Abstract
Porcine circovirus 2 (PCV2) is the primary etiological agent of postweaning multisystemic wasting syndrome (PMWS), one of the most economically important emerging swine diseases worldwide. Virulent PCV2 was first identified following nearly simultaneous outbreaks of PMWS in North America and Europe in the 1990s and has since achieved global distribution. However, the processes responsible for the emergence and spread of PCV2 remain poorly understood. Here, phylogenetic and cophylogenetic inferences were utilized to address key questions on the time scale, processes, and geographic diffusion of emerging PCV2. The results of these analyses suggest that the two genotypes of PCV2 (PCV2a and PCV2b) are likely to have emerged from a common ancestor approximately 100 years ago and have been on independent evolutionary trajectories since that time, despite cocirculating in the same host species and geographic regions. The patterns of geographic movement of PCV2 that we recovered appear to mimic those of the global pig trade and suggest that the movement of asymptomatic animals is likely to have facilitated the rapid spread of virulent PCV2 around the globe. We further estimated the rate of nucleotide substitution for PCV2 to be on the order of 1.2 × 10−3 substitutions/site/year, the highest yet recorded for a single-stranded DNA virus. This high rate of evolution may allow PCV2 to maintain evolutionary dynamics closer to those of single-stranded RNA viruses than to those of double-stranded DNA viruses, further facilitating the rapid emergence of PCV2 worldwide.
Livestock species are an increasingly important focus of emerging-disease research. Livestock are susceptible to infections from related wild species and can act as a conduit through which zoonotic diseases spill over into human populations. For example, contact between humans and livestock has been implicated in the emergence of pandemic influenza virus (25), Ebola-Reston virus (53), Nipah virus (8), and Chlamydia psittaci (4). Pigs in particular are potentially important reservoirs for emerging human disease and have been implicated in the recent emergence of pandemic H1N1 influenza A virus (66), among others (3, 8, 53). Characteristics of the modern pig industry include high-density farming and expanding global trade, a combination that favors increased transmission and spread for many infectious agents. The role of swine in emerging infectious diseases may warrant further scrutiny, considering the expansive array of pathogens to which pigs are host, including a wide range of viruses: arboviruses, circoviruses, flaviviruses, herpesviruses, nidoviruses, orthomyxoviruses, paramyxoviruses, and picornaviruses all cause common infections in swine (52). Given the number of emergent pathogens currently identified in swine, it is essential to understand the circumstances under which these pathogens emerge and evolve. In this study, we examine the evolutionary dynamics of one of the most economically important emerging pig pathogens, porcine circovirus 2 (PCV2), the primary etiological agent of postweaning multisystemic wasting syndrome (PMWS).
PCV2 is a single-stranded, nonenveloped, circular DNA virus with an ambisense genome of only ∼1.76 kb, making it the smallest autonomously replicating virus. The genome of PCV2 contains at least three open reading frames (ORFs) with known functions: ORF1 codes for two replicase proteins (rep gene products), ORF2 for the structural protein (cap gene product), and ORF3 for a protein implicated in viral pathogenesis (44, 45, 48). PCV2 is highly infectious, and transmission can occur through oronasal, fecal, urinary, sexual, and vertical routes (38, 40, 62, 64). The current prevalence of PCV2 in pigs has been estimated to be as high as 40 to 60%, with studies reporting up to 100% of farms infected in a given region (21, 58, 60, 65, 68, 73). PCV2 has been identified as the causative agent of PCV-associated diseases (PCVAD), a term recently used to describe all PCV2-associated clinical and subclinical manifestations, including those formerly considered to be PMWS (systemic infection) as well as porcine dermatitis and nephropathy syndrome and PCV2-associated pneumonia, enteritis, and reproductive failure (7, 55). However, clinical diagnosis of PCVAD remains rare, and the vast majority of research on PCV2 has centered on the causes and consequences of PMWS.
The majority of PCV2 infections (and subsequent PMWS disease symptoms) appear to occur shortly after weaning, when the protective effects of maternal immunity have waned (51, 59). The transition from infection with PCV2 (often asymptomatic) to a clinical diagnosis of PMWS is thought to depend on several factors, of which viral load and immune activation (potentially caused by early vaccination or infection with multiple pathogens) appear to be the most significant (35, 36, 37). It has been suggested that there is a viral genetic component to the observed variation in virulence of PCV2, although this is still controversial (15, 18, 70). It is possible that the emergence of a new genotype (PCV2b) may be related to the appearance of highly virulent PCV2, although this link has yet to be confirmed (55). Symptoms of PMWS most commonly include wasting/weight loss, histologic lesions (particularly in the lymphatic system or lungs), and early mortality or abortion (67). Morbidity due to PMWS on an affected farm is often 5 to 15%, with a case fatality rate of close to 100% (18, 31).
PMWS was initially identified in Canada in 1991, with the first large outbreaks occurring in Europe in the late 1990s (9, 20, 30). Retrospective studies have now identified both antibodies to PCV2 and (in some cases) viral RNA dating back as far as 1969, indicating that PCV2 was present at least 22 years before the emergence of PCVAD (21, 47, 60). Outbreaks of PMWS and other PCVAD have now been confirmed in nearly every region with industrial pig farming (2, 11, 12, 17, 18, 21), and the incidences of all PCVAD have been steadily rising, particularly since 2004 (9, 55, 58). It remains unclear what event(s) precipitated the recent worldwide emergence of PCVAD (and PMWS in particular) from PCV2, despite the high level of scientific and economic interest directed toward this cause.
To address some of the uncertainty surrounding the recent emergence of PCVAD and PCV2, the present study employed phylogenetic inference to address the following key questions. (i) Is PCV2 a newly emergent pig virus, or is it much older? (ii) What are the rate and time scale of the evolution of PCV2? (iii) Are there viral genetic factors that are associated with virulence in PCV2? (iv) How did severe PCVAD emerge in both North America and Europe within a 10-year interval?
MATERIALS AND METHODS
Evolutionary relationships and time scale.
For the PCV2-specific analyses, all available PCV2 full-genome, rep, or cap gene sequences for which information on the sampling year was available were downloaded from GenBank (Table 1). Sequences were manually aligned by gene or genome by using Se-Al (version 2.0a11 Carbon) (http://tree.bio.ed.ac.uk/software/seal/) and examined for evidence of recombination using the RDP, GENECONV, Bootscan, MaxChi, and LARD methods with default parameters, as implemented in RDP3 (49). All potential recombinants identified by two or more methods within RDP3 were removed from further analyses. Phylogenetic trees were created for both the PCV2 genomes and the cap genes by using a Bayesian Markov chain Monte Carlo (MCMC) method implemented in the BEAST package (version 1.4.8) (13), which incorporates time-of-sampling information and returns rooted trees. BEAST was also used to estimate the rate of evolution and the time to the most recent common ancestor (TMRCA) of PCV2 for both the full-genome data set and each ORF separately (as ORF3 is completely within ORF1, the substitution rates for ORF1 were analyzed both with and without inclusion of the ORF3 region). A relaxed molecular clock with an uncorrelated log-normal distribution of rates, a GTR+Γ4+I model of nucleotide substitution (determined by Modeltest version 3.7) (57), and a Bayesian skyline coalescent model were used in all PCV2 analyses. A minimum of four independent runs were performed for each data set, with sampling every 10,000 generations, until convergence of all parameters was reached. Statistical uncertainty is reflected in values of the 95% highest probability density (95% HPD). To confirm that the tip-dated circovirus sequences exhibited sufficient phylogenetic signal to estimate evolutionary rates and the TMRCA, the date-sequence relationships were randomized to create 10 unique data sets and a BEAST analysis was performed on each. To examine the influence of the prior distributions on the rate and TMRCA parameters, the MCMC chains were also run on three data sets with the same priors as in the original analysis, but without the inclusion of sequence data.
TABLE 1.
Bayesian MCMC estimates of rates of nucleotide substitution in PCV2
| Genomic region | No. of samples | Sequence length (nt) | Yr range | No. of nucleotide substitutions/site/yr (95% HPD) |
|---|---|---|---|---|
| Full genome | 160 | 1,780 | 1980-2007 | 1.21 × 10−3 (8.23 × 10−4 to 1.61 × 10−3) |
| Coat protein | 327 | 693 | 1980-2007 | 1.44 × 10−3 (1.01 × 10−3 to 1.90 × 10−3) |
| Replicase | 168 | 942 | 1980-2007 | 1.03 × 10−3 (7.37 × 10−4 to 1.32 × 10−3) |
| Replicase-ORF3 | 168 | 627 | 1980-2007 | 8.74 × 10−4 (5.94 × 10−4 to 1.15 × 10−3) |
| ORF3 | 168 | 312 | 1980-2007 | 1.32 × 10−3 (7.94 × 10−4 to 1.86 × 10−3) |
| Genome (big clade) | 122 | 1,780 | 1998-2007 | 1.15 × 10−3 (8.34 × 10−4 to 1.45 × 10−3) |
| Genome (small clade) | 38 | 1,780 | 1980-2005 | 1.64 × 10−3 (9.43 × 10−4 to 2.40 × 10−3) |
To infer the divergence times and probable origin of PCV2 in relation to other circoviruses, representative gene sequences were collected from GenBank for 10 other circovirus species and aligned with a random sample of the PCV2 data set. Due to the high levels of divergence between circovirus species, only an amino acid alignment for rep could reliably be constructed for the genus. Beak and feather disease virus was also excluded, as it could not be convincingly aligned with the other circoviruses. One representative from each circovirus was then used to estimate the TMRCA for each species with BEAST, using the WAG model of amino acid substitution and a birth-death (speciation) model. It has been suggested that circoviruses may not have recently emerged but instead have strictly codiverged with their avian and swine host species over millions of years (29). To test this hypothesis, complete mitochondrial cytochrome b gene sequences were collected from GenBank for those vertebrate species considered to be reservoir hosts of distinct circoviruses. Bayesian phylogenetic trees were constructed for both the circoviruses (n = 134) and hosts (n = 49) using MrBayes (version 3) (61). Trees were inferred using the WAG amino acid transition model for the circoviruses and the HKY+Γ4+I model of nucleotide substitution for the hosts, as determined by Modeltest. Each analysis consisted of a minimum of two independent runs with 10 million generations, using four chains each, sampling every 5,000 generations. After confirmation that each host or circovirus species was monophyletic, these trees were pruned to include one taxon per species for the codivergence analysis. The topologies of all trees were confirmed using maximum likelihood methods (see the supplemental material).
To test for topological congruence between the host and circovirus phylogenies, TreeMap was used (version 2.0β) (available at http://www.it.usyd.edu.au/∼mcharles/). TreeMap assumes a fixed host phylogeny and creates multiple potentially optimal (POpt) solutions by mapping the circovirus phylogeny into that of the host, using varying combinations of codivergence events (CEs), host switches, duplications and losses (collectively termed noncodivergence events). In this process, TreeMap simultaneously considers both the given circovirus phylogeny and the corresponding distribution of the virus over the host tree (27). In this study, all POpt solutions generated by TreeMap were considered as hypotheses of the evolution of circoviruses with respect to their vertebrate hosts (see the supplemental material). Significance testing was performed by creating 1,000 randomized circovirus trees and mapping each of those into the fixed host phylogeny. Significance was assessed using the number of CEs, where the proportion of randomized reconciliations with equal or greater numbers of CEs was compared to that determined for the actual circovirus tree (see the supplemental material for details). The level of congruence determined for the actual circovirus tree must be greater than that expected for the randomly generated trees to support a history of codivergence (α = 0.05).
PCV2 and the emergence of PCVAD.
Two separate estimates of selection pressure were undertaken with this data set to identify lineages or regions of the genome that may have been under positive selection during the recent emergence of PCVAD. Global estimates of the ratio of nonsynonymous to synonymous nucleotide changes (dN/dS ratio) per site were estimated for each of the following genomic fragments (coding regions only): cap, rep, rep-ORF3, and ORF3. The HyPhy package was used to fit a codon model to each alignment by using the MG94xHKY85_3x4 substitution model, and the global dN/dS ratio and corresponding 95% confidence interval (CI) were estimated (34). If there was strong evidence that dN was >dS for any genomic region, the per-site dN/dS ratio was then estimated using the fixed-effect-likelihood algorithm implemented in HyPhy (32). A dN/dS ratio of >1 for any given site was considered significant if the P value derived from the likelihood ratio test was greater than 0.1.
A lineage-specific genetic algorithm approach was also used to infer the presence of positive selection along the branches of the Bayesian maximum clade credibility tree (full genome only) derived from BEAST, implemented in HyPhy (GABranch) (33). This approach assigns each branch to an incrementally estimated number of classes of dN/dS ratios without requiring a specification of the branches a priori and is less parameterized than a fully local model. Due to the computational intensity of this approach, the genome tree was pruned from 160 taxa down to 98 by arbitrarily removing all but one sequence from any monophyletic clade containing samples from any one outbreak/study.
PCVAD were discovered in both North America and Europe within a very short time frame, potentially as the result of high levels of migration of PCV2-infected pigs between disparate geographic regions. Here, we inferred the strength of PCV2 movement between geographic locations by using a geographically explicit Bayesian MCMC method implemented in BEAST (version 1.5) (43). This method simultaneously estimates a reversible diffusion rate matrix between previously defined locations along with the evolutionary and coalescent parameters. The diffusion pathways were estimated on two levels: by continent (Asia, Australia, Europe, North America, and South America; 10 possible diffusion pathways) and by country (Australia, Brazil, Canada, China/Taiwan, Denmark, France/Spain, Germany, Greece, Hungary, The Netherlands, and the United States; 55 possible diffusion pathways). Bayesian stochastic variable search selection was used to identify links between these locations that were necessary to explain the geographic diffusion process along the posterior sets of trees. Summary statistics based on a Bayes factor (BF) test were used to determine which diffusion links were statistically significant over the whole tree.
RESULTS
Evolutionary relationships and time scale.
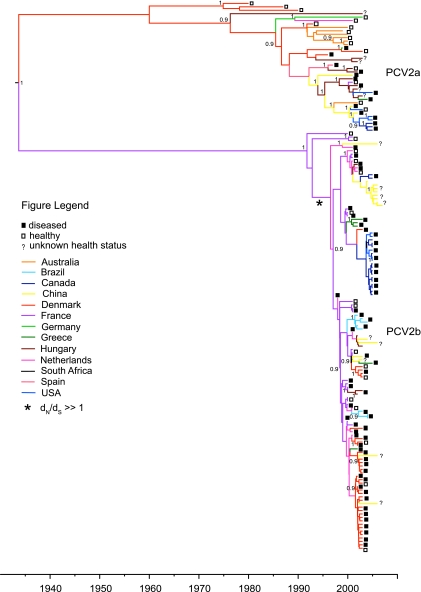
The phylogenetic trees inferred using tip-dated sequences resulted in very similar general topologies for the PCV2 genome and cap gene data sets, as previously noted (54). However, the increased sequence length in the genome data set (1,780 versus 693 bp) did result in a tree with substantially higher Bayesian posterior probabilities (BPPs) at the internal nodes (Fig. 1 for the genome tree; see Fig. S5 in the supplemental material for the cap tree). In both the cap and the genome trees, the two large clades (designated PCV2a and PCV2b) seen in previous phylogenetic analyses of PCV2 were recovered with high nodal support (7, 10, 42, 70). The majority of taxa fell within clade PCV2b, which may be replacing the older PCV2a genotype on a global scale (70). Notably, the PCV2b clade contained larger proportions of sequences both from recent outbreaks (post-1998) and from PMWS-affected animals (Fig. 1). Although both the PCV2a and the PCV2b clades contained taxa from Asia, Europe, and North America, the South American sequences were restricted to clade PCV2b and the Australian sequences to PCV2a. The majority of relationships within PCV2a had high nodal support, as did the deeper relationships within PCV2b (BPP > 0.9) (Fig. 1). The associations at the tips of the PCV2b clade were not significantly supported in the majority of cases, although small geographically structured clusters were resolved throughout the tree (Fig. 1).
FIG. 1.
Bayesian maximum clade credibility phylogenetic tree of the full-genome sequences of PCV2. Tips are colored by the country of sampling, and the internal branches are colored based on the reconstructed ancestral state (country) as estimated by the reversible diffusion model. Tip times reflect the sampling time of the sequence in question (x axis). Numbers at the nodes indicate BPPs greater than 0.9. Disease status (when reported) is associated with each tip. The location of the branch leading to the primary PCV2b clade with a dN/dS ratio of >1 is indicated with an asterisk (*).
The estimated age of the current diversity of PCV2 and the corresponding rate of nucleotide substitution were determined using a Bayesian MCMC analysis. The posterior distribution of the rate and TMRCA parameters from these analyses were compared with those from the data sets generated by randomizing the sequence-date relationships of all taxa as well as those from analyses using priors only. The parameter values estimated from the actual analyses had nonoverlapping 95% HPDs with all analyses from both the randomized data sets and the prior-only data sets. These results suggest that sufficient temporal structure was present in the data to estimate both the rates of nucleotide substitution and the TMRCA. After examining the distinct two-clade structure of the PCV2 phylogeny, rates were also estimated independently for the PCV2a and PCV2b clades. The mean substitution rate for the full-genome data set was 1.21 × 10−3 nucleotide substitutions per site per year, with a 95% HPD that ranged from 8.23 × 10−4 to 1.61 to 10−3 substitutions/site/year. This rate was consistent with those estimated independently for each genomic segment (Table 1). The TMRCA estimated for the current diversity of all PCV2 sampled viruses was 70 years before present (ybp) (95% HPD = 36 to 115 ybp), while the TMRCA values for the PCV2a and PCV2b genotypes were estimated at 41 ybp (95% HPD = 24 to 62 ybp) and 18 ybp (95% HPD = 12 to 27 ybp), respectively. Analyzing the population dynamics of PCV2a versus those of PCV2b also revealed genotype-specific differences, although neither Bayesian skyline plot demonstrated a significant departure from a constant level of genetic diversity (see Fig. S6 in the supplemental material). Under a model of neutral evolution, this is equivalent to a constant population size through time. However, there was a trend toward a recent increase in genetic diversity in PCV2b that was not mirrored in the Bayesian skyline plot of PCV2a. In addition, the relative genetic diversity estimated for both genotypes was extremely low (see Fig. S6 in the supplemental material).
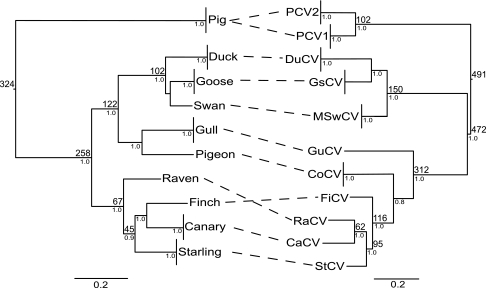
Two competing hypotheses exist regarding the origin and emergence of PCV2 and PCVAD: either PCV2 is part of a family of viruses that have codiverged with their vertebrate hosts over millions of years (29) or it has recently emerged through cross-species transmission and/or host expansion. To distinguish between these hypotheses, we assessed the topological congruence between the circovirus and associated host phylogenies. The inferred phylogenies for both the circovirus and the host phylogenies were well resolved with high nodal support (Fig. 2). Phylogenetic reconciliation analysis of the host and virus trees returned 15 POpt solutions, containing three, four, or five codivergent nodes (6, 8 or 10 CEs) (see Fig. S7 in the supplemental material). Significance testing based on the maximum number of CEs revealed that 74% of randomly generated trees could be mapped onto the host tree with at least 10 CEs, revealing no statistically significant evidence for congruence by this measure. It was possible to generate significant congruence between the host and virus phylogenies by invoking a CE at the root of the vertebrate tree (see the supplemental material). However, the presence of a CE between circoviruses and the last common ancestor of birds and mammals is highly implausible, given the age of this split in the vertebrate tree, estimated to have occurred over 300 million years ago (6, 56). To test for consistency between the divergence times of each circovirus species with those of their hosts, the TMRCA values for a range of avian circoviruses and PCVs were estimated based on the rate of amino acid substitution (Fig. 2). The TMRCA for PCV2 was estimated to be more recent under the amino acid substitution model than when nucleotides were used (38 ybp; 95% HPD = 14 to 69 ybp); and as tip-dated estimates of divergence times may be too recent as a whole (23), we considered only the upper 95% HPD of these TMRCA estimates for a conservative comparison with the host divergence times (Fig. 2). Despite this, our estimated divergence times for the circoviruses were very recent, with a mean TMRCA for PCV1 and PCV2 of 102 ybp and the mean divergence of pig and avian circoviruses estimated to have occurred 491 ybp.
FIG. 2.
Tanglegram indicating the associations between each circovirus and its reservoir host. The numbers in smaller print at the nodes are BPPs, while the numbers in larger print indicate the divergence time for that node, as estimated from the literature (in millions of years ago, for hosts) or directly from the data (in ybp, for circoviruses). Values are shown for duck (DuCV), goose (GsCV), swan (MSwCV), gull (GuCV), columbid (CoCV), finch (FiCV), raven (RaCV), canary (CaCV), and starling (StCV) circoviruses in addition to PCVs.
PCV2 and the emergence of PCVAD.
The dN/dS ratios for cap, rep, and rep-ORF3 were relatively low (dN/dS = 0.230, 0.193, and 0.173, respectively), indicating that most sites within these regions are under strong purifying selection. This is in accord with previous analyses of dN/dS in PCV2, where purifying selection was found in both rep and cap (26, 54). Including the region of rep that overlaps with ORF3 did not increase the strength of purifying selection measured for the rep gene. However, the dN/dS ratio for ORF3 was 1.363 (95% CI = 1.114 to 1.651), which might indicate some positive selection in this region (although the presence of overlapping reading frames complicates this analysis). A site-by-site analysis of the dN/dS ratio within ORF3 revealed 37 amino acid sites where dN was >dS, and five of these were significant (P <0.10) (sites 41, 57, 76, 87, 94, and 100). However, a more thorough analysis of the effect(s) of overlapping reading frames on measures of dN/dS is needed before the significance of these results can be interpreted.
To look for the presence of selection events that may have occurred only once in the evolutionary history of PCV2, dN/dS was also calculated along individual branches of the genome tree. Four dN/dS categories optimally described the selection pressure throughout the tree (based on Akaike Information Criterion scores; data not shown). Three of these had dN/dS ratios of substantially less than 1, which accounted for 95% of the branches (see Fig. S8 in the supplemental material). The remaining dN/dS category had a dN/dS ratio of 15.4 and best fit 5% of the branches. Critically, while many of the branches associated with dN/dS of >1 were near the tips of the tree (see Fig. S8 in the supplemental material) and therefore of little interest due to the influence of transient deleterious mutations, there was 99% support for dN/dS of >1 on one of the primary branches leading to the PCV2b clade (Fig. 1). This branch was associated with an amino acid transition from threonine to proline at position 151 of the cap gene, present in the majority of taxa in the PCV2b clade.
Two phenomena may explain the nearly synchronous emergences of PCVAD in North America and Europe: either the emergence of PCVAD occurred independently in multiple regions or virulent PCV2 emerged only once and spread rapidly throughout the globe via the pig trade. When the diffusion pathways for PCV2 were estimated between continents, three were found to be significant: (in order of increasing significance) those between South America and Europe, Asia and North America, and Europe and Asia (BF > 12) (see Table S2 in the supplemental material). When the diffusion pathways were estimated between countries, three pathways were again significant: those between France/Spain and Hungary, Canada and the United States, and France and The Netherlands (BF > 17) (see Table S2 in the supplemental material). The reconstruction of the ancestral locations (by country) of the internal nodes on the PCV2 genome tree is shown in Fig. 1.
DISCUSSION
Macroevolution of PCV2.
This study provides a unique insight into the evolutionary context of the recent emergence of PCV2-associated diseases in swine. PMWS and other PCVAD are rapidly increasing in prevalence worldwide and are often associated with severe economic impacts. The origin and time scale of the emergence of PCV2 have remained unclear, as have questions surrounding a possible viral genetic component to recent reported increases in virulence of PCV2 (15, 18, 70). The outcome of our study indicates that PCV2 has recently emerged, potentially as the result of a cross-species jump from birds into swine, most likely through intermediate contact with wild boars (39, 50). Our results corroborate previous suggestions that a new genotype (PCV2b) may have contributed to the recent expansion of highly virulent PCV2, although it is clear from our analyses that PMWS is also associated with recent PCV2a infections (Fig. 1). We suggest that the high levels of movement of (asymptomatic) PCV2-infected pigs that occur as a result of the swine trade may have assisted in the rapid spread of the virulent PCV2 genotype around the globe.
Our PCV2 phylogeny shows the division of this virus into two genotypes, PCV2a and PCV2b, congruent with the results of previous studies (Fig. 1) (18, 54). The PCV2b clade contained the majority of recently sampled sequences and was more often associated with disease symptoms than PCV2a (e.g., all sequences from the PMWS-free country of Australia fell within the PCV2a clade). While previous work has indicated that the global emergence of PCVAD is coupled to recent increases in the prevalence of PCV2b, it is clear that this genotype is not exclusively associated with the presence of disease (Fig. 1) (1, 15, 18). However, our analysis of the population dynamics of PCV2 did reveal a trend toward increasing genetic diversity (and population size) in PCV2b that was not reflected in PCV2a, which could be the result of a recent population expansion that occurred in PCV2b only (see Fig. S6 in the supplemental material). As these results are not significant, a larger sample size is needed to confirm this trend. Notably, this analysis also indicates that the relative genetic diversity for both genotypes is extremely low, a feature that could be indicative of continuous selective sweeps over time or repeated population bottleneck events, both of which act to purge variation from the population.
The development of PMWS is often (but not exclusively) thought to be the result of multiple factors, one of which must be the presence of PCV2. Factors often linked to the development of a PCVAD from an asymptomatic infection include the presence of a coinfecting pathogen (such as porcine respiratory and reproductive syndrome virus or porcine parvovirus) and the stimulation of the neonatal immune system from early vaccination (46, 55). The combination of factors often associated with the development of PCVAD may help explain the presence of diseased animals in both the PCV2a and the PCV2b clades. If PCV2b is selectively favored over PCV2a due to properties such as increased replication rate, transmissibility, or virulence, we might expect to see evidence of selection along the lineage leading to the PCV2b clade. While the results of our branch-specific selection analysis did find evidence of positive selection (dN/dS of > 1) along a branch leading to the primary PCV2b clade, no obvious directional evolution of multiple nucleotide or amino acid sites was identified (Fig. 1; see also Fig. S8 in the supplemental material). This contrasts with the clear signature of positive selection and directional evolution seen in the lineage leading to another newly emergent single-stranded DNA (ssDNA) virus (canine parvovirus) (63). However, the lack of a similarly strong signal in our data could be the result of an insufficient sample size.
The lack of strong evidence for recent positive selection could alternatively suggest that PCV2 is not a virus that has recently emerged in pigs but instead has perhaps only recently begun to cause disease. Based on similarities in the branching patterns of the Circoviridae phylogeny and those of their corresponding hosts, Johne et al. (29) suggested that circoviruses have been coevolving with vertebrates for millions of years. However, our analysis of the temporal and topological congruence between the host and circovirus phylogenies indicated that the circovirus phylogeny was no more topologically congruent with that of the host than would be expected by chance (Fig. 2). In addition, a comparison of the respective divergence times of host and circovirus species revealed substantial incompatibilities between the divergence times of these groups, which conflicts strongly with the hypothesis of codivergence. The last shared ancestor between mammals and birds is thought to have lived approximately 300 million years ago, whereas the results of our Bayesian MCMC analyses indicate that the common ancestor of all sampled circovirus diversity existed only 500 years ago (Fig. 2). In contrast to the results reported by Johne et al. (29), the lack of both temporal and topological congruence between the host and circovirus phylogenies suggests strongly that circoviruses have not shared a long history with their hosts.
Given that circoviruses are likely to be of relatively recent origin, we further estimated the TMRCA for the sampled genetic diversity of the PCV2 lineage. The results of our analyses, using both nucleotide and amino acid substitution rates, indicated that PCV2 originated within approximately the last 100 years. The TMRCA for PCV2b (∼18 ybp; 95% HPD, 12 to 27 ybp) was more recent than that for PCV2a (∼41 ypb; 95% HPD, 24 to 62 ybp), with almost nonoverlapping 95% HPDs. This result is consistent with the hypothesis that the PCV2b is associated with the emergence of PCVAD worldwide. The first PMWS case was identified in Canada in 1991, identical to our mean estimated date of origin for PCV2b. Interestingly, the deep bifurcation and long branch lengths leading to the PCV2a and PCV2b clades reveal that PCV2b, while more recent, did not arise from PCV2a. Rather, these two groups shared a common ancestor approximately 100 years ago and have been independently evolving, while cocirculating, since that time. How PCV2a and PCV2b have remained genetically distinct while coinfecting the same species in the same geographic area is unknown but is an interesting avenue for further research.
It has been a challenge to explain the cause of the nearly simultaneous emergences of PCVAD in Europe and North America and the rapid global spread that followed. The results of the geographic diffusion model used in this study indicate that significant global movement of PCV2 exists, with links detected between countries in North America, Europe, and Asia as well as potentially between South America and Europe (Fig. 1; see also Table S2 in the supplemental material). France (Spain), Hungary, and The Netherlands were identified as being particularly important foci of PCV2 movement in Europe, while high levels of movement also appear to exist between Canada and the United States. Critically, the high levels of diffusion of PCV2 around the globe detected in this analysis are similar to those reported for the trade of live swine between nations (UN database, COMTRADE; available at http://comtrade.un.org/db/; accessed on 6 January 2009). Since the worldwide emergence of PCV2 was reported in 1997, more than 18 billion kilograms of live pigs have been exported from the countries for which PCV2 sequence data were available (based on reported data only). The largest exporters of swine since 1997 have been Canada, China, Denmark, and The Netherlands, while the majority of these exports went to France, Germany, Spain, and the United States. While both the COMTRADE database and the results of our geographic diffusion model are likely skewed by uneven reporting between countries, the similarities in movement patterns that were revealed through these two disparate types of data are striking. The high levels of swine trafficking that occur due to the globalization of the pig/pork industry, along with the high prevalence of asymptomatic PCV2 infections, combine to create an ideal circumstance for the emergence of a highly virulent strain of PCV2 (55). Epidemiological links between infected farms have been determined for outbreaks in the United Kingdom and New Zealand, among others, highlighting the potential for PCV2 transmission through the pig trade (41, 71).
Microevolution of PCV2.
Virulence in PCV2 is likely determined by two genomic regions, those encoding the capsid protein (ORF2), potentially involved in tissue tropism (39), and ORF3, implicated in the apoptotic action of the virus (44, 45). While our selection analyses did not recover evidence for positive selection in the cap protein as a whole, we found tentative evidence for positive selection in ORF3, with individually selected sites throughout the protein. Previous in vitro and in vivo work has demonstrated that ORF3 is directly involved in viral pathogenesis and acts by inducing the apoptosis of T cells (44, 45). This destruction of T cells is thought to lead to immune suppression and, in turn, to a higher viral load in infected animals (44). Critically, we could not identify specific amino acids in ORF3 that were associated with the emergence of PCV2b or the presence of PCVAD in general; therefore, it is unlikely that strong directional selection is occurring within ORF3. Instead, the evolutionary pressures on this protein may be diversifying, indicative of a rough fitness surface favoring a range of phenotypes. However, that ORF3 is completely within ORF1 (and translated in the opposite direction) greatly complicates this analysis.
Another notable result of our analysis is that the estimated rate of nucleotide substitution for PCV2 (∼1.2 × 10−3 substitutions/site/year) (Table 1) is higher than that measured for any ssDNA virus to date. This places PCV2 within the range of evolutionary rates estimated for most ssRNA viruses and is particularly striking for a DNA virus that replicates using the host polymerase with its associated proofreading capabilities (19, 22, 28, 69). Although such a high substitution rate could have been driven by rapid, strong positive selection, we found little evidence for this process. A variety of factors may therefore explain our anomalously high rate estimates. First, as PCV2 is the smallest known autonomously replicating virus, it is possible that it is able to tolerate a higher mutation rate per site (in keeping with error threshold theory) (5). A rapid rate of viral replication may also contribute to a high error, as replication speed and mutational fidelity may be inversely correlated in RNA viruses (16). Unfortunately there are no available experimental measures of the replication rate of PCV2 or other ssDNA viruses by which to test this hypothesis. Alternatively, it is possible that our rate estimation is strongly skewed toward the mutation rate rather than the long-term substitution rate, a result of the bias in sampling toward the recent past (23, 24). However, this effect is unlikely to have inflated substitution rates by the orders of magnitude needed to reconcile it with those observed in double-stranded DNA viruses. More generally, the mechanism(s) by which ssDNA viruses can achieve high mutation rates while using host DNA polymerases to replicate is still unknown and represents a clear area for future study (14). Despite these uncertainties, our analysis further confirms that ssDNA viruses are closer to RNA than double-stranded DNA viruses in their evolutionary dynamics, helping to create the biological conditions necessary for rapid emergence (72).
Taken together, the results of our analyses strongly suggest that PCV2 is a virus of relatively recent origin in swine. We speculate that the emergence of this virus was likely the result of a recent host switch from birds into pigs, perhaps with an intermediate jump into wild boars (40, 50). High levels of pig movement due to the global trade market may have created a situation resembling a standing network, whereby any highly infectious pathogen, such as (but not limited to) PCV2, would be able to rapidly infect pigs around the world.
Supplementary Material
Acknowledgments
Funding to C.F. was provided by the Natural Sciences and Engineering Research Council (Canada). E.C.H. was funded in part by NIH grant R01 GM080533.
We thank Andrew Read, Anton Nekrutenko, and Bryan Grenfell for their insight into PCV2 evolution and emergence; Sergei Kosakovsky Pond for his technical advice and support; and Maia Rabaa for her editorial assistance and invaluable contribution to all things swine.
Footnotes
Published ahead of print on 7 October 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Allan, G. M., F. McNeilly, M. McMenany, I. McNair, S. G. Krakowka, S. Trimmusk, D. Walls, M. Donnely, D. Minachim, J. Ellis, P. Wallgren, and C. Fossum. 2007. Temporal distribution of porcine circovirus 2 genogroups recovered from postweaning multisystemic wasting syndrome-affected and nonaffected farms in Ireland and Northern Ireland. J. Vet. Diagn. Investig. 19:668-673. [DOI] [PubMed] [Google Scholar]
- 2.An, D. J., I. S. Roh, D. S. Song, C. K. Park, and B. K. Park. 2007. Phylogenetic characterization of porcine circovirus type 2 in PMWS and PDNS Korean pigs between 1999 and 2006. Virus Res. 129:115-122. [DOI] [PubMed] [Google Scholar]
- 3.Barrette, R. W., S. A. Metwally, J. M. Rowland, L. Xu, S. R. Zaki, S. T. Nichol, P. E. Rollin, J. S. Towner, W.-J. Shieh, B. Batten, T. K. Sealy, C. Carrillo, K. E. Moran, A. J. Bracht, G. A. Mayr, M. Sirios-Cruz, D. P. Catbagan, E. A. Lautner, T. G. Ksiazek, W. R. White, and M. T. McIntosh. 2009. Discovery of swine as a host for the Reston ebolavirus. Science 325:204-206. [DOI] [PubMed] [Google Scholar]
- 4.Beeckman, D. S., and D. C. Vanrompay. 2009. Zoonotic Chlamydophila psittaci infections from a clinical perspective. Clin. Microbiol. Infect. 15:11-17. [DOI] [PubMed] [Google Scholar]
- 5.Belshaw, R., A. Gardner, A. Rambaut, and O. G. Pybus. 2008. Pacing a small cage: mutation and RNA viruses. Trends Ecol. Evol. 23:188-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, J. W., J. S. Rest, J. Garcia-Moreno, M. D. Sorenson, and D. P. Mindell. 2008. Strong mitochondrial DNA support for a Cretaceous origin of modern avian lineages. BMC Biol. 6:6. doi: 10.1186/1741-7007-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carman, S., H. Y. Cai, J. DeLay, S. A. Youssef, B. J. McEwen, C. A. Gagnon, D. Tremblay, M. Hazlett, P. Lusis, J. Fairles, H. S. Alexander, and T. van Dreumel. 2008. The emergence of a new strain of porcine circovirus-2 in Ontario and Quebec swine and its association with severe porcine circovirus associated disease—2004-2660. Can. J. Vet. Res. 72:259-268. [PMC free article] [PubMed] [Google Scholar]
- 8.Chua, K. B., W. J. Bellini, P. A. Rota, B. H. Harcourt, A. Tamin, S. K. Lam, T. G. Ksiazek, P. E. Rollin, S. R. Zaki, W.-J. Shieh, C. S. Goldsmith, D. J. Gubler, J. T. Roehrig, B. Eaton, A. R. Gould, J. Olson, H. Field, P. Daniels, A. E. Ling, C. J. Peters, L. J. Anderson, and B. W. Mahy. 2000. Nipah virus: a recently emergent deadly paramyxovirus. Science 288:1432-1435. [DOI] [PubMed] [Google Scholar]
- 9.Clark, E. G. 1997. Post-weaning multisystemic wasting syndrome. Proc. Am. Assoc. Swine Pract. 28:499-501. [Google Scholar]
- 10.de Boisseson, C., V. Beven, L. Bigarre, R. Thiery, N. Rose, E. Eveno, F. Madec, and A. Jestin. 2004. Molecular characterization of porcine circovirus type 2 isolates from post-weaning multisystemic wasting syndrome-affected and non-affected pigs. J. Gen. Virol. 85:293-304. [DOI] [PubMed] [Google Scholar]
- 11.de Castro, A. M., A. Cortez, M. B. Heinemann, P. E. Brandão, and L. J. Richtzenhain. 2008. Molecular diversity of Brazilian strains of porcine circovirus type 2 (PCV-2). Res. Vet. Sci. 85:197-200. [DOI] [PubMed] [Google Scholar]
- 12.Drew, T. W., S. S. Grierson, D. P. King, D. Hicks, S. Done, J. A. Neser, D. P. B. Evans, P. Grimbeek, and M. Banks. 2004. Genetic similarity between porcine circovirus type 2 isolated from the first reported case of PMWS in South Africa and North American isolates. Vet. Rec. 155:149-151. [DOI] [PubMed] [Google Scholar]
- 13.Drummond, A. J., and A. Rambaut. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duffy, S., L. A. Shackelton, and E. C. Holmes. 2008. Rates of evolutionary change in viruses: patterns and determinants. Nat. Rev. Genet. 9:267-276. [DOI] [PubMed] [Google Scholar]
- 15.Dupont, K., E. O. Nielsen, P. Baekbo, and L. E. Larsen. 2008. The genomic analysis of PCV2 from Danish archives and a current PMWS case-control study supports a shift in genotypes with time. Vet. Microbiol. 128:56-64. [DOI] [PubMed] [Google Scholar]
- 16.Elena, S. F., and F. Sanjuán. 2007. Virus evolution: insights from an experimental approach. Annu. Rev. Ecol. Evol. Syst. 38:27-52. [Google Scholar]
- 17.Finlaison, D., P. Kirkland, R. Luong, and A. Ross. 2007. Survey of porcine circovirus 2 and postweaning multisystemic wasting syndrome in New South Wales piggeries. Aust. Vet. J. 85:304-310. [DOI] [PubMed] [Google Scholar]
- 18.Gagnon, C. A., D. Tremblay, P. Tijssen, M.-H. Venne, A. Houde, and S. M. Elahi. 2007. The emergence of porcine circovirus 2b genotype (PCV-2b) in swine in Canada. Can. Vet. J. 48:811-819. [PMC free article] [PubMed] [Google Scholar]
- 19.Ge, L. M., J. T. Zhang, X. P. Zhou, and H. Y. Li. 2007. Genetic structure and population variability of Tomato yellow leaf curl China virus. J. Virol. 81:5902-5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gresham, A., G. Jackson, N. Giles, G. Allan, F. McNeilly, and S. Kennedy. 2000. PMWS and porcine dermatitis nephropathy syndrome in Great Britain. Vet. Rec. 146:143. [PubMed] [Google Scholar]
- 21.Grierson, S. S., D. P. King, T. Sandvik, D. Hicks, Y. Spencer, T. W. Drew, and M. Banks. 2004. Detection and genetic typing of type 2 porcine circoviruses in archived pig tissues from the UK. Arch. Virol. 149:1171-1183. [DOI] [PubMed] [Google Scholar]
- 22.Hanada, K., Y. Suzuki, and T. Gojobori. 2004. A large variation in the rates of synonymous substitution for RNA viruses and its relationship to a diversity of viral infection and transmission. Mol. Biol. Evol. 21:1074-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ho, S. Y., B. Shapiro, M. J. Phillips, A. Cooper, and A. J. Drummond. 2007. Evidence for time dependency of molecular rate estimates. Syst. Biol. 56:515-522. [DOI] [PubMed] [Google Scholar]
- 24.Holmes, E. C. 2003. Patterns of intra- and interhost nonsynonymous variation reveal strong purifying selection in dengue virus. J. Virol. 77:11296-11298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horimoto, T., and Y. Kawaoka. 2005. Influenza: lessons from past pandemics, warnings from current incidents. Nat. Rev. Microbiol. 3:591-600. [DOI] [PubMed] [Google Scholar]
- 26.Hughes, A. L., and H. Piontkivska. 2008. Nucleotide sequence polymorphism in circoviruses. Infect. Genet. Evol. 8:130-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jackson, A. P., and M. A. Charleston. 2004. A cophylogenetic perspective of RNA-virus evolution. Mol. Biol. Evol. 21:45-57. [DOI] [PubMed] [Google Scholar]
- 28.Jenkins, G. M., A. Rambaut, O. G. Pybus, and E. C. Holmes. 2002. Rates of molecular evolution in RNA viruses: a quantitative phylogenetic analysis. J. Mol. Evol. 54:156-165. [DOI] [PubMed] [Google Scholar]
- 29.Johne, R., D. Fernandez-de-Luco, U. Hofle, and H. Muller. 2006. Genome of a novel circovirus of starlings, amplified by multiply primed rolling-circle amplification. J. Gen. Virol. 87:1189-1195. [DOI] [PubMed] [Google Scholar]
- 30.Kennedy, S., G. Allan, F. McNeilly, B. M. Adair, A. Hughes, and P. Spillane. 1998. Porcine circovirus infection in Northern Ireland. Vet. Rec. 142:495-496. [PubMed] [Google Scholar]
- 31.Kim, J., H.-K. Chung, T. Jung, W.-S. Cho, C. Choi, and C. Cahe. 2002. Postweaning multisystemic wasting syndrome of pigs in Korea: prevalence, microscopic lesions and coexisting microorganisms. J. Vet. Med. Sci. 64:57-62. [DOI] [PubMed] [Google Scholar]
- 32.Kosakovsky Pond, S. L., and S. D. W. Frost. 2005. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol. 22:1208-1222. [DOI] [PubMed] [Google Scholar]
- 33.Kosakovsky Pond, S. L., and S. D. W. Frost. 2005. A genetic algorithm approach to detecting lineage-specific variation in selection pressure. Mol. Biol. Evol. 22:478-485. [DOI] [PubMed] [Google Scholar]
- 34.Kosakovsky Pond, S. L., S. D. W. Frost, and S. V. Muse. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21:676-679. [DOI] [PubMed] [Google Scholar]
- 35.Krakowka, S., J. Ellis, F. McNeilly, S. Ringler, D. M. Rings, and G. Allan. 2001. Activation of the immune system is the pivotal event in the production of wasting disease in pigs infected with porcine circovirus-2 (PCV-2). Vet. Pathol. 38:31-42. [DOI] [PubMed] [Google Scholar]
- 36.Krakowka, S., J. Ellis, F. McNeilly, C. Waldner, and G. Allan. 2005. Features of porcine circovirus-2 disease: correlations between lesions, amount and distribution of virus, and clinical outcome. J. Vet. Diagn. Investig. 17:213-222. [DOI] [PubMed] [Google Scholar]
- 37.Kyriakis, S. C., K. Saoulidis, S. Lekkas, C. C. Miliotis, P. A. Papoutsis, and S. Kennedy. 2002. The effects of immuno-modulation on the clinical and pathological expression of postweaning multisystemic wasting syndrome. J. Comp. Pathol. 126:38-46. [DOI] [PubMed] [Google Scholar]
- 38.Ladekjaer-Mikkelsen, A. S., J. Nielsen, T. Storgaard, A. Bøtner, G. Allan, and F. McNeilly. 2001. Tranplacental infection with PCV-2 associated with reproductive failure in a gilt. Vet. Rec. 148:759-760. [PubMed] [Google Scholar]
- 39.Larochelle, R., R. Magar, and S. D'Allaire. 2000. PCR detection and evidence of shedding of porcine circovirus type 2 in boar semen. J. Clin. Microbiol. 38:4629-4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Larochelle, R., R. Magar, and S. D'Allaire. 2002. Genetic characterization and phylogenetic analysis of porcine circovirus type 2 (PCV2) strains from cases presenting various clinical conditions. Virus Res. 90:101-112. [DOI] [PubMed] [Google Scholar]
- 41.Lawton, D. E. B., T. Rawdon, S. S. Bastianello, A. C. Johnstone, and R. S. Morris. 2004. PMWS in New Zealand, part 1: diagnosis and investigation. Proceedings of the 18th Congress. International Pig Veterinary Society, Hamburg, Germany.
- 42.Lefebvre, D. J., S. Costers, J. Van Doorsselaere, G. Misinzo, P. L. Delputte, and H. J. Nauwynck. 2008. Anitgenic differences among porcine circovirus type 2 strains, as demonstrated by the use of monoclonal antibodies. J. Gen. Virol. 89:177-187. [DOI] [PubMed] [Google Scholar]
- 43.Lemey, P., A. Rambaut, A. J. Drummond, and M. A. Suchard. Bayesian phylogeography finds its roots. PLoS Comp. Biol, in press. [DOI] [PMC free article] [PubMed]
- 44.Liu, J., I. Chen, Q. Du, H. Chua, and J. Kwang. 2006. The ORF3 protein of porcine circovirus type 2 is involved in viral pathogenesis in vivo. J. Virol. 80:5065-5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu, J., I. Chen, and J. Kwang. 2005. Characterization of a previously unidentified viral protein in porcine circovirus type 2-infected cells and its role in virus-induced apoptosis. J. Virol. 79:8262-8274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Madec, F., N. Rose, B. Grasland, R. Cariolet, and A. Jestin. 2008. Post-weaning multisystemic wasting syndrome and other PCV2-related problems in pigs: a 12-year experience. Transbound. Emerg. Dis. 55:273-283. [DOI] [PubMed] [Google Scholar]
- 47.Magar, R., P. Muller, and R. Larochelle. 2000. Retrospective serological survey of antibodies to porcine circovirus type 1 and type 2. Can. J. Vet. Res. 64:184-186. [PMC free article] [PubMed] [Google Scholar]
- 48.Mankertz, A., R. Caliskan, K. Hattermann, B. Hillenbrand, P. Kurzendoerfer, B. Mueller, C. Schmitt, T. Steinfeldt, and T. Finsterbusch. 2004. Molecular biology of porcine circovirus: analyses of gene expression and viral replication. Vet. Microbiol. 98:81-88. [DOI] [PubMed] [Google Scholar]
- 49.Martin, D. P., C. Williamson, and D. Posada. 2005. RDP3: recombination detection and analysis from sequence alignments. Bioinformatics 21:260-262. [DOI] [PubMed] [Google Scholar]
- 50.McIntosh, K. A., J. C. S. Harding, S. Parker, J. A. Ellis, and G. D. Appleyard. 2006. Nested PCR detection and duration of porcine circovirus type 2 in semen with sperm morphological analysis from naturally infected boars. J. Vet. Diagn. Investig. 18:380-384. [DOI] [PubMed] [Google Scholar]
- 51.McKeown, N. E., T. Opriessnig, P. Thomas, D. K. Guenette, F. Elvinger, M. Fenaux, P. G. Halbur, and X. J. Meng. 2005. Effects of porcine circovirus 2 (PCV2) maternal antibodies on experimental infection of piglets with PCV2. Clin. Diagn. Lab Immunol. 12:1347-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morilla, A., K.-J. Yoon, and J. L. Zimmerman (ed.). 2002. Trends in emerging viral infections of swine, p. 387. Iowa State Press, Ames, IA.
- 53.Morris, K. 2009. First pig-to-human transmission of Ebola-Reston virus. Lancet Infect. Dis. 9:148. [Google Scholar]
- 54.Olvera, A., M. Cortey, and J. Segales. 2007. Molecular evolution of porcine circovirus type 2 genomes: phylogeny and clonality. Virology 357:175-185. [DOI] [PubMed] [Google Scholar]
- 55.Opriessnig, T., N. E. McKeown, E. M. Zhou, X. J. Meng, and P. G. Halbur. 2006. Genetic and experimental comparison of porcine circovirus type 2 (PCV2) isolates from cases with and without PCV2-associated lesions provides evidence for differences in virulence. J. Gen. Virol. 87:2923-2932. [DOI] [PubMed] [Google Scholar]
- 56.Pereira, S. L., D. P. Johnson, D. H. Clayton, and A. J. Baker. 2007. Mitochondrial and nuclear DNA sequences support a Cretaceous origin of Columbiformes and a dispersal-driven radiation in the Paleocene. Syst. Biol. 56:656-672. [DOI] [PubMed] [Google Scholar]
- 57.Posada, D., and K. A. Crandall. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 58.Ramirez-Mendoza, H., H. Castillo-Juarez, J. Hernandez, P. Correa, and J. Segales. 2009. Retrospective serological survey of porcine circovirus-2 infection in Mexico. Can. J. Vet. Res. 73:21-24. [PMC free article] [PubMed] [Google Scholar]
- 59.Rodriguez-Arrioja, G. M., J. Segales, M. Calsamiglia, A. R. Resendes, M. Balasch, J. Plana-Duran, J. Casal, and M. Domingo. 2002. Dynamics of porcine circovirus type 2 infection in a herd of pigs with postweaning multisystemic wasting syndrome. Am. J. Vet. Res. 63:354-357. [DOI] [PubMed] [Google Scholar]
- 60.Rodriguez-Arrioja, G. M., J. Segales, C. Rosell, A. Rovira, J. Pujols, J. Plana-Duran, and M. Domingo. 2003. Retrospective study on porcine circovirus type 2 infection in pigs from 1985 to 1997 in Spain. J. Vet. Med. 50:99-101. [DOI] [PubMed] [Google Scholar]
- 61.Ronquist, F., and J. P. Huelsenbeck. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572-1574. [DOI] [PubMed] [Google Scholar]
- 62.Segales, J., M. Calsamiglia, A. Olvera, M. Sibila, L. Badiella, and M. Domingo. 2005. Quantification of porcine circovirus type 2 (PCV2) DNA in serum and tonsillar, nasal, tracheo-bronchial, urinary and faecal swabs of pigs with and without postweaning multisystemic wasting syndrome (PMWS). Vet. Microbiol. 111:223-229. [DOI] [PubMed] [Google Scholar]
- 63.Shackelton, L. A., C. R. Parrish, U. Truyen, and E. C. Holmes. 2005. High rate of viral evolution associated with the emergence of carnivore parvovirus. Proc. Natl. Acad. Sci. USA 102:379-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shibata, I., Y. Okuda, S. Yazawa, M. Ono, T. Sasaki, M. Itagaki, N. Nakajima, Y. Okabe, and I. Hidejima. 2003. PCR detection of porcine circovirus type 2 DNA in whole blood, serum, oropharyngeal swab, nasal swab, and feces from experimentally infected pigs and field cases. J. Vet. Med. Sci. 65:405-408. [DOI] [PubMed] [Google Scholar]
- 65.Shuai, J., W. Wei, X. Li, N. Chen, Z. Zhang, X. Chen, and W. Fang. 2007. Genetic characterization of porcine circovirus type 2 (PCV2) from pigs in high-seroprevalence areas in southeastern China. Virus Genes 35:619-627. [DOI] [PubMed] [Google Scholar]
- 66.Smith, G. J., D. Vijaykrishna, J. Bahl, S. J. Lycette, M. Worobey, O. G. Pybus, S. K. Ma, C. L. Cheung, J. Raghwani, J. Bhatt, J. S. Peiris, Y. Guan, and A. Rambaut. 2009. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 25:1122-1125. [DOI] [PubMed] [Google Scholar]
- 67.Sorden, D. 2000. Update on porcine circovirus and postweaning multisystemic wasting syndrome (PMWS). Swine Health Prod. 8:133-136. [Google Scholar]
- 68.Turner, M. J., G. F. Medley, K. A. Woodbine, J. A. Slevin, and L. E. Green. 2009. The relationship between porcine circovirus 2 antigen score and antibody titre and histology of lymph nodes with 375 euthanased sick and healthy pigs from 113 British pig farms with and without postweaning multisystemic wasting syndrome. Prev. Vet. Med. 88:213-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Urbino, C., G. Thébaud, M. Granier, S. Blanc, and M. Peterschmitt. 2008. A novel cloning strategy for isolating, genotyping and phenotyping genetic variants of geminiviruses. Virol. J. 5:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wiederkehr, D. D., T. Sydler, E. Buergi, M. Haessig, D. Zimmermann, A. Paspischil, E. Brugner, and X. Sidler. 2009. A new emerging genotype subgroup within PCV-2b dominates the PMWS epizooty in Switzerland. Vet. Microbiol. 136:27-35. [DOI] [PubMed] [Google Scholar]
- 71.Woodbine, K. A., G. F. Medley, J. Slevin, A. L. Kilbride, E. F. Novell, M. J. Turner, M. J. Keeling, and L. E. Green. 2007. Spatiotemporal patterns and risks of herd breakdowns in pigs with postweaning multisystemic wasting syndrome. Vet. Rec. 160:751-762. [DOI] [PubMed] [Google Scholar]
- 72.Woolhouse, M. E., L. H. Taylor, and D. T. Haydon. 2001. Population biology of multihost pathogens. Science 292:1109-1112. [DOI] [PubMed] [Google Scholar]
- 73.Zhou, J. Y., Q. X. Chen, J. X. Ye, H. G. Shen, T. F. Chen, and S. B. Shang. 2006. Serological investigation and genomic characterization of PCV2 isolates from different geographic regions of Zhejiang province in China. Vet. Res. Commun. 30:205-220. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.