Introduction
Left-ventricular hypertrophy (LVH) is one of the strongest independent predictors of cardiovascular morbidity and mortality(1-3) in the general population. Although both hypertension and obesity are well-established independent risk factors for the development of LVH, they explain less than 25-50% of the variance of left ventricular mass (LVM) in humans(4,5). A substantial body of evidence suggests that there is a genetic basis to the observed inter-individual variability in the susceptibility to the development of LVH(6-9). Given the continuous relationship between LVM and cardiovascular morbidity and mortality (10), elucidating the genetic determinants of inter-individual differences in the susceptibility to LVH is of considerable public health importance. It promises the opportunity to identify high-risk individuals for targeted intervention and may identify novel therapeutic targets for improved prevention and treatment strategies.
Evidence of a genetic contribution to the development of LVH
The Framingham investigators examined the genomics of left-ventricular remodeling in a Framingham study sample that included adults in the original Framingham study and the Framingham Offspring Study who were free of coronary artery disease, congestive heart failure, diabetes mellitus, renal insufficiency, heart disease related to cardiac valve disease, and severe ventricular hypertrophy. Intra-class correlations for left ventricular mass (LVM) among first degree relatives, second-degree relatives and unrelated spouse pairs were calculated to determine the contribution of heredity to the variability in LVM. After adjustments for age, height, weight and systolic blood pressure, the intra-class correlations between first-degree relatives were 0.15 (parent-child, P<0.0001), 0.16 (siblings, P<0.001); between second-degree relatives the contribution was 0.06 (P=NS) and between spouses it was 0.05 (P=NS). Overall, the estimated heritability of adjusted LVM was between 0.24 to 0.32 (9). Similar findings were reported from the Strong Heart Study (SHS), a population-based cohort survey of cardiovascular risk factors and prevalent cardiovascular disease in 13 American Indian tribes in the Southwestern United States. Baseline echocardiograms were analyzed in 1373 American Indian participants from 445 families to determine the heritability of LVM. After adjustment for sex, age, body weight, height, heart rate, medications and diabetes, the heritability of LVM was estimated to be 0.17 (P<0.05)(11). In another study of 376 normotensive Caucasian twin pairs (182 monozygotic and 194 dizygotic) aged 25-79 years, the adjusted estimated heritability of LVM was estimated at 0.59 (95% CI 0.50-0.67) suggesting a significant polygenic basis to the determination of LVM(12).
The natriuretic peptide system as a candidate gene system for susceptibility to LVH
Our research has focused upon genes critical to the signaling pathways of the natriuretic peptide system and explored the general hypothesis that genetic variation within these genes contributes to inter-individual susceptibility to hypertensive heart disease. In response to increases in cardiac volume or pressure, cardiomyocytes and fibroblasts synthesize and secrete atrial natriuretic peptide (ANP) and brain natriuretic peptide (BNP)(13,14). ANP and BNP are peptide hormones that circulate and activate the type A natriuretic peptide receptor (NPR-A) leading to the generation of cGMP pool(15) by activation of the particulate gunaylate cyclase domain of the NPR-A receptor. The NPR-A derived cGMP pool is compartmentalized and contributes to unique signaling pathways within the cardiomyocyte, in contradistinction to the signaling pthways activated by the cGMP pool derived from soluble guanylate cyclase activity, stimulated by NO-donors (16). The NPR-A receptor is abundantly expressed by cardiomyocytes, pulmonary and systemic vasculature, kidney, and the zona glomerulosa of the adrenal glands. The multiple biological actions of ANP and BNP within target tissues are best described as “compensatory” in nature and include: vasodilation of arteries and veins(17), opposing activation of the renin-angiotensin-aldosterone system (RAAS) (18) , enhancement of natriuresis by opposing distal tubule sodium reabsorption(19-21) and direct inhibition of endothelin release by the renal vascular endothelium (22). The vascular and neurohormonal actions of the NPS oppose elevations of systemic blood pressure, and murine gene knockout models have clearly demonstrated the demonstrating the importance of the NPS to blood-pressure regulation (15,23).
The NPS attenuated cardiac hypertrophy independent of its effect upon blood pressure
The natriuretic peptide system also functions as an autocrine and paracrine system within the heart that opposes the development of cardiac hypertrophy independent of the actions of the NPS on systemic blood pressure. The murine NPR-A knockout model demonstrated that mice with NPR-A deletion develop hypertension with an exaggerated cardiac hypertrophic response seemingly “out of proportion” to the increase in blood pressure(24). Subsequent experiments using sophisticated tissue-targeted deletion strategies have demonstrated that the NPS opposes cardiac hypertrophy in a “pressure-independent” manner. Kishimoto and colleagues demonstrated that over-expression of an NPR-A transgene in cardiomyocytes in wild-type or NPR-A knockout mice protected against cardiac hypertrophy despite no alteration in blood pressure(25). Holtwick and colleagues then demonstrated that cardiomyocyte-restricted disruption of the NPR-A receptor results in mice with a lower systemic blood pressure compared to wild-type littermates, however, despite the lower systemic blood pressure, the cardiomyocyte-restricted NPR-A deletion mice demonstrated increased cardiac hypertrophy as compared to wild-type littermates(26). Moreover, upon pressure-overload introduced by aortic banding, the cardiomyocyte-restricted NPR-A deletion mice demonstrated even greater increases in LVM compared to wild-type controls and significantly greater activation of the hypertrophic gene cascade. More recently, cardiac specific attenuation of the activity of NPR-A in transgenic mice produced cardiomyocyte restricted expression of a dominant negative mutation of the NPR-A receptor resulted in reduced myocardial cGMP levels, enhanced cardiac fibrosis and hypertrophy, and increased mortality when compared to the wild-type littermates(27). Thus, murine genetic models have clearly demonstrated that the local autocrine/paracrine actions of the NPS function to oppose the development of cardiac hypertrophy and fibrosis.
Molecular mechanisms of antihypertrophic and antifibrotic actions
The molecular mechanisms that underlie the cardiac antihypertrophic actions of NPS are multifactorial. The anti-hypertrophic actions of the NPS are dependent upon adequate stimulation of the NPR-A receptor expressed by cardiomyocytes, resulting in cGMP production by the particulate cyclase domain and secondary activation of cGMP-dependent protein kinase G (28). The downstream signaling effects are multifactorial and include interference with MAP kinase signaling cascades. In one study, ANP strongly induced expression of MAPK phosphatase-1 (MKP-1) and over expression of MKP-1 inhibited angiotensin II or endothelin 1-induced hypertrophic responses. More recent data has demonstrated that one of the major molecular mechanisms of the antihypertrophic actions of the NPS include the ability of the NPR-A to activate regulator of G-protein signaling, type 4, (RGS-4) via protein kinase G-mediated phosphorylation. This results in an attenuation of Galpha(q) signaling and its downstream hypertrophic signaling cascades, in particular, upstream components of the calcineurin-mediated hypertrophic gene cascade. (29). In addition to the NPS's attenuation of cardiomyocyte hypertrophic gene activation cascades, the NPS also exerts anti-fibrotic actions by interfering with TGF-beta signaling pathways(30), and enhancing the expression and activation of metalloproteinases from cardiac fibroblasts, while simultaneously inhibiting cardiac fibroblast collagen synthesis (31).
Corin as a candidate gene within the NPS signaling system
In order for the NPR-A receptor to become activated, ANP and BNP must first be adequately processed into biologically active peptides. For example, BNP is produced as a 126 amino acid precursor molecule, and after the secretion signal is removed, proBNP exists as an 108 amino acid molecule. ProBNP (BNP1-108) is cleaved in a highly sequence-specific manner into a 32 amino acid, carboxyl fragment (BNP-32) and a 76 amino acid amino terminal fragment (NT-BNP).
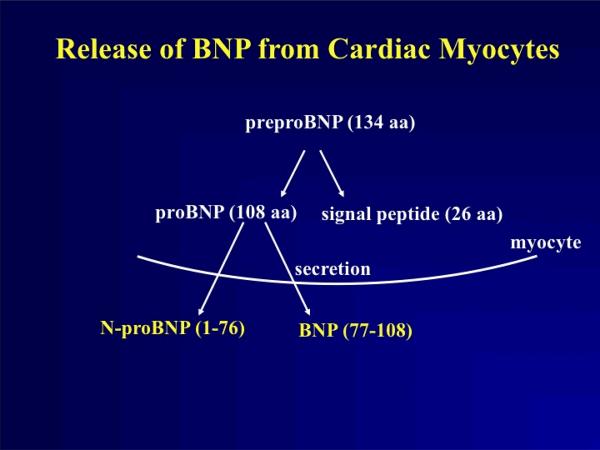
Recently the major enzyme responsible for the processing of proANP and proBNP has been identified to be corin, a type II trans-membrane serine protease predominantly expressed by cardiomyocytes(32); therefore, corin appears to be the long sought after “NP convertase”. The current paradigm suggests that after synthesis in ventricular cardiomyocytes, BNP is not stored, but rather targeted for secretion using the large dense-core vesicle (LDCV) pathway characteristic of neurotransmittor secretion and secretion of other peptide hormones(33). According to the present hypothesis, BNP 1-108 is then released from cardiomyocytes and interacts with corin's extracellular catalytic domain whereupon it is cleaved into BNP-32 and NT-BNP. (Figure 1) Even in the case of ANP that is stored in atrial secretory granules, the ANP stored in the secretory granules is thought to be unprocessed proANP, and corin is thiught to participates in extracellular proANP cleavage upon exocytosis from atrial secretory granules. Some investigators have also suggested that furin, a ubiquitously expressed serine endopeptidase, may participate in intracellular BNP processing, but this remains controversial and an active area of research(34,35). However, corin is clearly one of the most significant contributors to natriuretic peptide processing as evidenced by the murine corin knockout model(36). The corin knockout mice as compared to controls develop salt-sensitive hypertension and cardiac hypertrophy, with evidence of circulating unprocessed proANP. Administration of activated soluble corin results in a rapid decline in proANP, increase in plasma cGMP and lowering of systemic blood pressure.
Figure 1.
ProBNP is produced in cardiomyocytes where it is not stored but rapidly targeted for secretion after removal of the targeting sequence. Our current paradigm suggests that BNP 1-108 is released from ventricular cardiomyocytes and interacts with the extracellular protease fragment of membrane-bound corin, a type II transmembrane serine protease. This interaction cleaves corin in a sequence-dependent manner resulting in a 32 amino acid carboxyl fragment, termed BNP-32, and a 76 amino acid amino terminal fragment, termed NT-BNP.
Adequate processing of natriuretic peptides required fir biological activity
The importance of corin and adequate natriuretic peptide processing to the function of the NPS is related to the fact that unprocessed BNP (BNP 1-108) is unable to effectively serve as a ligand for NPR-A. In one study(37) , investigators reported that recombinant proBNP was completely devoid of biological activity as assessed by its ability to stimulate cGMP production in cardiomyocytes and cardiac fibroblasts cell cultures. The use of E.Coli produced recombinant BNP was problematic because this proBNP was not glycosylated, and human BNP is known to be glycosylated(38). Another group of investigators(38) used the same cell-based system to explore the ability of recombinant BNP 1-108 made in mammalian cells to ensure adequate glycosylation. In contrast to the previous report, this recombinant proBNP possessed some biological activity, however, as compared to BNP-32 the activity was 6-fold lower. Therefore, abnormalities in corin-mediated natriuretic peptide processing would be expected to interfere with the autocrine/paracrine anti-hypertrophic actions of the NPS, especially in conditions of pressure-overload.
Corin as a candidate gene
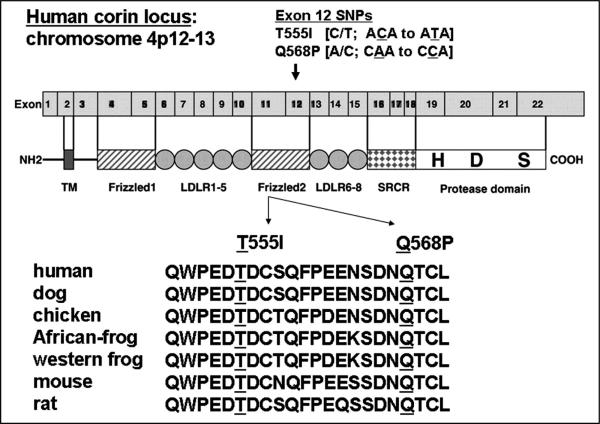
We decided to focus attention on corin as a candidate gene based upon the hypothesis that impaired corin activity would reduce natriuretic peptide processing and reduce the in vivo function of the endogenous NPS. The human corin gene was sequenced in 33 African-Americans with dilated cardiomyopathy leading to the identification of two previously unreported non-synonymous, non-conservative single nucleotide polymorphisms, T555I and Q568P, each located in exon 11 of the human corin gene. The T555I and Q568P amino acid substitutions are located in the second cysteine-rich, frizzled-like domain of corin, a domain demonstrated to be essential to the bioactivity of corin (39). Moreover, these amino acid changes are biochemically non-conservative and change highly conserved amino acid residues (Figure 2) increasing the apriori probability that they might alter corin protein function.
Figure 2.
The corin I555(P568) minor allele is comprised of two single nucleotide polymorphisms that are in complete linkage disequilibrium in African-Americans with an allelic prevalence of 6%. The SNPs result in a T555I and Q568P amino acid change within the second cysteine-rich, frizzled domain in corin. These amino acid changes are biochemically non-conservative and demonstrate strong conservation. Because of the linkage disequilibrium, the SNPs travel on the same parental chromosome are the two resulting amino acid changes will be transcribed into the same corin molecule in heterozygous carriers of the corin I555(P568) minor allele.
Our initial genotyping efforts in the Dallas Heart Study(40), a large, population-based longitudinal cohort, demonstrated that these two SNPs were exclusive to persons of African ancestry and in complete linkage disequilibrium. Thus, the T555I and Q568P mutations travel on the same chromosome and comprise, in essence, what we term the minor corin I555(P568) allele with an ~ 6% allelic prevalence in the African-American population. Genotyping within the African-American population has demonstrated that the minor corin I555(P568) allele is primarily present in the heterozygous state with approximately 12% of African-Americans found to be heterozygous for the corin I555(P568) allele.
The corin I555(P568) allele and prevalent hypertension in African-Americans
We genotyped self-identified African-American subjects in three cohorts, the Dallas Heart Study (DHS), the Multi-Ethnic Study of Atherosclerosis (MESA) and the Chicago Genetics Study, for the corin I555(P568) allele and studied its association with blood pressure parameters and prevalent hypertension defined according to JNC VII criteria(41). In our primary population sample, the Dallas Heart Study, after adjustment for potential confounders, including population stratification, the corin I555 (P568) allele remained independently associated with increased risk for prevalent hypertension (odds ratio, 1.63; 95% CI, 1.11 to 2.38; P=0.013). The corin I555 (P568) allele also was associated with higher systolic blood pressure in subjects not using antihypertensive medication in unadjusted (133.7+/−20.7 versus 129.4+/−17.4 mm Hg; P=0.029) and adjusted (132.5+/−1.6 versus 128.9+/−0.6 mm Hg; P=0.029) analyses. The independent association of the minor corin allele with increased risk for prevalent hypertension was confirmed in the Multi-Ethnic Study of Atherosclerosis (odds ratio, 1.50; 95% CI, 1.09 to 2.06; P=0.014). In addition, the association of the minor corin I555 (P568) allele with higher systolic blood pressure was confirmed in adjusted analysis in the Chicago Genetics of Hypertension Study (125.8+/−1.9 versus 121.4+/−0.7 mm Hg; P=0.03).
Evidence for causation: T555I and Q568P reduce corin in vitro activity
Recently, in vitro experiments have demonstrated that the presence of both the T555I and Q568P amino acid substitutions significantly reduce the natriuretic processing activity of corin(42). In both human embryonic kidney (HEK)293 cells and murine HL-1 cardiomyocytes, the corin variant T555I/Q568P had a reduced (38+/−7%, P<0.01) pro-ANP processing activity compared to that of wild type. The variant also exhibited a low activity (44+/−15%, P<0.05) in processing pro-brain natriuretic peptide (BNP). Interestingly, the presence of each mutation individually did not significantly reduce the biological activity of corin. This is intriguing because in the African-American populations we have studied, the T555I and Q568P SNPs are indeed in complete linkage disequilibrium, therefore, both amino acid changes are transcribed into the same corin molecule in subjects that are heterozygous for the corin I555(P568) allele. In additional experiments, the biochemical basis for the loss of activity in T555I/Q568P variant was studied. It was demonstrated that the I555/P568 substitutions significantly impair corin zymogen activation that is required for catalytic activity. This appears to account for the impaired natriuretic peptide processing, since soluble, pre-activated corin containing the I555/P568 substitutions processes proBNP normally. Therefore, the corin I555(P568) minor allele associated with hypertension and cardiac hypertrophy in untreated hypertensive patients impair corin zymogen activation and natriuretic peptide processing activity in vitro, suggesting a causal molecular basis to the observed phenotypic associations within African-Americans.
Corin I555(P568) allele and susceptibility to pressure-overload concentric LVH
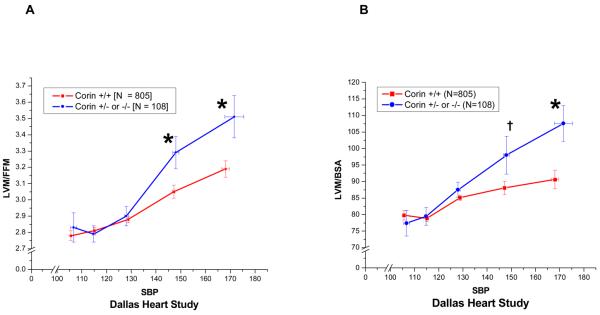
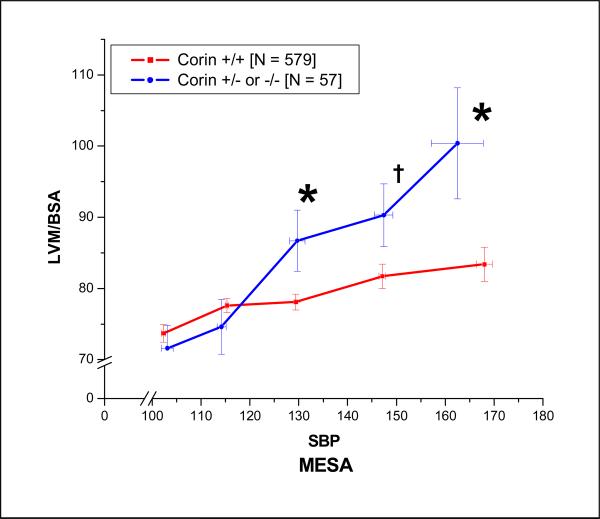
We next determined the association of the minor corin I555(P568) allele with concentric cardiac remodeling in untreated (no reported use of any anti-hypertensive medication) African-American hypertensive subjects in the DHS and MESA. In the DHS, the relationship of left ventricular mass index (LVMI), indexed to either DEXA-determined fat-free mass or body-surface area, and systolic blood pressure demonstrated increased LVMI in the corin I555(P568) group as compared to wild-type participants in the higher ranges of SBP (Figure 3) The ratio of left-ventricular mass (LVM) to left-ventricular end-diastolic dimension (LVM/EDV), a measure of concentric cardiac remodeling, also increased as SBP increased consistent with concentric cardiac hypertrophy (Figure 4). The increased LVMI at higher ranges of SBP that was demonstrated in the DHS was replicated in the untreated African-American participants in the MESA study (Figure 5), despite the fact that MESA excluded subjects with known cardiovascular disease other than hypertension.
Figure 3.
The relationship of left-ventricular mass indexed (LVMI) to either fat-free mass or body-surface area and systolic blood pressure as a function of corin I555(P568) carrier status was examined in untreated (no antihypertensive medication) African-Americans participating in the Dallas Heart Study. Participants heterozygous for the corin I555(P568) allele demonstrated increased LVMI as compared to wild-type corin carriers in the higher ranges of systolic blood pressure. († P < 0.1; * P<0.5 ).
Figure 4.
An increase in the ratio of left ventricular mass (LVM) (grams) to left ventricular end diastolic volume (EDV) (cc) is a measure of concentric left ventricular hypertrophy. The untreated self-identified African-American Dallas Heart Study participants heterozygous for the corin I555(P568) allele as compared to wild-type participants demonstrated increasing degrees of concentric cardiac hypertrophy as systolic blood pressure increased.
Figure 5.
The relationship of left-ventricular mass indexed (LVMI) to body-surface area and systolic blood pressure as a function of corin I555(P568) carrier status was examined in untreated (no antihypertensive medication) African-Americans participating in the Multi-Ethnic Study of Atherosclerosis (MESA). Similar to findings in the Dallas Heart Study, untreated African-American participants heterozygous for the corin I555(P568) allele demonstrated increased LVMI compared to corin wild-type participants in the higher ranges of systolic blood pressure. († P < 0.1; * P<0.5 ).
We hypothesize that the non-linear disparities in the LVMI and SBP relationship as a function of the presence of absence of the corin I555(P568) allele may relate to the fact that the majority of African-Americans carrying the corin I555(P568) allele are heterozygous for the corin allele associated with reduced corin function. Assuming equal allelic expression, approximately 50% of the corin protein in the heterozygotes would be expected to be fully functional. Therefore, corin's natriuretic peptide processing ability may become rate-limiting in heterozygotes only under conditions of marked increases in ANP or BNP expression, such as occurs under conditions of pressure-overload caused by increases in systolic blood pressure.
Corin I555(P568) allele and prevalent LVH
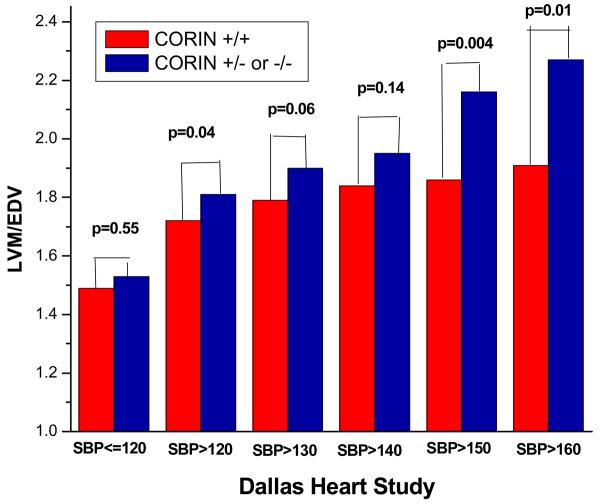
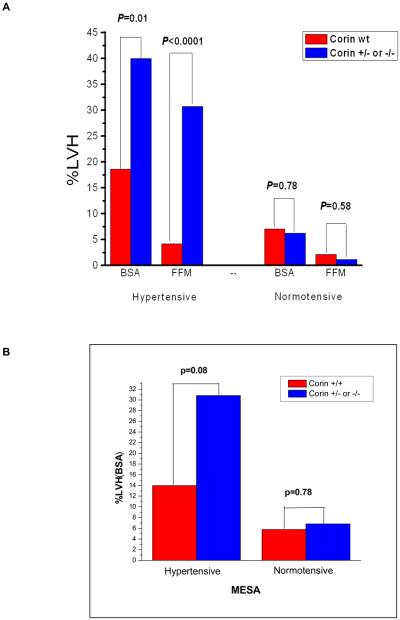
We next examined whether corin variant status affected the prevalence of LVH defined as a dichotomous trait. In the DHS (Figure 6A), the prevalence of LVH, whether defined by FFM or BSA indexation, was significantly higher in the corin variant compared to non-variant group among those untreated African-American participants with SBP≥140 (hypertensive). In the MESA study, a similar trend was discovered (Figure 6B). As mentioned, it is important to note that MESA may have been biased against replication of this result because it excluded from enrollment patients with known pre-existing cardiovascular disease excluding hypertension. Given the independent association of LVH with cardiovascular disease, such as heart failure and atrial fibrillation, the demonstration of a similar trend in MESA, albeit not statistically significant, was nonetheless reassuring. In the Dallas Heart Study, upon multivariate logistic regression analysis, the corin I555(P568) allele remained independently associated with increased odds for prevalent LVH despite adjusting for differences in age, gender, diabetes, SBP, BMI, and estimated African-ancestry. Moreover, the odds ratio for prevalent LVH increased in magnitude in the higher ranges of SBP (Table 1).
Figure 6A and B.
In the Dallas Heart Study, the prevalence of LVH, defined by indexing left-ventricular mass to either fat-free mass (FFM) or body-surface area (BSA) was significantly greater in untreated hypertensive African-American participants that were heterozygous for the corin I555(P568) allele as compared to wild-type. In the Multi-Ethnic Study of Atherosclerosis, there was a trend for a higher prevalence of LVH, defined by indexing left-ventricular mass to body-surface area, in untreated African-American participants that were heterozygous for the corin I555(P568) allele as compared to wild-type
Table 1.
Association between minor corin I555(P568) allele and prevalent left ventricular hypertrophy in subjects from the Dallas Heart Study with no history of antihypertensive medication, grouped according to increasing systolic blood pressure.
| OR | Adjusted 95% CI |
P | OR | Ancestry adjusted ** 95% CI |
P | |
|---|---|---|---|---|---|---|
| All untreated (N=983) |
2.86 | (1.26, 6.08) | 0.008 | 2.89 | (1.27, 6.19) | 0.008 |
| SBP > 120 mmHg (N=600) |
3.52 | (1.52, 7.84) | 0.002 | 3.60 | (1.54, 8.05) | 0.002 |
| SBP > 130 mmHg (N=338) |
4.37 | (1.74, 10.34) | 0.001 | 4.90 | (1.91, 12.25) | 0.0007 |
| SBP >140 mmHg (N=177) |
9.89 | (3.31, 31.52) | 0.0001 | 9.73 | (3.24, 31.02) | <0.0001 |
| SBP > 150 mmHg (N=96) |
11.45 | (1.82, 102.86) | 0.013 | 12.69 | (1.82, 139.69) | 0.014 |
LVH is defined as LVM/FFM > 3.7 g/kg for both men and women, where LVM=left ventricular mass and FFM=fat free mass derived from dual X-ray absorptiometry. The odds ratios are adjusted for age, gender, diabetes, body mass index, systolic blood pressure, and a comparison is made with and without adjustment for genetic ancestry.
also adjusted for estimated African ancestry in addition to age, gender, diabetes, body mass index and systolic blood pressure.
LVH and genetic variation in other natriuretic peptide candidate genes
Other investigators have reported associations between LVH and genetic variation within other components of the natriuretic peptide system. Nakayama and colleagues investigators isolated the 5′-flanking region of the type A human NP receptor gene and identified an insertion/deletion mutation in this region in Japanese patients with essential hypertension that was also associated with LVH(43). The deletion allele lacks 8 nucleotides and alters binding sites for the activator protein-2 (AP-2) and Zeste transcriptional factors; reporter gene assays demonstrate that transcriptional activity of the deletion allele is 30% lower than wild-type. The investigators genotyped 200 subjects with essential hypertension and 200 non-hypertensive controls, and found 9 subjects with the deletion (8 in the EH group and 1 in the NT group). All 9 individuals were heterozygous. The normotensive subject with the mutation had left ventricular hypertrophy without hypertension. The plasma levels of BNP in the essential hypertension brain patients with the deleted allele were significantly higher than the levels in the essential hypertension patients with the wild-type allele, and plasma brain NP levels were significantly higher in subjects with the deleted allele than in subjects with the wild-type allele, despite comparable blood pressures. The investigators concluded that the human NPR-A 5′flanking region insertion mutation is associated with reduced receptor activity and increased susceptibility to the development of hypertension and LVH.
In another recent study, Rubattu and colleagues(44) studied 203 hypertensive patients that underwent two-dimensional echocardiography. Three markers of the ANP gene (−C664G, G1837A, and T2238C polymorphisms) and a microsatellite marker of both the NPRA and BNP genes were characterized. They reported that patients carrying the ANP gene promoter allelic variant had increased left ventricular mass index, and other measures of cardiac concentric remodeling, as compared with the wild-type genotype. These associations were independent from other potential confounders and were replicated in a large subgroup of never-treated hypertensive patients . Carrier status of the ANP gene promoter allelic variant was associated with significantly lower plasma proANP levels, and a significant association for NPRA gene variants with left ventricular mass index and left ventricular septal thickness was found. The investigators concluded that genetic variation in the NPS significantly contributes to ventricular remodeling in human essential hypertension.
Most recently, Xue and colleagues (45) tested for any association between two single nucleotide polymorphisms in the ANP gene (one in the promoter and one exonic) with cardiac hypertrophy in 2118 hypertensive patients, including 945 with LVH and 1173 without LVH, as well as 816 healthy control subjects. All subjects were genotyped for the −A2843G and A188G polymorphisms. The investigators reported that the GG genotype at position −2843 conferred a 2.2-fold risk for LVH defined by left-ventricular mass index and other measures of concentric cardiac remodeling as compared with the AA or AG genotypes. In addition, plasma levels of ANP were significantly lower in the hypertensive patients with LVH carrying the GG genotypes compared with those carrying the AA or AG genotypes (P<0.01). The investigators concluded that the −A2843G polymorphism in the ANP gene promoter might be a genetic risk factor for the development of LVH in patients with hypertension.
Conclusions
In summary, using a candidate gene system approach, we have identified a minor corin allele that is associated with increased risk for concentric LVH in untreated hypertensive African-Americans. Based on the recently identified functional effects of these two mutations on corin activity, coupled with the known anti-fibrotic and anti-hypertrophic actions of the NPS, we believe that the corin I555(P568) minor allele is an “LVH-sensitizing” gene variant. Future studies will utilize more advanced genomic methodologies to explore these hypotheses in greater detail. We believe that this is the first example of a population-specific LVH sensitizing gene variant. Given the importance of the natriuretic peptide system to cardiac and hemodynamic homeostasis in a variety of conditions, we anticipate that modern genomic technologies and methodologies will continue to elucidate the importance of genetic variation in this system and its relationship to inter-individual susceptibility to cardiovascular disease and drug response.
References
- 1.Levy D, Garrison RJ, Savage DD, Kannel WB, Castelli WP. Prognostic implications of echocardiographically determined left ventricular mass in the Framingham Heart Study. N Engl J Med. 1990;322:1561–6. doi: 10.1056/NEJM199005313222203. [DOI] [PubMed] [Google Scholar]
- 2.Levy DGR, Savage DD, Kannel WB, Castelli WP. Left ventricular mass and incidence of coronary heart disease in an elderly cohort. The Framingham Heart Study. Ann Intern Med. 1989;110:101–107. doi: 10.7326/0003-4819-110-2-101. [DOI] [PubMed] [Google Scholar]
- 3.Okin PM, Devereux RB, Jern S, et al. Regression of electrocardiographic left ventricular hypertrophy during antihypertensive treatment and the prediction of major cardiovascular events. Jama. 2004;292:2343–9. doi: 10.1001/jama.292.19.2343. [DOI] [PubMed] [Google Scholar]
- 4.Devereux RBRM, de Simone G, O'Grady MJ, Paranicas M, Yeh JL, Fabsitz RR, Howard BV. Relations of left ventricular mass to demographic and hemodynamic variables in American Indians: the Strong Heart Study. Circulation. 1997;96:1416–1423. doi: 10.1161/01.cir.96.5.1416. [DOI] [PubMed] [Google Scholar]
- 5.Gardin JMWL, Anton-Culver H, Flack J, Gidding S, Kurosaki T, Wong ND, Manolia TA. Relationship of cardiovascular risk factors to echocardiographic left ventricular mass in healthy young black and white adult men and women. The CARDIA Study. Circulation. 1995;92:380–387. doi: 10.1161/01.cir.92.3.380. [DOI] [PubMed] [Google Scholar]
- 6.Adams TDYF, Fisher AG, Ridges JD, Nelson AG, Hagan AD, Williams RR, Hunt SC. Heritability of cardiac size: an echocardiographic and electrocardiographic study of monozygotic and dizygotic twins. Circulation. 1985;71:39–44. doi: 10.1161/01.cir.71.1.39. [DOI] [PubMed] [Google Scholar]
- 7.Benjamin EDA, Larson MG, O'Donnell J, Vasan RS, Levy D. Genetic linkage analyses for left ventricular mass phenotypes in the Framingham Heart Study. Circulation. 2000;102:II–860. [Google Scholar]
- 8.Harshfield GAGC, Hwang C, Savage DD, Anderson SJ. Genetic and environmental influences on echocardiographically determined left ventricular mass in black twins. Am J Hypertens. 1990;3:538–543. doi: 10.1093/ajh/3.7.538. [DOI] [PubMed] [Google Scholar]
- 9.Post WSLM, Myears RH, Galderisi M, Levy D. Heritability of left ventricular mass: the Framingham Heart Study. Hypertension. 1997;30:1025–1028. doi: 10.1161/01.hyp.30.5.1025. [DOI] [PubMed] [Google Scholar]
- 10.Schillaci GVP, Porcellati C, Cuccurullo O, Cosco C, Perticone F. Continuous relation between left ventricular mass and cardiovascular risk in essential hypertension. Hypertension. 2000;35:580–586. doi: 10.1161/01.hyp.35.2.580. [DOI] [PubMed] [Google Scholar]
- 11.Bella JN, MacCluer JW, Roman MJ, et al. Heritability of left ventricular dimensions and mass in American Indians: The Strong Heart Study. J Hypertens. 2004;22:281–6. doi: 10.1097/00004872-200402000-00011. [DOI] [PubMed] [Google Scholar]
- 12.Sharma P, Middelberg RP, Andrew T, Johnson MR, Christley H, Brown MJ. Heritability of left ventricular mass in a large cohort of twins. J Hypertens. 2006;24:321–4. doi: 10.1097/01.hjh.0000202815.18083.03. [DOI] [PubMed] [Google Scholar]
- 13.Brandt RRWR, Redfield MM, Burnett JC. Atrial natriuretic peptide in heart failure. J Am Coll Cardiol. 1993;22:86A–92A. doi: 10.1016/0735-1097(93)90468-g. [DOI] [PubMed] [Google Scholar]
- 14.Burnett JC., Jr The atrial peptide system in cardiac disease. Am J Hypertens. 1988;1:410S–420S. doi: 10.1093/ajh/1.4.410s. [DOI] [PubMed] [Google Scholar]
- 15.Potter LR, Abbey-Hosch S, Dickey DM. Natriuretic peptides, their receptors, and cyclic guanosine monophosphate-dependent signaling functions. Endocr Rev. 2006;27:47–72. doi: 10.1210/er.2005-0014. [DOI] [PubMed] [Google Scholar]
- 16.Castro LR, Verde I, Cooper DM, Fischmeister R. Cyclic guanosine monophosphate compartmentation in rat cardiac myocytes. Circulation. 2006;113:2221–8. doi: 10.1161/CIRCULATIONAHA.105.599241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moritoki H, Yoshikawa T, Hisayama T, Takeuchi S. Possible mechanisms of age-associated reduction of vascular relaxation caused by atrial natriuretic peptide. Eur J Pharmacol. 1992;210:61–8. doi: 10.1016/0014-2999(92)90652-k. [DOI] [PubMed] [Google Scholar]
- 18.Kurtz A, Della Bruna R, Pfeilschifter J, Taugner R, Bauer C. Atrial natriuretic peptide inhibits renin release from juxtaglomerular cells by a cGMP-mediated process. Proc Natl Acad Sci U S A. 1986;83:4769–73. doi: 10.1073/pnas.83.13.4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gunning ME, Brady HR, Otuechere G, Brenner BM, Zeidel ML. Atrial natriuretic peptide(31-67) inhibits Na+ transport in rabbit inner medullary collecting duct cells. Role of prostaglandin E2. J Clin Invest. 1992;89:1411–7. doi: 10.1172/JCI115730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeidel ML. Renal actions of atrial natriuretic peptide: regulation of collecting duct sodium and water transport. Annu Rev Physiol. 1990;52:747–59. doi: 10.1146/annurev.ph.52.030190.003531. [DOI] [PubMed] [Google Scholar]
- 21.Zeidel ML. Regulation of collecting duct Na+ reabsorption by ANP 31-67. Clin Exp Pharmacol Physiol. 1995;22:121–4. doi: 10.1111/j.1440-1681.1995.tb01967.x. [DOI] [PubMed] [Google Scholar]
- 22.Isono M, Haneda M, Maeda S, Omatsu-Kanbe M, Kikkawa R. Atrial natriuretic peptide inhibits endothelin-1-induced activation of JNK in glomerular mesangial cells. Kidney Int. 1998;53:1133–42. doi: 10.1046/j.1523-1755.1998.00869.x. [DOI] [PubMed] [Google Scholar]
- 23.Melo LG, Steinhelper ME, Pang SC, Tse Y, Ackermann U. ANP in regulation of arterial pressure and fluid-electrolyte balance: lessons from genetic mouse models. Physiol Genomics. 2000;3:45–58. doi: 10.1152/physiolgenomics.2000.3.1.45. [DOI] [PubMed] [Google Scholar]
- 24.Kuhn M, Holtwick R, Baba HA, Perriard JC, Schmitz W, Ehler E. Progressive cardiac hypertrophy and dysfunction in atrial natriuretic peptide receptor (GC-A) deficient mice. Heart. 2002;87:368–74. doi: 10.1136/heart.87.4.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kishimoto IIRK, Garbers DL. A genetic model provides evidence that the receptor for atrial natriuretic peptide (guanylyl cyclase-A) inhibitis cardiac hypertrophy in natriuretic peptide receptor-A-deficient mice. Proc Natl Acad Sci U S A. 2001;98:2703–2706. doi: 10.1073/pnas.051625598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holtwick REM, Skryavin BV, et al. Pressure independent cardiac hypertrophy in mice with cardiomyocyte-restricted inactivation of the atrial natriuretic peptide receptor guanylyl cyclase Á. J Clin Invest. 2003;111:1399–1407. doi: 10.1172/JCI17061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patel JB, Valencik ML, Pritchett AM, Burnett JC, Jr., McDonald JA, Redfield MM. Cardiac-specific attenuation of natriuretic peptide A receptor activity accentuates adverse cardiac remodeling and mortality in response to pressure overload. Am J Physiol Heart Circ Physiol. 2005;289:H777–84. doi: 10.1152/ajpheart.00117.2005. [DOI] [PubMed] [Google Scholar]
- 28.Nishikimi T, Maeda N, Matsuoka H. The role of natriuretic peptides in cardioprotection. Cardiovasc Res. 2006;69:318–28. doi: 10.1016/j.cardiores.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 29.Tokudome T, Kishimoto I, Horio T, et al. Regulator of G-protein signaling subtype 4 mediates antihypertrophic effect of locally secreted natriuretic peptides in the heart. Circulation. 2008;117:2329–39. doi: 10.1161/CIRCULATIONAHA.107.732990. [DOI] [PubMed] [Google Scholar]
- 30.Kapoun AM, Liang F, O'Young G, et al. B-type natriuretic peptide exerts broad functional opposition to transforming growth factor-beta in primary human cardiac fibroblasts: fibrosis, myofibroblast conversion, proliferation, and inflammation. Circ Res. 2004;94:453–61. doi: 10.1161/01.RES.0000117070.86556.9F. [DOI] [PubMed] [Google Scholar]
- 31.Tsuruda T, Boerrigter G, Huntley BK, et al. Brain natriuretic Peptide is produced in cardiac fibroblasts and induces matrix metalloproteinases. Circ Res. 2002;91:1127–34. doi: 10.1161/01.res.0000046234.73401.70. [DOI] [PubMed] [Google Scholar]
- 32.Yan W, Sheng N, Seto M, Morser J, Wu Q. Corin, a mosaic transmembrane serine protease encoded by a novel cDNA from human heart. J Biol Chem. 1999;274:14926–35. doi: 10.1074/jbc.274.21.14926. [DOI] [PubMed] [Google Scholar]
- 33.Rybkin II, Kim MS, Bezprozvannaya S, et al. Regulation of atrial natriuretic peptide secretion by a novel Ras-like protein. J Cell Biol. 2007;179:527–37. doi: 10.1083/jcb.200707101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sawada Y, Inoue M, Kanda T, et al. Co-elevation of brain natriuretic peptide and proprotein-processing endoprotease furin after myocardial infarction in rats. FEBS Lett. 1997;400:177–82. doi: 10.1016/s0014-5793(96)01385-3. [DOI] [PubMed] [Google Scholar]
- 35.Sawada Y, Suda M, Yokoyama H, et al. Stretch-induced hypertrophic growth of cardiocytes and processing of brain-type natriuretic peptide are controlled by proprotein-processing endoprotease furin. J Biol Chem. 1997;272:20545–54. doi: 10.1074/jbc.272.33.20545. [DOI] [PubMed] [Google Scholar]
- 36.Chan JC, Knudson O, Wu F, Morser J, Dole WP, Wu Q. Hypertension in mice lacking the proatrial natriuretic peptide convertase corin. Proc Natl Acad Sci U S A. 2005;102:785–90. doi: 10.1073/pnas.0407234102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heublein DM. Immunoreactivity and cyclic GMP activating actions of various molecular forms of human B-type natriuretic peptide. Hypertension. 2007 doi: 10.1161/HYPERTENSIONAHA.106.081083. [DOI] [PubMed] [Google Scholar]
- 38.Liang F, O'Rear J, Schellenberger U, et al. Evidence for functional heterogeneity of circulating B-type natriuretic peptide. J Am Coll Cardiol. 2007;49:1071–8. doi: 10.1016/j.jacc.2006.10.063. [DOI] [PubMed] [Google Scholar]
- 39.Knappe S, Wu F, Madlansacay MR, Wu Q. Identification of domain structures in the propeptide of corin essential for the processing of proatrial natriuretic peptide. J Biol Chem. 2004;279:34464–71. doi: 10.1074/jbc.M405041200. [DOI] [PubMed] [Google Scholar]
- 40.Victor RG, Haley RW, Willett DL, et al. The Dallas Heart Study: a population-based probability sample for the multidisciplinary study of ethnic differences in cardiovascular health. Am J Cardiol. 2004;93:1473–80. doi: 10.1016/j.amjcard.2004.02.058. [DOI] [PubMed] [Google Scholar]
- 41.Dries DL, Victor RG, Rame JE, et al. Corin gene minor allele defined by 2 missense mutations is common in blacks and associated with high blood pressure and hypertension. Circulation. 2005;112:2403–10. doi: 10.1161/CIRCULATIONAHA.105.568881. [DOI] [PubMed] [Google Scholar]
- 42.Wang W, Liao X, Fukuda K, et al. Corin variant associated with hypertension and cardiac hypertrophy exhibits impaired zymogen activation and natriuretic peptide processing activity. Circ Res. 2008;103:502–8. doi: 10.1161/CIRCRESAHA.108.177352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakayama T, Soma M, Takahashi Y, Rehemudula D, Kanmatsuse K, Furuya K. Functional deletion mutation of the 5′-flanking region of type A human natriuretic peptide receptor gene and its association with essential hypertension and left ventricular hypertrophy in the Japanese. Circ Res. 2000;86:841–5. doi: 10.1161/01.res.86.8.841. [DOI] [PubMed] [Google Scholar]
- 44.Rubattu S, Bigatti G, Evangelista A, et al. Association of atrial natriuretic peptide and type a natriuretic peptide receptor gene polymorphisms with left ventricular mass in human essential hypertension. J Am Coll Cardiol. 2006;48:499–505. doi: 10.1016/j.jacc.2005.12.081. [DOI] [PubMed] [Google Scholar]
- 45.Xue H, Wang S, Wang H, et al. Atrial natriuretic peptide gene promoter polymorphism is associated with left ventricular hypertrophy in hypertension. Clin Sci (Lond) 2008;114:131–7. doi: 10.1042/CS20070109. [DOI] [PubMed] [Google Scholar]