Abstract
African green monkeys (AGMs) infected with the AGM type of SIV (SIVagm) do not develop chronic immune activation and AIDS, despite viral loads similar to those detected in humans infected with HIV-1 and rhesus macaques (RMs) infected with the RM type of SIV (SIVmac). Because chronic immune activation drives progressive CD4+ T cell depletion and immune cell dysfunctions, factors that characterize disease progression, we sought to understand the molecular basis of this AGM phenotype. To this end, we longitudinally assessed the gene expression profiles of blood- and lymph node–derived CD4+ cells from AGMs and RMs in response to SIVagm and SIVmac infection, respectively, using a genomic microarray platform. The molecular signature of acute infection was characterized, in both species, by strong upregulation of type I IFN–stimulated genes (ISGs). ISG expression returned to basal levels after postinfection day 28 in AGMs but was sustained in RMs, especially in the lymph node–derived cells. We also found that SIVagm induced IFN-α production by AGM cells in vitro and that low IFN-α levels were sufficient to induce strong ISG responses. In conclusion, SIV infection triggered a rapid and strong IFN-α response in vivo in both AGMs and RMs, with this response being efficiently controlled only in AGMs, possibly as a result of active regulatory mechanisms.
Introduction
The molecular mechanisms underlying the pathogenesis of HIV infection are still not fully understood. HIV infection is characterized by progressive CD4+ T cell depletion and immune cells dysfunctions that are driven by chronic immune activation (1, 2). Interestingly, the level of immune activation in the early phase of HIV-1 infection is predictive of the disease outcome (3–5). In the early phase of infection, innate immune responses to the virus play a crucial role. One of the principal actors of the innate immune system during viral infections is the plasmacytoid dendritic cell (PDC). PDCs are professional IFN-I producers (6). IFN-I is of critical importance not only for its direct antiviral effect, but also because it triggers adaptive immunity (7–9). Upon exposure to HIV-1 in vitro, PDCs from healthy donors produce IFN-α and TNF-α, which, in turn, induce bystander myeloid dendritic cell maturation (10). In acute HIV-1 and SIVmac infections of humans and macaques, respectively, IFN-α is detected at high levels (11, 12). It is unclear, however, whether IFN-α has a beneficial or harmful effect in HIV-1 infection. On the one hand, IFN-α could be beneficial, as it has been shown to directly decrease HIV-1 replication in vitro, although only partially (13). Moreover, the IFN-α effect on promoting antiviral adaptive immune responses may also result in reduced virus replication in vivo. A beneficial effect of type I IFN is also suggested by the observation of higher levels of PDCs and IFN-α production by TLR9-stimulated PDCs in HIV-infected long-term nonprogressors (14). On the other hand, IFN-α levels and interferon-stimulated gene (ISG) expression are increased in progressor as compared with nonprogressor HIV-infected individuals (15, 16). IFN-α might exert deleterious effects through various mechanisms, including the upregulation of TNF-related apoptosis-inducing ligand (TRAIL), which may induce apoptosis of uninfected CD4+ T lymphocytes (17, 18). In addition, chronic high levels of IFN-α could also induce defects in the thymopoiesis and bias in T cell selection and contribute to the generalized immune activation (19).
It is of crucial importance to understand the role of IFN-α in the outcome of HIV infection. In this context, natural hosts of SIV, such as African green monkeys (AGMs) and sooty mangabeys (SMs), are of particular interest, since they do not display CD4+ T cell apoptosis and exhibit no chronic immune activation when infected with SIV, despite persistently high virus replication (20). Reminiscent of what has been reported for humans, contrasting data have been published on the role of IFN-I responses in the natural SIV hosts. Some studies reported substantially reduced innate immune responses to the virus that were hypothesized to be responsible for the lack of chronic T cell activation. For instance, PDCs of noninfected AGMs and SMs displayed only low levels of in vitro IFN-α production in response to TLR7 and/or TLR9 stimulation (21, 22). Moreover, plasma IFN-α levels during chronic infection were most often lower than in pathogenic SIVmac infection in macaques (18, 21, 23). In contrast, other studies provided evidence for strong innate responses during acute SIV infection of natural hosts. For example, PDCs from acutely infected AGMs produced high levels of IFN-α in response to TLR9 stimulation (21), and ISGs were highly expressed at day 10 after infection (p.i.) of AGMs with SIVagm (23).
To explain these discordant results, we have now conducted a comparative, longitudinal assessment of the regulation of IFN-α and ISGs in nonpathogenic (AGM) and pathogenic (rhesus macaque [RM]) SIV infection by using whole-genome expression analysis. Of note, we included in our study very early time points p.i. in order to gain important insight into the innate response. Since CD4+ cells are central to AIDS pathogenesis, we focused our attention on these cells and studied their gene expression profile in both peripheral blood and LNs, i.e., the site where the immune responses are induced, shaped, and synchronized. In particular, we assessed IFN-α production in SIVagm-infected AGMs and SIVmac-infected RMs. We used multiple blood samples collected at very short intervals in order to be able to identify time of peak IFN-α production. Since IFN-α responses are reflected at the molecular level by induction of ISGs, we assessed the expression profiles longitudinally and analyzed whether there is a correlation between the IFN-α and type I ISG levels in SIVagm infection. In addition, we developed functional assays to study the in vitro induction of ISGs by SIVagm and IFN-α in AGM cells, and, for the first time to our knowledge in a natural host species, cells from infected animals were tested for their capacity to produce IFN-α after in vitro stimulation with SIV. Taken together, our results revealed that SIVagm induces IFN-α production both in vitro and in vivo and that the lack of sustained ISG response in AGMs during chronic SIV infection is not due to a genetic inability to produce IFN-α but rather occurs as a result of negative regulatory mechanisms.
Results
Viral replication and disease outcome.
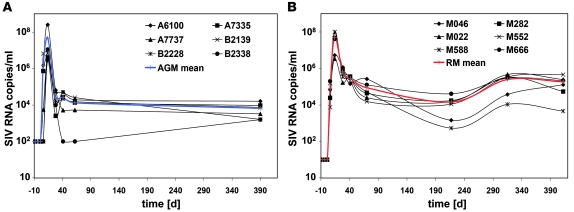
Six SIVagm-infected AGMs and 6 SIVmac-infected RMs were sampled between days 1 and 592 p.i. (Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI40093DS1). As expected, based on previous studies (20, 24), plasma viral loads (PVLs) were similar in AGMs and RMs (Figure 1), and the peak of PVL coincided with a decline in CD4+ T cell levels in both species (Supplemental Figure 2). In RMs, peripheral blood CD4+ T cell counts remained low after acute infection and progressively declined to less than 200 cells/μl within 1.5 years after SIVmac infection (Supplemental Figure 2B). We next monitored T cell proliferation by using Ki-67 as a marker and found that, consistent with previous observations (25), acute SIVagm infection of AGMs is associated with a transient increase in the percentages of CD4+Ki-67+ and CD8+Ki-67+ T cells (Supplemental Figure 3, A and C) in blood and a modest and nonsignificant increase in LN (Supplemental Figure 4, A and C). RMs manifested more intense increases of CD4+Ki-67+ and CD8+Ki-67+ T cell percentages in blood (Supplemental Figure 3, B and D) and LN (Supplemental Figure 4, B and D).
Figure 1. Plasma viral loads.
Plasma SIV RNA copy numbers (cut off value: 0.5 × 102 to 1 × 102 copies/ml of plasma) in (A) AGMs and (B) RMs. Designations for individual animals are shown. The mean is indicated in blue for AGMs and in red for RMs.
Whole-genome expression analysis.
To investigate gene expression patterns for the whole genome, we have previously identified a human genomic microarray platform that is highly sensitive for simian RNA (26). We previously reported the expression of more than 13,000 genes in in vitro activated CD4+ cells of AGMs and RMs, i.e., a number similar to what we observed for human cells (26). Here, we analyzed gene expression profiles ex vivo. In order to obtain a robust baseline for each animal, blood and LNs were collected respectively at 4 and 1 or 2 time points before infection (Supplemental Figure 1). We analyzed gene expression in purified CD4+ cells and considered a change in the gene profile significant when at least 1 time point p.i. was significantly different (P < 0.05) from baseline. We found that, in LNs, more genes were significantly modulated in RMs (4,022 genes) as compared with AGMs (2,082 genes), results consistent with the notion that SIV-infected RMs have higher levels of immune activation than AGMs. Surprisingly, the opposite was observed in blood (3,053 genes in AGMs and 1,187 in RMs).
We then performed gene ontology analysis to identify the biological processes that were significantly modulated during acute infection (days 1–28 p.i.). Similar to what has been described for total cells (27, 28), most changes in the CD4+ cells from blood and LNs were associated with immune responses, cell cycle, cell death, signal transduction, and nucleic acid or protein metabolism (Supplemental Figure 5A). The number of differentially regulated signaling pathways in blood was increased in both species, but during the course of the infection, it increased further in RMs (16 pathways at day 115 p.i.), while in AGMs it decreased after acute infection (i.e., down to 9 pathways at day 115 p.i.; data not shown).
We next compared the gene expression profiles of IL10 and IFNG, as measured using the current microarray platform, to the profile of the same 2 genes as analyzed in a previous study by RT-PCR (24, 29). Consistent with the presence of an attenuated inflammatory profile in acute SIVagm infection (24), IL10 was increased to a greater degree in AGMs than in RMs (Supplemental Figure 6, A and B), while IFNG was upregulated only transiently in AGMs when compared with the more persistent upregulation in RMs (Supplemental Figure 6, E and F). These gene expression data were consistent with the plasma levels of the same proteins (Supplemental Figure 6, C, D, G, and H). We also compared the microarray data with the published findings on CD4+ T cell activation as assessed by immunophenotypic analysis. Consistent with the lack of CD4+ T cell apoptosis reported in nonpathogenic SIVagm infection of AGMs (30, 31), we found that proapoptotic genes, such as caspase-7 (CASP7), were induced only in RM CD4+ cells, whereas antiapoptotic genes such as BCL2 were found transiently upregulated during acute infection in AGM and not modulated in RM CD4+ cells (Supplemental Figure 7).
SIV infection induces ISG expression in AGMs and RMs.
The genes most intensely modulated by SIV infection were those associated with IFN-mediated immunity (Supplemental Figure 5B). For both RMs and AGMs, the strongest increases in gene expression were detected for the type I ISGs, such as MX1 and IP-10 (CXCL10) (Figure 2). To validate the microarray data, we performed RT-PCR, and a strong correlation was found between the 2 measurements, as shown for MX1 gene expression (Supplemental Figure 8A). To determine whether the upregulation of type I ISG transcripts correlated with their expression at the protein level, we next quantified proteins that are known to be induced by IFN-α, such as IP-10, in the plasma. The IP-10 gene transcription profile indeed correlated with the plasma IP-10 levels in AGMs and RMs (Supplemental Figure 8B).
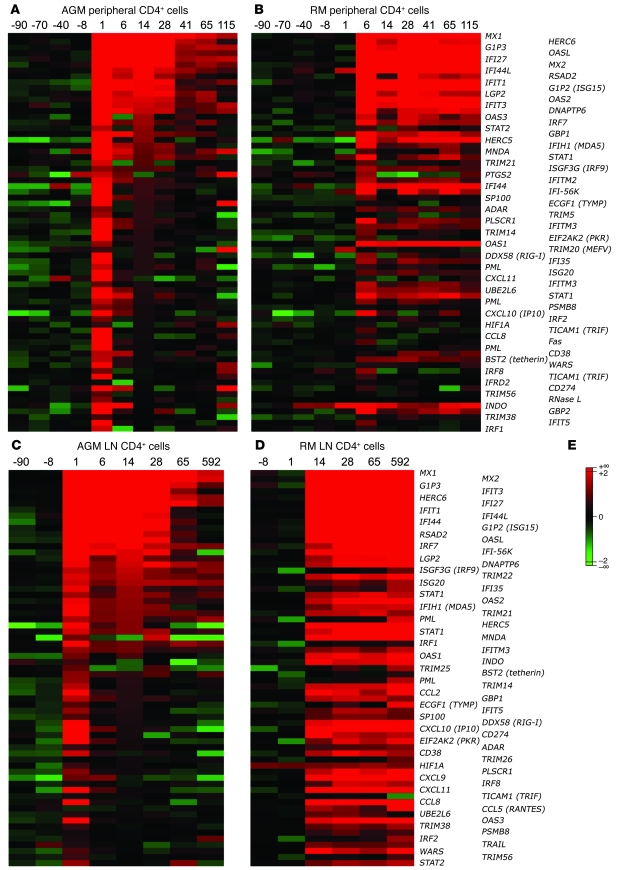
Figure 2. Type I ISG expression in blood and LN CD4+ cells.
The genes of the ISG cluster that were significantly regulated (P < 0.05) in at least 1 of the 2 species are represented here as heatmaps. Mean values of the log2Q of type I ISG expression in peripheral (A and B, respectively) and LN CD4+ cells (C and D, respectively) from 6 AGMs and 6 RMs are shown. Many of these genes can also be induced by IFN-II. (E) The color scheme indicates the log2Q. Gene expressions shown in red were upregulated, and those shown in green were downregulated.
The heatmaps shown in Figure 2 illustrate the high number of type I ISGs that are upregulated in response to SIV infection in both AGMs and RMs. Of note, the number of ISGs induced during acute infection was higher in AGMs’ blood CD4+ cells (67 in AGMs versus 43 in RMs), while it was higher in RMs within the LN CD4+ cells (63 in RMs versus 44 in AGMs). The most striking difference between the 2 species was observed during the transition from the acute to the chronic phase. Indeed, after day 28 p.i., the signal of most ISGs became progressively weaker in AGMs or even returned to baseline levels, while expression of the same genes remained sustained in RMs (Figure 2 and Supplemental Figure 9). In the RM LNs, most signals remained as high as during acute infection (Figure 2D). Thus, both species showed significant increases in type I ISG expression during acute infection, but after day 28 p.i., the expression of many ISGs returned to baseline levels in AGMs in contrast to RMs.
We also examined other genes that are involved in pattern recognition receptor (PRR) signaling or IFN-I signaling or are known to exert a positive or negative feedback on these pathways. The expression profiles observed for those genes were not different in peripheral CD4+ cells between the 2 species (Supplemental Figure 10, A and B), but it was again striking to observe that their expression was sustained in RM in contrast to AGM LN CD4+ cells (Supplemental Figure 10, C and D). Still, most type I ISGs induced by SIV infection were found in both species. For instance, IRF7, MDA5 (IFIH1), RIG-I (DDX58), and TRIF (TICAM1), which are all known to be involved in the transcription of type I ISGs (32), were similarly upregulated in AGMs and RMs (Figures 2 and 3 and Supplemental Table 1). Furthermore, the upregulation of a series of IFN-I signal transduction genes, such as STAT1, STAT2, and IRF9 (ISGF3), was also observed. Many other ISGs with known antiviral activity, such as PKR (EIF2AK2), ISG20, the OAS (2′5′-oligoadenylate synthetases) genes, the MX genes, ADAR1, TRIM5, and TRIM22 were also markedly induced, as were genes with known inhibitory activity on the IFN-I pathway, such as LGP2 and ISG15 (Figure 2). Importantly, there were, however, a few exceptions. For instance, TRAIL, CCL5 (RANTES), and IRF8 were upregulated only in the LNs of RMs. Moreover, the expression of several other chemokines was induced in LNs of AGMs but immediately controlled in contrast to RMs (CCL2, CCL8, CXCL9, CXCL11).
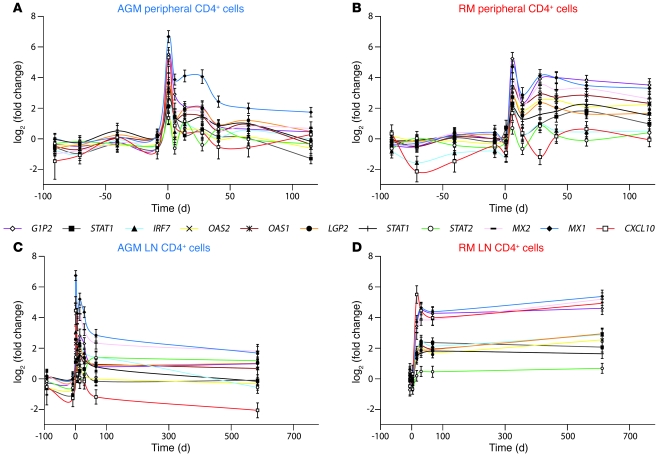
Figure 3. Expression profiles of representative type I ISGs.
Microarray gene expression profiles of 11 ISGs are shown for CD4+ cells in (A and B) blood and (C and D) LN from (A and C) AGMs and (B and D) RMs. Statistics associated with these profiles are provided in Supplemental Table 1.
Together, these data show that there was a strong induction of type I ISG during acute SIVagm infection, followed by an efficient downregulation. The kinetic analysis revealed that this downregulation was achieved after day 28 p.i. The tissue analysis demonstrated that in LNs, fewer genes were induced during nonpathogenic compared with pathogenic SIV infection.
Systemic IFN-I production in AGMs.
To determine whether this ISG upregulation associated with acute SIV infection was caused by IFN-I, we first quantified IFN-α levels in plasma by using a highly sensitive functional assay (21). IFN-α was detected at day 1 p.i. in 4 of the 6 AGMs (>100 IU/ml) and at day 14 p.i. in 5 of them (12–63 IU/ml) (data not shown). In RMs, IFN-α was detected in 5 of 6 animals at day 6 p.i. (15–250 IU/ml) and in 3 animals at day 14 p.i. (15–150 IU/ml) (data not shown). The IFN-α levels detected at days 6 and 14 p.i. were relatively low in both species. Of note, the earlier detection of IFN-α in AGMs is consistent with the earlier induction of ISGs in these animals.
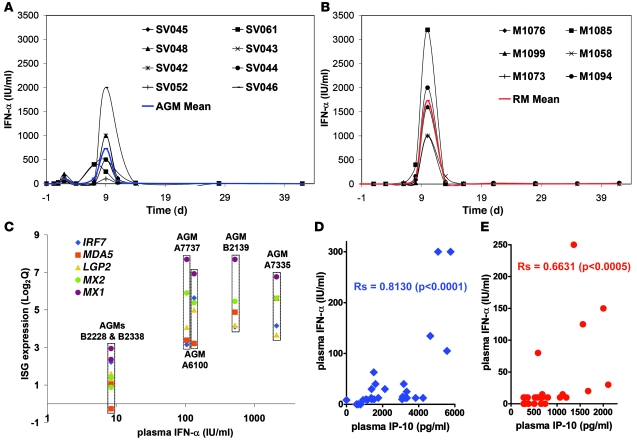
We then reasoned that by sampling only every 6–8 days, we probably missed the peak of IFN-α production, since we and others have shown that the peak of plasma IFN-α is close to the time of peak PVL, which in these experimental infections usually occurs between days 8 and 12 p.i. (12, 21, 33). We therefore decided to include additional animals in the study (8 AGMs, 6 RMs) and measured IFN-α on a tighter schedule including multiple time points close to the peak PVL. The animals were of the same origin and were infected according to the same protocol used for the animals included in the microarray study. In AGMs, a first small increase in IFN-α plasma levels was detected at day 2 p.i. (50–200 IU/ml) and a second, stronger increase at day 9 p.i. (100–2,000 IU/ml; median, 500 IU/ml), corresponding to the time of peak PVL (Figure 4A). In RMs, a clear peak IFN-I was observed at day 10 p.i., i.e., also close to the peak PVL (1,000–3,200 IU/ml; median, 1,600 IU/ml) (Figure 4B). Importantly, the fact that some AGMs showed levels similar to those in some of the RMs indicates that these natural SIV hosts have a preserved ability to produce this cytokine in response to SIVagm infection in vivo (21). On average, the IFN-α plasma levels at the peak were significantly lower in AGMs than in RMs (P < 0.05). This observation is to be interpreted with caution, since the lower levels of IFN-α production observed in AGMs were still associated with an upregulation of the ISGs that was as strong as that in RMs.
Figure 4. Systemic IFN-α levels and correlation with type I ISG expression and plasma IP-10 quantities.
Levels of bioactive IFN-α in plasma of (A) 8 additional AGMs and (B) 6 additional RMs. The detection level of the assay was 2 IU/ml. (C) Expression levels of 5 upregulated ISGs at day 1 p.i. were plotted against respective plasma IFN-α levels for the 6 AGMs included in the microarray analysis. Correlation between IFN-α and IP-10 plasma levels in (D) AGMs (blue) and (E) RMs (red) during acute infection (days 1–28). The Spearman coefficient (Rs) is indicated.
The IFN-α levels in plasma of the SIV-infected AGMs included in the microarray study correlated with the type I ISG expression levels observed in both blood and LNs during acute SIVagm infection (Supplemental Table 2). In particular, the 4 AGMs (A7737, A6100, B2139, A7335) that displayed the highest plasma IFN-α levels were those with the highest ISG expression ratios, in both blood (Figure 4C) and LN (data not shown). Moreover, the plasma levels of IP-10 correlated positively with plasma levels of IFN-α in both species (Figure 4D).
Production of IFN-α and ISGs by AGM peripheral mononuclear cells.
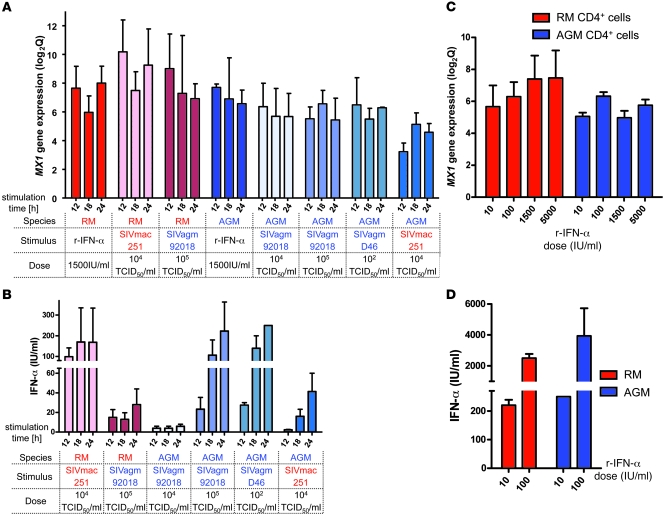
We then developed a functional assay to study whether SIVagm is capable of inducing type I ISGs in AGM cells via IFN-α. First, we stimulated PBMCs from AGMs and RMs with SIVagm and SIVmac and quantified ISG expression in CD4+ cells, thus enabling us to analyze the same type of cells that were studied in the microarray experiments. To this end, CD4+ and CD4– cells from healthy AGMs and RMs were cultured in 2 distinct chambers. RNA was extracted from the CD4+ cells, and ISGs, such as MX1, were quantified. As a positive control for stimulation, we used recombinant IFN-α. This cytokine induced MX1 gene expression in AGM and RM CD4+ cells to similar levels (Figure 5A). In these experiments, AGM cells were then stimulated with 2 species-specific isolates: the isolate used here for the in vivo infections (SIVagm.sab92018) and a nonrelated isolate, SIVagm.sabD46 (34). RM cells were stimulated with the same virus used for the in vivo infections (SIVmac251). Importantly, MX1 gene expression was induced in CD4+ cells of both species after stimulation with SIV (Figure 5A). This was also true for other ISGs tested, such as TRAIL (Supplemental Figure 11 and data not shown).
Figure 5. In vitro induction of type I ISG and IFN-α.
Peripheral CD4+ cells of uninfected AGMs (blue) and RMs (red) were cultured in the presence of the CD4– cell fraction in a 2-chamber system. (A and B) Cells were incubated for 12, 18, and 24 hours with SIV or 1,500 IU/ml of human recombinant IFN-α. Viral infectious titers used are indicated. (C and D) Cells were incubated with a range of doses of recombinant IFN-α. (A and C) For MX1 gene expression, the mean ± SEM of log2Q is shown. (B and D) Means of the amounts of bioactive IFN-α produced in supernatants are represented (±SEM). The means were calculated from 3 independent experiments with cells from 3 different animals of each species.
This experiment demonstrates that SIV induces type I ISG expression in both RM and AGM cells. To study whether this ISG upregulation could be mediated by IFN-α, we next determined IFN-α concentration in the supernatants. No IFN-α was found in the supernatants of nonstimulated cells, while it was detected in all supernatants from SIV-stimulated cells, albeit at variable levels (Figure 5B). Of note, the levels of IFN-α produced by AGM PBMCs depended on the viral dose, as well as on the viral isolate (Figure 5B). A higher IFN-α production was observed when AGM PBMCs were stimulated with the autologous virus (SIVagm.sab92018) compared with a heterologous virus (SIVmac251), and, analogously, IFN-α production by RM cells was more efficient after stimulation with SIVmac251 than with SIVagm.sab92018. The quantities of IFN-α produced were thus higher when cells were stimulated with their own species-specific virus rather than with a heterologous virus.
Low quantities of IFN-α induce strong ISG expression in AGM cells.
The difference in IFN-α production after cross-species stimulations with SIV was not observed at the level of MX1 expression (Figure 5A), and, in fact, strong MX1 gene expression was observed even in cultures in which low levels of IFN-α were detected (4 IU/ml or less). While we cannot exclude that another soluble factor is contributing to type I ISG induction in this in vitro assay, it is possible that ISG expression is actually very sensitive to IFN-I and that even low quantities of IFN-α efficiently induce ISG expression. In order to test this hypothesis, we performed a dose response assay in which PBMCs were stimulated with different concentrations of IFN-α. Already at the lowest dose of IFN-α (10 IU/ml), we observed a strong induction of MX1 gene expression in both AGM and RM CD4+ cells (Figure 5C). Thus, low levels of IFN-α were sufficient to induce high ISG expression.
Interestingly, the exposure of AGM and RM PBMCs to exogenous IFN-α also enhanced IFN-α production (Figure 5D). Indeed, IFN-α production increased more than 20-fold in supernatants of cells cultured with 100 IU/ml or 10 IU/ml of exogenous IFN-α (160–250 IU/ml after stimulation with 10 IU/ml and 1,800–7,500 IU/ml after stimulation with 100 IU/ml after 12–24 hours of culture). In the mock-treated cultures, we did not detect any IFN-α. This finding demonstrates the existence of a positive feedback loop, suggesting that low levels of IFN-α can significantly amplify the IFN-α response in vivo.
Repeated stimulations with SIV or IFN-α induce ISG expression.
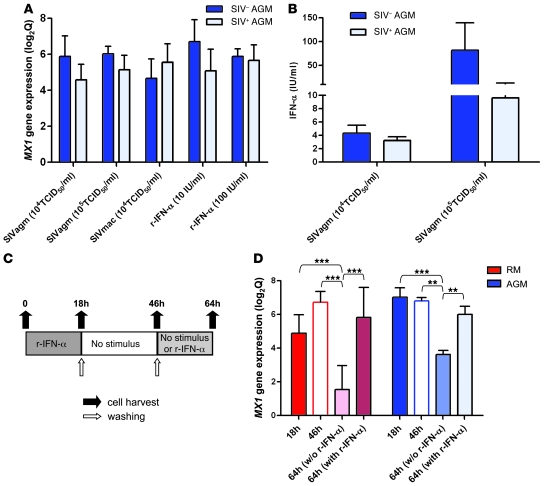
We next wondered which potential mechanisms responsible for downregulation of type I ISG response may be active in AGMs during the transition from acute to chronic SIV infection. We thus investigated whether the more effective control of the IFN-I activation that we observed in AGMs is related to an intrinsic resistance to repeated stimulation. To this end, we first stimulated cells of chronically SIV-infected AGMs, using the same protocol that was previously applied to samples from SIV-uninfected AGMs, and tested whether previous in vivo exposure to SIV or other factors may have desensitized the cells. Of note, all used stimuli (SIVagm, SIVmac, or IFN-α) induced MX1 gene expression to levels similar to those observed in cells of noninfected AGMs (Figure 6A). In addition, IFN-α was also produced by cells of chronically infected AGMs (Figure 6B). We then used a second approach that consisted of quantification of ISG expression in PBMCs from healthy AGMs and RMs stimulated 2 times with IFN-α, as depicted in Figure 6C. As in previous experiments, MX1 gene expression was induced by IFN-α in both species after 18 hours of stimulation (Figure 6D). After an additional 28-hour interval without stimulation, the expression level of MX1 remained stable, and it was only after another 18 hours without stimulus (for a total of 64 hours) that the expression of MX1 was significantly decreased. However, restimulation with IFN-α resulted in a significant increase in MX1 gene expression at 64 hours compared with the un-restimulated cells in both species. While these experiments cannot rule out the possibility that AGM cells that were prestimulated are less sensitive to a second stimulus than nonexposed cells, they clearly suggest that AGM PBMCs are not more refractory to restimulation than RM cells.
Figure 6. In vitro restimulation of AGM cells.
(A and B) As in Figure 5, peripheral CD4+ cells of 5 uninfected and 6 chronically infected AGMs were cultured in the presence of the CD4– cell fraction for 18 hours with SIV or IFN-α. (A) MX1 gene expression (mean ± SEM of log2Q). (B) Mean ± SEM of bioactive IFN-α produced in supernatants. The difference in IFN-α production between SIV+ and SIV– AGMs was not significant. (C and D) Repeated treatments with recombinant IFN-α were conducted on PBMCs from 3 healthy AGMs and 2 healthy RMs as described in Methods and shown in C. (D) MX1 gene expression. **P < 0.01, ***P < 0.001.
Immunosuppressive genes.
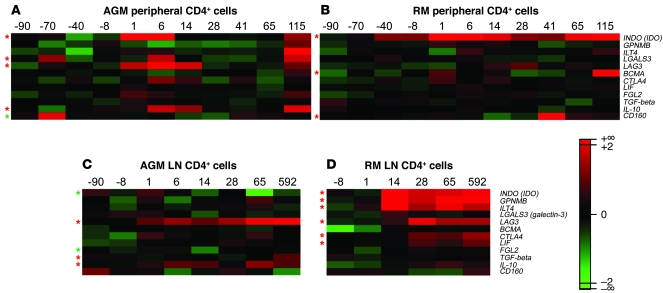
In an attempt to identify alternative mechanisms involved in the efficient control of ISG responses in chronically SIV-infected AGMs, we searched for differences in the expression profiles of genes coding for immunosuppressive proteins. In blood, AGM, but not RM, CD4+ cells showed increases in LAG3, LGALS3 (galectin-3), and IL10 expression. In LNs, LAG3 and IL10 were also increased in AGMs, while RM CD4+ cells displayed chronic increases of other genes, such as CTLA4 (Figure 7).
Figure 7. Microarray results for immunosuppressive genes in blood and LN CD4+ cells of SIVagm-infected AGMs and SIVmac-infected RMs.
Heatmaps of expression profiles of immunosuppressive genes are shown for AGM and RM peripheral CD4+ cells (A and B, respectively) and LN CD4+ cells (C and D, respectively). The color scheme indicates the log2Q. Gene expressions shown in red were upregulated and those shown in green were downregulated with respect to baseline levels. Red asterisks indicate probes found to be significantly increased, and green asterisks those significantly decreased after infection as compared with before infection (P < 0.05).
Discussion
The outcome of HIV/SIV infections is critically dependent on the early events following the initial encounter of the virus with its host cells (35). Innate immune responses are initiated immediately after viral infections. To investigate the events that take place during the first days and weeks p.i., we took advantage of our validated comparative nonhuman primate models comparing pathogenic (RM) with nonpathogenic (AGM) SIV infection. While previous studies reported gene expression data on total cells (23, 27, 28), we focused our current study on CD4+ cells, which are the target cells of the virus and play a central role in AIDS pathophysiology. Crucially, we sampled the animals at short time intervals during the acute and post-acute phase of SIV infection, which allowed us to identify the time point at which regulatory mechanisms become active. We were particularly interested in the regulation of the IFN-I response in the major site of immune education, i.e., secondary lymphoid organs (LNs).
Our assessment of the transcriptional profile indicated that type I ISGs (44–67 genes) were strongly induced in CD4+ cells of blood and LNs during acute SIV infection of both AGMs and RMs. Among these genes were those involved in IFN-α production (such as IRF7), IFN signaling (such as STAT1), and antiviral factors (such as MX1 and members of the TRIM family).
ISGs are excellent surrogate markers of IFN activity. Our study revealed that ISGs can be induced in AGM and RM CD4+ cells in vitro by very low levels of IFN-α. This is an interesting finding, since it reconciles previous reports on low in vitro IFN-α production by healthy AGMs and SMs (21, 22). Our data demonstrate that low levels are not incompatible with a strong ISG induction.
Although in both species, the in vivo induction of ISGs was strong and broad in terms of the number of genes that were upregulated, we also detected some important species-specific differences. In particular, we noticed differences related to the tissue analyzed: in blood, more genes were induced in AGMs than in RMs, while the opposite was the case in LN, where more genes were significantly modulated in RMs. The reasons underlying this pattern of tissue-specific gene expression are still unclear but may be related to the higher viral loads in the LN during pathogenic versus nonpathogenic SIV infection (36, 37) and/or to a sequestration of activated CD4+ T cells in RM LN. Indeed, activated T cells are drastically increased in RM LNs while rare in AGM LNs (24, 31). Accordingly, we found that only CD4+ cells from RM LN permanently upregulated the expression of the IFN-I–inducible chemokines IP-10, CCL5, and CXCL11, which are known to be involved in the homing of memory T cells to LN or inflammatory sites (38). This could contribute, in RMs, to a prolonged retention of CD4+ T cells in the LN. Another gene that was also differentially expressed between the 2 species was TRAIL, which was only increased in RM LN CD4+ cells. TRAIL has been associated specifically with CD4+ T cell apoptosis (17, 18), and, in line with this notion, AGM LNs are characterized by a lack of CD4+ T cell apoptosis (31). Since in vitro IFN-α–stimulated AGM PBMCs upregulate TRAIL expression, AGMs seem to control TRAIL expression in vivo by an as-yet-unknown mechanism. Together, our results revealed that many type I ISGs were induced in both AGMs and RMs during acute SIV infection, even though the tissue distribution, as well as the expression profiles of some specific genes, was different in the 2 species.
Another major difference between AGMs and RMs was that most type I ISGs that were induced during acute SIV infection in AGMs were downmodulated during the transition toward the chronic phase, while in RMs, the same genes remained upregulated throughout the course of infection. Indeed, our kinetic analysis revealed that the downregulation in AGMs started to be significant after day 28 p.i. and was then persistent throughout the follow-up (until day 115 p.i. in blood and day 592 p.i. in LNs).
Importantly, a similarly strong but transient upregulation of many ISGs was found during SIV infection of another natural host, the SM (39), suggesting that the activation of effective immunomodulatory mechanisms that downregulate the initial type I IFN response to SIV is a common feature of these 2 species, and possibly an even more common phenomenon among monkeys that experience a nonpathogenic infection. Instead, the observation that the upregulation of ISGs upon SIV infection was more rapid in AGMs than in RMs was not repeated in the parallel comparative study of SMs and RMs, suggesting that the rapidity of ISG induction is unlikely to play a key role in causing the nonpathogenic phenotype of natural SIV infections. The fact that, in both AGMs and SMs, the acute phase was associated with a strong upregulation of ISGs is consistent with our previous study showing that upon TLR9 stimulation, blood and LN PDCs collected during acute SIVagm infection were able to produce high amounts of IFN-α (21). In addition, our functional assays demonstrate that SIV can induce IFN-α and ISGs in AGM cells. Although the IFN-α levels were somewhat lower than those observed in RMs, our data demonstrate that the IFN-I pathway is fully functional in natural hosts. The data here and the results of the parallel study conducted in SIV-infected SMs contradict the hypothesis that an inborn defect in the innate immunity, and particularly PDCs, in responding to viral infections is the key driver of the different levels of immune activation in pathogenic versus nonpathogenic SIV infections (22). In fact, both AGMs and SMs are clearly able to mount an in vivo innate immune response to SIV, with one key difference: that this IFN-I response is transient in the 2 natural SIV hosts but persistent in RMs. While the mechanisms underlying this divergent phenotype during chronic SIV infection remain unclear, it is highly unlikely that they simply reflect a genetic inability of AGM or SM PDCs to produce IFN-I in response to viral TLR ligands (22).
We show here that IFN-α enhances IFN-α production in AGM cells in vitro. It is not clear which cells produced the IFN-α in our functional assays. Nonetheless, this positive feedback loop could explain our previous data showing that PDCs collected during acute SIVagm infection have the highest capacity to produce IFN-α shortly after the in vivo peak IFN-α (21).
Elucidation of the mechanisms responsible for the downregulation of the IFN-I pathway in the post-acute phase of SIV infection of natural hosts is the focus of intense studies, as these mechanisms of active downregulation might play a major role in protection against harmful chronic immune activation observed in pathogenic HIV and SIV infections. As discussed above, in the context of a normal ability to produce IFN-α, and similar PVL in both species, the downregulation of the IFN-α response in AGMs is very likely the result of negative control mechanisms. Of note, the fact that AGM cells from SIV-infected animals can be repeatedly stimulated in vitro to produce IFN-α and upregulate ISGs suggests that the downregulation of type I ISG expression during chronic SIVagm infection is not related to a stronger resistance to restimulation in AGM compared with RM cells.
In the course of a normal antiviral immune response, negative regulatory mechanisms are generally induced in order to return to the preinfection state and avoid the harmful effects of chronic immune activation (40). In blood-derived CD4+ cells of AGMs, we detected early increases in IDO, LAG3, IL10, and LGALS3 gene expression upon SIVagm infection. On the other hand, we also observed that more immunosuppressive genes were persistently induced in RM LN CD4+ cells. This apparently paradoxical finding is actually consistent with the possibility that a migration of regulatory T cells to LNs of SIV-infected RMs occurs as a consequence of ongoing inflammation (41). The key question, however, is why these negative regulatory mechanisms would be successful in downmodulating immune activation in AGMs but not in RMs. In this regard, it was interesting that RMs upregulated the expression of IRF8, which can further enhance IFN-I production as part of a positive feedback regulation mechanism (42) in LN CD4+ cells of RMs. Indeed, the ISG profiles in RMs recall the exaggerated inflammation seen in highly pathogenic influenza strains (43). In line with this observation, a recent report demonstrated that the inflammation in early HIV-1 infection is considerably stronger than in HCV and HBV infections (11). Collectively, these data support the hypothesis that there is an exacerbated activation of innate immunity in pathogenic HIV/SIVmac infections (44), which might then overwhelm and inhibit normal control mechanisms (45). In contrast, in AGMs, a differential expression of genes in the LNs was observed. In particular, several chemokines were not induced or were immediately controlled in AGM LNs, while upregulation of negative regulators, such as ISG15 and other immunosuppressive genes, was observed. Thus, in AGM, the gene expression pattern between the tissue compartments might indicate a more coordinated activation of the inflammatory and antiinflammatory responses that leads to the efficient control of the immune activation in the post-acute phase.
In conclusion, our study indicates that SIV triggers a strong and rapid innate response in vivo in AGMs. Since AGM cells are able to produce IFN-I, the transient ISG response in these animals is likely the result of active regulatory mechanisms that are lacking or are inhibited in SIV-infected RMs. Our study highlights the need to decipher the factors causing a better coordination of immune responses in AGMs. Elucidation of these mechanisms may lead to the identification of novel candidates for strategies aimed at blocking the harmful chronic immune activation that is associated with progression to AIDS in HIV-infected individuals.
Methods
Animals and infections.
The Central Committee for Animals at Institut Pasteur reviewed and approved the use and care of animals. Chinese RMs (Macaca mulatta) were intravenously infected with 50 AID50 (50% animal infectious dose) of the cell-free SIVmac251 stock provided by A.M. Aubertin (Université Louis Pasteur, Strasbourg, France) and AGMs (Chlorocebus sabaeus) with 250 TCID50 (50% tissue culture infectious dose) of SIVagm.sab92018 as previously described (37). These doses were chosen as they give the same PVL during acute infection in both models.
Sample processing.
Whole blood was collected from 6 AGMs and 6 RMs before infection (days –90, –70, –40, and –8), during acute infection (days 1, 6, 14, and 28), and during the chronic phase (days 41, 65, and 115) (Supplemental Figure 1). Peripheral LNs were obtained by excision before (days –90, –8 for AGMs and –8 for RMs) and after infection (days 1, 6, 14, 28, 65, and 592 for AGMs and days 1, 14, 28, 65 and day of euthanasia for RMs). The CD4+ cells were isolated from PBMCs and LN mononuclear cells using magnetic beads (Miltenyi Biotec). The fractions routinely had a purity of greater than 95% CD3+CD4+ T cells as described previously (26).
Viral load.
PVL was determined by real-time PCR based on amplification of LTR RNA (24).
Cytokine quantifications.
Titers of bioactive IFN-α were determined as described previously (21, 33). Other cytokines were quantified using Luminex (46) and ELISA technologies (24).
Flow cytometry.
We used a panel of mAbs that were originally designed to detect human molecules but that we and others have shown to be cross-reactive with AGMs and RMs (31, 47, 48): CD3–Alexa Fluor 488 (SP34-2; BD Biosciences), CD4-PerCP (L200; BD), CD8-PeCy7 (Sk1; BD), Ki-67–FITC (Ki-67 [Dako] for RMs and B56 [BD] for AGMs), as well as isotype controls.
RNA extraction.
Total RNA was extracted using the RNeasy Mini extraction Kit (QIAGEN), and the quality and concentration were assessed as before (26).
Microarray proceedings.
RNA was analyzed on the ABI Human Whole Genome Arrays v2.0 (Applied Biosystems) as previously described (26). One single round of linear amplification was performed from 600 ng of total RNA. cDNA synthesis, in vitro transcription, amplification, fragmentation, hybridization, staining, and scanning were performed as directed by the suppliers. To avoid any bias, the order of the 216 sample hybridizations was calculated by random assignment.
Data analysis.
We previously described acute data analysis of ABI arrays (26, 49). Briefly, normalization was achieved using the NeONORM method (50). Significance of log2 (fold change) (designated “log2Q”) was determined based on a mixture lognormal distribution hypothesis of signal intensities using mixture ANOVA methodology (51). Changes in gene expression profiles were considered significant when at least 1 time-point was different from the baseline (P < 0.05). Heatmaps were visualized using MultiExperiment Viewer software (TM4) (52). Gene ontology (GO) annotations were analyzed using the Panther Protein Classification System (53). Methods used are explained in more detail in Supplemental Methods. Correlations were determined by using the Spearman rank method.
Quantification of cellular mRNA.
Quantification of ISG transcripts was performed by real-time RT-PCR in triplicate using TaqMan gene expression assays (Applied Biosystems). The expression of each gene was normalized against that of 18S rRNA (24).
Stimulation assays.
Cells were cultured in a 2-chamber system (Transwell; Corning): 4.5 × 105 CD4– cells were placed in the bottom wells and 1.8 × 105 CD4+ cells in the top wells in the presence of SIV, human recombinant IFN-α2 (IntronA, Schering-Plough), or a mock agent. After 12, 18, and 24 hours of culture, each cell fraction was collected and total RNA extracted from CD4+ cells as described above. In restimulation assays, PBMCs were incubated with human recombinant IFN-α2 or a mock agent for 18 hours. After 18 hours, the cells were washed, cultured for another 28 hours, and then treated again or not with IFN-α2 for 18 hours. Cells were harvested at time points shown in Figure 6C, and total RNA was extracted. MX1 transcripts were quantified by real-time PCR as described above. Transcript levels were normalized against those found in mock-treated cells for the same time point. The fold changes were then calculated relative to the MX1 transcript levels before any treatment.
Accession numbers.
The transcriptome data have been annotated following MIAME standards I and II and were deposited in the transcriptome data repository MACE (http://mace.ihes.fr). The 2 datasets are available under accession numbers 3070984318 (AGM) and 2932572286 (RM).
Statistics.
The methods used for analysis of microarray data are described above in Data analysis. To compare the peak IFN-γ levels in the 2 species, a 2-tailed Mann-Whitney U test was used (P < 0.05). To compare the gene expression levels within the same species and same animals after distinct in vitro stimulations, a 2-way ANOVA methodology was used. Correlations were estimated with a Spearman test. P values are indicated in the respective figures.
Supplementary Material
Acknowledgments
We are grateful to Roger Le Grand, Benoît Delache, Christophe Joubert, Xavier Montagutelli, Pascal Lavedan, and the staff of the CEA and Institut Pasteur animal facilities for excellent work in care of the animals used in this study. We thank Mickaël Ploquin and Bruno Vaslin for helpful discussions. This study was supported by grants from Institut Pasteur (GPH no. 2) and ANRS (no. 03/172). Work in the Benecke group was funded by the “Institut des Hautes ωtudes Scientifiques,” the CNRS, the Genopole Evry, and the ANRS. Nonhuman cytokine detection was supported by NIH grant P51 RR013986. D. Kunkel and G. Petitjean received fellowships from ANRS and A-S. Liovat received a fellowship from the “Ministère de l’Enseignement Supérieur et de la Recherche.”
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 119:3544–3555 (2009). doi:10.1172/JCI40093.
See the related Commentary beginning on page 3512.
References
- 1.Giorgi J.V., et al. Shorter survival in advanced human immunodeficiency virus type 1 infection is more closely associated with T lymphocyte activation than with plasma virus burden or virus chemokine coreceptor usage. J. Infect. Dis. 1999;179:859–870. doi: 10.1086/314660. [DOI] [PubMed] [Google Scholar]
- 2.Sousa A.E., Carneiro J., Meier-Schellersheim M., Grossman Z., Victorino R.M. CD4 T cell depletion is linked directly to immune activation in the pathogenesis of HIV-1 and HIV-2 but only indirectly to the viral load. J. Immunol. 2002;169:3400–3406. doi: 10.4049/jimmunol.169.6.3400. [DOI] [PubMed] [Google Scholar]
- 3.Deeks S.G., et al. Immune activation set point during early HIV infection predicts subsequent CD4+ T-cell changes independent of viral load. Blood. 2004;104:942–947. doi: 10.1182/blood-2003-09-3333. [DOI] [PubMed] [Google Scholar]
- 4.Hazenberg M.D., et al. Persistent immune activation in HIV-1 infection is associated with progression to AIDS. AIDS. 2003;17:1881–1888. doi: 10.1097/00002030-200309050-00006. [DOI] [PubMed] [Google Scholar]
- 5.van Asten L., et al. Pre-seroconversion immune status predicts the rate of CD4 T cell decline following HIV infection. AIDS. 2004;18:1885–1893. doi: 10.1097/00002030-200409240-00004. [DOI] [PubMed] [Google Scholar]
- 6.Siegal F.P., et al. The nature of the principal type 1 interferon-producing cells in human blood. Science. 1999;284:1835–1837. doi: 10.1126/science.284.5421.1835. [DOI] [PubMed] [Google Scholar]
- 7.Colonna M., Trinchieri G., Liu Y.-J. Plasmacytoid dendritic cells in immunity. Nat. Immunol. 2004;5:1219. doi: 10.1038/ni1141. [DOI] [PubMed] [Google Scholar]
- 8.Jaehn P.S., Zaenker K.S., Schmitz J., Dzionek A. Functional dichotomy of plasmacytoid dendritic cells: antigen-specific activation of T cells versus production of type I interferon. Eur. J. Immunol. 2008;38:1822–1832. doi: 10.1002/eji.200737552. [DOI] [PubMed] [Google Scholar]
- 9.McKenna K., Beignon A.-S., Bhardwaj N. Plasmacytoid dendritic cells: linking innate and adaptive immunity. J. Virol. 2005;79:17–27. doi: 10.1128/JVI.79.1.17-27.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fonteneau J.-F., et al. Human immunodeficiency virus type 1 activates plasmacytoid dendritic cells and concomitantly induces the bystander maturation of myeloid dendritic cells. J. Virol. 2004;78:5223–5232. doi: 10.1128/JVI.78.10.5223-5232.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stacey A.R., et al. Induction of a striking systemic cytokine cascade prior to peak viraemia in acute human immunodeficiency virus type 1 infection, in contrast to more modest and delayed responses in acute hepatitis B and C virus infections. J. Virol. 2009;83:3719–3733. doi: 10.1128/JVI.01844-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malleret B., et al. Primary infection with simian immunodeficiency virus: plasmacytoid dendritic cell homing to lymph nodes, type I interferon, and immune suppression. Blood. 2008;112:4598–4608. doi: 10.1182/blood-2008-06-162651. [DOI] [PubMed] [Google Scholar]
- 13.Audige A., et al. Anti-HIV state but not apoptosis depends on IFN signature in CD4+ T cells. J. Immunol. 2006;177:6227–6237. doi: 10.4049/jimmunol.177.9.6227. [DOI] [PubMed] [Google Scholar]
- 14.Hosmalin A., Lebon P. Type I interferon production in HIV-infected patients. J. Leukoc. Biol. 2006;80:984–993. doi: 10.1189/jlb.0306154. [DOI] [PubMed] [Google Scholar]
- 15.Bosinger S.E., et al. Gene expression profiling of host response in models of acute HIV infection. J. Immunol. 2004;173:6858–6863. doi: 10.4049/jimmunol.173.11.6858. [DOI] [PubMed] [Google Scholar]
- 16.Hyrcza M.D., et al. Distinct transcriptional profiles in ex vivo CD4+ and CD8+ T cells are established early in human immunodeficiency virus type 1 infection and are characterized by a chronic interferon response as well as extensive transcriptional changes in CD8+ T cells. J. Virol. 2007;81:3477–3486. doi: 10.1128/JVI.01552-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herbeuval J.-P., Shearer G.M. HIV-1 immunopathogenesis: how good interferon turns bad. Clin. Immunol. 2007;123:121–128. doi: 10.1016/j.clim.2006.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meythaler M., et al. Differential CD4+ T-lymphocyte apoptosis and bystander T-Cell activation in rhesus macaques and sooty mangabeys during acute simian immunodeficiency virus infection. J. Virol. 2009;83:572–583. doi: 10.1128/JVI.01715-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keir M.E., et al. Generation of CD3+CD8low thymocytes in the HIV Type 1-infected thymus. J. Immunol. 2002;169:2788–2796. doi: 10.4049/jimmunol.169.5.2788. [DOI] [PubMed] [Google Scholar]
- 20.Liovat A.S., Jacquelin B., Ploquin M.J., Barre-Sinoussi F., Muller-Trutwin M.C. African non human primates infected by SIV - why don’t they get sick? Lessons from studies on the early phase of non-pathogenic SIV infection. Curr. HIV Res. 2009;7:39–50. doi: 10.2174/157016209787048546. [DOI] [PubMed] [Google Scholar]
- 21.Diop O.M., et al. Plasmacytoid dendritic cell dynamics and alpha interferon production during Simian immunodeficiency virus infection with a nonpathogenic outcome. J. Virol. 2008;82:5145–5152. doi: 10.1128/JVI.02433-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mandl J.N., et al. Divergent TLR7 and TLR9 signaling and type I interferon production distinguish pathogenic and nonpathogenic AIDS virus infections. Nat. Med. 2008;14:1077. doi: 10.1038/nm.1871. [DOI] [PubMed] [Google Scholar]
- 23.Favre D., et al. Critical loss of the balance between Th17 and T regulatory cell populations in pathogenic SIV infection. PLoS Pathog. 2009;5:e1000295. doi: 10.1371/journal.ppat.1000295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kornfeld C., et al. Antiinflammatory profiles during primary SIV infection in African green monkeys are associated with protection against AIDS. J. Clin. Invest. 2005;115:1082–1091. doi: 10.1172/JCI23006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lozano Reina J.M., et al. Gag p27-specific B- and T-cell responses in Simian immunodeficiency virus SIVagm-infected African green monkeys. J. Virol. 2009;83:2770–2777. doi: 10.1128/JVI.01841-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacquelin B., et al. Long oligonucleotide microarrays for African green monkey gene expression profile analysis. FASEB J. 2007;21:3262–3271. doi: 10.1096/fj.07-8271com. [DOI] [PubMed] [Google Scholar]
- 27.Lederer S., et al. Transcriptional profiling in pathogenic and non-pathogenic SIV infections reveals significant distinctions in kinetics and tissue compartmentalization. PLoS Pathog. 2009;5:e1000296. doi: 10.1371/journal.ppat.1000296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.George M.D., Sankaran S., Reay E., Gelli A.C., Dandekar S. High-throughput gene expression profiling indicates dysregulation of intestinal cell cycle mediators and growth factors during primary simian immunodeficiency virus infection. Virology. 2003;312:84–94. doi: 10.1016/S0042-6822(03)00207-1. [DOI] [PubMed] [Google Scholar]
- 29.Ploquin M.J., et al. Distinct expression profiles of TGF-beta1 signaling mediators in pathogenic SIVmac and non-pathogenic SIVagm infections. Retrovirology. 2006;3:37. doi: 10.1186/1742-4690-3-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Estaquier J., et al. Programmed cell death and AIDS: significance of T-cell apoptosis in pathogenic and nonpathogenic primate lentiviral infections. Proc. Natl. Acad. Sci. U. S. A. 1994;91:9431–9435. doi: 10.1073/pnas.91.20.9431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cumont M.C., et al. Early divergence in lymphoid tissue apoptosis between pathogenic and nonpathogenic simian immunodeficiency virus infections of nonhuman primates. J. Virol. 2008;82:1175–1184. doi: 10.1128/JVI.00450-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bonjardim C.A., Ferreira P.C., Kroon E.G. Interferons: signaling, antiviral and viral evasion. Immunol. Lett. 2009;122:1–11. doi: 10.1016/j.imlet.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khatissian E., et al. The relationship between the interferon alpha response and viral burden in primary SIV infection. AIDS Res. Hum. Retroviruses. 1996;12:1273–1278. doi: 10.1089/aid.1996.12.1273. [DOI] [PubMed] [Google Scholar]
- 34.Muller M.C., et al. Simian immunodeficiency viruses from central and western Africa: evidence for a new species-specific lentivirus in tantalus monkeys. J. Virol. 1993;67:1227–1235. doi: 10.1128/jvi.67.3.1227-1235.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Q., et al. Visualizing antigen-specific and infected cells in situ predicts outcomes in early viral infection. Science. 2009;323:1726–1729. doi: 10.1126/science.1168676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beer B., et al. Lack of dichotomy between virus load of peripheral blood and lymph nodes during long-term simian immunodeficiency virus infection of African green monkeys. Virology. 1996;219:367. doi: 10.1006/viro.1996.0262. [DOI] [PubMed] [Google Scholar]
- 37.Diop O.M., et al. High levels of viral replication during primary simian immunodeficiency virus SIVagm infection are rapidly and strongly controlled in African green monkeys. J. Virol. 2000;74:7538–7547. doi: 10.1128/JVI.74.16.7538-7547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Foley J.F., et al. Roles for CXC chemokine ligands 10 and 11 in recruiting CD4+ T cells to HIV-1-infected monocyte-derived macrophages, dendritic cells, and lymph nodes. J. Immunol. 2005;174:4892–4900. doi: 10.4049/jimmunol.174.8.4892. [DOI] [PubMed] [Google Scholar]
- 39.Bosinger S.E., et al. Global genomic analysis reveals rapid control of a robust innate response in SIV-infected sooty mangabeys. J. Clin. Invest. 2009;119:3556–3572. doi: 10.1172/JCI40115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O’Garra A., Vieira P.L., Vieira P., Goldfeld A.E. IL-10-producing and naturally occurring CD4+ Tregs: limiting collateral damage. J. Clin. Invest. 2004;114:1372–1378. doi: 10.1172/JCI23215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Estes J.D., et al. Premature induction of an immunosuppressive regulatory T cell response during acute simian immunodeficiency virus infection. J. Infect. Dis. 2006;193:703–712. doi: 10.1086/500368. [DOI] [PubMed] [Google Scholar]
- 42.Tailor P., et al. The feedback phase of type I interferon induction in dendritic cells requires interferon regulatory factor 8. Immunity. 2007;27:228–239. doi: 10.1016/j.immuni.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baskin C.R., et al. Early and sustained innate immune response defines pathology and death in nonhuman primates infected by highly pathogenic influenza virus. Proc. Natl. Acad. Sci. U. S. A. 2009;106:3455–3460. doi: 10.1073/pnas.0813234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boasso A., Shearer G.M. Chronic innate immune activation as a cause of HIV-1 immunopathogenesis. Clin. Immunol. 2008;126:235–242. doi: 10.1016/j.clim.2007.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Korn T., et al. IL-6 controls Th17 immunity in vivo by inhibiting the conversion of conventional T cells into Foxp3+ regulatory T cells. Proc. Natl. Acad. Sci. U. S. A. 2008;105:18460–18465. doi: 10.1073/pnas.0809850105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Giavedoni L.D. Simultaneous detection of multiple cytokines and chemokines from nonhuman primates using luminex technology. J. Immunol. Methods. 2005;301:89. doi: 10.1016/j.jim.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 47.Gordon S.N., et al. Severe depletion of mucosal CD4+ T cells in AIDS-free simian immunodeficiency virus-infected sooty mangabeys. J. Immunol. 2007;179:3026–3034. doi: 10.4049/jimmunol.179.5.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mortara L., et al. Phenotype and function of myeloid dendritic cells derived from African green monkey blood monocytes. J. Immunol. Methods. 2006;308:138. doi: 10.1016/j.jim.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 49.Noth S., Benecke A. Avoiding inconsistencies over time and tracking difficulties in Applied Biosystems AB1700/Panther probe-to-gene annotations. BMC Bioinformatics. 2005;6:307. doi: 10.1186/1471-2105-6-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Noth S., Brysbaert G., Benecke A. Normalization using weighted negative second order exponential error functions (NeONORM) provides robustness against asymmetries in comparative transcriptome profiles and avoids false calls. Genomics Proteomics Bioinformatics. 2006;4:90–109. doi: 10.1016/S1672-0229(06)60021-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Noth S., Brysbaert G., Pellay F., Benecke A. High-sensitivity transcriptome profiles exhibit distinct signal distribution properties having significant implications for data analysis and biologic interpretation. Genomics Proteomics Bioinformatics. 2006;4:212–229. doi: 10.1016/S1672-0229(07)60002-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saeed A.I., et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 53.Thomas P.D., et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.