Abstract
There are two protein primers involved in picornavirus RNA replication, VPg, the viral protein of the genome, and VPgpUpUOH. A cis-acting replication element (CRE) within the open reading frame of poliovirus (PV) RNA allows the viral RNA-dependent RNA polymerase 3DPol to catalyze the conversion of VPg into VPgpUpUOH. In this study, we used preinitiation RNA replication complexes (PIRCs) to determine when CRE-dependent VPg uridylylation occurs relative to the sequential synthesis of negative- and positive-strand RNA. Guanidine HCl (2 mM), a reversible inhibitor of PV 2CATPase, prevented CRE-dependent VPgpUpUOH synthesis and the initiation of negative-strand RNA synthesis. VPgpUpUOH and nascent negative-strand RNA molecules were synthesized coincident in time following the removal of guanidine, consistent with PV RNA functioning simultaneously as a template for CRE-dependent VPgpUpUOH synthesis and negative-strand RNA synthesis. The amounts of [32P]UMP incorporated into VPgpUpUOH and negative-strand RNA products indicated that 100 to 400 VPgpUpUOH molecules were made coincident in time with each negative-strand RNA. 3′-dCTP inhibited the elongation of nascent negative-strand RNAs without affecting CRE-dependent VPg uridylylation. A 3′ nontranslated region mutation which inhibited negative-strand RNA synthesis did not inhibit CRE-dependent VPg uridylylation. Together, the data implicate 2CATPase in the mechanisms whereby PV RNA functions as a template for reiterative CRE-dependent VPg uridylylation before and during negative-strand RNA synthesis.
A common feature of positive-strand RNA viruses is the asymmetric replication of viral RNA. Poliovirus (PV) RNA replication has been studied extensively; however, it remains to be determined exactly how the synthesis of negative-strand RNA and that of positive-strand RNA are mechanistically distinct, culminating in the synthesis of greater amounts of positive-strand than negative-strand RNA (2). A cis-acting replication element (CRE) within the 2C open reading frame of PV RNA functions as a template for the conversion of the viral protein of the genome (VPg) into VPgpUpUOH (24, 26, 37). 3D polymerase (3DPol), in concert with other viral proteins, catalyzes the conversion of VPg into VPgpUpUOH on CRE RNA templates (22). It remains to be determined whether the tyrosine hydroxyl of VPg (14, 20, 21), the 3′ hydroxyl of VPgpUpUOH (22, 23, 43), or both (38) are used to prime negative-strand RNA synthesis. It would be informative to know whether VPg is converted into VPgpUpUOH before, during, and/or after the initiation of viral negative-strand RNA synthesis. Conversion of VPg into VPgpUpUOH before the initiation of negative-strand RNA synthesis would be consistent with the possibility that it primes the initiation of negative-strand RNA synthesis. Conversely, if VPg were not converted into VPgpUpUOH until after the initiation of negative-strand RNA synthesis, VPgpUpUOH could not possibly participate in the initiation of negative-strand RNA synthesis. Also, because multiple copies of VPgpUpUOH are necessary to prime reiterative initiation of positive-strand RNA synthesis (35), VPg is most likely converted into abundant amounts of VPgpUpUOH before the initiation of positive-strand RNA synthesis.
PV preinitiation RNA replication complexes (PIRCs) were used in this study to examine when VPg is converted into VPgpUpUOH. PIRCs assemble and accumulate when PV mRNA is translated in reaction mixtures containing cytoplasmic extracts from uninfected HeLa cells and 2 mM guanidine HCl, a reversible inhibitor of viral RNA replication (5). The viral replication proteins expressed from the viral mRNA interact with lipid membranes in the cytoplasmic extracts to form RNA replication complexes (RCs) similar to those in infected cells (12). PIRCs convert VPg into VPgpUpUOH and initiate viral RNA replication when they are isolated from reaction mixtures containing guanidine and resuspended in reaction mixtures lacking guanidine (6, 19). Guanidine HCl functions as a reversible inhibitor of PV RNA replication, both in cells (11) and in cell-free translation-replication reactions (6). In cells, PV RNA RCs fail to immediately initiate RNA replication following the removal of guanidine HCl (11). Rather, PV RCs formed in the presence of guanidine in cells appear to be translocated to a region of the cytoplasm where the RCs and their contents may be recycled and/or destroyed (11), possibly by autophagic vesicles (17). Recycling and/or destruction of RCs by autophagic vesicles would preclude their function upon the removal of guanidine. PIRCs, which form in the presence of guanidine during the translation of PV mRNA in cytoplasmic extracts of HeLa cells, immediately initiate both negative-strand RNA synthesis and CRE-dependent VPg uridylylation upon the removal of guanidine (6, 19). Viral RNA replication and VPgpUpUOH synthesis are monitored by the incorporation of radiolabeled UTP (19-21). It is important to note that RNA replication in the context of PIRCs is artificial in that the PIRCs are stalled with guanidine and purified and then the guanidine block is removed. Despite this artificiality, the mechanisms of RNA replication within PIRCs appear to reliably represent the mechanisms of RNA replication in cells. There are several advantageous features of the PIRC experimental system: viral RNA replication is synchronous and sequential, with negative-strand RNA being made before positive-strand RNA (6); viral RNA replication is asymmetric, with an excess of positive-strand RNA being made from each negative-strand template; VPg is converted into VPgpUpUOH in a CRE-dependent manner (20, 21); and the reaction conditions, including nucleoside triphosphate concentrations, are easily manipulated (38). Importantly, PIRCs contain all of the viral proteins associated with RNA replication and RNA replication by PIRCs faithfully mimics the asymmetric replication of PV RNA observed in cells.
PV protein 2C, the target of guanidine HCl (30), is a critical but poorly understood component of PIRCs and RNA RCs in cells. PV protein 2C has an NH-terminal amphipathic helix which interacts with cellular membranes (40), a central ATPase domain where guanidine-resistant and guanidine-dependent mutations arise (31, 32), a cysteine-rich zinc binding motif (29), and a COOH-terminal RNA binding domain (34) which appears to work in concert with amino acid residues at the NH terminus to bind RNA. 2CATPase can oligomerize (1, 41), anchoring viral replication proteins and RNA templates within membranous RCs (4). The ability of guanidine HCl to reversibly inhibit both CRE-dependent VPg uridylylation and negative-strand RNA synthesis implicates 2CATPase in the mechanisms by which PV RNA functions coordinately as a template for both RNA replication and CRE-dependent VPgpUpUOH synthesis (19, 21).
In this study, we found that VPg was converted into VPgpUpUOH before and during negative-strand RNA synthesis and that 2CATPase activity, in the context of membranous PIRCs, allowed PV RNA to function simultaneously as a template for CRE-dependent VPg uridylylation and as a template for negative-strand RNA synthesis. We discuss how picornaviruses coordinate the synthesis of nucleotidylylated protein primers with other steps of viral RNA replication.
MATERIALS AND METHODS
PV cDNAs.
(i) pRNA2 was kindly provided by James B. Flanegan (University of Florida College of Medicine, Gainesville). pRNA2 encodes a PV subgenomic RNA replicon containing an in-frame deletion of nucleotides (nt) 1175 to 2956 within the capsid genes (10). T7 transcription of MluI-linearized pRNA2 cDNA produces PV RNA2 replicon RNA with two nonviral guanosine residues at its 5′ terminus that prevent positive-strand RNA synthesis (8).
(ii) pDNVR27.
pDNVR27 is identical to pRNA2, except for a 5′-terminal hammerhead ribozyme. T7 transcription of MluI-linearized pDNVR27, in concert with ribozyme cleavage, produces a PV RNA2 replicon possessing an authentic PV 5′ terminus (21).
(iii) p3′ΔGUA3 is a derivative of pRNA2 with a deletion of the conserved GUAAA sequence in the 3′ nontranslated region (NTR; nt 5636 to 5640 in RNA2, corresponding to nt 7418 to 7422 in PV RNA) (9).
HeLa S10 translation-replication reactions.
HeLa cell S10 extracts (S10) and HeLa cell translation initiation factors were prepared as previously described (7). HeLa S10 translation-replication reaction mixtures contained 50% (by volume) S10, 20% (by volume) initiation factors, 10% (by volume) 10× nucleotide reaction mixture (10 mM ATP, 2.5 mM GTP, 2.5 mM CTP, 600 mM KCH3CO2, 300 mM creatine phosphate, 4 mg/ml creatine kinase, 155 mM HEPES-KOH [pH 7.4]), and T7 transcripts of PV replicon RNA at 45 μg/ml. PIRCs formed during 4 h of PV RNA translation at 34°C in the presence of 2 mM guanidine HCl as previously described (6).
Translation of PV replicon mRNA was monitored by including [35S]methionine (1.2 mCi/ml; Amersham) in HeLa S10 translation-replication reaction mixtures. Amounts of protein synthesis were measured by the incorporation of [35S]methionine into acid-precipitable material and/or sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). For SDS-PAGE, [35S]methionine-labeled proteins were solubilized in sample buffer (2% SDS [Sigma], 62.5 mM Tris-HCl [pH 6.8], 0.5% 2-mercaptoethanol, 0.1% bromophenol blue, 20% glycerol). The samples were heated at 100°C for 5 min and separated by electrophoresis in SDS-9 to 18% polyacrylamide gels. [35S]methionine-labeled proteins were detected by phosphorimaging (Bio-Rad).
PV RNA replication and VPg uridylylation.
PV RNA replication and VPg uridylylation were assayed by using PIRCs containing PV RNAs as described previously (21). PIRCs were isolated from HeLa S10 translation-replication reaction mixtures containing 2 mM guanidine (described above) by centrifugation at 17,000 × g for 15 min at 4°C (6). PIRCs were resuspended in 40- to 50-μl reaction mixtures containing 27 mM HEPES KOH (pH 7.4), 60 mM KCH3CO2, 2.3 mM Mg(CH3CO2)2, 2.6 mM dithiothreitol, 2.3 mM KCl, 400 μg/ml creatine kinase, 30 mM creatine phosphate, 1 mM ATP, 250 μM GTP, 250 μM CTP, 50 μg/ml puromycin, and 1.25 to 100 μM UTP with or without guanidine HCl, as indicated in the figure legends. [α-32P]UTP (1 μCi/μl of reaction mixture, 800 Ci/mmol; MP Biomedical) was included in the reaction mixtures in order to radiolabel viral RNA and VPgpUpUOH. Reaction mixtures were incubated at 37°C with continuous labeling or pulse-labeling for the periods of time indicated in the figure legends and then quenched in ice water.
Analyses of viral RNA products by 1% agarose gel electrophoresis.
After incubation in reaction mixtures containing radiolabel, RNA RCs were reisolated by centrifugation at 17,000 × g for 15 min at 4°C and the supernatant containing unincorporated radiolabel was discarded. The isolated RCs containing radiolabeled products were solubilized in SDS buffer (0.5% SDS, 10 mM Tris HCl [pH 7.5], 1 mM EDTA, 100 mM NaCl). Radiolabeled viral RNA products were phenol-chloroform extracted, ethanol precipitated, and separated by electrophoresis on a nondenaturing 1% agarose-Tris-borate-EDTA (TBE) gel. Radiolabeled RNAs within the gels were detected and quantified by phosphorimaging.
RNase T1 fingerprints of viral RNA products.
Radiolabeled viral RNAs were isolated by phenol-chloroform extraction and ethanol precipitation as described above. The viral RNAs were then resuspended in 10 μl methylmercury hydroxide (MeHgOH) sample buffer (50 mM boric acid, 5 mM sodium borate, 10 mM sodium sulfate). MeHgOH was not added to the sample until a 1-μl portion of the sample was placed in 5 ml of trichloroacetic acid (TCA) solution (428 mM TCA, 44.8 mM sodium pyrophosphate) to acid precipitate the labeled RNA products, which were then collected on nitrocellulose filters and quantified by scintillation counting. One microliter of 0.5 M MeHgOH was then added to the remaining 9 μl of the RNA sample, and the mixture was incubated at room temperature for 10 min to denature double-stranded RNAs. The MeHgOH in the samples was then inactivated by adding 1 μl of 1.4 M β-mercaptoethanol. One microliter of RNase T1 (300 U/μl) was then added to each reaction mixture, and the reaction mixtures were incubated at 37°C for 30 min. When indicated, proteinase K (200 μg/ml) was added to the reaction mixtures, which were incubated at 37°C for another 30 min to remove VPg from VPg-linked poly(U) products of RNA replication. Reactions were terminated by the addition of an equal volume of 2× urea sample buffer (18 M urea, 8.9 mM Tris base, 8.9 mM boric acid [pH 8.3], 0.2 mM EDTA, 20% [wt/vol] sucrose, 0.05% [wt/vol] bromophenol blue, 0.05% xylene cyanol). Radiolabeled T1 oligonucleotides in the urea sample buffer were denatured at 100°C for 3 min and separated by electrophoresis in 7 M urea-18% polyacrylamide gels in TBE buffer (89 mM Tris base, 89 mM boric acid [pH 8.3], 2 mM EDTA) for 5 h at 25 W. Radiolabeled RNAs within the gels were detected and quantified by phosphorimaging (Bio-Rad).
Analyses of VPgpUpUOH by Tris-Tricine SDS-PAGE.
After incubation in reaction mixtures containing radiolabel, RNA RCs were reisolated by centrifugation at 17,000 × g for 15 min at 4°C and the supernatant containing unincorporated radiolabel was discarded. The isolated RCs containing radiolabeled viral RNA and VPgpUpUOH products were solubilized in Tris-Tricine sample buffer (3% SDS, 62.5 mM Tris-hydrochloride [pH 6.8], 5% β-mercaptoethanol, 10% glycerol, 0.1% bromophenol blue). The samples were fractionated by electrophoresis (70 mA of constant current for 15 min, followed by 17 mA of constant current for 12 h) in a 0.75-mm-thick polyacrylamide (29:1 ratio of acrylamide to bisacrylamide)-Tris-Tricine gel system consisting of a 4% polyacrylamide stacking gel (4% polyacrylamide, 1.0 M Tris [pH 8.45], 0.1% SDS) and a 12% polyacrylamide separating gel (1.0 M Tris [pH 8.45], 0.1% SDS) by using a Tris-Tricine running buffer (0.1 M Tris-Tricine [pH 8.25], 0.1% SDS). Gels were fixed in 50% TCA and then dried. Radiolabeled VPgpUOH and VPgpUpUOH in the gel were detected by phosphorimaging. The mobility of VPgpUpUOH is indicated in each figure.
3′-dCTP.
3′-dCTP was obtained from TriLink BioTechnologies, San Diego, CA.
RESULTS
CRE-dependent VPg uridylylation during negative-strand RNA synthesis.
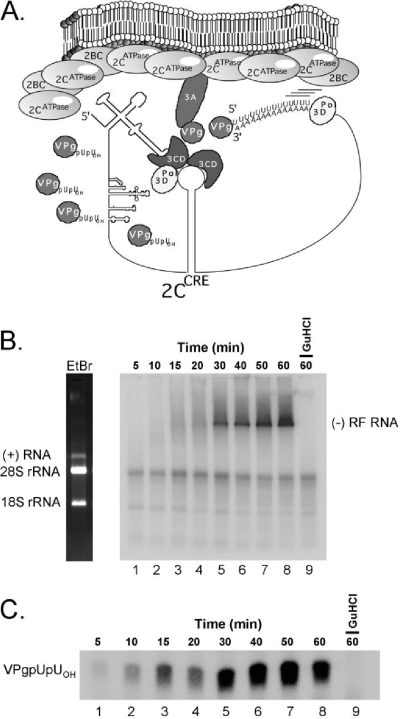
We (21) and others (9, 16, 43) postulate that the 5′ cloverleaf RNA, the 3′ NTR/poly(A) tail, and CRE RNA interact in concert with viral replication proteins to mediate CRE-dependent VPg uridylylation and the initiation of negative-strand RNA synthesis (Fig. 1A, illustration of preinitiation RNA RC moments after the removal of guanidine HCl). As illustrated in this model, PV RNA would function simultaneously as a template for both negative-strand RNA synthesis and CRE-dependent VPg uridylylation. Reiterative CRE-dependent VPg uridylylation during negative-strand RNA synthesis could make the molar excess of VPgpUpUOH primers necessary for positive-strand RNA synthesis.
FIG. 1.
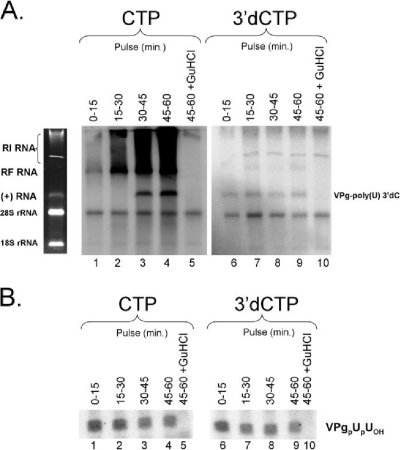
CRE-dependent VPg uridylylation during negative-strand RNA synthesis. (A) Model illustrating CRE-dependent VPg uridylylation coincident in time with negative-strand RNA synthesis. (B) PIRCs containing PV RNA2 templates were incubated for 5 to 60 min as indicated in 40-μl reaction mixtures containing 40 μCi [α-32P]UTP in the absence (lanes 1 to 8) or presence (lane 9) of 2 mM guanidine HCl (GuHCl). Reaction mixtures contained 1 mM ATP, 250 μM CTP, 250 μM GTP, 5 μM UTP, and 1× S10 buffer. Forty microcuries of [α-32P]UTP in a 40-μl reaction mixture containing 5 μM UTP corresponds to a specific radioactivity of 200 Ci/mmol UTP. RNA products of the reactions were separated by electrophoresis in a nondenaturing 1% agarose-TBE gel. PV RNA2 template and rRNA were detected when the gel was stained with ethidium bromide and visualized with UV light (EtBr panel). Phosphorimaging revealed the incorporation of radiolabel into RF RNA along with faint labeling of rRNAs. (C) Radiolabeled VPgpUOH and VPgpUpUOH made within the reaction mixtures was separated by electrophoresis in a Tris-Tricine-polyacrylamide gel and detected by phosphorimaging. The mobility of VPgpUpUOH is indicated. VPgpUOH products migrate slightly faster than VPgpUpUOH products in the gel.
To test these hypotheses, we examined whether [α-32P]UTP was incorporated into both VPgpUpUOH and negative-strand RNA products following the removal of guanidine HCl from PIRCs. [α-32P]UTP was incorporated into both negative-strand RNA (Fig. 1B) and VPgpUpUOH (Fig. 1C) when PIRCs containing PV RNA2 templates were resuspended in reaction mixtures without guanidine. The ongoing presence of 2 mM guanidine prevented the incorporation of radiolabel into both negative-strand RNA (Fig. 1B, lane 9) and VPgpUpUOH (Fig. 1C, lane 9). Ethidium bromide staining revealed the mobility of viral template RNAs and rRNAs within the reaction products (Fig. 1B, left panel). Replicative-form (RF) RNA containing radiolabeled negative-strand RNA was clearly evident after 30 min of incubation (Fig. 1B, lane 5). At earlier time points, nascent negative-strand RNAs were smaller, more heterogeneous in mobility, and less prominent (Fig. 1B, lanes 1 to 4). Radiolabeled VPgpUpUOH was detected at the earliest time points examined (Fig. 1C, lane 1, 5 min postremoval of guanidine). Substantial amounts of radiolabeled VPgpUpUOH accumulated over the initial 20 to 30 min of incubation, coincident in time with the elongation of nascent negative-strand RNAs and their accumulation within RF RNA. These results, as in previous investigations (6, 20, 21), implicate 2CATPase in the mechanisms by which PV RNA functions simultaneously as a template for both negative-strand RNA synthesis and VPgpUpUOH synthesis.
Molar ratios of VPgpUpUOH and negative-strand RNA.
Both VPgpUpUOH and negative-strand RNA were made in common reaction mixtures containing PIRCs and the same amount of [32P]UTP (1 μCi/μl) (Fig. 1B and C). In order to compare the molar amounts of VPgpUpUOH and negative-strand RNA made during the incubation period, the amounts of [32P]UMP incorporated into VPgpUpUOH (detected in Tris-Tricine gels) and negative-strand RNA products (detected in agarose gels) were quantified by phosphorimaging. Thirty-minute reaction products (Fig. 1B and C, lanes 5) contained 67,204 phosphorimage U of VPgpUpUOH and 505,550 phosphorimage U of radiolabel in RF RNA (Table 1). VPgpUpUOH has 2 mol of UMP per molecule, while negative-strand RNA from PV RNA2 templates has 1,792 mol of UMP per molecule (Table 1). The mole-to-mole comparisons were made by correcting phosphorimage units to moles of UMP per molecule (Table 1). The amounts of [32P]UMP in VPgpUpUOH and negative-strand RNA after 30 min of incubation indicated that 119 VPgpUpUOH molecules were made coincident in time with each negative-strand RNA (Table 1). This molar excess of VPgpUpUOH made during negative-strand RNA synthesis is consistent with the abundant amounts of VPgpUpUOH needed to prime subsequent positive-strand RNA synthesis (20, 21, 35).
TABLE 1.
Calculation of the VPgpUpUOH/negative-strand RNA molar ratioa
| Product | [32P]UMP countb | Total mol of UMP/molecule | Counts/molecule | Molar ratio of VPgpUpUOH/negative-strand RNA |
|---|---|---|---|---|
| VPgpUpUOH | 67,204 | 2 | 33,602 | 119:1 |
| Negative-strand RNA | 505,550 | 1,792 | 282 |
The values shown were from the 30-min time point in Fig. 1.
The values shown are phosphorimage units.
The amounts of [32P]UMP in VPgpUpUOH and negative-strand RNA products varied between 100 and 400 VPgpUpUOH molecules per negative-strand RNA, depending on the experiment (data not shown). Variability in the molar ratios from experiment to experiment may be due to slight variations in the magnitudes of translation from one experiment to another (thereby altering the amounts of VPg precursors available for conversion to VPgpUpUOH), to slight variations in the recovery of reaction products from experiment to experiment (note processing of reaction mixtures necessary for gel electrophoresis), or to other considerations. Because PV mRNA appears to function simultaneously as a template for both negative-strand RNA synthesis and CRE-dependent VPg uridylylation (Fig. 1), we predicted that the elongation of 3DPol through CRE ribonucleoprotein (RNP) complexes during negative-strand RNA synthesis might inhibit ongoing VPg uridylylation, thereby affecting the amounts of VPgpUpUOH made during negative-strand RNA synthesis (21). To test this possibility, we used 3′-dCTP to inhibit the elongation of 3DPol during negative-strand RNA synthesis (Fig. 2), we validated the ability of 3′-dCTP to block the elongation of 3DPol into the heteropolymeric region of negative-strand RNA (Fig. 3), and we reexamined the amounts of VPgpUpUOH made under conditions where 3DPol could not elongate through CRE RNP complexes (Fig. 4, 5, and 6).
FIG. 2.
3′-dCTP inhibited the elongation of nascent negative-strand RNA. (A) Model depicting termination of negative-strand RNA elongation by 3′-dCTP. (B) Structure of 3′-dCTP. (C and D) PIRCs containing PV RNA2 were incubated for 60 min in 50-μl reaction mixtures containing 50 μCi [α-32P]UTP (1.25 μM UTP, 800 Ci/mmol UTP) and 250 μM CTP (lane 1) or 50 to 200 μM 3′-dCTP (lanes 2 to 4). Two millimolar guanidine HCl (GuHCl) was included in a control reaction mixture containing 250 μM CTP (lane 5). RNA products of the reactions were separated by electrophoresis in a nondenaturing 1% agarose-TBE gel. (C) PV RNA2 template (asterisk) and rRNA were detected when the gel was stained with ethidium bromide and visualized with UV light (Ethidium panel). (D) Phosphorimaging revealed the incorporation of radiolabel into RF RNA along with faint labeling of RNA products comigrating with PV RNA2 template [lane 4, VPg-poly(U) 3′-dCMP]. Background labeling of 28S rRNA is evident in each lane (1 to 5). ssRNA size markers (in kilobases) are indicated on the left in panel C.
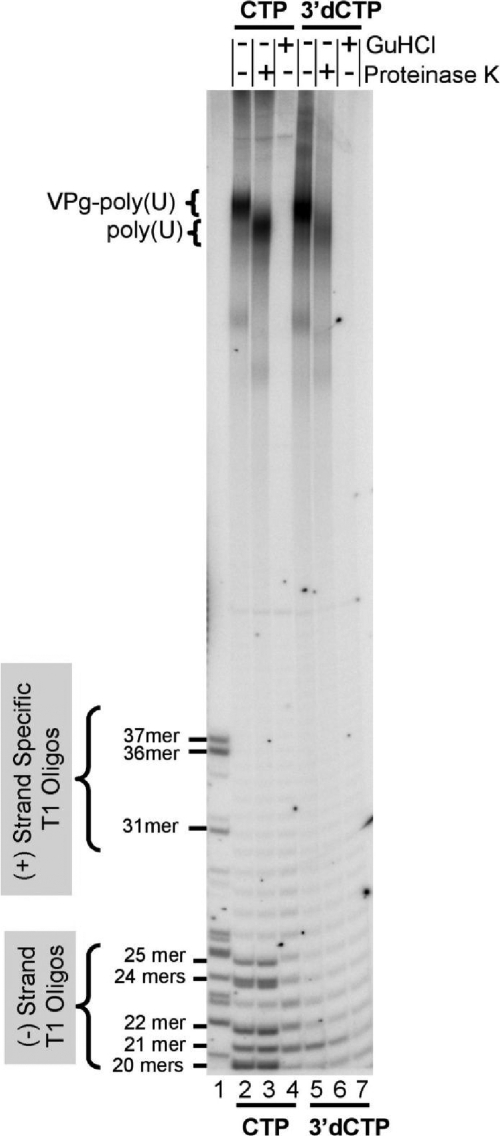
FIG. 3.
3′-dCTP blocks negative-strand RNA elongation immediately after copying the poly(A) tail into VPg-linked poly(U). Denaturing 7 M urea-20% polyacrylamide gel of negative-strand RNA products from 40-μl reaction mixtures containing 150 μCi of [α-32P]UTP (final concentration of 4.7 μM UTP; 800 Ci/mmol UTP) without (lanes 2 to 4) and with (lanes 5 to 7) 3′-dCTP. RNA products were digested with 30 U/μl RNase T1 only (lanes 2, 4, 5, and 7) or RNase T1 and 200 μg/ml proteinase K (lanes 3 and 6). RNA2 template labeled with [α-32P]UTP digested with RNase T1 is in lane 1. Sizes of positive-strand RNA template fragments, negative-strand RNA fragments, and poly(U) size with or without VPg are labeled. GuHCl, guanidine HCl.
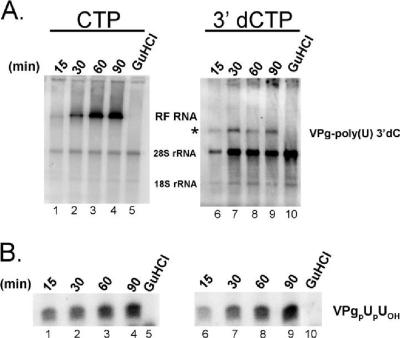
FIG. 4.
Kinetics and amounts of negative-strand RNA synthesis and VPgpUpUOH synthesis measured by continuous labeling. (A) Negative-strand RNA synthesis. PIRCs containing PV RNA2 templates were incubated for the indicated periods of time in 45-μl reaction mixtures containing 45 μCi [α-32P]UTP in the absence (lanes 1 to 4 and 6 to 9) or presence (lanes 5 and 10) of 2 mM guanidine HCl (GuHCl). Reaction mixtures contained 1× S10 buffer along with 1 mM ATP, 250 μM GTP, 10 μM UTP, and endogenous CTP (lanes 1 to 5) or 200 μM 3′-dCTP (lanes 6 to 10). RNA products of the reactions were separated by electrophoresis in a nondenaturing 1% agarose-TBE gel. Phosphorimaging revealed the incorporation of radiolabel into RF RNA (lanes 1 to 4) along with faint labeling of RNA products comigrating with PV RNA2 template [lanes 6 to 9, VPg-poly(U) 3′-dCMP]. The asterisk indicates mobility of the PV RNA2 template. (B) VPgpUpUOH synthesis. Reactions were performed in duplicate with those in panel A. VPgpUpUOH products were separated on a polyacrylamide-Tris-Tricine gel and detected by phosphorimaging. The mobility of VPgpUpUOH is indicated. VPgpUOH products migrate slightly faster than VPgpUpUOH products in the gel.
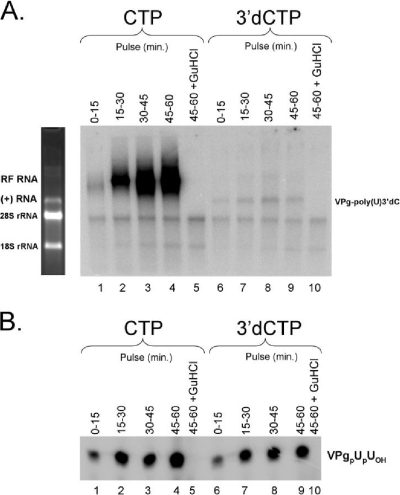
FIG. 5.
Kinetics and amounts of negative-strand RNA synthesis and VPgpUpUOH synthesis measured by pulse-labeling. (A) Negative-strand RNA synthesis. PIRCs containing PV RNA2 templates were incubated in the absence (lanes 1 to 4 and 6 to 9) or presence (lanes 5 and 10) of 2 mM guanidine HCl (GuHCl) in 40-μl reaction mixtures containing 1× S10 buffer along with 1 mM ATP, 250 μM GTP, 10 μM UTP, and endogenous CTP (lanes 1 to 5) or 200 μM 3′-dCTP (lanes 6 to 10). Forty microcuries of [α-32P]UTP was added to each 40-μl reaction mixture for 15-min periods of time as indicated. Reactions were terminated after the indicated pulse-labeling period, and RNA products of the reactions were separated by electrophoresis in a nondenaturing 1% agarose-TBE gel. Phosphorimaging revealed the incorporation of radiolabel into RF RNA (lanes 1 to 4) along with faint labeling of RNA products comigrating with PV RNA2 template [lanes 6 to 9, VPg-poly(U) 3′-dCMP]. (B) VPgpUpUOH synthesis. Reactions were performed in duplicate with those in panel A. VPgpUOH and VPgpUpUOH products were separated on a polyacrylamide-Tris-Tricine gel and detected by phosphorimaging. The mobility of VPgpUpUOH is indicated. VPgpUOH products migrate slightly faster than VPgpUpUOH products in the gel.
FIG. 6.
RNA replication and VPg uridylylation measured by pulse-labeling. (A) Negative- and positive-strand RNA synthesis. PIRCs containing PV DNVR27 RNA templates were incubated in the absence (lanes 1 to 4 and 6 to 9) or presence (lanes 5 and 10) of 2 mM guanidine HCl (GuHCl) in 40-μl reaction mixtures containing 1× S10 buffer along with 1 mM ATP, 250 μM GTP, 100 μM UTP, and endogenous CTP (lanes 1 to 5) or 200 μM 3′-dCTP (lanes 6 to 10). Forty microcuries of [α-32P]UTP was added to the 40-μl reaction mixtures for 15-min periods of time as indicated. Reactions were terminated after the indicated pulse-labeling period, and RNA products of the reactions were separated by electrophoresis in a nondenaturing 1% agarose-TBE gel. Phosphorimaging revealed the incorporation of radiolabel into RF RNA, replicative intermediate RNA, and positive-strand RNA (lanes 1 to 4), along with faint labeling of RNA products comigrating with the PV DNVR27 template [lanes 6 to 9, VPg-poly(U) 3′-dCMP]. (B) VPgpUpUOH synthesis. Reactions were performed in duplicate with those in panel A. VPgpUOH and VPgpUpUOH products were separated on a polyacrylamide-Tris-Tricine gel and detected by phosphorimaging. The mobility of VPgpUpUOH is indicated. VPgpUOH products migrate slightly faster than VPgpUpUOH products in the gel.
3′-dCTP inhibited 3DPol elongation during negative-strand RNA synthesis.
Negative-strand RNA synthesis initiates on the 3′ poly(A) tail of positive-strand RNA templates (Fig. 2A). Incorporation of 3′-dCTP into negative-strand RNA products should prevent further elongation of 3DPol due to the absence of the 3′ hydroxyl in 3′-dCMP (36), thereby inhibiting negative-strand RNA synthesis (Fig. 2A and B). When increasing concentrations of 3′-dCTP were included in the reaction mixtures containing PIRCs, the amounts of [32P]UMP incorporated into RF RNA decreased and the mobility of radiolabeled product RNAs increased to coincide with the mobility of template RNAs (Fig. 2C, lanes 2 to 4). Ethidium bromide staining and UV light were used to detect PV RNA templates within the cell-free reaction mixtures (Fig. 1B, left panel, and 2C). RF RNA was not detected by ethidium bromide and UV light (Fig. 2C and D, compare lanes 1; note that ethidium bromide reveals input PV RNA templates but does not reveal RF RNA), indicating that the majority of PV RNA templates within the reaction mixtures were not copied into negative-strand RNA products.
Because the sequence of PV RNA adjacent to the poly(A) tail is 5′-GGAGpoly(A), we predicted that 3′-dCTP would be incorporated into negative-strand RNA immediately after 3DPol copied the 3′ poly(A) tail (Fig. 2A). The ability of 3′-dCTP to prevent the incorporation of [32P]UMP into RF RNA is consistent with this possibility, the comigration of the putative VPg-poly(U) 3′-dCMP product with PV RNA templates is consistent with this possibility, and the reduced amount of [32P]UMP incorporated into the putative VPg-poly(U) 3′-dCMP is consistent with this possibility (Fig. 2C and D, lanes 1 and 4). The amount of [32P]UMP incorporated into VPg-poly(U) 3′-dCMP [∼80 mol of UMP per mol of VPg-poly(u)] was about 1/20 of that incorporated into RF RNA (∼1,792 mol of UMP per mol of negative strand) [Fig. 2D, compare RF RNA in lane 1 with the VPg-poly(U) 3′-dCMP RNA product in lane 4]. This is consistent with 3′-dCTP blocking 3DPol elongation soon after the synthesis of heteropolymeric negative-strand RNA.
VPg-poly(U) 3′-dCMP products analyzed by RNase T1 fingerprinting.
In order to confirm that 3′-dCTP prevented the elongation of 3DPol into the heteropolymeric portion of PV RNA templates, we examined the RNase T1 oligonucleotides in PV negative-strand RNA (Fig. 3). PV negative-strand RNA has a distinct RNase T1 fingerprint where the largest fragment is a 25-mer, followed by two 24-mers, no 23-mers, a 22-mer, a 21-mer, and six 20-mers. The 21-mer, the 25-mer, and one of the 24-mers are all within the 5′ half of the negative-strand RNA. The 21-mer is only 35 nt after the poly(U) sequence at the 5′ end of the negative-strand RNA.
PV RF RNA digested with RNase T1 yielded RNase T1 oligonucleotides characteristic of full-length negative-strand RNA, including the 25-mer, 24-mers, and 21-mer (Fig. 3, lanes 2 and 3). VPg-linked poly(U) from the 5′ end of negative-strand RNA was detected near the top of the polyacrylamide gel (Fig. 3, lane 2). Proteinase K treatment increased the mobility of VPg-poly(U) products without affecting the mobilities of other T1 oligonucleotides (Fig. 3, compare lanes 2 and 3). RNase T1 digestion of radiolabeled products from reaction mixtures containing 3′-dCTP revealed the presence of VPg-poly(U) from the 5′ end of the negative-strand RNA (Fig. 3, lanes 5 and 6); however, RNase T1 oligonucleotides from the heteropolymeric portion of the negative-strand RNA were not detected (Fig. 3, lanes 5 and 6, note the absence of the PV 25-mer, 24-mers, 22-mer, 21-mer, and 20-mers). The absence of the 21-mer in T1-digested products from reaction mixtures containing 3′-dCTP indicated that 3′-dCTP prevented elongation of 3DPol somewhere within the 3′ NTR of the template. Nonviral (background) oligonucleotides between 20 and 30 bases long were detected in the products of reaction mixtures containing 2 mM guanidine HCl (Fig. 3, lanes 4 and 7). Similar background bands were seen in this region of the gel from 3′-dCTP reaction products (Fig. 3, compare lanes 5 and 6 with lanes 4 and 7). These data confirm that 3′-dCTP effectively inhibited the elongation of 3DPol into the heteropolymeric portion of PV RNA templates without inhibiting the initiation of negative-strand RNA synthesis, as illustrated in Fig. 2A. Furthermore, VPg-poly(U) 3′-dCMP products comigrated with PV RNA templates (Fig. 2C and D) due to the complementarity of VPg-poly(U) 3′-dCMP products with the poly(A) template (Fig. 2A). We monitored the synthesis of VPg-poly(U) 3′-dCMP as a marker of the initiation of negative-strand RNA synthesis in subsequent experiments.
VPgpUpUOH synthesis in the presence and absence of 3′-dCTP.
Because PV mRNA appears to function simultaneously as a template for both negative-strand RNA synthesis and CRE-dependent VPg uridylylation (Fig. 1), we predicted that the elongation of 3DPol through CRE RNP complexes during negative-strand RNA synthesis might inhibit ongoing VPg uridylylation. To test this possibility, we compared the amounts of VPgpUpUOH made in reaction mixtures with and without 3′-dCTP (Fig. 4). As established above, 3′-dCTP prevented the elongation of 3DPol into the heteropolymeric region of PV RNA templates (Fig. 4A). VPg-poly(U) 3′-dCMP was synthesized in reaction mixtures containing 3′-dCTP (Fig. 4A, right panel), whereas RF RNA was synthesized in reaction mixtures without 3′-dCTP (Fig. 4A, left panel). Contrary to our predictions, VPgpUpUOH synthesis was unaffected when 3′-dCTP was used to inhibit the elongation of 3DPol through the CRE structure (Fig. 4B, compare the left and right panels). Thus, inhibiting the elongation of 3DPol with 3′-dCTP did not increase or decrease the amounts of VPg uridylylation observed during negative-strand RNA synthesis.
To more comprehensively examine when VPg is converted into VPgpUpUOH during viral RNA replication, we used 15 min of pulse-labeling to examine VPgpUpUOH synthesis in reaction mixtures containing PIRCs, as described below.
Pulse-labeling to determine when VPg uridylylation occurs relative to negative- and positive-strand RNA synthesis.
PIRCs synchronously and sequentially synthesize negative- and positive-strand RNA (6). Under the conditions used in Fig. 1, it took approximately 20 min to synthesize full-length negative-strand RNA on the 5,741-base-long PV RNA2 template (Fig. 1). This corresponds to an elongation rate of ∼287 bases/min. Under these conditions, it would take approximately 10 min for 3DPol elongating nascent negative-strand RNAs to reach and copy through the CRE RNA 3,000 bases upstream from the 3′ poly(A) tail of PV RNA templates. Therefore, we used pulse-labeling to determine whether VPgpUpUOH was made before and/or after the elongation of 3DPol through CRE RNA structures within template RNAs.
[32P]UTP was incorporated into nascent negative-strand RNA from 0 to 15 min following the removal of guanidine HCl from replication reaction mixtures (Fig. 5A, lane 1). Full-length negative-strand RNA was radiolabeled with pulse-labels from 15 to 30, 30 to 45, and 45 to 60 min (Fig. 5A, lanes 2 to 4). The incorporation of radiolabel into negative-strand RNA from 45 to 60 min after the removal of guanidine was unexpected because it only takes 20 to 30 min to make a full-length negative-strand RNA molecule under these conditions. The synthesis of negative-strand RNA after 30 min of incubation could be due to delayed initiation of negative-strand RNA synthesis in a subpopulation of PIRCs within reaction mixtures following the removal of guanidine and/or to paused 3DPol elongation on some negative-strand RNAs in the reaction mixtures.
Pulse-labeling with [α-32P]UTP in reaction mixtures containing 3′-dCTP indicated that VPg-poly(U) 3′-dCMP was made during the initial pulse from 0 to 15 min, as well as during pulses from 15 to 30 min, 30 to 45 min, and 45 to 60 min (Fig. 5A, lanes 6 to 9). The ongoing presence of 2 mM guanidine prevented the incorporation of radiolabel into VPg-poly(U) 3′-dCMP (Fig. 5A, lane 10). The incorporation of [α-32P]UTP into VPg-poly(U) 3′-dCMP long after the synthesis of RF RNA is most consistent with delayed initiation of negative-strand RNA synthesis. RNase T1 fingerprints revealed that T1 oligonucleotides corresponding to the 3′ end of negative-strand RNAs were present at a one-to-one molar ratio with T1 oligonucleotides from the 5′ end of negative-strand RNAs (Fig. 3; unpublished data). Therefore, we see no quantitative evidence of paused elongation between VPg-poly(U) at the 5′ end of negative-strand RNAs and T1 oligonucleotides at the 3′ end of negative-strand RNA. This does not exclude the possibility of paused elongation on a small fraction of templates, but paused elongation on a small number of templates would not affect our interpretations or conclusions from data representing the majority of the RNA population. VPgpUpUOH was synthesized in each 15-min pulse during the 60-min period following the removal of guanidine HCl (Fig. 5B, lanes 1 to 4 and 6 to 9). The ongoing presence of 2 mM guanidine prevented the incorporation of radiolabel into VPgpUpUOH (Fig. 5B, lanes 5 and 10). Thus, VPgpUpUOH synthesis was detected both before and after the synthesis of full-length negative-strand RNAs; however, it appeared as if a subpopulation of PIRCs within the reaction mixtures was delaying initiation, contributing to the synthesis of VPgpUpUOH coordinate in time with ongoing negative-strand RNA synthesis.
To further characterize the timing of VPg uridylylation relative to negative- and positive-strand RNA synthesis, we performed the experiment described above with a template capable of both negative- and positive-strand RNA synthesis (Fig. 6). RF RNA was radiolabeled during the 0- to 15-min and 15- to 30-min periods, consistent with negative-strand RNA synthesis after the removal of guanidine HCl (Fig. 6A, lanes 1 to 2). RF, replicative intermediate, and progeny positive-strand RNAs were radiolabeled during the 30- to 45-min and 45- to 60-min periods (Fig. 6A, lanes 3 and 4). As before, VPg-poly(U) 3′-dCMP was detected in each 15-min pulse during the 60-min period following the removal of guanidine HCl (Fig. 6A, lanes 6 to 9). In addition, VPgpUpUOH was synthesized in each 15-min pulse during the 60-min period, including during positive-strand RNA synthesis (Fig. 6B, lanes 1 to 4).
VPg-poly(U) 3′-dCMP synthesis throughout the time course and VPgpUpUOH synthesis during positive-strand RNA synthesis were consistent with the possibility that some RCs were delaying initiation, thereby confounding our interpretations of when VPg was converted into VPgpUpUOH relative to negative- and positive-strand RNA synthesis. Therefore, we used guanidine readdition to block delayed initiation of RNA replication and reexamined VPg uridylylation under such conditions (Fig. 7).
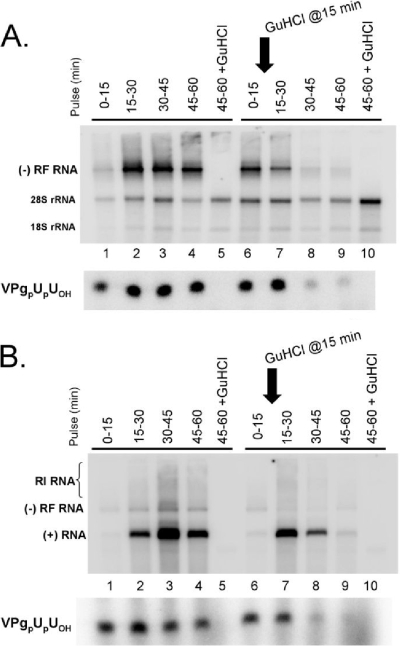
FIG. 7.
Guanidine inhibited asynchronous initiation of negative-strand RNA synthesis and CRE-dependent VPg uridylylation. PIRCs containing PV RNA2 templates (A) or DNVR27 RNA templates (B) were incubated in the absence (lanes 1 to 4 and 6 to 9) or presence (lanes 5 and 10) of 2 mM guanidine HCl (GuHCl) in 40-μl reaction mixtures containing 1× S10 buffer, 1 mM ATP, 250 μM GTP, 250 μM CTP, and 100 μM UTP. Forty microcuries of [α-32P]UTP was added to the 40-μl reaction mixtures for 15-min periods of time as indicated. Two millimolar guanidine HCl was readded to reaction mixtures after being absent for 15 min (lanes 7 to 9). Reactions were terminated after the indicated pulse-labeling period, and RNA products of the reactions were separated by electrophoresis in a nondenaturing 1% agarose-TBE gel to detect viral RNAs (top of panels A and B) or a Tris-Tricine-polyacrylamide gel to detect VPgpUOH and VPgpUpUOH (bottom of panels A and B). The mobility of VPgpUpUOH is indicated. VPgpUOH products migrate slightly faster than VPgpUpUOH products in the gel.
Guanidine HCl blocks delayed negative-strand RNA synthesis and delayed VPg uridylylation.
Fifteen-minute pulse-labeling was used to detect the replication of viral RNA and the synthesis of VPgpUpUOH in reaction mixtures containing PV RNA2 templates capable of negative-strand RNA replication (Fig. 7A) and in reaction mixtures containing PV RNA2 ribozyme templates capable of both negative- and positive-strand RNA replication (Fig. 7B). Based on the amounts of [32P]UMP incorporated into VPgpUpUOH and RF RNA, 300 to 400 molecules of VPgpUpUOH were made for every negative-strand RNA molecule in this experiment (Fig. 7A). Readdition of guanidine HCl after 15 min of incubation in the absence of guanidine did not immediately inhibit ongoing negative-strand RNA synthesis or ongoing VPgpUpUOH synthesis, as has been shown previously (Fig. 7A, lane 7, 15- to 30-min pulse) (6). However, after being present for 15 min, guanidine did inhibit delayed negative-strand RNA synthesis and delayed VPgpUpUOH synthesis (Fig. 7A, compare lanes 8 and 9 with lanes 3 and 4). Thus, ongoing negative-strand RNA elongation and ongoing VPg uridylylation continued for up to 15 min in the presence of 2 mM guanidine (Fig. 7A, lane 7) but ceased thereafter (Fig. 7A, lanes 8 and 9). Comparable results were obtained with templates capable of both negative- and positive-strand RNA replication (Fig. 7B). Thus, consistent with previously published results (6), 2 mM guanidine HCl does not inhibit either ongoing negative- or positive-strand RNA synthesis but does inhibit the initiation of negative-strand RNA synthesis. Likewise, 2 mM guanidine HCl inhibited the initiation of VPg uridylylation rather than ongoing VPg uridylylation.
Importantly, following the readdition of guanidine, ongoing VPg uridylylation ceased coordinate in time with completion of ongoing negative-strand RNA synthesis (Fig. 7A, compare lanes 7 and 8). This tight correlation between the timing of VPg uridylylation and negative-strand RNA synthesis suggested that the process of VPg uridylylation occurred coordinately in time with negative-strand RNA synthesis (Fig. 7A). Furthermore, the completion of most negative-strand RNA synthesis within 15 min after the readdition of guanidine indicated that 3DPol completed negative-strand RNA elongation without significant pausing during elongation (Fig. 7A, compare lanes 8 and 9 with lanes 3 and 4). These data reinforce our conclusion that the incorporation of [32P]UMP into VPg-poly(U) 3′-dCMP coincides with delayed initiation of negative-strand RNA synthesis rather than paused elongation by 3DPol following initiation.
Conversion of VPg into VPgpUpUOH before negative-strand RNA synthesis.
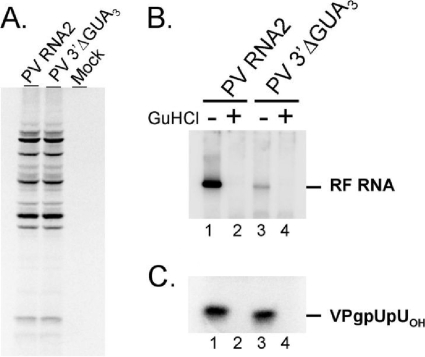
Based on the results described above, especially those in Fig. 7A, we concluded that VPg was converted into VPgpUpUOH coincident in time with negative-strand RNA elongation. Nonetheless, such results do not exclude the possibility that VPg can be converted into VPgpUpUOH before (and independent from) negative-strand RNA elongation. To test whether the initiation of negative-strand RNA synthesis is required for CRE-dependent VPg uridylylation within PIRCs, we used a 3′ NTR mutation to inhibit negative-strand RNA synthesis while simultaneously examining the amount of VPgpUpUOH synthesis (Fig. 8). A deletion of residues within the 3′ NTR (3′ΔGUA3) did not affect the amount of PV mRNA translation or polyprotein processing (Fig. 8A). The 3′ NTR mutation inhibited negative-strand RNA synthesis, as expected (Fig. 8B), without affecting the amount of VPg uridylylation (Fig. 8C). Negative-strand RNA synthesis was reduced to 8.4% of the wild-type level, while VPgpUpUOH synthesis was 99% of the wild-type level (Fig. 8B and C). 3′ NTR mutations that inhibit RNA replication are commonly thought to block the initiation of negative-strand RNA synthesis (46). The 3′ΔGUAAA mutation diminished incorporation of radiolabel into RF RNA, and radiolabel corresponding to VPg-poly(U) was not detected (Fig. 8B). Thus, the 3′ΔGUAAA mutation appeared to inhibit the initiation of negative-strand RNA synthesis without affecting VPg uridylylation. These results indicate that CRE-dependent VPg uridylylation can occur independent of negative-strand RNA synthesis just as negative-strand RNA synthesis can occur independent of CRE-dependent VPg uridylylation (14, 20, 21). Thus, CRE-dependent VPg uridylylation converted VPg into VPgpUpUOH both before and during negative-strand RNA synthesis.
FIG. 8.
Initiation of negative-strand RNA synthesis does not necessarily precede CRE-dependent VPg uridylylation. (A) PV RNA2 and 3′ΔGUA3 protein synthesis. [35S]methionine-labeled proteins were separated by electrophoresis in an SDS-9 to 18% polyacrylamide gel. The mock-treated control lacked PV RNA. (B) Negative-strand RNA synthesis. PV RNA2 (lanes 1 and 2) and PV 3′ΔGUA3 (lanes 3 and 4) RNA products labeled with 40 μCi of [α-32P]UTP in 40-μl reaction mixtures (final concentration of 1.25 μM UTP [800 Ci/mmol UTP]) in the presence (+) or absence (−) of 2 mM guanidine HCl (GuHCl) were separated by electrophoresis in a 1% agarose-TBE gel. (C) VPgpUpUOH synthesis. VPgpUOH and VPgpUpUOH products from PV RNA2 and PV 3′ΔGUA3 were separated by electrophoresis in a Tris-Tricine gel. All gels were dried, and radiolabel was detected with a PhosphorImager. The mobility of VPgpUpUOH is indicated. VPgpUOH products migrate slightly faster than VPgpUpUOH products in the gel.
DISCUSSION
The conversion of VPg into VPgpUpUOH is a critical step of picornavirus RNA replication because VPgpUpUOH is a primer for viral RNA replication (26). A CRE RNA structure at various positions within picornavirus genomes functions as a template for 3DPol-catalyzed addition of UTP to the tyrosine hydroxyl of VPg, resulting in the synthesis of VPgpUpUOH (reviewed in reference 37). It is important to determine when VPg is converted into VPgpUpUOH relative to negative- and positive-strand RNA synthesis to help understand how picornaviruses coordinate the synthesis of primers with the other steps of viral RNA replication. Therefore, we used [α-32P]UTP to detect the synthesis of VPgpUpUOH during the sequential synthesis of PV negative- and positive-strand RNAs. We found that VPg can be converted into VPgpUpUOH both before (Fig. 8) and during (Fig. 1 and 4 to 7) negative-strand RNA synthesis. These data are consistent with the ability of VPgpUpUOH to prime both negative- and positive-strand RNA synthesis (43). While VPg can prime negative-strand RNA synthesis in a CRE- and VPgpUpUOH-independent manner (14, 20, 21), accumulating evidence is consistent with the possibility that VPgpUpUOH made on CRE RNA templates functions as the predominant primer of negative-strand RNA synthesis (38). It is already clear that VPgpUpUOH made on CRE RNA templates functions as the primer for positive-strand RNA synthesis (20, 21). The synthesis of abundant amounts of VPgpUpUOH (100 to 400 VPgpUpUOH molecules per negative-strand RNA, Table 1 and Fig. 1 and 7A) before and during negative-strand RNA synthesis provides the primers needed for asymmetric replication of positive-strand RNA. It remains to be determined how VPgpUpUOH molecules and 3DPol are translocated from CRE RNAs to the 3′ termini of positive- and negative-strand RNA templates (23, 37).
The removal of guanidine from PIRCs allowed PV RNA to function simultaneously as a template for both VPg uridylylation and negative-strand RNA synthesis (Fig. 1). While it is possible that VPg uridylylation and negative-strand RNA synthesis occurred on distinct subpopulations of PV RNA templates, several lines of evidence suggest that VPg uridylylation and negative-strand RNA synthesis occur simultaneously on PV RNA templates within membrane-associated RNA RCs (as illustrated in Fig. 1A). First, under the conditions of our experiments, guanidine inhibited both VPg uridylylation and negative-strand RNA synthesis, indicating that the PV RNA templates used for these two processes are associated with 2CATPase membrane-anchored RNA RCs. Second, the 5′ cloverleaf of PV RNA templates is required in cis for both CRE-dependent VPg uridylylation and negative-strand RNA synthesis (9, 19), indicating that cis-active structures at the 5′ and 3′ termini of PV RNA templates function coordinately with CRE RNA sequences within PIRCs. Third, CRE-dependent VPg uridylylation increases the amounts of negative-strand RNA synthesis within PIRCs, suggesting that the VPgpUpUOH products of CRE-dependent VPg uridylylation can prime the initiation of negative-strand RNA synthesis (38). In addition, CRE-dependent VPg uridylylation is strictly required for positive-strand RNA synthesis within PIRCs (14, 20, 21). VPgpUpUOH products of CRE-dependent VPg uridylylation prime the initiation of positive-strand RNA synthesis via their complementarity to adenosine residues at the 3′ end of negative-strand RNA intermediates (35). Taken together, these data indicate that CRE-dependent VPg uridylylation and PV negative- and positive-strand RNA synthesis occur in concert with one another within PIRCs.
Pulse-labeling experiments in which the pulse-labeling period corresponded to the time required by 3DPol to traverse the viral RNA templates established that the conversion of VPg into VPgpUpUOH commenced and concluded coincident in time with negative-strand RNA synthesis (Fig. 7). Because PV RNA was functioning simultaneously for two synthetic events (CRE-dependent VPg uridylylation and negative-strand RNA synthesis), 3DPol must move through CRE RNPs (22) during the elongation of negative-strand RNA (Fig. 1). Therefore, we tested whether 3DPol elongation through CRE RNPs affected the kinetics or magnitude of CRE-dependent VPg uridylylation. 3′-dCTP inhibited the elongation of 3DPol through CRE RNPs in PV RNA templates (Fig. 2 and 3) but did not affect the amount of VPgpUpUOH made over time (Fig. 4 to 6) or the synthesis of VPg-poly(U) corresponding to the 5′ end of negative-strand RNA (Fig. 3). These data indicated that the elongation of 3DPol during negative-strand RNA synthesis had no effect on CRE-dependent VPg uridylylation.
The ability of 2 mM guanidine to inhibit both VPgpUpUOH synthesis and negative-strand RNA synthesis implicates 2CATPase in the mechanisms by which PV RNA functions as a template for both CRE-dependent VPg uridylylation and negative-strand RNA synthesis (Fig. 1 to 8). The requirement of 2CATPase activity suggests that the conversion of VPg into VPgpUpUOH and the synthesis of negative-strand RNA both occurred within membrane-based RCs because 2CATPase is a membrane-associated protein that anchors viral RNA templates and replication intermediates within RCs (34, 40). While purified 3DPol can convert VPg into VPgpUpUOH in reaction mixtures containing CRE RNA templates (22, 26), in the absence of membranes and the absence of 2CATPase, such reaction mixtures do not support asymmetric RNA replication. CRE RNAs function coordinately with the other cis-active RNA sequences and structures at the 5′ and 3′ termini of picornavirus RNA templates and with the full complement of viral RNA replication proteins during normal RNA replication. 2CATPase is critical in normal membranous RNA RCs; however, the mechanistic function of 2CATPase activity is unclear. While it has long been considered a putative helicase (18), diligent efforts failed to detect 2C-mediated helicase activity (29, 30). We found that 2 mM guanidine inhibited the synthesis of VPg-poly(U) corresponding to the 5′ end of negative-strand RNA (Fig. 3). Furthermore, 2 mM guanidine needed to be removed from PIRCs to allow for CRE-dependent VPg uridylylation; however, readdition of 2 mM guanidine did not immediately inhibit ongoing VPg uridylylation (Fig. 7). Thus, there is a requirement for 2CATPase activity before and/or at the moment of initiation of RNA replication (Fig. 7); however, guanidine-inhibited 2CATPase activity does not appear to be required for ongoing negative-strand RNA elongation, as previously established (6), or for ongoing CRE-dependent VPg uridylylation (Fig. 7). These data and previous studies (29, 30) made us consider potential mechanistic roles for 2CATPase other than helicase activity. We currently favor the possibility that 2CATPase functions as an RNP chaperone to mediate the formation of functional RNP complexes on viral cis-active RNA structures, a mechanism previously suggested by Pfister and Wimmer. We consider this function to be similar to the RNP chaperone activities of SM-like proteins, which are required to assemble functional RNP complexes involved in RNA splicing (28).
snRNP complexes can self-assemble by combining individual protein and RNA subunits in vitro (in the absence of survival of motor neurons [SMN] complexes), but the ATPase activity of SMN complexes is required for the assembly and activity of snRNPs in cells (28). snRNPs can self-assemble inappropriately in vitro on U-rich RNAs other than snRNAs, leading to nonfunctional RNPs. SMN complexes are machines capable of preventing the assembly of snRNPs on U-rich RNAs other than snRNAs (28). The ATPase activity of particular subunits within SMN complexes is necessary for this ability to prevent assembly of illicit nonfunctional snRNPs (3, 13, 15, 27, 28, 44, 45). Thus, SMN complexes, through ATPase activity, function as RNP chaperones to prevent assembly of illicit nonfunctional snRNPs (28). We hypothesize that ATPases within viral RCs may function in a similar manner. In the case of PV, we speculate that 2CATPase may function as an RNP chaperone within membranous RCs to coordinately assemble 5′ cloverleaf, 2C-CRE, and 3′NTR RNPs. Such 2CATPase activities could coordinate CRE-dependent VPg uridylylation with VPgpUpUOH priming at the 3′ termini of viral RNA templates. This hypothesis accounts for some of the apparent paradoxes observed between experiments using purified subunits (reconstitution experiments) and work using PIRCs (2). Purified 3DPol can be used to make negative-strand RNA in vitro; however, 3DPol will use both viral and nonviral template RNAs and oligonucleotide primers rather than VPg or VPgpUpUOH (42). Using purified 3DPol, VPg can be uridylylated in vitro in the absence of membranes or 2CATPase (24, 25, 33); however, within PIRCs, VPg uridylylation requires the 5′ cloverleaf and 2CATPase activity and VPg uridylylation occurs predominantly on CRE RNA sequences within PV RNA templates rather than on 3′ poly(A) sequences of PV RNA templates (20, 21). VPg uridylylation requires membranes when natural PV RCs are isolated from infected cells (39).
In the population of RCs within our reaction mixtures, negative-strand RNA synthesis did not begin and end in the time needed to synthesize full-length negative-strand RNA (Fig. 5A and 7A). Rather, a significant amount of delayed initiation of negative-strand RNA synthesis was detected [Fig. 5A and 6A, note ongoing VPg-poly(U) 3′-dCMP synthesis during each 15-min pulse]. Substantial amounts of VPg-poly(U) 3′-dCMP, corresponding to the 5′ end of nascent negative-strand RNAs (illustration in Fig. 2) were made as late as 45 to 60 min after the removal of guanidine (Fig. 5A, lane 9, and 6A, lane 9). The reason for substantial amounts of delayed initiation is unclear; however, the ability of guanidine to inhibit delayed initiation (Fig. 7A, lanes 8 and 9) invokes a role for 2CATPase. It is possible that the interactions between 5′ and 3′ RNP complexes involved in the initiation of RNA replication (Fig. 1A) are not properly oriented in a portion of RCs and that when they arise (perhaps due to 2CATPase-mediated reorientation), they initiate replication.
Thus, we found that 2CATPase activity within PIRCs allowed PV RNA to function simultaneously as a template for both CRE-dependent VPg uridylylation and negative-strand RNA synthesis. VPg was converted into VPgpUpUOH both before and during negative-strand RNA synthesis, with 100 to 400 VPgpUpUOH molecules synthesized per negative-strand RNA.
Acknowledgments
This work was supported by NIH grants AI042189 and T32AI052066.
Footnotes
Published ahead of print on 7 October 2009.
REFERENCES
- 1.Adams, P., E. Kandiah, G. Effantin, A. C. Steven, and E. Ehrenfeld. 2009. Poliovirus 2C protein forms homo-oligomeric structures required for ATPase activity. J. Biol. Chem. 284:22012-22021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agol, V. I., A. V. Paul, and E. Wimmer. 1999. Paradoxes of the replication of picornaviral genomes. Virus Res. 62:129-147. [DOI] [PubMed] [Google Scholar]
- 3.Baccon, J., L. Pellizzoni, J. Rappsilber, M. Mann, and G. Dreyfuss. 2002. Identification and characterization of Gemin7, a novel component of the survival of motor neuron complex. J. Biol. Chem. 277:31957-31962. [DOI] [PubMed] [Google Scholar]
- 4.Banerjee, R., and A. Dasgupta. 2001. Interaction of picornavirus 2C polypeptide with the viral negative-strand RNA. J. Gen. Virol. 82:2621-2627. [DOI] [PubMed] [Google Scholar]
- 5.Barton, D. J., E. P. Black, and J. B. Flanegan. 1995. Complete replication of poliovirus in vitro: preinitiation RNA replication complexes require soluble cellular factors for the synthesis of VPg-linked RNA. J. Virol. 69:5516-5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barton, D. J., and J. B. Flanegan. 1997. Synchronous replication of poliovirus RNA: initiation of negative-strand RNA synthesis requires the guanidine-inhibited activity of protein 2C. J. Virol. 71:8482-8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barton, D. J., B. J. Morasco, and J. B. Flanegan. 1996. Assays for poliovirus polymerase, 3D(Pol), and authentic RNA replication in HeLa S10 extracts. Methods Enzymol. 275:35-57. [DOI] [PubMed] [Google Scholar]
- 8.Barton, D. J., B. J. Morasco, and J. B. Flanegan. 1999. Translating ribosomes inhibit poliovirus negative-strand RNA synthesis. J. Virol. 73:10104-10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barton, D. J., B. J. O'Donnell, and J. B. Flanegan. 2001. 5′ cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO J. 20:1439-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collis, P. S., B. J. O'Donnell, D. J. Barton, J. A. Rogers, and J. B. Flanegan. 1992. Replication of poliovirus RNA and subgenomic RNA transcripts in transfected cells. J. Virol. 66:6480-6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egger, D., and K. Bienz. 2005. Intracellular location and translocation of silent and active poliovirus replication complexes. J. Gen. Virol. 86:707-718. [DOI] [PubMed] [Google Scholar]
- 12.Fogg, M. H., N. L. Teterina, and E. Ehrenfeld. 2003. Membrane requirements for uridylylation of the poliovirus VPg protein and viral RNA synthesis in vitro. J. Virol. 77:11408-11416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friesen, W. J., A. Wyce, S. Paushkin, L. Abel, J. Rappsilber, M. Mann, and G. Dreyfuss. 2002. A novel WD repeat protein component of the methylosome binds Sm proteins. J. Biol. Chem. 277:8243-8247. [DOI] [PubMed] [Google Scholar]
- 14.Goodfellow, I. G., C. Polacek, R. Andino, and D. J. Evans. 2003. The poliovirus 2C cis-acting replication element-mediated uridylylation of VPg is not required for synthesis of negative-sense genomes. J. Gen. Virol. 84:2359-2363. [DOI] [PubMed] [Google Scholar]
- 15.Gubitz, A. K., Z. Mourelatos, L. Abel, J. Rappsilber, M. Mann, and G. Dreyfuss. 2002. Gemin5, a novel WD repeat protein component of the SMN complex that binds Sm proteins. J. Biol. Chem. 277:5631-5636. [DOI] [PubMed] [Google Scholar]
- 16.Herold, J., and R. Andino. 2001. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol. Cell 7:581-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jackson, W. T., T. H. Giddings, Jr., M. P. Taylor, S. Mulinyawe, M. Rabinovitch, R. R. Kopito, and K. Kirkegaard. 2005. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 3:e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kadaré, G., and A. L. Haenni. 1997. Virus-encoded RNA helicases. J. Virol. 71:2583-2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lyons, T., K. E. Murray, A. W. Roberts, and D. J. Barton. 2001. Poliovirus 5′-terminal cloverleaf RNA is required in cis for VPg uridylylation and the initiation of negative-strand RNA synthesis. J. Virol. 75:10696-10708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morasco, B. J., N. Sharma, J. Parilla, and J. B. Flanegan. 2003. Poliovirus cre(2C)-dependent synthesis of VPgpUpU is required for positive- but not negative-strand RNA synthesis. J. Virol. 77:5136-5144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murray, K. E., and D. J. Barton. 2003. Poliovirus CRE-dependent VPg uridylylation is required for positive-strand RNA synthesis but not for negative-strand RNA synthesis. J. Virol. 77:4739-4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pathak, H. B., J. J. Arnold, P. N. Wiegand, M. R. Hargittai, and C. E. Cameron. 2007. Picornavirus genome replication: assembly and organization of the VPg uridylylation ribonucleoprotein (initiation) complex. J. Biol. Chem. 282:16202-16213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paul, A. 2002. Possible unifying mechanism of picornavirus genome replication, p. 227-246. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, DC.
- 24.Paul, A. V., E. Rieder, D. W. Kim, J. H. van Boom, and E. Wimmer. 2000. Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg. J. Virol. 74:10359-10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paul, A. V., J. H. van Boom, D. Filippov, and E. Wimmer. 1998. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature 393:280-284. [DOI] [PubMed] [Google Scholar]
- 26.Paul, A. V., J. Yin, J. Mugavero, E. Rieder, Y. Liu, and E. Wimmer. 2003. A “slide-back” mechanism for the initiation of protein-primed RNA synthesis by the RNA polymerase of poliovirus. J. Biol. Chem. 278:43951-43960. [DOI] [PubMed] [Google Scholar]
- 27.Pellizzoni, L., J. Baccon, J. Rappsilber, M. Mann, and G. Dreyfuss. 2002. Purification of native survival of motor neurons complexes and identification of Gemin6 as a novel component. J. Biol. Chem. 277:7540-7545. [DOI] [PubMed] [Google Scholar]
- 28.Pellizzoni, L., J. Yong, and G. Dreyfuss. 2002. Essential role for the SMN complex in the specificity of snRNP assembly. Science 298:1775-1779. [DOI] [PubMed] [Google Scholar]
- 29.Pfister, T., K. W. Jones, and E. Wimmer. 2000. A cysteine-rich motif in poliovirus protein 2C(ATPase) is involved in RNA replication and binds zinc in vitro. J. Virol. 74:334-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pfister, T., and E. Wimmer. 1999. Characterization of the nucleoside triphosphatase activity of poliovirus protein 2C reveals a mechanism by which guanidine inhibits poliovirus replication. J. Biol. Chem. 274:6992-7001. [DOI] [PubMed] [Google Scholar]
- 31.Pincus, S. E., D. C. Diamond, E. A. Emini, and E. Wimmer. 1986. Guanidine-selected mutants of poliovirus: mapping of point mutations to polypeptide 2C. J. Virol. 57:638-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pincus, S. E., and E. Wimmer. 1986. Production of guanidine-resistant and -dependent poliovirus mutants from cloned cDNA: mutations in polypeptide 2C are directly responsible for altered guanidine sensitivity. J. Virol. 60:793-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rieder, E., A. V. Paul, D. W. Kim, J. H. van Boom, and E. Wimmer. 2000. Genetic and biochemical studies of poliovirus cis-acting replication element cre in relation to VPg uridylylation. J. Virol. 74:10371-10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodríguez, P. L., and L. Carrasco. 1995. Poliovirus protein 2C contains two regions involved in RNA binding activity. J. Biol. Chem. 270:10105-10112. [DOI] [PubMed] [Google Scholar]
- 35.Sharma, N., B. J. O'Donnell, and J. B. Flanegan. 2005. 3′-terminal sequence in poliovirus negative-strand templates is the primary cis-acting element required for VPgpUpU-primed positive-strand initiation. J. Virol. 79:3565-3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shim, J., G. Larson, V. Lai, S. Naim, and J. Z. Wu. 2003. Canonical 3′-deoxyribonucleotides as a chain terminator for HCV NS5B RNA-dependent RNA polymerase. Antivir. Res. 58:243-251. [DOI] [PubMed] [Google Scholar]
- 37.Steil, B. P., and D. J. Barton. 2009. cis-active RNA elements (CREs) and picornavirus RNA replication. Virus Res. 139:240-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steil, B. P., and D. J. Barton. 2008. Poliovirus cis-acting replication element-dependent VPg uridylylation lowers the Km of the initiating nucleoside triphosphate for viral RNA replication. J. Virol. 82:9400-9408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takegami, T., R. J. Kuhn, C. W. Anderson, and E. Wimmer. 1983. Membrane-dependent uridylylation of the genome-linked protein VPg of poliovirus. Proc. Natl. Acad. Sci. USA 80:7447-7451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Teterina, N. L., A. E. Gorbalenya, D. Egger, K. Bienz, and E. Ehrenfeld. 1997. Poliovirus 2C protein determinants of membrane binding and rearrangements in mammalian cells. J. Virol. 71:8962-8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tolskaya, E. A., L. I. Romanova, M. S. Kolesnikova, A. P. Gmyl, A. E. Gorbalenya, and V. I. Agol. 1994. Genetic studies on the poliovirus 2C protein, an NTPase. A plausible mechanism of guanidine effect on the 2C function and evidence for the importance of 2C oligomerization. J. Mol. Biol. 236:1310-1323. [DOI] [PubMed] [Google Scholar]
- 42.Tuschall, D. M., E. Hiebert, and J. B. Flanegan. 1982. Poliovirus RNA-dependent RNA polymerase synthesizes full-length copies of poliovirion RNA, cellular mRNA, and several plant virus RNAs in vitro. J. Virol. 44:209-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Ooij, M. J., D. A. Vogt, A. Paul, C. Castro, J. Kuijpers, F. J. van Kuppeveld, C. E. Cameron, E. Wimmer, R. Andino, and W. J. Melchers. 2006. Structural and functional characterization of the coxsackievirus B3 CRE(2C): role of CRE(2C) in negative- and positive-strand RNA synthesis. J. Gen. Virol. 87:103-113. [DOI] [PubMed] [Google Scholar]
- 44.Wang, J., and G. Dreyfuss. 2001. A cell system with targeted disruption of the SMN gene: functional conservation of the SMN protein and dependence of Gemin2 on SMN. J. Biol. Chem. 276:9599-9605. [DOI] [PubMed] [Google Scholar]
- 45.Wang, J., and G. Dreyfuss. 2001. Characterization of functional domains of the SMN protein in vivo. J. Biol. Chem. 276:45387-45393. [DOI] [PubMed] [Google Scholar]
- 46.Zoll, J., H. A. Heus, F. J. van Kuppeveld, and W. J. Melchers. 2009. The structure-function relationship of the enterovirus 3′-UTR. Virus Res. 139:209-216. [DOI] [PubMed] [Google Scholar]