Abstract
Most single-gene diseases, including muscular dystrophy, display a nonuniform phenotype. Phenotypic variability arises, in part, due to the presence of genetic modifiers that enhance or suppress the disease process. We employed an unbiased mapping approach to search for genes that modify muscular dystrophy in mice. In a genome-wide scan, we identified a single strong locus on chromosome 7 that influenced two pathological features of muscular dystrophy, muscle membrane permeability and muscle fibrosis. Within this genomic interval, an insertion/deletion polymorphism of 36 bp in the coding region of the latent TGF-β–binding protein 4 gene (Ltbp4) was found. Ltbp4 encodes a latent TGF-β–binding protein that sequesters TGF-β and regulates its availability for binding to the TGF-β receptor. Insertion of 12 amino acids into the proline-rich region of LTBP4 reduced proteolytic cleavage and was associated with reduced TGF-β signaling, decreased fibrosis, and improved muscle pathology in a mouse model of muscular dystrophy. In contrast, a 12-amino-acid deletion in LTBP4 was associated with increased proteolysis, SMAD signaling, and fibrosis. These data identify Ltbp4 as a target gene to regulate TGF-β signaling and modify outcomes in muscular dystrophy.
Introduction
In muscular dystrophy, there is progressive loss of muscle fibers and replacement by fibroblasts and adipocytes, resulting in muscle weakness. Mutations in the genes encoding dystrophin or its associated proteins, the sarcoglycans, lead to a classical form of disease with early onset and a severe course. Mutations that disrupt dystrophin or the sarcoglycans render the muscle plasma membrane fragile and lead to increased permeability. A leaky plasma membrane produces elevated intracellular calcium and initiates a cascade of proteolysis, cellular dysfunction, and, ultimately, death of myofibers. As with most monogenetic disorders, there is phenotypic variability even with an identical gene mutation. The precise dystrophin gene mutation in Duchenne muscular dystrophy (DMD) explains aspects of the phenotypic spectrum. Age of onset and ambulatory loss, as well as targeted muscle groups, are influenced by the precise genetic mutation in the dystrophin gene. However, even within families where the same primary mutation occurs, there is a range of disease severity.
The related disorder limb girdle muscular dystrophy (LGMD) type 2C, arises from mutations in the gene encoding the dystrophin-associated protein γ-sarcoglycan (Sgcg). There is a single common Sgcg mutation that is widely prevalent among LGMD patients. The frameshifting Δ521-T Sgcg mutation causes the loss of γ-sarcoglycan protein (1) and has been associated with variable outcome, ranging from a severe, early-onset, Duchenne-like phenotype to a milder disease course, with individuals remaining ambulatory into their third and fourth decades (2). This same mutation produces heterogeneous phenotypes among siblings in 75% of families (3). The uniform genetic underpinning of this muscular dystrophy underscores the presence of modifying factors as major determinants of outcome. It is likely that modifiers are equally important in DMD, but the large number of mutations complicates the analysis of genetic modifier loci.
Phenotypic variability may relate to environmental or genetic factors, and in the muscular dystrophies, genetic modifiers have been studied using candidate gene approaches in animal models (4). Genetic mapping strategies using models of monogenetic disorders offer an unbiased approach to mapping genes that influence outcome. The mdx mouse represents the mouse model for DMD but displays a milder phenotype than that of human DMD patients (reviewed in ref. 5). The Sgcg-null mouse model of muscular dystrophy and cardiomyopathy displays a phenotype similar to that of the mdx mouse (6, 7). We selected mice lacking γ-sarcoglycan (Sgcg-null mice), as the model in which to map genetic modifiers, since the phenotype is more severe and there was ample evidence for genetic modifiers in the comparable human form of muscular dystrophy.
We previously introduced the Sgcg-null allele into 4 distinct genetic backgrounds, finding that the DBA/2J background conferred a more severe muscular dystrophy phenotype than the 129T2/SvEmsJ strain, demonstrating the presence of genetic modifier loci (7). In the present study, we interbred the mice from the two parental backgrounds to generate mice homozygous for the Sgcg-null allele in a mixed genetic background. We examined SNPs across the genome that differed between the parental strains and found that there was a single strong genetic region on chromosome 7 that was strongly associated with a suppressed or enhanced phenotype. We investigated the chromosome 7 region and found an insertion/deletion of 36 bp associated with an internal repeat structure within an exon of Ltbp4. Latent transforming growth factor–binding proteins (LTBPs) are members of a superfamily that binds TGF-β directly and regulates the release of TGF-β from the extracellular matrix. We examined the functional consequences of inserting 12 amino acids into or deleting them from the proline-rich region of LTBP4 and found altered proteolytic susceptibility. Moreover, cleavage of LTBP4 was associated with increased TGF-β signaling and provides a mechanism to explain how this genetic polymorphism alters the disease process.
Results
F2 mice display an intermediate phenotype between the two parental strains.
We generated F2 mice homozygous for the Sgcg-null allele in a mixed genetic background of 129T2/SvEmsJ and DBA/2J. The phenotypic range was determined using two different measures of the disease progress, membrane permeability and fibrosis. Disruption of the dystrophin glycoprotein complex (DGC) enhances membrane leak and promotes abnormal membrane permeability measured as Evans blue dye content in myofibers in vivo (7). The second assay quantifies the hydroxyproline (HOP) content of muscle as a direct reflection of the degree of fibrosis (7). All analyses were conducted in 8-week-old animals, since this time frame corresponds to an early point in the pathologic process but one at which the muscle phenotype is evident.
The interbred F2 generation (γ-sarcoglycan–null in the F2 generation [F2-Sgcg]) displayed a phenotype intermediate between the two parental backgrounds with the γ-sarcoglycan–null allele (Figure 1). 129-Sgcg mice had a milder phenotype, with decreased dye uptake and fibrosis compared with D2-Sgcg mice. The mean dye uptake for the F2-Sgcg mice (n = 186, or n = 372 quadriceps muscles) was 29.9 ± 13.1, close to the midpoint between the 129-Sgcg (15.8 ± 8.9) and the D2-Sgcg (35.5 ± 20.2) strains. We determined fibrosis by measuring HOP content and also observed an intermediate phenotype for the F2-Sgcg animals. The 129-Sgcg quadriceps muscles had 9.6 ± 1.7 nM HOP/mg, while the D2-Sgcg had 24.3 ± 7.7, and the value for the F2-Sgcg was 12.2 ± 3.3.
Figure 1. Mice lacking γ-sarcoglycan (Sgcg-null mice) serve as a model for muscular dystrophy.
The Sgcg-null allele displays a mild phenotype in the 129 background strain (129-Sgcg) and a severe phenotype in the D2 strain (D2-Sgcg). Sgcg mice from the two backgrounds were interbred (F2-Sgcg), and the F2-Sgcg animals displayed an intermediate phenotype. (A) Muscle membrane leak in the quadriceps muscle was measured by Evans blue dye uptake in the 2 parental strains (129-Sgcg and D2-Sgcg) and the intercrossed F2-Sgcg generation. (B) Quadriceps muscle fibrosis was measured by determining HOP content in the two parental strains; the F2-Sgcg mice also revealed an intermediate phenotype. (C) An example of the phenotypic range is shown from diaphragm muscles from 2 F2-Sgcg animals (the two left panels) and 1 wild-type control (right panel). Dye uptake (blue) and fibrotic replacement (white) are shown. (D) Grip strength was measured in 8-week-old animals of the 129 and D2 backgrounds with and without the Sgcg-null allele. The D2-Sgcg mice were weaker than 129-Sgcg mice, while the parental strains without the muscular dystrophy gene were not significantly different.
To evaluate whether these pathological features correlated with muscle function, we evaluated grip strength of 8-week Sgcg-null mice in both genetic backgrounds. We found that there was no significant difference between the two wild-type parental strains in grip strength performance. Sgcg-null mice in the D2 background were weaker than Sgcg-null mice in the 129 background (Figure 1D), confirming that these pathological differences also influenced muscle function. For mapping studies, we relied on the pathological quantitative traits, since these traits could be measured more reliably and allowed the assay of multiple muscle groups individually.
Genome-wide mapping identifies a single strong locus, dMOD1, that modifies both membrane leak and fibrosis.
We used whole-genome SNP analysis to map genetic regions that segregated with enhanced or suppressed phenotype. SNP genotyping was performed using 328 SNPs that differ between the parental 129 and D2 strains. The whole-genome scan was conducted on F2-Sgcg animals. For genotyping, we selected animals with dye uptake and HOP values at least 1 SD above or below the mean in at least two muscle groups, and we chose animals with concordance in two or more muscle groups to minimize intra-animal variability. In addition, we discarded data from animals with very large intramuscular variability, defined by an SD of greater than 20% of the mean for a given muscle pair. Based on these criteria, DNA samples from 80 of 186 F2-Sgcg animals were analyzed. For skeletal muscle dye uptake measurements, 49 animals were genotyped (25 animals had increased dye uptake and 24 animals had decreased dye uptake). For skeletal muscle HOP, 31 animals were genotyped (18 animals chosen for increased HOP and 13 for decreased HOP).
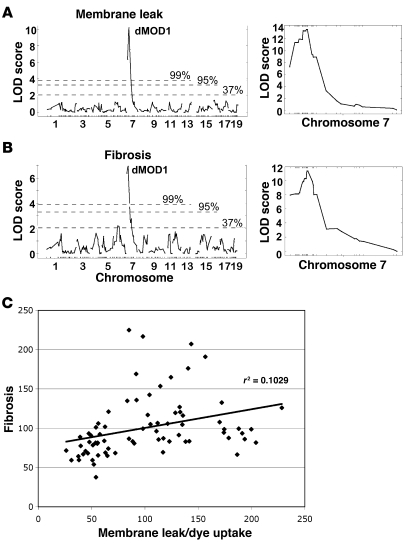
We identified one major locus, dMOD1, on chromosome 7 that influenced both membrane permeability and fibrosis (Figure 2). For dMOD1, the peak lod score for dye uptake was 10.17 at SNP marker rs3156053 at 28.01 Mb (build 36.1). This same SNP was also significantly associated with fibrosis in the quadriceps muscles, yielding a lod score of 6.95. Thus, the same SNP was highly linked to both features of muscular dystrophy, membrane permeability and replacement fibrosis. Using a denser chromosome 7 SNP panel, we further refined dMOD1 using the same cohort of F2-Sgcg mice. The peak lod scores increased to 13.59 for membrane leak and 11.32 for fibrosis (Figure 2, A and B, right).
Figure 2. The pathological muscular dystrophy traits of membrane leak and fibrosis independently map to the dMOD1 locus on chromosome 7.
(A) lod scores are shown for 328 SNPs for the trait of membrane leak (dye uptake) in the quadriceps muscles. dMOD1 maps to chromosome 7 with a peak lod of 10.17. (B) lod scores across the genome correlated to fibrosis (HOP content) also map to the dMOD1 on chromosome 7 with a peak lod score of 6.95. Chromosomes and SNP positions are indicated on the x axis, and lod scores are on the y axis. The percentages indicate the chance that this event occurred non-randomly. The right panels show lod scores derived from fine mapping of chromosome 7 near dMOD1, with a peak lod score for dye uptake and fibrosis of 13.52 and 11.32, respectively. (C) The degree of membrane leak and fibrosis correlate. Measurements from quadriceps muscle of membrane leak (dye uptake) and fibrosis (HOP content) were plotted (r2 = 0.1029; P < 0.005).
We analyzed the correlation between membrane leak and fibrosis in the same animals. Although the correlation was weak (r2 = 0.1029), a t test showed that the correlation was significant (P < 0.005; Figure 2C). In our previous analysis, we were unable to demonstrate a clear correlation between these two aspects of pathology, membrane leak and fibrosis (7), and assumed that they may be influenced by different modifier loci. This current study was performed with a larger sample size and thus likely accounts for the weak, but statistically evident, correlation. The finding that both traits were influenced by the same strong modifier region on chromosome 7 indicates that the responsible gene can affect both aspects of disease, membrane leak and fibrosis, or that the two aspects of pathology are interdependent on each other.
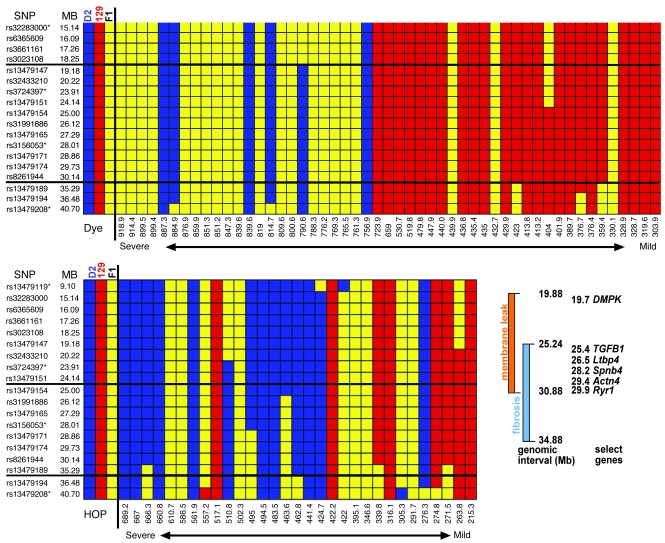
Given the unexpected result of a single strong locus affecting muscular dystrophy, we analyzed data from individual animals separately for both pathological traits to more fully dissect the genomic interval associated with dMOD1 (Figure 3). We aligned the dye uptake values from mild to severe and analyzed the SNP data, including those genotypes from the high-resolution genotyping. Using only the dye uptake values, the 95% confidence interval for dMOD1 extended from 19.88 Mb to 30.88 Mb. This data set was larger because more animals met criteria of mild or severe disease. This analysis highlights a comparative absence of the homozygous DBA allele, consistent with a dominant effect of the modifier gene on membrane leak. When the HOP values were considered similarly, the mode of genetic inheritance was less clear and was consistent with a potential semidominant mode of inheritance. The 95% confidence interval for dMOD1 HOP content extended from 25.24 Mb to 34.88 Mb. Since these intervals overlap, we assume that a single gene mediates both membrane leak and fibrosis and therefore refine dMOD1 to the interval between 25.24 Mb and 30.88 Mb on chromosome 7 (Figure 3).
Figure 3. Refining the chromosome 7 dMOD1 interval.
The upper panel shows SNP data arranged in rank order of membrane leak (dye uptake) in all muscle groups. The lower panel shows SNP data arranged in rank order of fibrosis (HOP uptake) in all muscle groups. Animals on the left were the most severely affected, having high levels of membrane leak and fibrosis, while those on the right were less severely affected. D2 homozygous chromosomes are shown in blue, 129 in red, and the heterozygous chromosomes in yellow. The thick horizontal lines indicate the 95% confidence interval for each trait. Shown in the box are the genetic regions associated with membrane leak and with fibrosis, demonstrating overlap and listing several candidate genes including TGFB1 and Ltbp4. Asterisks indicate markers also used for the whole-genome scan.
Candidate gene analysis of dMOD1.
Muscular dystrophy is characterized by ongoing degeneration and active regeneration within the same muscle group. Given the complexity of this process, finding a single genomic modifier locus of very high significance was unexpected. The D2 strain is known to be associated with dystrophic calcification. Notably, dMOD1 falls near the Dyscalc locus, a region previously linked to vascular and myocardial calcinosis (8). Dyscalc1 is a major contributor to dystrophic cardiovascular calcification in susceptible strains, affecting both the vessels and myocardium (8). Two genes have been independently implicated in Dyscalc, Abcc6 and Emp3 (9, 10). Both genes were excluded as accounting for dMOD1 gene using known disease-associated polymorphisms and finding that both parental strains had the susceptible alleles of each gene (data not shown).
TGF-β signaling has been extensively implicated in fibrosis in many different cell and tissue types, including muscle, and enhanced fibrosis was a feature we used to map dMOD1. Increased TGF-β signaling has been observed in muscular dystrophy (11). Moreover, inhibition of TGF-β signaling improves muscle pathology and function in the mdx mouse model of muscular dystrophy (12). Two genes involved in the TGF-β pathway are within the dMOD1 interval, TGFB1 and Ltbp4. We fully sequenced the coding regions, promoter, and flanking intronic regions of TGFB1. However, there were no polymorphisms in the TGFB1 gene between the parental strains D2 and 129.
An insertion/deletion polymorphism in Ltbp4 segregates with severity of muscular dystrophy pathology.
TGF-β1 is a secreted growth factor that is held latent in the extracellular matrix (13). LTBP4 is a member of the latent TGF-β–binding protein multigene family that contains 4 LTBPs and 3 fibrillins (14, 15). Ltbp4 is preferentially expressed in cardiac, skeletal, and smooth muscle (14, 15). We evaluated gene expression profiles for Ltbp4 and found that Ltbp4 expression is induced upon myoblast differentiation and with regeneration after injury. In the mouse, a hypomorphic, loss-of-function allele of Ltbp4 develops pulmonary fibrosis, cardiomyopathy, and colon cancer (16). Like other LTBPs, LTBP4 is part of the large latent complex that sequesters TGF-β in the extracellular matrix, limiting its availability to TGF-β receptors (17, 18).
Because of the reported involvement of TGF-β1 in muscular dystrophy and its general role in fibrosis, we evaluated Ltbp4 in both parental strains. We found a 36-bp polymorphism within exon 12 of Ltbp4 (Figure 4). The protected 129 strain had 36 additional bp (Ltbp4+36), and the severely affected D2 strain was deleted for these 36 bp (Ltbp4Δ36). Closer examination revealed a repeating DNA sequence that is duplicated in the 129 strain, and this repeating structure may have predisposed to the apparent insertion into 129. This repeated sequence and the deletion/insertion are unrelated to the intron-exon boundary, since the deletion occurs wholly within an exon and not at the intron-exon border.
Figure 4. An insertion/deletion polymorphism in Ltbp4 predicts the phenotype in muscular dystrophy.
Ltbp4 encodes a TGF-β–binding protein expressed in skeletal and cardiac muscle. (A) The gene structure is shown for Ltbp4. Exons 11, 12, 13 (red bars) of Ltbp4 encode a proline-rich region. (B) 129-Sgcg mice, with a milder phenotype, have a 36-bp insertion that encodes an extended proline-rich region, while severely affected D2-Sgcg mice have a deletion of 36 bp. The insertion/deletion occurs wholly within exon 12. (C) Ltbp4+36 correlates with reduced membrane permeability and reduced fibrosis in F2-Sgcg mice. Congenic Sgcg-null mice in the C57BL/6J background or in the CD1 background have a mild phenotype comparable to that of 129-Sgcg mice (7), and these mice also have the protective insertion Ltbp4+36 allele. mdx mice, the model for DMD, in the C57BL/10 background, also have the protective Ltbp4+36 allele.
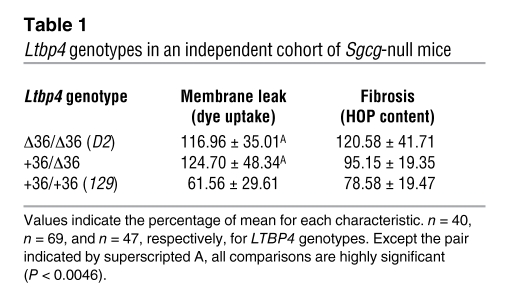
We confirmed that the Ltbp4 insertion/deletion segregates with phenotypic variability in muscular dystrophy by examining mixed-background γ-sarcoglycan–null animals with a range of values for membrane permeability defects and fibrosis. Those animals with the lowest values for dye uptake and fibrosis carried the Ltbp4+36 allele (Figure 4C). We generated congenic strains for the γ-sarcoglycan–null allele in the C57BL/6J and CD1 VAF+ backgrounds. Membrane permeability and fibrosis in mice of the congenic strains B6-Sgcg and CD1-Sgcg were comparable to what was seen in 129-Sgcg mice (7), and these strains were also found to harbor the protective allele (Ltbp4+36) (Figure 4C). The mdx model was also found to carry the protective Ltbp4+36 allele (Figure 4C). To ensure that the segregation of Ltbp4 with disease phenotype was not unique to the restricted cohort of animals used for mapping, we replicated these results in an independently bred cohort of additional F2-Sgcg mice from 129-Sgcg and D2-Sgcg parental strains. We genotyped the Ltbp4 allele in this new cohort and found a highly significant correlation between the Ltbp4Δ36 allele and the degree of membrane permeability and fibrosis (Table 1).
Table 1 .
Ltbp4 genotypes in an independent cohort of Sgcg-null mice
Insertion of 12 amino acids in the proline-rich region protects against proteolysis.
LTBP4 interacts with the matrix via its aminoterminal segment (19). At the carboxyl terminus, the third cysteine-rich region binds TGF-β1 (17, 18). In between these two domains is a proline-rich region thought to be a target for proteases (14, 15), although the range of proteases responsible for this cleavage is not well defined. Cleavage releases TGF-β from the matrix and renders it accessible to TGF-β receptors, where the ligand can engage the receptor and mediate TGF-β effects (20, 21). The severe phenotype in the D2 background was associated with a deletion of 12 amino acids in the proline-rich region of LTBP4.
To evaluate differential proteolytic sensitivity of LTBP4 from the D2 and 129 strains, we cultured muscle fibroblasts from the parental strains as well as those carrying the Sgcg-null allele. Exposure to either pancreatin or plasmin resulted in more digestion of full-length LTBP4 from the D2 background compared with the 129 background, irrespective of the presence of the Sgcg-null allele (Figure 5, A and B). To attribute this differential proteolytic susceptibility to the insertion or deletion of 12 amino acids in the proline-rich region, we expressed portions of LTBP4 in vitro. The regions expressed are depicted in Figure 5C; the in vitro products were radiolabeled with [35S]cysteine found only at the carboxyl terminus of the fragments. The 129 and D2 proline-rich regions of LTBP4 were expressed and treated with plasmin, an enzyme implicated in the cleavage of LTBP4 and release of TGF-β (15). Prior to proteolysis, the expressed regions displayed distinct electrophoretic patterns, with the smaller protein migrating more slowly, consistent with altered conformation. The LTBP4 region from the 129 strain (+12 amino acids) was minimally susceptible to proteolysis even at high doses of plasmin (Figure 5E). In contrast, the region of LTBP4 from the D2 strain (deleted for 12 amino acids) displayed a greater susceptibility to plasmin degradation (Figure 5A). Enhanced susceptibility to proteolysis should be associated with increased TGF-β release from the matrix and increased availability to engage cell-surface TGF-β receptors and activate canonical TGF-β signaling.
Figure 5. The 12-amino-acid deletion increases LTBP4 protease susceptibility.
(A and B) Total protein was extracted from D2 and 129 fibroblasts with and without the Sgcg-null allele, digested, and immunoblotted with an antibody directed toward the amino terminus of LTBP4. (A) Digests with 12 μg of pancreatin. Each pair represents Sgcg mutant, then wild-type. (B) Digestion with 25 μg of plasmin. Each set of 4 represents 3 mutants, then a wild-type. LTBP4 from the D2 strain was digested more easily than LTBP4 from the 129 strain. The percent digested refers to the proteolyzed lower product. On average, plasmin digested 38% of 129-derived LTBP4 versus 57% of D2-derived LTBP4. (C and D) In vitro expression constructs. D2- and 129-derived LTBP4 proline-rich sequences were expressed in vitro. The LTBP4 sequence is underlined; the insertion is shown in gray; and the deletion is represented by dashed lines. The position of the cysteine residues in the expressed sequences is indicated by the asterisks. (E) The constructs were expressed in vitro with [35S]cysteine and then exposed to plasmin, a protease implicated in cleavage of LTBP4 (15). The D2-derived sequences are digested at low levels of plasmin, while only the highest level of plasmin can begin to produce the fully digested product from the 129-derived sequence. The slower migration of the 12-amino-acid-deleted proline-rich region, prior to digestion, is consistent with an altered conformational state that is more susceptible to proteolysis. The arrow indicates the cleavage product.
TGF-β signaling is increased by deletion of 12 amino acids in LTBP4.
Upon ligand interaction, TGF-β receptors activate SMAD signaling, whereby SMAD2/3 is phosphorylated and translocated to the nucleus to drive gene expression (13). The Ltbp4 alleles from 129 and D2 backgrounds should reflect a variable capacity to sequester inactive TGF-β and therefore should elicit differential TGF-β–induced signaling because more TGF-β is available for the ligand-receptor interaction. To evaluate this possibility, we isolated muscle fibroblasts from 129-Sgcg and D2-Sgcg mice and then exposed these cells to identical doses of exogenously added TGF-β. We chose fibroblasts for this assay because γ-sarcoglycan is not expressed in fibroblasts, and therefore TGF-β signaling in this assay is isolated from the ongoing disease process itself which can itself activate TGF-β signaling. We maintained the fibroblast cultures at confluence for 5 days, since this condition is shown to be associated with increased LTBP4 expression and deposition into the matrix (22). TGF-β signaling in this assay is dependent on the presence of intact LTBP4 (23). After exposure to TGF-β, fibroblasts from D2-Sgcg mice displayed significantly more SMAD2/3 phosphorylation than those from 129-Sgcg mice (Figure 6A). This increased signaling was not dependent on the presence of the Sgcg-null allele, since wild-type DBA also had increased p-SMAD2/3 for the same dose of TGF-β. Increased TGF-β signaling did not arise from altered LTBP4 protein levels, since LTBP4 protein levels were similar in the 2 strains (Figure 6B). Therefore, these data are consistent with a model wherein increased TGF-β–induced signaling in the D2 strain derives from enhanced proteolytic susceptibility of LTBP4 and release of TGF-β, leading to SMAD activation.
Figure 6. Increased proteolytic cleavage is associated with enhanced TGF-β availability and SMAD signaling, accounting for the more severe phenotype.
(A) Fibroblasts were cultured from D2-Sgcg and 129-Sgcg muscle. Fibroblasts were exposed to TGF-β, and the amount of phosphorylated SMAD was determined. Fibroblasts from the severely affected D2-Sgcg muscle respond to TGF-β with enhanced p-SMAD signaling. Coomassie-stained actin is the loading control. (B) The amount of LTBP4 protein is similar in D2 and 129 fibroblasts. Muscle fibroblasts were isolated and subjected to immunoblotting with an anti-LTBP4 antibody. The graph represents the densitometer readings of p-SMAD normalized to actin, and the highest value of the 8 animals was set to 100%.
TGF-β signaling is enhanced in the presence of the Ltbp4Δ36 allele.
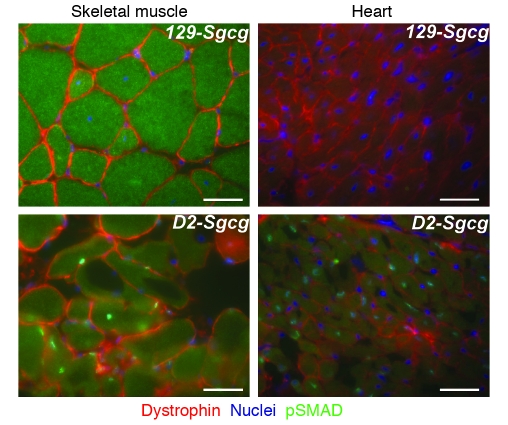
We examined SMAD signaling within the heart and muscle of mutant animals in the enhanced and suppressed genetic backgrounds. We found that TGF-β signaling was enhanced within muscle nuclei, consistent with a TGF-β effect on both fibroblasts and the neighboring myofibers. Within skeletal muscle, the myofiber nuclei of the severe D2-Sgcg strain had increased p-SMAD staining (Figure 7, green nuclei). The centrally placed nuclei within dystrophic skeletal muscle indicate newly regenerated fibers, and these fibers consistently displayed the most intense signaling compared with other nuclei within the same sections. Within cardiac muscle, the D2-Sgcg hearts also showed p-SMAD–positive nuclei (Figure 7). Within the hearts, p-SMAD–positive nuclei were clustered in regions of damage and were more often seen in the right ventricle. Neither wild-type strain showed activation of SMAD signaling (data not shown). These data support a model whereby LTBP4 is preferentially cleaved in the D2-Sgcg background, leading to enhanced TGF-β availability and increased SMAD signaling.
Figure 7. Increased TGF-β signaling within skeletal myofibers and cardiomyocytes in the D2 background.
p-SMAD2 (green) is increased in the myonuclei of D2-Sgcg compared with 129-Sgcg skeletal and cardiac muscle. In skeletal muscle, the centrally placed nuclei, indicative of recent regeneration, show the most intense staining (green). Scale bars: 10 μm.
Discussion
We used a genome-wide mapping strategy to identify genetic loci that modified two different pathological features of muscular dystrophy. Using this approach, we found evidence for a single genomic region that was highly significantly linked to these two different features, dye uptake and fibrosis. The finding that one genomic locus linked so strongly to both dye uptake and fibrosis suggests that these two pathological features may be more tightly coupled than previously appreciated. The role for TGF-β signaling in fibrosis is well established, but the role of TGF-β signaling in mediating dye uptake, a reflection of membrane leakage, was less anticipated. Dye uptake is thought to be an early aspect of muscle pathology deriving from disruption of the dystrophin protein complex (24, 25). In contrast, fibrosis has been considered a more typical “scar” response to myofiber loss in muscular dystrophy. Serum creatine kinase (CK) levels are elevated in muscular dystrophy patients and in this model (6), and serum CK is thought to similarly reflect membrane leak. CK values were not used in this study as a quantitative trait, since we have found this trait to be too highly variable to be useful for genetic mapping. In the absence of the dystrophin protein complex, the muscle plasma membrane is fragile, because dystrophin is known to participate in a mechanically strong link between the plasma membrane and the extracellular matrix (26–28). Blocking TGF-β signaling using anti–TGF-β antibodies reduced SMAD signaling and improved outcome in the mdx mouse without a significant improvement in CK (12). The failure of TGF-β inhibition to detectably reduce CK levels in the Cohn et al. study (12) may indicate that TGF-β exerts more of an effect on matrix remodeling than plasma membrane stability. However, this current genetic study and the tight linkage of the Ltbp4 allele to dye uptake support a model whereby there is a close interplay between membrane leak and surrounding fibrosis.
Ltbp4 is highly expressed in muscle and therefore positioned to be a determinant of damage and fibrosis in muscle diseases (14, 15). It is also notable that there was a functional correlate of these pathological features in that the D2-Sgcg mice had muscle weakness. The expression of LTBP4 in muscle is not restricted to fibroblasts, since Ltbp4 mRNA levels are increased in the C2C12 cell culture model of muscle differentiation (29). Therefore, multiple cell types in skeletal muscle mediate the deposition of LTBP4 into the matrix and sequestration of TGF-β (Figure 8). We found that insertion of 12 amino acids into the proline-rich region of LTBP4 resulted in reduced susceptibility to proteolysis associated with reduced SMAD signaling. These data are consistent with recent results generated from fibroblasts deficient for LTBP4 (16). Fibroblasts deficient for LTBP4 are impaired in their ability to activate TGF-β, consistent with a requirement of LTBP4 for normal TGF-β activation. In LTBP4-deficient fibroblasts, reexpression of LTBP4 restored TGF-β activation, and this activation depended on the presence of both the TGF-β–binding and matrix-binding domains (23).
Figure 8. Model of LTBP4 action.
TGF-β forms the small latent complex with its inactive domain. The small latent complex binds to LTBP4 to form the large latent complex, where TGF-β is held inactive in the extracellular matrix. Proteolytic cleavage of LTBP4 releases TGF-β from the large latent complex, where it is available to bind TGF-β receptors and activated phosphorylation and nuclear translocation of SMADs and activate gene expression.
It is presumed that increased SMAD signaling within myofibers is pathogenic, although this remains to be formally demonstrated. Deficiency of Ltbp4 leads to activation of bone morphogenetic protein–4 (BMP-4) (30). Although this study examined TGF-β, it is possible that LTBP4 mediates its effects by regulating BMP or even myostatin activity. Cohn et al. examined the effect of TGF-β blockade on muscle regeneration and found that regeneration was enhanced in the mdx mouse model after inhibiting TGF-β (12). TGF-β blockade promoted regeneration and factors associated with regenerating muscle such as periostin and neonatal myosin (12). Periostin is a known downstream target of TGF-β signaling. Matrix remodeling in muscular dystrophy may explain impaired regeneration and contribute to plasma membrane defects (31). The current study strongly supports a pathogenic role for TGF-β signaling beyond the effect on regeneration, since we noted increased nuclear SMAD signaling within cardiomyocytes, a cell type that lacks the robust regenerative response of skeletal muscle. Interestingly, p-SMAD signaling was not uniformly increased in all myofibers or cardiomyocytes but instead exhibited regional variation, similar to what is seen with dye uptake into muscle and heart.
Given the general role of TGF-β in regulating fibrosis, it is possible that Ltbp4 mediates membrane leak and fibrosis beyond this single genetic disease. Muscular dystrophy deriving from a similar pathogenic process, such as the loss of dystrophin, may be also regulated by Ltbp4 alleles. We tested multiple mouse strains for the presence of the Ltbp4+36/+36 allele and found that the protective allele is the most common form of the allele (Supplemental Figure 1). The presence of the deletion allele correlates well with phylogenetic maps derived from SNP data (32). However, when the genetic relationship between mouse strains was determined based on Y chromosome or mitochondrial DNA, the Ltbp4 insertion/deletion allele did not segregate as well (data not shown). The repetitive nature of the sequence underlying the Ltbp4 insertion/deletion allele may have promoted the polymorphism having arisen more than once.
The dMOD1 region is sizable and contains a number of genes that could modulate phenotypic variability in muscular dystrophy. It is also possible that more than one gene within this interval contributes to the modifier effect, and additional genetic experiments will be required to conclude that only Ltbp4 modifies muscular dystrophy. We compared the dMOD1 region in the 129 and D2 strains using the sequence data of the Mouse Phenome project (http://phenome.jax.org/pub-cgi/phenome/mpdcgi?rtn=snps/door). With this analysis, there are approximately 45 nonsynonymous SNPs in this interval, and only 29 SNPs fall into predicted or known proteins (Supplemental Table 1). One SNP falls within a gene important for muscle function, the RYR1 gene encoding the ryanodine receptor of skeletal muscle. Mutations in RYR1 cause malignant hyperthermia or central core disease (33), and recently, pharmacologic targeting of nitrosylation of the ryanodine receptor was shown to be effective in mitigating some of the damage in muscular dystrophy (34). Between the D2 and 129 strains, there is a single coding SNP, RYR1 Q2028R. However, it is unlikely that this SNP contributes significantly to modifying muscular dystrophy, since both the 129 and C57BL/6 background are protective for muscular dystrophy, and the RYR1 SNP differs between these two strains. The SNP list in the Phenome project is not exhaustive, and therefore, additional differences likely exist within dMOD1 between the D2 and 129 strains. The finding of only a single strong modifier locus remains surprising given the complexity of this disease process.
The ability of LTBP4 to regulate fibrosis in human muscle remains an open question. It is striking that the related proline-rich region of human LTBP4 contains a much larger deletion. The canine genome also supports a larger deletion within the proline-rich region of LTBP4. Both human and dog have more fibrosis associated with muscular dystrophy compared with the murine model, including the severe D2 strain. The smaller proline-rich regions encoded by the human and dog LTBP4 genes may mediate more TGF-β signaling. The human gene demonstrates a similar pattern of gene expression, whereby LTBP4 is expressed in a number of tissues but more highly in heart, muscle, and smooth muscle (14). Therefore, LTBP4 may be well positioned to preferentially mediate TGF-β signaling in these tissues. Genetic disruption of murine Ltbp4 produces pulmonary fibrosis, cardiomyopathy, and colon cancer (16) consistent with the expression pattern of LTBP4.
Intriguingly, in a study of 304 human patients with chronic obstructive pulmonary disease, 3 nonsynonymous SNPs within LTBP4 associated with outcome (35). Close analysis of the data reveals that the LTBP4 SNPs were most significantly associated with exercise-related traits, including maximum work, increased exercise capacity, and an improved 6-minute walk distance (35). The LTBP4 SNPs in the study by Hersh et al. (35) encode alternative amino acids that fall within exons 5, 17, and 18 in regions involved in matrix association (Supplemental Figure 2). The association in this study requires replication, but it is nonetheless noteworthy in that it provides evidence that LTBP4 is important in human disease phenotypes that may involve muscle function.
Genetic mapping of modifiers for single-gene disorders is often successful in identifying genetic regions that predict altered phenotype. The nature of this type of mapping, where quantitative traits are linked to genetic regions in the absence of a clear pattern of Mendelian inheritance, makes it difficult to demonstrate genetic causation. Therefore, identification of the precise genes responsible for a genetic effect can be problematic. For example, genetic regions that modify cystic fibrosis have been identified in this same region of chromosome 7 in mice and the orthologous region of chromosome 19 in humans (36, 37). This same genomic interval, as noted in this study, also includes the TGFB1 gene itself. Specific haplotypes of the TGFB1 gene have been linked to outcome in cystic fibrosis (36, 38), confirming that this interval is important for additional phenotypes. Given the proximity of TGFB1 to LTBP4, it is possible that LTBP4 alleles may also contribute to the phenotypic variability ascribed to this genomic interval, chromosome 19 in humans and chromosome 7 in mice. TGF-β signaling has both beneficial and detrimental effects, and thus, exploiting TGF-β for therapy has a complicated side-effect profile. Our findings suggest that the large latent complex of TGF-β, including LTBPs, offers a mechanism to target this TGF-β signaling preferentially within tissues or cell types.
Methods
Animals.
Sgcg-null mice were generated by deleting exon 2 of γ-sarcoglycan and express no γ-sarcoglycan protein in the homozygous state (6). Sgcg-heterozygous mice were bred over 10 generations onto the 129/SVemst/J (129) or DBA/2J (D2) strain (The Jackson Laboratory). Breeding was conducted in heterozygous animals to avoid any selection for fitness that may occur with homozygous null mice. Homozygous null 129-Sgcg and D2-Sgcg mice were interbred and analyzed at 8 weeks of age. All animals used in this study were uniformly housed in a single, pathogen-free barrier facility and treated under a protocol approved by the University of Chicago Animal Care and Use Committee.
Mice were sacrificed at 8 weeks of age; the muscles were harvested and weighed. Evans blue dye (dye uptake) assays were conducted immediately, while tissues for HOP assay were frozen at –80°C. Dye uptake assays were preformed on triceps, gastrocnemius/soleus, hamstrings, abdominal muscles, and one-half of each quadriceps as 9 separate muscle groups. HOP assays were performed on diaphragm, heart ventricles, abdominal muscles, and one-half of each quadriceps as 5 separate muscle groups.
Grip strength.
Grip strength was measured in 8-week-old animals of the genotypes D2, 129, D2-Sgcg, and 129-Sgcg, with comparable numbers of males and females in each group (n = at least 8 in each group) as described previously (39).
Quantitative dye uptake.
Prior to sacrifice, mice were injected intraperitoneally with Evans blue dye at 5 μl/g body weight (Sigma-Aldrich, E2129) dissolved in PBS at 10 mg/ml. Tissues were harvested 40 hours later, finely minced, weighed, and incubated at 55°C in 1 ml formamide for 2 hours before spectrophotometric absorbance was measured at 620 nm as described previously (40). Results are reported as arbitrary OD units/mg tissue or were normalized for a particular muscle group.
HOP measurement.
The HOP assay was performed as described previously (41). Muscle was minced, weighed, and hydrolyzed overnight in 2 ml of 6 M hydrochloric acid at 110°C. Hydrolysate (10 μl) was mixed with 150 μl isopropanol, then 75 μl of 1.4% chloramine-T (Sigma-Aldrich) in citrate buffer and oxidized at room temperature for 10 minutes. One milliliter of Ehrlich’s reagent [3 g of 4-(dimethylamino) benzaldehyde (Sigma-Aldrich), 10 ml ethanol, 675 μl sulfuric acid] was added, mixed vigorously, and incubated for 30 minutes at 55°C, followed by extinction measurement at 558 nm. A standard curve (0–5,000 nM, trans-4-hydroxy-l-proline; Sigma-Aldrich) was included in each assay. Therefore, results are reported as nM HOP/mg tissue.
Genetic analysis.
Autosomal SNPs (n = 328) were selected from the fixed panel of 768 SNPs of the Illumina GoldenGate platform because they differed between the 129 and D2 parental strains, with an average distance between SNPs of 8.35 Mb and a maximum distance between any two SNPs of 60.81 Mb. Higher-resolution mapping was accomplished by genotyping of a denser SNP panel on chromosome 7 with the Sequenom iPLEX platform (Harvard Partners Center for Genomics and Genetics; Sequenom). The dense chromosome 7 panel consisted of 16 SNPs between 15.14 and 40.70 Mb. All chromosome distances were established using NCBI build 36.1.
The Gene Expression Omnibus database was queried for LTBP4 expression profiles (http://www.ncbi.nlm.nih.gov/geo/, gene expression profiles for LTBP4 and C2C12). Expression of C2C12, a muscle cell line that lacks other cell types, was queried so as to avoid fibroblast contamination. Records GDS4212 and GDS586 were examined.
Sequence analysis and PCR.
Exons and exon-intron boundaries of genes were sequenced in both 129-Sgcg and D2-Sgcg strains. At least 50 bp of intronic sequence was acquired and analyzed at each exon-intron boundary by selecting primers approximately 70 bp within intronic regions flanking exons.
Cell culture.
Primary muscle fibroblast cultures were generated following the primary myoblasts isolation protocol (42), except that in the preplating step, the fibroblasts were collected. Cells were grown in DMEM with 10% FBS media to 100% confluence. The cultures were maintained for 5 additional days in culture to allow adequate LTBP4 protein expression. Cultures were placed in Opti-MEM (Invitrogen) media for 1 day and then treated with 1 ng/ml TGF-β1 (TGF-β1 including the LAP domain; R&D Systems, 240-B) for 30 minutes. Supernatants were collected for analysis of LTBP4 protein (Santa Cruz Biotechnology Inc., SC33144 directed at the LTBP amino terminus residues 30–300). Cell lysates were collected and analyzed for phosphorylated SMAD (anti–p-SMAD2; Cell Signaling Technology, 3101).
Immunofluorescence microscopy.
Tissues were sectioned as described previously (6). The anti–p-SMAD2 and anti-dystrophin (Dys2; Novocastra Laboratories) antibodies were incubated on the sections for 2 hours at room temperature. Secondary antibodies (Invitrogen) were also diluted in the block solution and incubated at room temperature for 1 hour. Nuclei were illuminated using Vectashield with DAPI (Vector Laboratories). Images were acquired on a Zeiss Axiocam and recorded and processed using AxioVision 2.0.
In vitro expression and proteolysis.
Constructs representing the proline-rich domain (amino acids 482–608, NM_175641) were generated by PCR from both the 129 and D2 backgrounds and ligated into pcDNA3.1. In vitro transcription/translation was carried out using the reticulocyte lysate system (Promega) in the presence of [35S]cysteine. Three microliters of the 50-μl reaction was exposed to plasmin (2.5 μg to 25 μg) at 37°C for 15 minutes. The products were analyzed on a 12% gel, dried, and autoradiographed.
Statistics.
J/QTL software (http://churchill.jax.org/software/jqtl.shtml) was utilized to calculate lod scores for the whole-genome SNP analysis. The software assembled a genetic map from the SNP data. All of the animals for which we had both genotype and phenotype data were included in the J/QTL analyses. The settings of nonparametric model, EM scan method, and 1,000 permutations were selected. The 95% confidence interval was generated using estimation maximum. Values were compared using InStat and Prism (GraphPad), where P values less than 0.05 were considered significant. Statistical tests were chosen based on appropriateness of the distribution and comparison to be made, and thus ANOVA with Bonferroni post test analyses was performed. Error bars indicate the mean ± SD.
Supplementary Material
Acknowledgments
This work was supported by NIH grants HL 61322 to E.M. McNally and U01HD43430 to D.R. Beier.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 119:3703–3712 (2009). doi:10.1172/JCI39845.
Jennifer L. Moran’s present address is: Genetic Analysis Platform, Broad Institute of MIT and Harvard, Cambridge, Massachusetts, USA.
References
- 1.Noguchi S., et al. Mutations in the dystrophin-associated protein gamma-sarcoglycan in chromosome 13 muscular dystrophy. Science. 1995;270:819–822. doi: 10.1126/science.270.5237.819. [DOI] [PubMed] [Google Scholar]
- 2.McNally E.M., et al. Mild and severe muscular dystrophy caused by a single gamma-sarcoglycan mutation. Am. J. Hum. Genet. 1996;59:1040–1047. [PMC free article] [PubMed] [Google Scholar]
- 3.Kefi M., et al. Phenotype and sarcoglycan expression in Tunisian LGMD 2C patients sharing the same del521-T mutation. Neuromuscul. Disord. 2003;13:779–787. doi: 10.1016/S0960-8966(03)00136-6. [DOI] [PubMed] [Google Scholar]
- 4.Heydemann A., Doherty K.R., McNally E.M. Genetic modifiers of muscular dystrophy: implications for therapy. Biochim. Biophys. Acta. 2007;1772:216–228. doi: 10.1016/j.bbadis.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 5.Banks G.B., Chamberlain J.S. The value of mammalian models for Duchenne muscular dystrophy in developing therapeutic strategies. Curr. Top. Dev. Biol. 2008;84:431–453. doi: 10.1016/S0070-2153(08)00609-1. [DOI] [PubMed] [Google Scholar]
- 6.Hack A.A., et al. Gamma-sarcoglycan deficiency leads to muscle membrane defects and apoptosis independent of dystrophin. J. Cell Biol. 1998;142:1279–1287. doi: 10.1083/jcb.142.5.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heydemann A., Huber J.M., Demonbreun A., Hadhazy M., McNally E.M. Genetic background influences muscular dystrophy. Neuromuscul. Disord. 2005;15:601–609. doi: 10.1016/j.nmd.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 8.Ivandic B.T., et al. A locus on chromosome 7 determines myocardial cell necrosis and calcification (dystrophic cardiac calcinosis) in mice. Proc. Natl. Acad. Sci. U. S. A. 1996;93:5483–5488. doi: 10.1073/pnas.93.11.5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meng H., et al. Identification of Abcc6 as the major causal gene for dystrophic cardiac calcification in mice through integrative genomics. Proc. Natl. Acad. Sci. U. S. A. 2007;104:4530–4535. doi: 10.1073/pnas.0607620104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aherrahrou Z., et al. Ultrafine mapping of Dyscalc1 to an 80-kb chromosomal segment on chromosome 7 in mice susceptible for dystrophic calcification. Physiol. Genomics. 2007;28:203–212. doi: 10.1152/physiolgenomics.00133.2006. [DOI] [PubMed] [Google Scholar]
- 11.Bernasconi P., et al. Expression of transforming growth factor-beta 1 in dystrophic patient muscles correlates with fibrosis. Pathogenetic role of a fibrogenic cytokine. J. Clin. Invest. 1995;96:1137–1144. doi: 10.1172/JCI118101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohn R.D., Liang H.Y., Shetty R., Abraham T., Wagner K.R. Myostatin does not regulate cardiac hypertrophy or fibrosis. Neuromuscul. Disord. 2007;17:290–296. doi: 10.1016/j.nmd.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Massague J., Seoane J., Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- 14.Giltay R., Kostka G., Timpl R. Sequence and expression of a novel member (LTBP-4) of the family of latent transforming growth factor-beta binding proteins. FEBS Lett. 1997;411:164–168. doi: 10.1016/S0014-5793(97)00685-6. [DOI] [PubMed] [Google Scholar]
- 15.Saharinen J., Taipale J., Monni O., Keski-Oja J. Identification and characterization of a new latent transforming growth factor-beta-binding protein, LTBP-4. J. Biol. Chem. 1998;273:18459–18469. doi: 10.1074/jbc.273.29.18459. [DOI] [PubMed] [Google Scholar]
- 16.Sterner-Kock A., et al. Disruption of the gene encoding the latent transforming growth factor-beta binding protein 4 (LTBP-4) causes abnormal lung development, cardiomyopathy, and colorectal cancer. Genes Dev. 2002;16:2264–2273. doi: 10.1101/gad.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Y., et al. Amino acid requirements for formation of the TGF-beta-latent TGF-beta binding protein complexes. J. Mol. Biol. 2005;345:175–186. doi: 10.1016/j.jmb.2004.10.039. [DOI] [PubMed] [Google Scholar]
- 18.Saharinen J., Keski-Oja J. Specific sequence motif of 8-Cys repeats of TGF-beta binding proteins, LTBPs, creates a hydrophobic interaction surface for binding of small latent TGF-beta. Mol. Biol. Cell. 2000;11:2691–2704. doi: 10.1091/mbc.11.8.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koli K., Saharinen J., Hyytiainen M., Penttinen C., Keski-Oja J. Latency, activation, and binding proteins of TGF-beta. Microsc. Res. Tech. 2001;52:354–362. doi: 10.1002/1097-0029(20010215)52:4<354::AID-JEMT1020>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 20.Flaumenhaft R., et al. Role of the latent TGF-beta binding protein in the activation of latent TGF-beta by co-cultures of endothelial and smooth muscle cells. J. Cell Biol. 1993;120:995–1002. doi: 10.1083/jcb.120.4.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyazono K., Olofsson A., Colosetti P., Heldin C.H. A role of the latent TGF-beta 1-binding protein in the assembly and secretion of TGF-beta 1. EMBO J. 1991;10:1091–1101. doi: 10.1002/j.1460-2075.1991.tb08049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koli K., Hyytiainen M., Ryynanen M.J., Keski-Oja J. Sequential deposition of latent TGF-beta binding proteins (LTBPs) during formation of the extracellular matrix in human lung fibroblasts. Exp. Cell Res. 2005;310:370–382. doi: 10.1016/j.yexcr.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 23.Zhou Y., et al. Latent transforming growth factor-beta-binding protein-4 regulates transforming growth factor-beta1 bioavailability for activation by fibrogenic lung fibroblasts in response to bleomycin. Am. J. Pathol. 2009;174:21–33. doi: 10.2353/ajpath.2009.080620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuda R., Nishikawa A., Tanaka H. Visualization of dystrophic muscle fibers in mdx mouse by vital staining with Evans blue: evidence of apoptosis in dystrophin-deficient muscle. J. Biochem. (Tokyo). 1995;118:959–964. doi: 10.1093/jb/118.5.959. [DOI] [PubMed] [Google Scholar]
- 25.Straub V., Rafael J.A., Chamberlain J.S., Campbell K.P. Animal models for muscular dystrophy show different patterns of sarcolemmal disruption. J. Cell Biol. 1997;139:375–385. doi: 10.1083/jcb.139.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Menke A., Jockusch H. Decreased osmotic stability of dystrophin-less muscle cells from the mdx mouse. Nature. 1991;349:69–71. doi: 10.1038/349069a0. [DOI] [PubMed] [Google Scholar]
- 27.Pasternak C., Wong S., Elson E.L. Mechanical function of dystrophin in muscle cells. J. Cell Biol. 1995;128:355–361. doi: 10.1083/jcb.128.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rybakova I.N., Patel J.R., Ervasti J.M. The dystrophin complex forms a mechanically strong link between the sarcolemma and costameric actin. J. Cell Biol. 2000;150:1209–1214. doi: 10.1083/jcb.150.5.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tomczak K.K., et al. Expression profiling and identification of novel genes involved in myogenic differentiation. FASEB J. 2004;18:403–405. doi: 10.1096/fj.03-0568fje. [DOI] [PubMed] [Google Scholar]
- 30.Koli K., et al. Disruption of LTBP-4 function reduces TGF-beta activation and enhances BMP-4 signaling in the lung. J. Cell Biol. 2004;167:123–133. doi: 10.1083/jcb.200403067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alexakis C., Partridge T., Bou-Gharios G. Implication of the satellite cell in dystrophic muscle fibrosis: a self-perpetuating mechanism of collagen overproduction. Am. J. Physiol. Cell Physiol. 2007;293:C661–669. doi: 10.1152/ajpcell.00061.2007. [DOI] [PubMed] [Google Scholar]
- 32.Frazer K.A., et al. A sequence-based variation map of 8.27 million SNPs in inbred mouse strains. Nature. 2007;448:1050–1053. doi: 10.1038/nature06067. [DOI] [PubMed] [Google Scholar]
- 33.Durham W.J., Wehrens X.H., Sood S., Hamilton S.L. Diseases associated with altered ryanodine receptor activity. Subcell. Biochem. 2007;45:273–321. doi: 10.1007/978-1-4020-6191-2_10. [DOI] [PubMed] [Google Scholar]
- 34.Bellinger A.M., et al. Hypernitrosylated ryanodine receptor calcium release channels are leaky in dystrophic muscle. Nat. Med. 2009;15:325–330. doi: 10.1038/nm.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hersh C.P., et al. Genetic association analysis of functional impairment in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2006;173:977–984. doi: 10.1164/rccm.200509-1452OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dorfman R., et al. Complex two-gene modulation of lung disease severity in children with cystic fibrosis. J. Clin. Invest. 2008;118:1040–1049. doi: 10.1172/JCI33754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rozmahel R., et al. Modulation of disease severity in cystic fibrosis transmembrane conductance regulator deficient mice by a secondary genetic factor. Nat. Genet. 1996;12:280–287. doi: 10.1038/ng0396-280. [DOI] [PubMed] [Google Scholar]
- 38.Drumm M.L., et al. Genetic modifiers of lung disease in cystic fibrosis. N. Engl. J. Med. 2005;353:1443–1453. doi: 10.1056/NEJMoa051469. [DOI] [PubMed] [Google Scholar]
- 39.Meyer O.A., Tilson H.A., Byrd W.C., Riley M.T. A method for the routine assessment of fore- and hindlimb grip strength of rats and mice. Neurobehav. Toxicol. 1979;1:233–236. [PubMed] [Google Scholar]
- 40.Thurston G., et al. Leakage-resistant blood vessels in mice transgenically overexpressing angiopoietin-1. Science. 1999;286:2511–2514. doi: 10.1126/science.286.5449.2511. [DOI] [PubMed] [Google Scholar]
- 41.Flesch M., et al. Contractile systolic and diastolic dysfunction in renin-induced hypertensive cardiomyopathy. Hypertension. 1997;30:383–391. doi: 10.1161/01.hyp.30.3.383. [DOI] [PubMed] [Google Scholar]
- 42.Rando T.A., Blau H.M. Methods for myoblast transplantation. Methods Cell Biol. 1997;52:261–272. doi: 10.1016/s0091-679x(08)60382-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.