Abstract
The bovine immunodeficiency virus (BIV) Rev protein (186 amino acids [aa] in length) is involved in the nuclear exportation of partially spliced and unspliced viral RNAs. Previous studies have shown that BIV Rev localizes in the nucleus and nucleolus of infected cells. Here we report the characterization of the nuclear/nucleolar localization signals (NLS/NoLS) of this protein. Through transfection of a series of deletion mutants of BIV Rev fused to enhanced green fluorescent protein and fluorescence microscopy analyses, we were able to map the NLS region between aa 71 and 110 of the protein. Remarkably, by conducting alanine substitution of basic residues within the aa 71 to 110 sequence, we demonstrated that the BIV Rev NLS is bipartite, maps to aa 71 to 74 and 95 to 101, and is predominantly composed of arginine residues. This is the first report of a bipartite Rev (or Rev-like) NLS in a lentivirus/retrovirus. Moreover, this NLS is atypical, as the length of the sequence between the motifs composing the bipartite NLS, e.g., the spacer sequence, is 20 aa. Further mutagenesis experiments also identified the NoLS region of BIV Rev. It localizes mainly within the NLS spacer sequence. In addition, the BIV Rev NoLS sequence differs from the consensus sequence reported for other viral and cellular nucleolar proteins. In summary, we conclude that the nucleolar and nuclear localizations of BIV Rev are mediated via novel NLS and NoLS motifs.
Bovine immunodeficiency virus (BIV) is a lentivirus of the Retroviridae family which shares morphological, genetic, antigenic, and/or biologic properties with human immunodeficiency virus type 1 (HIV-1) and other animal lentiviruses including equine infectious anemia virus (EIAV) (22, 23, 68). Although the association of clinical diseases with BIV is still controversial, persistent lymphocytosis, neurological disorders associated with central nervous system lesions, weight loss, diminished milk production, lymphoid hyperplasia, and the presence of opportunistic bacterial infections have been associated with BIV infection (5, 47, 57, 70). Interestingly, studies of rabbits experimentally infected with BIV have shown the development of a disease characterized by a fatal dysfunction of the immune system similar to that observed in humans, nonhuman primates, and cats infected with HIV-1 (or HIV-2), simian immunodeficiency virus, and feline immunodeficiency virus, respectively (34, 35).
BIV is categorized as a complex nonprimate lentivirus (68). The BIV provirus DNA is 8,960 nucleotides in length with a typical retroviral genomic structure containing the gag, pol, and env genes flanked by long terminal repeats of 589 nucleotides in length at the 5′ and 3′ termini (23, 68). In proximity to the pol/env junction, the BIV genome also contains additional open reading frames that may encode nonstructural regulatory/accessory proteins (23, 68). These open reading frames are designated vif (viral infectivity factor), tat (trans-activator factor of transcription), rev (regulator of virus expression), vpw, vpy, and tmx. Only the Tat and Rev proteins have been reported to regulate viral expression at the transcriptional and posttranscriptional levels, respectively (21-23, 68). Among the latter proteins, only the Tat protein and its transactivator response element (TAR) located within the long terminal repeat sequence have been intensively studied (3, 6, 10, 19, 58, 60, 72).
BIV Rev is a 23-kDa (186-aa-long) phosphoprotein produced from a multiply spliced mRNA that contains the untranslated leader (exon 1) and two encoding exons (exons 2 and 3) (55). It has been shown previously that BIV Rev localizes to the nucleus and nucleoli of BIV-infected cells (56). As reported for HIV-1 Rev, BIV Rev mediates the nuclear exportation of partially spliced viral RNAs encoding structural proteins and of unspliced RNAs that serve as genomic RNA by interacting with a stem-loop structure termed Rev-responsive element (RRE) present in these RNAs (59). The Rev proteins contain at least three central functional domains: a basic arginine-rich domain that mediates RNA binding (RBD) and contains the nuclear/nucleolar localization signal (NLS/NoLS), a multimerization domain, and a leucine-rich domain that is necessary for the nuclear exportation of Rev (51, 59).
In HIV-1, the Rev protein shuttles between the nucleus and the cytoplasm of the infected cells via the importin/exportin proteins or nucleoporin pathway (59). The shuttling of HIV-1 Rev into the nucleus is mediated by the direct binding of Rev NLS to the nuclear transport receptors, mainly importin β but also transportin, importin 5, and importin 7 (2). This mechanism differs from the classical protein importation α/β pathway where importin α serves as an adaptor that links the protein and importin β1 and recognizes NLSs within the proteins. Importin α recognizes two types of NLSs: the classical monopartite NLSs and the bipartite NLSs that are composed, respectively, of one or two clusters of basic amino acids (predominantly lysine residues) (40). Monopartite NLSs are currently categorized in five different classes on the basis of amino acid composition and of their interaction with importin α (38). One well-known example of a class 1 monopartite NLS is that of the simian virus 40 large T antigen (PKKKRRV) (33, 38). Bipartite NLSs are less diversified, as they are classified into two main types (long and short) according to the length of the so-called spacer region, which refers to the number of amino acids between the motifs that compose the NLS (63). An example of a bipartite NLS is the short-type NLS of the nucleoplasmin protein with a spacer region of 10 aa (62). The long-type bipartite NLS with a spacer of 30 aa is exemplified by the HnRNP1 protein (63).
The NLS motifs of numerous Rev or Rev-like proteins have been characterized so far including those of Rev HIV-1, feline immunodeficiency virus, simian immunodeficiency virus, and EIAV and of Rev-like family members such as the mouse mammary tumor virus Rem, the endogenous retrovirus human endogenous retrovirus K Rec, and the human T-cell leukemia virus type 1 (HTLV-1) Rex (31, 36, 41, 46, 51, 59, 73). These NLSs are all monopartite structures, and based on their amino acid composition, they are nonclassical because they contain clusters of basic amino acids, mainly arginine residues (28, 41, 45). Moreover, importin α, as mentioned above, is not involved in the recognition of HIV-1 Rev NLS (2, 52, 69). In addition to the NLS, HIV-1 and certain Rev-like proteins contain an NoLS that mediates the accumulation of Rev in nucleoli (7, 39). These NoLSs correspond to one or multiple copies of the R/K-R/K-X-R/K consensus sequence (17).
Little is known on the exact location and amino acid composition of the BIV Rev functional domains. This is undoubtedly of interest as the BIV Rev amino acid sequence does not display a high degree of homology with that of other lentiviral Rev proteins. Here, we have determined the localization of the NLS and NoLS regions of BIV Rev. By generating a series of deletion and single point mutants, we have mapped a bipartite NLS in a Rev protein for the first time in a lentivirus. We found that BIV Rev NLS is atypical, as it harbors a 20-aa spacer sequence between the motifs that compose the NLS. We also identified the BIV Rev NoLS, which also differs from the NoLSs previously described in any viral or cellular proteins in terms of amino acid composition and/or its location in the spacer region of the bipartite NLS.
MATERIALS AND METHODS
Cell cultures and transfections.
HEK 293T, Cf2Th, MDBK, and Vero cells were maintained at 37°C in a humidified atmosphere of 5% CO2 in Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (PAA Laboratories, Inc., Etobicoke, Ontario, Canada). Cell transfections were done in six-well cell culture plates seeded to a cell density of ∼50% confluence. The next day, plasmids were mixed with the FuGENE HD transfection reagent (Roche, Indianapolis, IN) and added to the cells according to the manufacturer's protocol.
Construction of Rev deletion mutants.
Rev mutants (M1 to M9) containing internal deletions were generated by PCR-ligation-PCR mutagenesis (1). Briefly, upstream and downstream blunt-ended cDNA fragments of each mutant were amplified from pcDNA3.1BIVRevWT (wild type [WT]) with appropriate phosphorylated primers (sequences available upon request). Fragments were gel purified and ligated to the corresponding upstream fragment. Ligation products were then amplified by using the forward Rev5′ (5′-ATGGATCAGGACCTAGACCGCGC-3′) and reverse Rev3′ (5′-TTTTGTTCCCTGGATCCATCC-3′) primers. The generated PCR products were cloned into pBluescript KS(+) (Stratagene, La Jolla, CA). The N-terminal (M1) and the C-terminal (M9) mutants were PCR amplified using a forward primer (containing an ATG initiation codon) and a reverse primer (containing a stop codon), respectively. All constructs were confirmed by DNA sequencing through the McGill University Sequencing Services (Montréal, Québec, Canada).
Rev fusion proteins.
To generate proteins containing c-Myc and six-histidine tags fused to the C terminus of Rev, BIV Rev WT and the deletion mutants were amplified by PCR from the pBluescript KS(+) constructs described above with primers that introduced 5′ NotI and 3′ ApaI restriction sites. Digested PCR products were cloned into the pcDNA3.1Myc/HisB expression vector (Invitrogen) and sequenced for validation. To generate enhanced green fluorescent protein (EGFP) fusion proteins, the Rev sequences were amplified by PCR from the WT and deletion constructs with primers that introduced 5′ XhoI and 3′ ApaI restriction sites. The PCR products were digested with XhoI and ApaI and subsequently cloned into the corresponding restriction sites in the pEGFP-C1 vector (Clontech, Palo Alto, CA) to generate pEGFP-BIVRevWT and pEGFP-BIVRev mutant constructs. To generate the EGFP-NLS BIV, the region containing the putative NLS of BIV (aa 71 to 110) of Rev WT was amplified by PCR and cloned in pEGFP-C1 as described above.
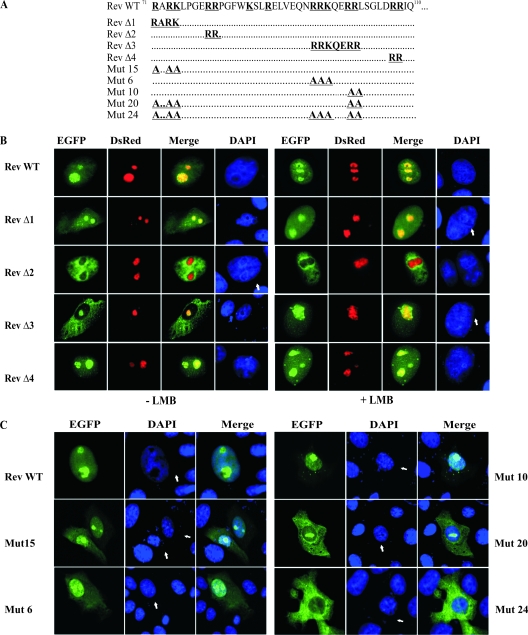
The Rev Δ constructs targeting selected regions of the BIV Rev WT were generated as described for the Rev deletion mutants by using appropriate phosphorylated primers (sequences available upon request). The resulting PCR products were directly cloned into pEGFP-C1. The Rev delta (Δ) constructs were as follow: Rev Δ1 (deletion of 71RARK74 residues), Rev Δ2 (deletion of 79RR80 residues), Rev Δ3 (deletion of 95RRKQERR101 residues), and Rev Δ4 (deletion of 107RR108 residues) (see Fig. 4A). Alanine substitution mutants were generated by site-directed mutagenesis using appropriate primers and were introduced into the EGFP-Rev protein (Table 1). All constructs were validated by DNA sequencing.
FIG. 4.
The NLS of the BIV Rev protein is of a bipartite type. (A) Several pEGFP-Rev deletion (Rev Δ1 to Rev Δ4) and alanine substitution (Mut 15, Mut 6, Mut 10, Mut 20, and Mut 24) mutant constructs targeting the aa 71 to 110 sequence of BIV Rev were generated from pEGFP-Rev WT. (B) Subcellular localization of EGFP-Rev deletion mutants by fluorescence microscopy. Vero cells were cotransfected for 24 h with pRed-C1Nucleolin and each of the pEGFP-Rev deletion constructs described for panel A in the presence or absence of 5 nM LMB and incubated for 24 h. The cells were fixed, counterstained with DAPI (in blue), and examined by fluorescence microscopy. The merge image represents the superposition of EGFP and DsRed-Nucleolin images. (C) Subcellular localization of EGFP-Rev alanine substitution mutants. Vero cells were transfected with each of the indicated pEGFP-Rev mutant constructs. The merge image represents the superposition of EGFP and DAPI images. The images shown are representative of the expression patterns (three independent experiments) observed in >70% of the cells.
TABLE 1.
Listing of the BIV Rev mutant proteinsa
| Mutant | Mutation at residue: |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 71R | 73R | 74K | 79R | 80R | 85K | 88R | 95R | 96R | 97K | 100R | 101R | 107R | 108R | 154K | 156K | 157R | |
| Mut 1 | A | ||||||||||||||||
| Mut 2 | A | A | |||||||||||||||
| Mut 3 | A | A | |||||||||||||||
| Mut 4 | A | ||||||||||||||||
| Mut 5 | A | ||||||||||||||||
| Mut 6 | A | A | A | ||||||||||||||
| Mut 7 | A | ||||||||||||||||
| Mut 8 | A | ||||||||||||||||
| Mut 9 | A | ||||||||||||||||
| Mut 10 | A | A | |||||||||||||||
| Mut 11 | A | A | |||||||||||||||
| Mut 12 | A | ||||||||||||||||
| Mut 13 | A | ||||||||||||||||
| Mut 14 | A | ||||||||||||||||
| Mut 15 | A | A | A | ||||||||||||||
| Mut 16 | A | A | A | A | A | ||||||||||||
| Mut 17 | A | A | |||||||||||||||
| Mut 18 | A | A | |||||||||||||||
| Mut 19 | A | A | A | A | A | A | A | ||||||||||
| Mut 20 | A | A | A | A | A | ||||||||||||
| Mut 21 | A | A | A | ||||||||||||||
| Mut 22 | A | A | A | A | A | A | A | ||||||||||
| Mut 23 | A | A | A | A | A | ||||||||||||
| Mut 24 | A | A | A | A | A | A | A | A | |||||||||
| Mut 25 | A | A | A | ||||||||||||||
| Mut 26 | A | A | A | A | |||||||||||||
The indicated arginine (R) and lysine (K) residues in the BIV Rev sequence were changed to alanine (A) residues.
Red fluorescence fusion proteins.
To generate the red fluorescent protein (RFP) in fusion with other proteins, the sequence of RFP cDNA was amplified by PCR from pDSRED2-N1 (Clontech) by using primers that introduced 5′ NheI and 3′ XhoI restriction sites. To clone the RFP, the pEGFP-C1 vector (Clontech) was digested with NheI and XhoI to replace the EGFP sequence with RFP. The resulting plasmid was designated pRED-C1. To generate the nucleolin, fibrillarin, and B23.1 versions of pRED-C1, the corresponding cDNAs were obtained by using KpnI and BamHI restriction enzymes from plasmids pEGFP-C1Nucleolin, pEGFP-C1Fibrillarin, and pEGFP-C1B23.1 (16), which were kindly provided by Julian A. Hiscox (University of Leeds, Leeds, United Kingdom). The nucleolin, fibrillarin, and B23.1 fragments were then subcloned into pRED-C1 previously digested with KpnI and BamHI restriction enzymes. All constructs were validated by DNA sequencing.
Indirect immunofluorescence assay.
Cells cultured on coverslips in six-well cell culture plates were transfected with the respective expression vectors. After an incubation of 48 h, the transfected cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS, pH 7.3) solution for 15 min, permeabilized with 0.2% Triton X-100 for 10 min, blocked with 4% bovine serum albumin in PBS for 1 h at 37°C, and then incubated with mouse primary anti-Myc antibodies (1 μg/ml; Roche) for 1 h at 37°C. After three washes with PBS containing 0.2% Triton X-100, the cells were incubated with Alexa 488- or Cy3-labeled anti-mouse secondary antibodies (Invitrogen) for 1 h at 37°C. Nuclei were counterstained with DAPI (4′,6-diamidino-2-phenylindole; Sigma-Aldrich, St. Louis, MO). The coverslips were then mounted on glass slides using ProLong Gold antifade reagent (Invitrogen). The fluorescence images were taken with an Eclipse Ti fluorescent microscope (Nikon) equipped with a 60× 0.75-numerical-aperture objective lens. Images were captured as 8-bit tagged image file format files with a Scion CFW-1608C camera. All images were analyzed with the Image-J software (61). For each protein examined, 50 positive cells were observed. The data shown below were representative, from three independent experiments, of expression patterns observed in >70% of the cells.
Fluorescence and confocal microscopy.
Cells cultured on coverslips in six-well cell culture plates were transfected with each of the respective pEGFP and pRed expression constructs. After 24 h of incubation, for certain samples, 5 nM of leptomycin B (LMB; Sigma-Aldrich) was added to the cell culture medium for 4 h. The cells were fixed for nuclear staining and mounted as described above. The fluorescence images were captured as described above. For confocal microscopy analysis, the cells were cultured on glass chamber slides (Thermo Fisher, Ottawa, Ontario, Canada). Unfixed cells were examined 24 h after transfection. The images were taken using an inverse microscope (Nikon TE-300) coupled to the confocal Bio-Rad MRC-1024ES system. All images were analyzed with the Image-J software. For each protein examined, 50 expressing cells were observed. The data shown below were representative, from three independent experiments, of expression patterns observed in >70% of cells.
CAT assay.
Rev nuclear export activity was quantified in transient-transfection assays using a pDM138-based BIV Rev chloramphenicol acetyltransferase (CAT) reporter construct. The minimal region of BIV RRE (50) was cloned into the claI site of plasmid pDM138 (30) (a generous gift of Yuying Liang from Emory University) to generate pRRE-BIV, the sequence of which was validated by sequencing. 293T cells were seeded in 12-well cell culture plates and then cotransfected the next day with 0.5 μg of empty pcDNA3.1 Myc/HisB or pEGFP-C1 or each of the pcDNA3.1 Myc/HisB or pEGFP constructs encoding either Rev WT or each of the Rev mutants, 0.5 μg of pRRE-BIV, and 0.2 μg of Rous sarcoma virus β-galactosidase plasmid (Fisher Scientific, Nepean, Ontario, Canada). The Rous sarcoma virus β-galactosidase plasmid was used as a control for transfection efficiency and normalization of data. Cells were harvested at 48 h following transfection and lysed with the lysis buffer (CAT enzyme-linked immunosorbent assay kit; Roche). The amount of CAT in 50 μg of total cellular proteins was determined using the CAT enzyme-linked immunosorbent assay kit. β-Galactosidase activity of each cell extract was determined by incubating 50 μg of proteins with 50 μl of β-galactosidase assay buffer (100 mM sodium phosphate buffer, pH 7.3, 2 mM MgCl2, 10 mM β-mercaptoethanol, and 133 mg/ml o-nitrophenyl-β-d-galactopyranoside) for 30 min at 37°C. The reaction was stopped by the addition of 150 μl of 1 M sodium carbonate. Colorimetric reaction was measured at 410 nm in a microplate reader (model 550; Bio-Rad). CAT expression data were normalized through the establishment of a ratio using β-galactosidase activity for each sample. Rev activity was defined as the ratio of CAT quantity obtained in cells transfected with the plasmids containing BIV RRE and Rev WT- or Rev mutant-encoding sequences to that obtained in cells transfected with the BIV RRE-containing plasmid alone. All cell transfections were performed in triplicate, and the experiments were repeated three times. Statistical analysis was performed using a two-tailed t test.
SDS-polyacrylamide gel electrophoresis and Western blot analyses.
Cell extracts were prepared as described previously (67). Total cell protein concentrations were quantified with the DC protein assay (Bio-Rad). For each sample, 50 μg of total cell extract was electrophoretically separated onto 12 or 15% sodium dodecyl sulfate (SDS)-polyacrylamide gels and transferred to nitrocellulose membranes. The membranes were blocked in PBS-T (PBS containing 0.05% Tween 20) in the presence of 5% nonfat dry milk powder for 1 h at room temperature. The membranes were incubated for 1 h at room temperature with mouse monoclonal primary antibodies specific to either GFP (Roche), Myc (Roche), or glyceraldehyde-3-phosphate dehydrogenase (Sigma-Aldrich). Following incubation with the antibodies, membranes were washed three times with PBS-T and incubated for 1 h (at room temperature) with anti-mouse horseradish peroxidase-conjugated immunoglobulin Gs (Bethyl, Montgomery, TX) used as secondary antibodies. The signal was detected by enhanced chemiluminescence (Perkin-Elmer, Boston, MA). The membranes were then exposed to Kodak Biomax Light-1 films.
RESULTS
BIV Rev fused to EGFP predominantly localizes to the nucleoli.
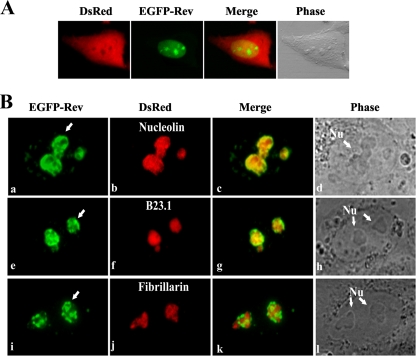
As reported in a previous study, BIV Rev localizes predominantly into the nucleus and nucleoli of infected cells as determined by immune gold staining (56). To confirm the intracellular distribution of BIV Rev, HEK 293 cells were cotransfected with plasmids pEGFP-RevWT, encoding EGFP in fusion with BIV Rev, and pDsRed, encoding the RFP used as a total cell marker. As shown in Fig. 1A, the EGFP-Rev fusion protein localized into the nucleus and more predominantly into the nucleoli of the transfected cells. About 5 to 10% of the transfected cells showed the presence of Rev within the cytoplasm, indicating a rapid nuclear transport and accumulation. This distribution pattern was also observed in all other cells tested. The predominant nucleus and nucleolus localization pattern observed here with pEGFP-Rev was also confirmed by immunofluorescence where Rev was fused to the c-Myc tag (Fig. 2C, WT panel). Finally, the main nucleolar localization of the EGFP-Rev fusion protein was also observed in BIV-permissive canine (Cf2Th) and bovine (MDBK) cells (not shown) and in simian (Vero) cells, suggesting that this localization is not species specific. Because Vero cells offer a small nucleus/cytoplasm volume ratio and contain a small number of well-defined and easy-to-observe nucleoli, subsequent experiments, unless otherwise specified, were conducted with these cells.
FIG. 1.
Nuclear and nucleolar localization of BIV Rev WT protein in fusion with EGFP (EGFP-Rev). (A) Confocal fluorescence microscopy analyses of EGFP-Rev (in green) and DsRed (in red) proteins transiently expressed in HEK 293 cells 24 h after cell transfection. DsRed protein was used as a whole-cell marker. The phase-contrast image of the cell is also shown. (B) Confirmation of the nucleolar localization of BIV EGFP-Rev. Vero cells were cotransfected for 24 h with pEGFP-RevWT and either pRed-C1Nucleolin (a to d), pRed-C1B23.1 (e to h), or pRed-C1Fibrillarin (i to l) vectors. The merge images represent the superposition of EGFP and either DsRed-Nucleolin, DsRed-B23.1, or DsRed-Fibrillarin images. The phase-contrast images of the cells are also shown. The arrows in panels a, e, and i indicate the accumulation of EGFP-Rev in the perinucleolar region of the cell. Nu, nucleoli. The images shown are representative of expression patterns (three independent experiments) observed in >70% of the cells.
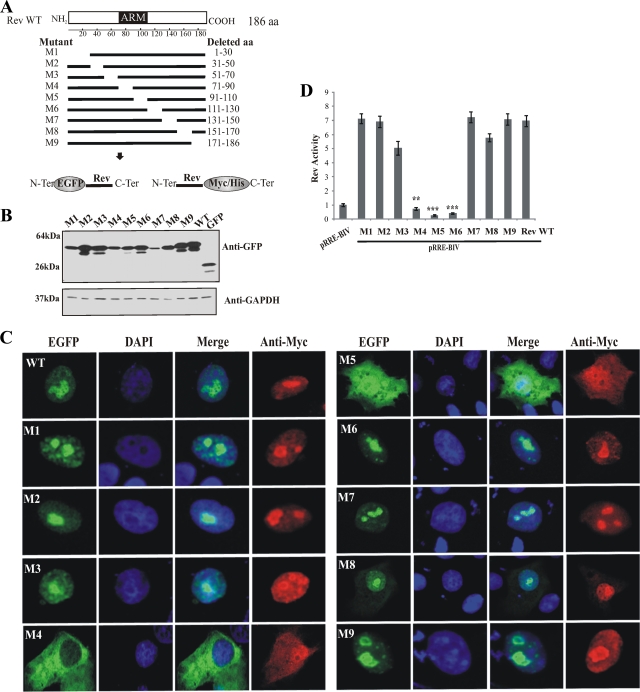
FIG. 2.
Analysis of BIV Rev protein deletion mutants reveals the importance of aa 71 to 110 for the intracellular localization of Rev. (A) The predicted functional motifs in BIV Rev are shown. Terminal and internal deletions were carried out from pcDNA3.1RevWT by PCR-ligation-PCR. The resulting constructs were cloned into the expression vectors pEGFP-C1 and pcDNA3.1Myc/HisB to generate the Rev amino acid deletion mutant proteins (M1 to M9). (B) Expression analyses of Rev WT and Rev mutant proteins in fusion with EGFP by Western blotting from Vero cells transfected with the appropriate plasmid constructs. Total cell proteins (50 μg) were separated on 15% SDS-polyacrylamide gels, electroblotted onto a nitrocellulose membrane, and probed with GFP-specific antibodies. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) immunostaining was used as a loading control. The molecular masses of the expressed proteins are indicated in the left margin. (C) Fluorescence microscopy analyses of Rev WT (WT) and Rev mutant (M1 to M9) proteins fused to EGFP or c-Myc/His tags. Vero cells were transfected with the respective plasmid constructs. After 24 h of incubation, the cells were fixed and counterstained with DAPI (in blue). The expression of EGFP fusion proteins is shown in green. The merge image represents the superposition of EGFP and DAPI images. The c-Myc/His-tagged proteins were visualized by indirect immunofluorescence using a Myc-specific primary antibody and an anti-mouse Cy3 secondary antibody. The images shown are representative of expression patterns (three independent experiments) observed in >70% of the cells. (D) Nuclear export activity of Rev-Myc/His WT and Rev-Myc/His mutants (M1 to M9) was determined using a CAT reporter assay. Fifty micrograms of cell lysate was used in the CAT assay. The CAT levels were normalized to the β-galactosidase activity. The results represent mean values of triplicate samples of at least three separate experiments. Rev activity is expressed as the ratio of Rev WT or mutant CAT expression to basal expression of pRRE-BIV alone. Vertical bars indicate the standard deviations about the means. The values significantly different from Rev WT, according to a two-tailed t test, are indicated by asterisks: **, P < 0.005; ***, P < 0.0005.
To identify more precisely the nucleolar distribution of BIV Rev, Vero cells were cotransfected with pEGFP-Rev and each of the plasmids containing well-known nucleolar protein markers in fusion with DsRed, namely, nucleolin (pDsRed-Nucleolin), fibrillarin (pDsRed-Fibrillarin), and B23.1 (pDsRed-B23.1). As shown in Fig. 1B, the nucleolar distribution of BIV Rev was similar to that of nucleolin (subpanels a to c) and of B23.1 (subpanels e to g). In contrast, the BIV Rev did not colocalize with fibrillarin (subpanels i to k). Finally the results also indicated that Rev accumulates, in a significant number of cells, at the periphery of the nucleoli (arrow in Fig. 1B, subpanels a, e, and i), an observation previously reported elsewhere (56).
Intracellular localization of BIV Rev deletion mutants.
In order to identify the region required for nuclear localization, we generated a series of 20- to 30-aa-long-deletion Rev mutants. The mutated Rev sequences were cloned in plasmid pEGFP or pcDNA3.1 Myc/HisB to produce two types of Rev-tagged proteins: the Rev-EGFP protein, in which Rev is fused to the C-terminal end of EGFP, and the Myc/His-Rev protein, in which Myc/His is fused to its C-terminal end (Fig. 2A). Western blot analyses revealed that all pEGFP constructs (Rev WT and mutated) were expressed at comparable levels upon transfection of Vero cells at the expected sizes (Fig. 2B). Similar results were obtained in cells transfected with the pcDNA3.1 Myc/His Rev WT and mutated constructs (data not shown).
The subcellular distribution of the deletion mutant proteins was analyzed by fluorescence microscopy. The results showed that six of the nine Rev mutants (M1, M2, M3, M6, M7, and M9) had a predominant nuclear/nucleolar localization similar to that of the WT version (Fig. 2C). In contrast, mutants M4 and M5, both with an aa 71 to 110 segment deletion, showed a diffuse cytoplasmic distribution with a slight M5 nuclear localization (Fig. 2B). Both mutant proteins did not accumulate in the nucleus or nucleoli as observed for the Rev WT (Fig. 2C). In contrast to the EGFP-tagged proteins, we could readily detect the Myc/His-tagged M4 and M5 mutants in the nucleus of transfected cells (Fig. 2B and data not shown). The nuclear localization of these mutated proteins was attributed to the low molecular weight of these proteins, conferring on them the ability to enter the nucleus through nuclear pores. No c-Myc or His signals were observed in cells transfected with empty plasmids (not shown). Interestingly, although mutant M8 (Δ151-170) displayed nuclear and nucleolar localizations similar to those of the WT, a higher proportion of cells (40%) showed the presence of the M8 mutant protein in the cytoplasm than that of cells expressing Rev WT (<10% of positive cells) (Fig. 2C). Although this finding was not addressed in this study, the presence of M8 in the cytoplasm is consistent with the dynamics of the cell nucleolus with the nucleoplasm, the alteration of which may result in either enrichment or exclusion of a particular protein in the nucleolus (17), and, ultimately, in the nucleus with a possible spillover in the cytoplasm.
After having established the subcellular localization of BIV Rev WT and each of the Rev mutant proteins, we wished to determine the impact of the mutations on the nuclear export activity of these proteins. This was assessed by a transient-transfection assay using the pRRE-BIV CAT reporter construct. This construct contains the CAT gene and a region containing the putative BIV RRE (50) located within an intron flanked by HIV-1 splice sites (30). By using this construct, unspliced transcripts of pRRE-BIV containing the CAT sequence enter in the cytoplasm only if Rev is present. In the absence of Rev, only traces of CAT activity are detected. As shown in Fig. 2D, M4, M5, and M6 mutated Rev proteins did not exert CAT activity, whereas all other mutants showed CAT activity comparable to that of Rev WT. The results obtained with M4 and M5 correlate with cytoplasmic localization of the proteins with no or negligible (for M5) distribution in the nucleus (Fig. 2C). Similar results for CAT activity were obtained with the EGFP-Rev WT and mutant fusion proteins (not shown). The lack of CAT activity obtained with the M6 mutant is associated with the fact that the amino acid sequence deleted in that protein contains the nuclear export signal of BIV Rev, which allows the exportation of unspliced transcripts in the cytoplasm where they are translated (A. Gomez Corredor and D. Archambault, unpublished data).
Two regions are essential for the nuclear localization of BIV Rev.
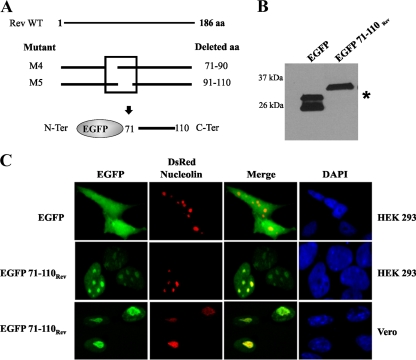
Nucleo-nucleolus-cytoplasmic trafficking of molecules is mediated by the NLS and NoLS (11, 16, 42). To determine whether the deleted region (aa 71 to 110) in M4 and M5 mutants functions as a bona fide NLS-NoLS, we directly fused this putative NLS-NoLS (40-aa) sequence to the C terminus of an EGFP (Fig. 3A). The expression of EGFP and of the EGFP fusion proteins in transfected Vero cells was confirmed by Western blot analysis (Fig. 3B). As expected, the control EGFP showed diffuse distribution in both the cytoplasm and the nucleus, whereas the EGFP-NLS-NoLS (EGFP 71-110Rev) fusion protein exclusively localized to the nucleus of HEK 293 and Vero cells (Fig. 3C). In addition, EGFP71-110Rev readily colocalized with the DsRed-Nucleolin into the nucleoli (Fig. 3C). These results clearly indicated that two regions within the M4 and M5 mutants that encompass aa 71 to 110 in BIV Rev contain functional NLS and NoLS that direct a cytoplasmic protein to the cell nucleus and nucleoli.
FIG. 3.
The region encompassing aa 71 to 100 of BIV Rev protein is associated with NLS and NoLS functions. (A) The region deleted in M4 and M5 Rev mutants was fused in the C-terminal end of EGFP, known to have diffuse localization in transfected cells. (B) Expression analysis of EGFP and EGFP 71-110Rev by Western blot assays of Vero cells transfected with the appropriate constructs. Total cell proteins (50 μg) were separated on 15% SDS-polyacrylamide gels, electroblotted onto a nitrocellulose membrane, and probed with GFP-specific antibodies. The molecular masses of the expressed proteins are indicated in the left margin. The asterisk indicates EGFP. (C) HEK 293 and Vero cells were cotransfected with pEGFP or pEGFP71-110Rev and with pRed-C1Nucleolin plasmids. After 24 h of incubation, the cells were fixed and counterstained with DAPI (in blue). Expression of the proteins was detected via the fluorescence of EGFP (in green) or RFP (in red). The merge image represents the superposition of EGFP and DsRed-Nucleolin images. The images shown are representative of the expression patterns (three independent experiments) observed in >70% of the cells.
The NLS motif in BIV Rev is bipartite.
NLSs and NoLSs are mainly composed of basic residues (17, 40). As shown in Fig. 4A, analysis of the sequence encompassing the aa 71 to 110 region of BIV Rev that contains the putative NLS and NoLS reveals four distinct clusters of basic amino acids (RARK, RR, RRKQERR, and RR) in addition to two separate K and R residues at positions 85 and 88, respectively. To determine which amino acids are necessary for the NLS-NoLS function of BIV Rev, we first generated a series of Rev mutants with deletions specific for each of the basic amino acid clusters. These mutants were then fused to EGFP (Fig. 4A). The constructs were then used to transfect Vero cells that were thereafter examined for subcellular localization of the mutated proteins. As Rev is a nucleocytoplasmic shuttling protein, experiments in cells were conducted in the presence or absence of LMB, an inhibitor of Rev nuclear export. Deletion of the first cluster (Rev Δ1; aa 71 to 74) slightly affected the subcellular distribution of the Rev-EGFP fusion protein in cells untreated with LMB, as demonstrated by a noticeable cytoplasmic localization (Fig. 4B). This result was not observed with EGFP-Rev WT (Fig. 1A). As expected, treatment of cells with LMB resulted in only nuclear and nucleolar localization of Rev Δ1. Deletion of the arginine residues at positions 79 and 80 in Rev Δ2 had no effect on the nuclear localization (Fig. 4B). However, these deletions completely abolished the nucleolar localization of the Rev protein in both LMB-untreated and LMB-treated cells. The deletion of the third cluster (Rev Δ3) (encompassing aa 95 to 101) disrupted intracellular distribution. In the absence of LMB, the Rev Δ3 protein was found not in the nucleus but rather in the cytoplasm and nucleoli. This nucleolus localization was confirmed by the colocalization of the mutated protein with DsRed-Nucleolin. In contrast, in the presence of LMB, the Rev Δ3 fusion protein was localized only into the nucleus and nucleoli (Fig. 4B). Taken together, these results indicate that the deleted residues in Rev Δ3 are necessary for the nuclear retention of BIV Rev and suggest that another region allows the entrance of the protein in the nucleus, indicating a possible bipartite NLS for BIV Rev. Finally, the last cluster of basic residues deleted in Rev Δ4 (aa 106 and 107) had no effect on the subcellular distribution of BIV Rev, indicating that these residues are not important for the nuclear localization of BIV Rev (Fig. 4B).
To confirm the results described above and to identify the amino acid residues of the NLS in BIV Rev, we generated a series of site-directed mutations in the basic residues present in the aa 71 to 110 sequence by replacing the arginine (R) and lysine (K) residues with alanine (A) residues (Table 1). The results obtained with the most relevant mutants are shown in Fig. 4. Single sets of substitutions of residues 71RARK74 to 71AAAA74 (Mut 15), 95RRK97 to 95AAA97 (Mut 6), or 100RR101 to 100AA101 (Mut 10) affected the proportion of the protein localized in the cytoplasm compared to the WT form of Rev (EGFP-Rev) (Fig. 4C and 1A). Interestingly, the combination of both 71RARK74 to 71AAAA74 and 100RR101 to 100AA101 (Mut 20) mutations disrupted the nuclear distribution of the mutated EGFP-Rev protein although the protein retained a nucleolar and cytoplasmic localization. Thus, cluster residues (95RRK97 and 100RR101) are necessary for the nuclear localization of EGFP-Rev. Finally, the combined mutations 71RARK74 to 71AAAA74, 95RRK97 to 95AAA97, and 100RR101 to 100AA101 (Mut 24) completely abolished the nuclear localization of the mutated EGFP-Rev, which was exclusively confined to the cytoplasm (Fig. 4C). Combined, the results indicate the presence of a bipartite NLS in BIV Rev that is composed of residues 71RARK74 and of 95RRK97/100RR101.
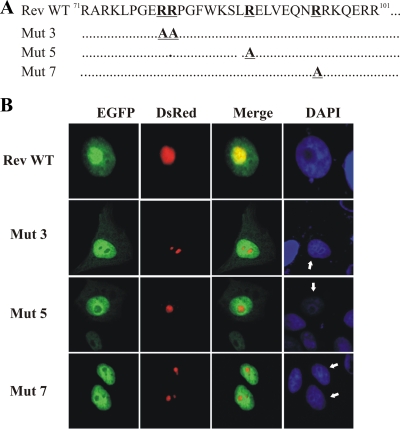
The NoLS of BIV Rev.
After having mapped the bipartite NLS in BIV Rev, we wished to identify the amino acid residues important for the nucleolar localization of the protein. To this end, we used the various mutated Rev versions described above in which the basic residues were changed to alanines in the region encompassing aa 71 to 110. In doing so, we were able to confirm the results obtained with the Rev Δ2 mutated protein (Fig. 4B). Indeed, alterations of 79RR80 to 79AA80 in EGFP-Rev (Mut 3) resulted in the localization of the protein to the nucleus but not in nucleoli (Fig. 5). Moreover, alterations of 88R to A (Mut 5) and 95R to A (Mut 7) in BIV EGFP-Rev disrupted the nucleolar localization of the protein with no effect on its nuclear localization (Fig. 5). Combined, the results demonstrated that residues 79RR80, 88R, and 95R are essential for the nucleolar targeting of BIV Rev.
FIG. 5.
Identification of amino acid residues necessary for the nucleolar localization of BIV Rev protein. (A) Alanine substitutions were introduced into the EGFP-Rev WT to generate the pEGFP-Rev Mut 3, Mut 5, and Mut 7 mutant constructs. (B) Vero cells were transfected with pRed-C1Nucleolin and each of the indicated pEGFP-Rev mutant constructs. After 24 h of incubation, the cells were fixed, counterstained with DAPI (in blue), and examined by fluorescence microscopy. The expression of EGFP-Rev mutant proteins and DsRed-Nucleolin is shown in green and red, respectively. The merge image represents the superposition of EGFP and DsRed-Nucleolin images. The image shown is a representative of the expression pattern (three independent experiments) observed in >70% of cells.
Functional analysis of BIV Rev NLS and NoLS mutants.
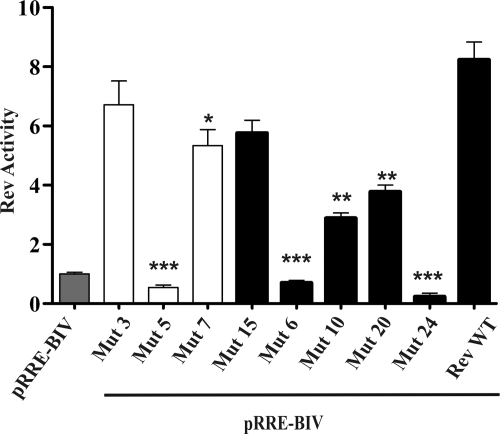
To determine whether mutations of BIV Rev NLS and NoLS amino acid residues had an impact in the nuclear export activity of the protein, Vero cells were cotransfected with the Rev-EGFP WT-encoding plasmid or each of the BIV Rev NLS and NoLS mutant plasmids and the pRRE-BIV CAT reporter construct as described above. The results showed that among all Rev NLS mutants tested (Fig. 6), only Mut 15 displayed an activity comparable to that of Rev WT. In contrast, the Mut 10 and Mut 20 mutants showed lower activity (at least 50%) than did Rev WT even though the proteins were present in the nucleus and the nucleoli, respectively (Fig. 4C). In contrast, the Mut 24 mutant, which was not localized in the nucleus and nucleoli, had no activity at all. Similarly, no activity was associated with the Mut 6 mutant despite its presence in the nucleus and nucleoli (Fig. 4C). When determining the activity of NoLS mutants (Fig. 6), Mut 3 and Mut 7 had activity comparable to or lower than that of Rev WT, respectively. In contrast, the Mut 5 mutant had no activity at all. As mentioned above, all of these mutants were localized only in the nucleoplasm.
FIG. 6.
Functional analysis of BIV Rev NLS and NoLS mutants. The nuclear export activities of EGFP-RevWT and EGFP-Rev NLS and NoLS mutants described for Fig. 4 and 5 were determined using a CAT reporter assay as described for Fig. 2C. Gray bar, pRRE-BIV alone; solid bars, NLS mutants; open bars, NoLS mutants. The results represent mean values of triplicate samples of at least three separate experiments. Vertical bars indicate the standard deviations about the means. The values significantly different from those for the Rev WT, according to a two-tailed t test, are indicated by asterisks: *, P < 0.05; **, P < 0.005; ***, P < 0.0005.
DISCUSSION
The Rev and Rev-like proteins of complex retroviruses are essential regulatory proteins that mediate nuclear export of partially spliced viral RNAs via functional domains that interact with cellular proteins and viral RNA. To exert their activity, lentivirus Rev proteins must enter the nucleus through NLSs (7, 39, 41, 69). Most nuclear proteins are targeted to the nucleus by mono- or bipartite basic amino acid sequences that constitute the NLS (11, 40, 62). In HIV-1 Rev, a 17-aa arginine-rich motif (Table 2) is located within the N-terminal half of the protein and serves as both the RRE-binding domain and the NLS (28). In addition to NLSs, HIV-1 and HTLV-1 target Rev and Rex proteins, respectively, to the nucleolus via NoLS (7, 39).
TABLE 2.
NLSs and NoLSsa
| Proteinc | NLS/NoLSb | Reference(s) |
|---|---|---|
| Bipartite NLS | ||
| Short-type NLSs | ||
| Nucleoplasmin | KRPAATKKAGQAKKKKLDK | 62 |
| CBP80 | RRRHSDENDGGQPHKRRK | 71 |
| hRAC3 | RKRKLPCDTPGQGLTCSGEKRRR | 74 |
| Sw15 | KKYENVVIKRSPRKRGRPRK | 63 |
| Consensus | [RK] (3), 8 to 16 aa [R/K] (4) | 63 |
| Long-type NLSs | ||
| HnRNP1 | KRGSDELFSTCVTNGPFIMSSNSASAANGNDSKKFKGDS | 63 |
| HIF-α | KRKMEHDGSLFQAVGIGTLLQQPDDHAATTSLSWKRVKG | 43 |
| Consensus | [KR] 30 to 32 aa [R/K] (4) | 63 |
| Unclassified-type NLSs | ||
| HCDA | KRPACTLKPECVQQLLVCSQEAKK | 65 |
| Rev BIV | RARKLPGERRPGFWKSLRELVEQNRRKQERR | This study |
| Viral and cellular monopartite NLSs and/or NoLSs | ||
| Rev, HIV-1 | RQARRNRRRRWRERQRQ | 7, 39 |
| Rev, EIAV | KRRRK | 41 |
| Rem, MMTV | ALRRKRRREMRK | 31 |
| Rex, HTLV-1 | PKTRRRPRRSQRKRPPTP | 39, 51, 53 |
| Rec, HERV-K | RRRRHRNRAP | 73 |
| Tat, HIV-1 | RKKRRQRRRAHQ | 8, 64 |
| hLa | SKGRRFKGKGK | 29 |
| MDM2 | KKLKKRNK | 42 |
| Consensus NoLS motif | R/K-R/K-X-R/K | 17, 29 |
| BIV Rev NoLS | ERRPGFWKSLRELVEQNR | This study |
Sequences of known short- and long-type bipartite NLSs and of NoLSs of various viral and cellular proteins with their respective consensus sequences in comparison with the novel type of bipartite NLS and NoLS sequences of BIV Rev protein.
The basic residues associated with NLSs are in bold, and the residues of NoLSs are underlined. The possible number of basic residues in the consensus sequences is indicated in parentheses.
Abbreviations: CBP80, CAP-binding protein 80; hRAC3, human receptor coactivator 3; Sw15, Saccharomyces cerevisiae transcription factor; HnRNP1, heterogeneous nuclear ribonucleoprotein; HIF-α, hypoxia-inducible factor α; HCDA, human cytidine deaminase; MMTV, mouse mammary tumor virus; HERV-K, human endogenous retrovirus K; Tat, trans activator factor of transcription; hLa, human La protein; MDM2, murine double minute 2 protein.
The present study is the first to directly characterize the cellular distribution of BIV Rev in details. We have demonstrated that the Rev protein of BIV localizes almost exclusively into the nucleus and the nucleoli of transiently transfected cells (Fig. 1), confirming a previous study conducted in BIV-infected cells (56). By using a series of Rev deletion mutants fused to the EGFP-encoding sequence, we thereafter identified the sequence necessary for the nuclear localization of BIV Rev. The subcellular localizations of both M4 and M5 mutants were disrupted as these proteins were distributed in the cytoplasm and the nucleus (Fig. 2C), a feature that contrasts with the predominant nuclear/nucleolar distribution of the Rev WT protein. Moreover, the biological activity of both M4 and M5 mutants was completely abolished in the CAT reporter assay (Fig. 2D). These results indicated that the regions deleted in M4 and M5 (aa 71 to 90 and aa 91 to 110, respectively) were important for the biological activity and intracellular localization of BIV Rev. The localization results also suggested the presence of a bipartite NLS in BIV Rev. To confirm the latter, we fused the aa 71 to 110 region to the C-terminal end of the EGFP protein and found that this sequence was sufficient to relocate the GFP into the nucleus and the nucleoli (Fig. 3).
The hallmark of bipartite NLSs is the presence of two clusters of basic amino acids whose integrity is essential for their function (62). As the amino acid deletion region in M4 and M5 contains several clusters of basic amino acid residues (Fig. 4A), we wished to identify which basic amino acid clusters were at play for the bipartite NLS function of BIV Rev. By using a series of deletions and point mutations within the Rev WT protein, we found that both the 71R-RK74 (Rev Δ1 mutant) and 95RRK-RR101 (Rev Δ3 mutant) basic residues were important for the nuclear localization of BIV Rev WT (Fig. 4). Interestingly, the 71R-RK74 cluster alone was able to transport the protein into the nucleus as shown by the result from the Rev Δ3 mutant, which contains an intact 71R-RK74 sequence. The latter mutant localized into the nucleoli and cytoplasm without nuclear accumulation in the absence of LMB (Fig. 4B). In contrast, in the presence of LMB, which blocks the putative exportation activity of Rev, the Rev Δ3 mutant was completely retained in the nucleus and nucleoli (Fig. 4B). Notably, in the 71RARK74 and 100RR101 to 71AAAA74 and 100AA101 mutant (Mut 20) (Fig. 4C), only the last two residues of the second cluster of basic residues (95RRK-RR101) are mutated to alanines. This mutant showed the same localization pattern as that of the Rev Δ3 mutant. Combined, these results suggest that the 71R-RK74 cluster mediates the transport of the protein into the nucleus and that the integrity of the 95RRK-RR101 cluster is necessary for both the nuclear localization and the retention of Rev. Our results unequivocally showed that the NLS of BIV Rev is bipartite and is composed of residues 71R-RK74 and 95RRK-RR101. This is the first report of a bipartite NLS for any Rev/Rex/Rev-like proteins in complex retroviruses. However, as shown in Table 2, the sequence of the two basic motifs in BIV Rev NLS differs from the consensus sequences reported for typical short- and long-type bipartite NLSs. Moreover, our results showed that the length of the spacer region (20 aa) separating the two basic clusters of BIV Rev NLS differs from the 8- to 16- and the 30- to 32-aa lengths described for the short- and long-type bipartite NLSs, respectively (32, 43, 62, 63, 71, 74). Although human cytidine deaminase and the integrase of HIV-1 have been reported to contain bipartite NLSs with spacer regions of 20 (Table 2) and 22 aa, respectively, the NLSs of these proteins were composed mainly of lysine residues (20, 65). This contrasts with the predominantly arginine residues described here. Combined, the results showed that BIV Rev contains a novel type of bona fide bipartite NLS.
The nucleus is a highly ordered structure containing several nonmembrane subcompartments, the largest subnuclear structure of which is the nucleolus. The nucleolus is morphologically divided into discrete regions including the fibrillar center, the dense fibrillar center (DFC), and the granular component (GC) (17). Beside the demonstrated role of the nucleolus in various steps of rRNA synthesis, a plurifunctional nucleolus hypothesis has been formulated and proposes that the nucleolus has multiple functions in health and disease (48, 66). In addition, the nucleolus can be a target for viral infection (26). For instance, changes in nucleolar structure can result from the coronavirus infectious bronchitis virus or from herpes simplex virus infection (4, 12, 44). Consistent with this and similar to what was previously reported for HIV-1 Rev (54), we also observed, in certain cell lines, altered nucleolar structure upon BIV Rev expression (not shown), an event that could be attributed to nucleolar accumulation of the protein (4, 44).
The nucleolar targeting of a protein depends on the presence of an NoLS or, in the absence of the latter, on interactions with nucleic acids or other NoLS-containing proteins that form a hub of proteins which, in turn, allows the binding of several protein partners (17). Here, we confirmed the nucleolar localization of the BIV Rev protein (56). We also showed that the BIV Rev protein did not colocalize with fibrillarin, which is known to localize in the DFC region of the nucleolus (Fig. 1B). In contrast, the BIV Rev protein colocalized with two other nucleolar markers, namely, nucleolin, known to localize in the GC and DFC region of the nucleolus, and B23.1, an isoform of B23 known to localize in the nucleolar GC (15, 16). As HIV-1 Rev was shown to colocalize and interact with B23 (13, 14, 18), it is possible that BIV also may interact with nucleolin and/or B23.1, an assumption that needs further studies.
We showed that BIV Rev contains an NoLS that mediates the nucleolar localization of this protein. We also showed that residues 79RR80, 88R, and 95R are essential for the nucleolar localization of BIV Rev (Fig. 4B and 5). Interestingly, none of the alanine substitutions within the NoLS disrupted the nuclear localization of BIV Rev (Fig. 5), thereby suggesting that the NoLS is independent of NLS function. HIV-1 Rev, HTLV Rex, and HIV-1 Tat proteins possess an NoLS that is intrinsically associated with the NLS (7, 39, 53, 64). These proteins contain an R/K-R/K-X-R/K consensus NoLS motif that is also present in a variety of other viral and cellular nucleolar proteins (9, 25, 29, 42) (Table 2). Remarkably, the BIV Rev does not harbor such a consensus motif, as several amino acid residues are present between the arginine residues associated with its NoLS function (Table 2). Like BIV Rev, the three NoLS sequences identified in parafibromin, a tumor suppressor protein, also differ from the consensus arginine/lysine motifs present in nucleolar proteins described so far (17, 24). In addition, the parafibromin NoLSs are localized in a region of the protein that is not intrinsically associated with the bipartite NLS (24). This contrasts with our results, where the 79RR80 and 88R residues of the Rev NoLS are localized between the two basic amino acid clusters that form the BIV Rev bipartite NLS and where the 95R residue is part of the second cluster of the bipartite NLS (95RRK-RR101). Thus, the BIV Rev protein harbors a novel type of NoLS different from NoLS motifs previously described for any viral or cellular proteins.
The NLS/NoLS and RBD motifs in several Rev/Rex/Rev-like proteins in complex retroviruses are closely associated with arginine-rich domains (27, 31, 45, 59, 73). Here we found that the nuclear export activity of the BIV Rev NLSs and NoLS mutants was diminished (Mut 7, Mut 10, and Mut 20) or absent (Mut 5 and Mut 6) even though they still localized in the nucleoplasm and/or nucleoli (Fig. 4C, 5, and 6). These results suggest that some of the amino acids that compose the NLS/NoLS are part of the RBD that mediates the binding of Rev to the RRE of the viral transcripts before their transport to the cytoplasm where they are translated. Finally, it has been suggested that the nucleolus plays a critical role in the nuclear export of HIV-1 Rev (8) and that the nucleoli “store” the protein (37). However, other investigators reported that HIV-1 Rev as well as HTLV-1 Rex exerts nuclear export activity with no nucleolar accumulation (49). A similar finding was observed here with the Mut 3 mutant that showed nuclear export activity like that of the Rev WT with no localization in the nucleoli (Fig. 6). The significance of this observation for the BIV life cycle has yet to be determined.
In conclusion, we demonstrated that the nucleolar and nuclear localizations of BIV Rev are mediated via novel NLS and NoLS motifs. BIV Rev NLS and NoLS may help in further understanding the dynamic network of proteins found in the nucleus and nucleoli.
Acknowledgments
A. Gomez Corredor was supported by a graduate studentship from La Fondation UQAM. This work was supported by an operating Discovery grant from the National Sciences and Engineering Research Council of Canada to D. Archambault.
We are grateful to Benoît Barbeau and Daniel Martineau for reviewing the manuscript and for helpful discussions.
Footnotes
Published ahead of print on 14 October 2009.
REFERENCES
- 1.Ali, S. A., and A. Steinkasserer. 1995. PCR-ligation-PCR mutagenesis: a protocol for creating gene fusions and mutations. BioTechniques 18:746-750. [PubMed] [Google Scholar]
- 2.Arnold, M., A. Nath, J. Hauber, and R. H. Kehlenbach. 2006. Multiple importins function as nuclear transport receptors for the Rev protein of human immunodeficiency virus type 1. J. Biol. Chem. 281:20883-20890. [DOI] [PubMed] [Google Scholar]
- 3.Barboric, M., R. Taube, N. Nekrep, K. Fujinaga, and B. M. Peterlin. 2000. Binding of Tat to TAR and recruitment of positive transcription elongation factor b occur independently in bovine immunodeficiency virus. J. Virol. 74:6039-6044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertrand, L., and A. Pearson. 2008. The conserved N-terminal domain of herpes simplex virus 1 UL24 protein is sufficient to induce the spatial redistribution of nucleolin. J. Gen. Virol. 89:1142-1151. [DOI] [PubMed] [Google Scholar]
- 5.Carpenter, S., L. D. Miller, S. Alexandersen, C. A. Whetstone, M. J. VanDerMaaten, B. Viuff, Y. Wannemuehler, J. M. Miller, and J. A. Roth. 1992. Characterization of early pathogenic effects after experimental infection of calves with bovine immunodeficiency-like virus. J. Virol. 66:1074-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, L., and A. D. Frankel. 1994. An RNA-binding peptide from bovine immunodeficiency virus Tat protein recognizes an unusual RNA structure. Biochemistry 33:2708-2715. [DOI] [PubMed] [Google Scholar]
- 7.Cochrane, A. W., A. Perkins, and C. A. Rosen. 1990. Identification of sequences important in the nucleolar localization of human immunodeficiency virus Rev: relevance of nucleolar localization to function. J. Virol. 64:881-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daelemans, D., S. V. Costes, E. H. Cho, R. A. Erwin-Cohen, S. Lockett, and G. N. Pavlakis. 2004. In vivo HIV-1 Rev multimerization in the nucleolus and cytoplasm identified by fluorescence resonance energy transfer. J. Biol. Chem. 279:50167-50175. [DOI] [PubMed] [Google Scholar]
- 9.Dang, C. V., and W. M. Lee. 1989. Nuclear and nucleolar targeting sequences of c-erb-A, c-myb, N-myc, p53, HSP70, and HIV tat proteins. J. Biol. Chem. 264:18019-18023. [PubMed] [Google Scholar]
- 10.Deng, G., Y. Su, J. Mu, R. Sha, Y. Geng, W. Qiao, and Q. Chen. 2008. Molecular basis of the internalization of bovine immunodeficiency virus Tat protein. Virus Genes 36:85-94. [DOI] [PubMed] [Google Scholar]
- 11.Dingwall, C., and R. A. Laskey. 1991. Nuclear targeting sequences—a consensus? Trends Biochem. Sci. 16:478-481. [DOI] [PubMed] [Google Scholar]
- 12.Dove, B. K., J. H. You, M. L. Reed, S. R. Emmett, G. Brooks, and J. A. Hiscox. 2006. Changes in nucleolar morphology and proteins during infection with the coronavirus infectious bronchitis virus. Cell. Microbiol. 8:1147-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dundr, M., G. H. Leno, M. L. Hammarskjold, D. Rekosh, C. Helga-Maria, and M. O. Olson. 1995. The roles of nucleolar structure and function in the subcellular location of the HIV-1 Rev protein. J. Cell Sci. 108:2811-2823. [DOI] [PubMed] [Google Scholar]
- 14.Dundr, M., G. H. Leno, N. Lewis, D. Rekosh, M. L. Hammarskjoid, and M. O. Olson. 1996. Location of the HIV-1 Rev protein during mitosis: inactivation of the nuclear export signal alters the pathway for postmitotic reentry into nucleoli. J. Cell Sci. 109:2239-2251. [DOI] [PubMed] [Google Scholar]
- 15.Dundr, M., T. Misteli, and M. O. Olson. 2000. The dynamics of postmitotic reassembly of the nucleolus. J. Cell Biol. 150:433-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emmott, E., B. K. Dove, G. Howell, L. A. Chappell, M. L. Reed, J. R. Boyne, J. H. You, G. Brooks, A. Whitehouse, and J. A. Hiscox. 2008. Viral nucleolar localisation signals determine dynamic trafficking within the nucleolus. Virology 380:191-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Emmott, E., and J. A. Hiscox. 2009. Nucleolar targeting: the hub of the matter. EMBO Rep. 10:231-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fankhauser, C., E. Izaurralde, Y. Adachi, P. Wingfield, and U. K. Laemmli. 1991. Specific complex of human immunodeficiency virus type 1 Rev and nucleolar B23 proteins: dissociation by the Rev response element. Mol. Cell. Biol. 11:2567-2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fong, S. E., J. D. Greenwood, J. C. Williamson, D. Derse, L. A. Pallansch, T. Copeland, L. Rasmussen, A. Mentzer, K. Nagashima, G. Tobin, and M. A. Gonda. 1997. Bovine immunodeficiency virus tat gene: cloning of two distinct cDNAs and identification, characterization, and immunolocalization of the tat gene products. Virology 233:339-357. [DOI] [PubMed] [Google Scholar]
- 20.Gallay, P., T. Hope, D. Chin, and D. Trono. 1997. HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc. Natl. Acad. Sci. USA 94:9825-9830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garvey, K. J., M. S. Oberste, J. E. Elser, M. J. Braun, and M. A. Gonda. 1990. Nucleotide sequence and genome organization of biologically active proviruses of the bovine immunodeficiency-like virus. Virology 175:391-409. [DOI] [PubMed] [Google Scholar]
- 22.Gonda, M. A., M. J. Braun, S. G. Carter, T. A. Kost, J. W. Bess, Jr., L. O. Arthur, and M. J. Van der Maaten. 1987. Characterization and molecular cloning of a bovine lentivirus related to human immunodeficiency virus. Nature 330:388-391. [DOI] [PubMed] [Google Scholar]
- 23.Gonda, M. A., D. G. Luther, S. E. Fong, and G. J. Tobin. 1994. Bovine immunodeficiency virus: molecular biology and virus-host interactions. Virus Res. 32:155-181. [DOI] [PubMed] [Google Scholar]
- 24.Hahn, M. A., and D. J. Marsh. 2007. Nucleolar localization of parafibromin is mediated by three nucleolar localization signals. FEBS Lett. 581:5070-5074. [DOI] [PubMed] [Google Scholar]
- 25.Hatanaka, M. 1990. Discovery of the nucleolar targeting signal. Bioessays 12:143-148. [DOI] [PubMed] [Google Scholar]
- 26.Hiscox, J. A. 2007. RNA viruses: hijacking the dynamic nucleolus. Nat. Rev. Microbiol. 5:119-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hope, T. J., B. L. Bond, D. McDonald, N. P. Klein, and T. G. Parslow. 1991. Effector domains of human immunodeficiency virus type 1 Rev and human T-cell leukemia virus type I Rex are functionally interchangeable and share an essential peptide motif. J. Virol. 65:6001-6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hope, T. J., X. J. Huang, D. McDonald, and T. G. Parslow. 1990. Steroid-receptor fusion of the human immunodeficiency virus type 1 Rev transactivator: mapping cryptic functions of the arginine-rich motif. Proc. Natl. Acad. Sci. USA 87:7787-7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horke, S., K. Reumann, M. Schweizer, H. Will, and T. Heise. 2004. Nuclear trafficking of La protein depends on a newly identified nucleolar localization signal and the ability to bind RNA. J. Biol. Chem. 279:26563-26570. [DOI] [PubMed] [Google Scholar]
- 30.Huang, X. J., T. J. Hope, B. L. Bond, D. McDonald, K. Grahl, and T. G. Parslow. 1991. Minimal Rev-response element for type 1 human immunodeficiency virus. J. Virol. 65:2131-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Indik, S., W. H. Gunzburg, B. Salmons, and F. Rouault. 2005. A novel, mouse mammary tumor virus encoded protein with Rev-like properties. Virology 337:1-6. [DOI] [PubMed] [Google Scholar]
- 32.Jans, D. A., T. Moll, K. Nasmyth, and P. Jans. 1995. Cyclin-dependent kinase site-regulated signal-dependent nuclear localization of the SW15 yeast transcription factor in mammalian cells. J. Biol. Chem. 270:17064-17067. [DOI] [PubMed] [Google Scholar]
- 33.Kalderon, D., B. L. Roberts, W. D. Richardson, and A. E. Smith. 1984. A short amino acid sequence able to specify nuclear location. Cell 39:499-509. [DOI] [PubMed] [Google Scholar]
- 34.Kalvatchev, Z., R. Walder, M. Barrios, and D. Garzaro. 1995. Acquired immune dysfunction in rabbits experimentally infected with an infectious molecular clone of the bovine immunodeficiency virus (BIV127). Viral Immunol. 8:159-164. [DOI] [PubMed] [Google Scholar]
- 35.Kalvatchev, Z., R. Walder, F. Perez, D. Garzaro, and M. Barrios. 1998. Infection of rabbits with R29 strain of bovine immunodeficiency virus: virulence, immunosuppression, and progressive mesenteric lymphadenopathy. Viral Immunol. 11:159-166. [DOI] [PubMed] [Google Scholar]
- 36.Kiyomasu, T., T. Miyazawa, T. Furuya, R. Shibata, H. Sakai, J. Sakuragi, M. Fukasawa, N. Maki, A. Hasegawa, T. Mikami, and A. Adachi. 1991. Identification of feline immunodeficiency virus rev gene activity. J. Virol. 65:4539-4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kjems, J., and P. Askjaer. 2000. Rev protein and its cellular partners. Adv. Pharmacol. 48:251-298. [DOI] [PubMed] [Google Scholar]
- 38.Kosugi, S., M. Hasebe, N. Matsumura, H. Takashima, E. Miyamoto-Sato, M. Tomita, and H. Yanagawa. 2009. Six classes of nuclear localization signals specific to different binding grooves of importin alpha. J. Biol. Chem. 284:478-485. [DOI] [PubMed] [Google Scholar]
- 39.Kubota, S., H. Siomi, T. Satoh, S. Endo, M. Maki, and M. Hatanaka. 1989. Functional similarity of HIV-I rev and HTLV-I rex proteins: identification of a new nucleolar-targeting signal in rev protein. Biochem. Biophys. Res. Commun. 162:963-970. [DOI] [PubMed] [Google Scholar]
- 40.Lange, A., R. E. Mills, C. J. Lange, M. Stewart, S. E. Devine, and A. H. Corbett. 2007. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J. Biol. Chem. 282:5101-5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee, J. H., S. C. Murphy, M. Belshan, W. O. Sparks, Y. Wannemuehler, S. Liu, T. J. Hope, D. Dobbs, and S. Carpenter. 2006. Characterization of functional domains of equine infectious anemia virus Rev suggests a bipartite RNA-binding domain. J. Virol. 80:3844-3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lohrum, M. A., M. Ashcroft, M. H. Kubbutat, and K. H. Vousden. 2000. Identification of a cryptic nucleolar-localization signal in MDM2. Nat. Cell Biol. 2:179-181. [DOI] [PubMed] [Google Scholar]
- 43.Luo, J. C., and M. Shibuya. 2001. A variant of nuclear localization signal of bipartite-type is required for the nuclear translocation of hypoxia inducible factors (1alpha, 2alpha and 3alpha). Oncogene 20:1435-1444. [DOI] [PubMed] [Google Scholar]
- 44.Lymberopoulos, M. H., and A. Pearson. 2007. Involvement of UL24 in herpes-simplex-virus-1-induced dispersal of nucleolin. Virology 363:397-409. [DOI] [PubMed] [Google Scholar]
- 45.Magin, C., J. Hesse, J. Lower, and R. Lower. 2000. Corf, the Rev/Rex homologue of HTDV/HERV-K, encodes an arginine-rich nuclear localization signal that exerts a trans-dominant phenotype when mutated. Virology 274:11-16. [DOI] [PubMed] [Google Scholar]
- 46.Malim, M. H., S. Bohnlein, R. Fenrick, S. Y. Le, J. V. Maizel, and B. R. Cullen. 1989. Functional comparison of the Rev trans-activators encoded by different primate immunodeficiency virus species. Proc. Natl. Acad. Sci. USA 86:8222-8226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martin, S. J., T. P. O'Neill, J. A. Bilello, and J. L. Eiseman. 1991. Lymphocyte transformation abnormalities in bovine immunodeficiency-like virus infected calves. Immunol. Lett. 27:81-84. [DOI] [PubMed] [Google Scholar]
- 48.Matthews, D. A., and M. O. Olson. 2006. What is new in the nucleolus?: workshop on the nucleolus: new perspectives. EMBO Rep. 7:870-873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McDonald, D., T. J. Hope, and T. G. Parslow. 1992. Posttranscriptional regulation by the human immunodeficiency virus type 1 Rev and human T-cell leukemia virus type I Rex proteins through a heterologous RNA binding site. J. Virol. 66:7232-7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Molina, R. P., M. Matukonis, B. Paszkiet, J. Zhang, M. Kaleko, and T. Luo. 2002. Mapping of the bovine immunodeficiency virus packaging signal and RRE and incorporation into a minimal gene transfer vector. Virology 304:10-23. [DOI] [PubMed] [Google Scholar]
- 51.Narayan, M., I. Younis, D. M. D'Agostino, and P. L. Green. 2003. Functional domain structure of human T-cell leukemia virus type 2 Rex. J. Virol. 77:12829-12840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Neville, M., F. Stutz, L. Lee, L. I. Davis, and M. Rosbash. 1997. The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr. Biol. 7:767-775. [DOI] [PubMed] [Google Scholar]
- 53.Nosaka, T., H. Siomi, Y. Adachi, M. Ishibashi, S. Kubota, M. Maki, and M. Hatanaka. 1989. Nucleolar targeting signal of human T-cell leukemia virus type I rex-encoded protein is essential for cytoplasmic accumulation of unspliced viral mRNA. Proc. Natl. Acad. Sci. USA 86:9798-9802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nosaka, T., T. Takamatsu, Y. Miyazaki, K. Sano, A. Sato, S. Kubota, M. Sakurai, Y. Ariumi, M. Nakai, S. Fujita, et al. 1993. Cytotoxic activity of rev protein of human immunodeficiency virus type 1 by nucleolar dysfunction. Exp. Cell Res. 209:89-102. [DOI] [PubMed] [Google Scholar]
- 55.Oberste, M. S., J. D. Greenwood, and M. A. Gonda. 1991. Analysis of the transcription pattern and mapping of the putative rev and env splice junctions of bovine immunodeficiency-like virus. J. Virol. 65:3932-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oberste, M. S., J. C. Williamson, J. D. Greenwood, K. Nagashima, T. D. Copeland, and M. A. Gonda. 1993. Characterization of bovine immunodeficiency virus rev cDNAs and identification and subcellular localization of the Rev protein. J. Virol. 67:6395-6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Onuma, M., E. Koomoto, H. Furuyama, Y. Yasutomi, H. Taniyama, H. Iwai, and Y. Kawakami. 1992. Infection and dysfunction of monocytes induced by experimental inoculation of calves with bovine immunodeficiency-like virus. J. Acquir. Immune Defic. Syndr. 5:1009-1015. [PubMed] [Google Scholar]
- 58.Pallansch, L. A., C. S. Lackman-Smith, and M. A. Gonda. 1992. Bovine immunodeficiency-like virus encodes factors which trans activate the long terminal repeat. J. Virol. 66:2647-2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pollard, V. W., and M. H. Malim. 1998. The HIV-1 Rev protein. Annu. Rev. Microbiol. 52:491-532. [DOI] [PubMed] [Google Scholar]
- 60.Puglisi, J. D., L. Chen, S. Blanchard, and A. D. Frankel. 1995. Solution structure of a bovine immunodeficiency virus Tat-TAR peptide-RNA complex. Science 270:1200-1203. [DOI] [PubMed] [Google Scholar]
- 61.Rasband, W. S. 1997-2004. ImageJ. National Institutes of Health, Bethesda, MD. http://rsb.info.nih.gov/ij/.
- 62.Robbins, J., S. M. Dilworth, R. A. Laskey, and C. Dingwall. 1991. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell 64:615-623. [DOI] [PubMed] [Google Scholar]
- 63.Romanelli, M. G., and C. Morandi. 2002. Importin alpha binds to an unusual bipartite nuclear localization signal in the heterogeneous ribonucleoprotein type I. Eur. J. Biochem. 269:2727-2734. [DOI] [PubMed] [Google Scholar]
- 64.Siomi, H., H. Shida, M. Maki, and M. Hatanaka. 1990. Effects of a highly basic region of human immunodeficiency virus Tat protein on nucleolar localization. J. Virol. 64:1803-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Somasekaram, A., A. Jarmuz, A. How, J. Scott, and N. Navaratnam. 1999. Intracellular localization of human cytidine deaminase. Identification of a functional nuclear localization signal. J. Biol. Chem. 274:28405-28412. [DOI] [PubMed] [Google Scholar]
- 66.Stark, L. A., and M. Taliansky. 2009. Old and new faces of the nucleolus. Workshop on the Nucleolus and Disease. EMBO Rep. 10:35-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.St-Louis, M. C., Y. Abed, and D. Archambault. 2005. The bovine immunodeficiency virus: cloning of a tat/rev cDNA encoding a novel Tat protein with enhanced transactivation activity. Arch. Virol. 150:1529-1547. [DOI] [PubMed] [Google Scholar]
- 68.St-Louis, M. C., M. Cojocariu, and D. Archambault. 2004. The molecular biology of bovine immunodeficiency virus: a comparison with other lentiviruses. Anim. Health Res. Rev. 5:125-143. [DOI] [PubMed] [Google Scholar]
- 69.Truant, R., and B. R. Cullen. 1999. The arginine-rich domains present in human immunodeficiency virus type 1 Tat and Rev function as direct importin beta-dependent nuclear localization signals. Mol. Cell. Biol. 19:1210-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Van der Maaten, M. J., W. A. Malmquist, and N. F. Cheville. 1972. Susceptibility of calves to bovine syncytial virus given by different inoculation routes. Am. J. Vet. Res. 33:1157-1160. [PubMed] [Google Scholar]
- 71.Weis, K., I. W. Mattaj, and A. I. Lamond. 1995. Identification of hSRP1 alpha as a functional receptor for nuclear localization sequences. Science 268:1049-1053. [DOI] [PubMed] [Google Scholar]
- 72.Xuan, C., W. Qiao, J. Li, G. Peng, M. Liu, Q. Chen, J. Zhou, and Y. Geng. 2008. BTat, a trans-acting regulatory protein, contributes to bovine immunodeficiency virus-induced apoptosis. Cell. Microbiol. 10:31-40. [DOI] [PubMed] [Google Scholar]
- 73.Yang, J., H. P. Bogerd, S. Peng, H. Wiegand, R. Truant, and B. R. Cullen. 1999. An ancient family of human endogenous retroviruses encodes a functional homolog of the HIV-1 Rev protein. Proc. Natl. Acad. Sci. USA 96:13404-13408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yeung, P. L., A. Zhang, and J. D. Chen. 2006. Nuclear localization of coactivator RAC3 is mediated by a bipartite NLS and importin alpha3. Biochem. Biophys. Res. Commun. 348:13-24. [DOI] [PubMed] [Google Scholar]