Abstract
We demonstrate the presence of a functional internal ribosome entry site (IRES) within the 5′ leader (designated 5L) from a variant of bicistronic mRNAs that encode the pp14 and RLORF9 proteins from Marek's disease virus (MDV) serotype 1. Transcribed as a 1.8-kb family of immediate-early genes, the mature bicistronic mRNAs have variable 5′ leader sequences due to alternative splicing or promoter usage. Consequently, the presence or absence of the 5L IRES in the mRNA dictates the mode of pp14 translation and leads to the production of two pp14 isoforms that differ in their N-terminal sequences. Real-time reverse transcription-quantitative PCR indicates that the mRNA variants with the 5L IRES is two to three times more abundant in MDV-infected and transformed cells than the mRNA variants lacking the 5L IRES. A common feature to all members of the 1.8-kb family of transcripts is the presence of an intercistronic IRES that we have previously shown to control the translation of the second open reading frame (i.e., RLORF9). Investigation of the two IRESs residing in the same bicistronic reporter mRNA revealed functional synergism for translation efficiency. In analogy with allosteric models in proteins, we propose IRES allostery to describe such a novel phenomenon. The functional implications of our findings are discussed in relation to host-virus interactions and translational control.
Marek's disease virus (MDV) is an oncogenic avian herpesvirus that induces malignant T-cell lymphomas and neurological disorders in its natural host, chicken (12, 32). Three MDV serotypes are recognized. Serotype 1 viruses (MDV-1) include the oncogenic MDVs and their cell culture-attenuated variants. Serotype 2 MDVs (MDV-2) include the naturally occurring nononcogenic chicken MDVs, while the nononcogenic turkey herpesviruses are classified as Meleagrid herpesvirus 1 (7, 8). The MDV genome is a double-stranded DNA with repeat structures that are characteristic of the Alphaherpesvirinae (10). The MDV genome consists of a unique long (UL) segment and a unique short (US) segment bracketed by inverted repeats. The genes located in the UL and US segments are largely homologous to, and arranged collinearly with, those of human herpesvirus 1 (herpes simplex virus type 1) and human herpesvirus 3 (varicella-zoster virus), whereas genus- and virus-specific genes are located in the inverted repeat regions (Fig. 1A) (10).
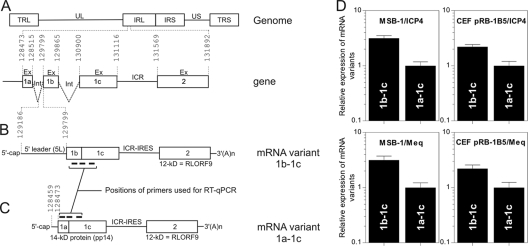
FIG. 1.
Genomic structure of MDV and expression of two variants of the 1.8-kb family of transcripts. (A) Schematic representation of the MDV genomic structure, consisting of UL and US regions, each bounded by a set of inverted repeats (TRL, IRL, IRS, and TRS). Introns (Int) and exons (Ex) are shown, as is the ICR region between exon 1c and exon 2. (B and C) Schematic representation of the bicistronic transcripts that we and others have cloned as cDNAs. The 5′ leader sequence in the mature transcript depicted in panel B is part of the intron between exon 1a and exon 1b. All genomic coordinates are according to MDV-1 strain Md5, accession number AF243438. (D) Relative expression level as measured by real-time RT-qPCR of the two transcript variants in MDV-transformed T-cell line MSB-1 and in primary CEFs infected with MDV-1 (pRB-1B5 bacterial artificial chromosome DNA). The level of each transcript was normalized to two MDV-1 genes, ICP4 and Meq. The means from triplicate qPCR assays with standard errors of the means are shown.
The repeat regions of MDV-1 have been the focus of intense investigations for several reasons. First, genes carried in herpesvirus repeat regions are virus specific (6). Second, abundant transcripts of immediate-early genes are derived from these regions. Third, and most importantly, transcripts derived from repeat regions may be associated with oncogenicity (3, 4). Transcripts originating from a bidirectional promoter located in the long internal repeat (IRL) in close proximity to the IRL/UL junction (40) are responsible for the expression of one of the major phosphoproteins, pp38, which appeared to be confined to the lytic phase (35). On the opposite strand, the same promoter seems to drive the transcription of a 1.8-kb family of transcripts (40). Several splice variants of the1.8-kb family of transcripts have been cloned as cDNAs, and their corresponding proteins have been identified (9, 18, 19). The role of the 1.8-kb family of transcripts in the maintenance of MDV latency in an MDV-1-transformed lymphoblastoid cell line was demonstrated by RNA interference experiments (23).
The importance of the 1.8-kb family of transcripts in the MDV-1 life cycle prompted us to investigate the translational control of a representative transcript that encodes the 14-kDa phosphoprotein (pp14), which is detected as an immediate-early protein in MDV-1-infected cells (18, 19). Being bicistronic, this transcript encodes the pp14 protein in two exons within the 5′ open reading frame (ORF) and the 107-amino-acid polypeptide (RLORF9) in the single exon of the 3′ ORF (18, 33). We have recently reported that the intercistronic (ICR) region between pp14 and RLORF9 contains a modular internal ribosome entry site (IRES) (41).
In the present study we demonstrated the presence of a functional IRES within the 5′ leader (designated 5L) of a subset of the mature bicistronic transcripts. The mRNA variants give rise to two pp14 protein variants that can be distinguished by the amino acid compositions of their N termini (19) (Fig. 1B and C) and by the mechanism underlying their translational control. Real-time reverse transcription-quantitative PCR (RT-qPCR) indicates that the mRNA variant with the 5L IRES is more abundant in MDV-infected and transformed cells than the mRNA variant lacking the 5L IRES. We hypothesize that the ability of the 5L IRES to mediate cap-independent translation initiation may enable this mRNA to overcome the interferon response and circumvent translation inhibition by the double-stranded-RNA-dependent protein kinase PKR, as has been reporter for the hepatitis C virus HCV IRES (42). Importantly, our findings revealed functional synergism between the 5L IRES and the ICR IRES within the same bicistronic RNA. In analogy with allosteric models of proteins, we propose IRES allostery to describe such a novel finding.
MATERIALS AND METHODS
Isolation of RNA and construction of cDNA library from MSB-1 cells.
RNA isolation from the MDV-transformed T-cell line MSB-1 and cDNA libraries were performed as previously described (41).
Real-time RT-qPCR.
Total RNA was extracted from MSB-1 cells and from chicken embryo fibroblasts (CEFs) infected with recombinant MDV-1 bacterial artificial chromosome DNA (pRB-1B5) (34) using Trizol (Invitrogen). The RNA was treated with DNase I (Promega) at 37°C for 1 h and reverse transcribed using Superscript II (Invitrogen) as previously described (43). PCR amplification was carried out in 20-μl reaction mixtures with 5 μl RT, 0.10 to 0.15 μM 6-carboxyfluorescein-labeled MGB probe (Applied Biosystems), 0.20 to 0.25 μM forward and reverse primers, and 10 μl of the Universal PCR master mix. The PCR conditions used were 95°C for 10 min, followed by 40 cycles of amplification at 95°C for 15 s and 60°C for 1 min. The primers used were F1a1c (5′-CGGCCGATCCCCGATA-3′), R1a1c (5′-AAAGTGGGTCCGCAGTCAAT-3′), F1b1c (5′-AAACCCTTCTCAGTTGTCGATTG-3′), and R1b1c (5′-GAAAGTGGGTCCGCAGTCAA-3′). The probes used were P1a1c (5′-TTTCCTGACCCCGGATG-3′), and P1b1c (5′-TGGGAACGACCCCGGA-3′). All real-time qPCRs were run in triplicate using the ABI Prism sequence detection system. The relative expression levels of the two MDV-1 mRNA variants 1a-1c and 1b-1c were measured by comparison to the MDV-1 mRNA transcripts ICP4 and Meq.
Cloning of the 5′ leader into monocistronic and bicistronic vectors.
All vectors used in this study to assess IRES activity were based on the psiCHECK-2 backbone (Promega). To generate a monocistronic vector, we first annealed two DNA oligonucleotides (sense, 5′-CTAGCCCTTAATTAAGGGGAGAGCTC CCGACTAGTC-3′; antisense, 5′-CTAGCGACTAGTCGGGAGCTCTCCCCTTAATTAAGG-3′) and then ligated the resulting DNA into the NheI site of the psiCHECK-2 vector. The resulting monocistronic vector was named pR and contains SpeI, SacI, and PacI restriction sites upstream from the Renilla luciferase (R-Luc) ORF. The 5′ leader sequence (5L) spanning nucleotides 129186 to 129799 according to the genomic coordinates of strain MDV-1 Md5 (accession number AF243438) was cloned into the monocistronic vector pR as follows. The 5L sequence was amplified by PCR using cDNA as a template and cloned into the pR vector in both sense and antisense orientations using SacI and PacI restriction sites; the resulting plasmids were designated p5L-R and p5L(AS)-R, respectively. To construct the promoterless vector, we digested the plasmids p5L-R, pR, and p5L(AS)-R with BglII and NheI to remove the simian virus 40 (SV40) promoter. The digested plasmids were then blunt ended using T4 DNA polymerase and religated. A bicistronic vector, designated pR/MCS-F, was constructed as previously described (41). In our previously published paper (41), the pR/MCS-F construct was named psiRF. The 5L sequence was cloned into the bicistronic vector pR/MCS-F using the PmeI and SpeI restriction sites, resulting in the construct pR/5L-F. The encephalomyocarditis virus (EMCV) IRES was PCR amplified from the pIRES2-AcGFP1 vector (Clontech) and cloned into the bicistronic plasmid using the PmeI and SpeI restriction sites to generate plasmid pR/EMCV-F. All constructs were verified by sequencing.
Construction of plasmids with dual IRES sequences.
We have previously reported the construction of plasmid pR/ICR-F (41), which contains an ICR IRES from the bicistronic MDV-1 mRNA and controls the translation of the RLORF9 protein. In our previously published paper (41), the pR/ICR-F was named psiRF-ICR. In order to construct a reporter plasmid that mimics the naturally occurring bicistronic viral mRNA, we first digested plasmid pR/ICR-F with NheI, which is located upstream of the R-Luc ORF. We then cloned into this site an oligonucleotide which contained unique SacI and PacI restriction sites. Finally, we cloned the 5′ leader sequence (designated 5L) as a SacI-PacI PCR-fragment into this plasmid to generate plasmid p5L-R/ICR-F.
Promoter prediction and validation.
We used different web-based programs for promoter predictions. These included the FPROM program (Softberry, Inc., Mt. Kisco, NY) and the Neural Network Promoter Prediction program (http://www.fruitfly.org/seq_tools/promoter). The target sequence spanned nucleotides 127801 to 129799 according to the genomic coordinates of MDV-1 strain Md5, accession number AF243438. Two promoters were predicted. The first promoter was located within the region that contained the bidirectional promoter that was previously identified by Shigekane et al. (40). This promoter is designated Pro-1 and was used as positive control for this study. The Pro-1 promoter was PCR amplified using pRB-1B5 DNA (34) as the template, forward primer 5′-GGGGTACCGGAGGGAGGGTGCCATCTGTGATGCCGAGA-3′, and reverse primer 5′-CATGCCATGGCGCGAAGAGAGAAGGAACCTCGCAACCGCCGC-3′. The second promoter (Pro-2) that was predicted was located within the first intron between exon1a and exon1b (Fig. 1). The region that contained the Pro-2 promoter was PCR amplified using forward primer 5′-GGGGTACCCGCTACGCTAGGCGACGAACGAGCTGAATTTCTCCC-3′ and reverse primer 5′-CCCAAGCTTGGCGAGGCTCGTGTGAAGAACCCTAGCAAGG-3′. The resulting PCR products were cloned into the pGL3-Basic vector (Promega) for validation. Pro-1 was cloned between KpnI and NcoI sites, whereas Pro-2 was cloned between KpnI and HindIII sites. Restriction sites are underlined in the primer sequences.
Site-directed mutagenesis.
We used the QuikChange II XL site-directed mutagenesis kit from Stratagene to mutate the multiple upstream AUG codons within the 5L sequence. All 11 upstream AUGs were individually mutated to AUA using the protocol provided by the manufacturer. The mutated clones were verified by sequencing.
shRNA.
Short interfering RNA (siRNA) sequences were cloned and expressed as small hairpin RNA (shRNA) from the pU6-shRNA vector using a previously described method (26, 41). The mRNA target sequences of the siRNAs were selected using http://genomics.jp/sidirect. The siRNA sequence (5′-CGATTATGGTTATACTTTACC-3′) targeted the 5L sequence within the MDV-1 genome at nucleotides 129700 to 129721 (accession number AF243438). As positive control, we used shRNA that was designed against the EMCV IRES (5′-GTCGCTGTTTGCACATTATCA-3′) and shRNA against the R-Luc ORF (5′-GCTGGACTCCTTCATCAAC-3′). We used pU6-shEMCV as a nonsilencing shRNA control for p5L-R/ICR-F and pR/5L-F and, conversely, pU6-sh5L as a nonsilencing shRNA control for pR/EMCV-F. All constructs were verified by sequencing.
Transient-expression assays.
Cell cultures of avian and mammalian cells and transfection with DNA constructs for analysis of IRES activity were carried out as described before (41). Firefly luciferase (F-Luc) and R-Luc were measured at 24 h posttransfection using the Dual-Glo luciferase assay system (Promega) and an Anthos Lucy1 microplate luminometer (Anthos Labtec Instruments, Austria). Cotransfection experiments for shRNA knockdown were performed as described before (41). Cotransfection experiments using the hypophosphorylated version of 4E-BP1 (14) and pACTAG null constructs in combination with p5L-R/ICR-F or pR/ICR-F were performed as described before (41).
Northern blotting analysis.
Northern blotting was performed on total RNA extracted from cells transfected with biscistronic vectors pR/EMCV-F and pR/5L-F, using 1.2% agarose-formaldehyde gels followed by vacuum blotting onto nylon membrane and hybridization with a random-primed 32P-labeled DNA fragment corresponding to the 5′ end of the F-Luc ORF. Detection was performed using storage phosphor screens and a Typhoon Trio scanner (GE Healthcare).
In vitro transcription and translation.
The plasmid constructs were linearized with BamHI. Capped transcripts were synthesized by using T7 mMessage mMachine (Ambion) according to the manufacturer's instructions. Transcripts were polyadenylated using the poly(A) tailing kit from Ambion. The transcripts were translated using the Retic Lysat IVT (Ambion) as previously described (41) except that we used a low-salt/high-salt ratio of 3:2 to obtain a concentration of 75 mM potassium acetate in the translation reaction mixtures. Assays supplemented with m7GpppG were carried out as previously described (1). In vitro translations with increasing amount of capped and polyadenylated transcripts (0.4 to 4.0 μM) were carried out in 25-μl reaction mixtures at 30°C for 90 min. The F-Luc and R-Luc activities were measured from in vitro translation reactions by using the Dual-Glo luciferase assay system (Promega) according to the manufacturer's instructions.
Analysis of in vitro translation reactions.
The F-Luc and R-Luc data from the in vitro translation reactions in the presence of increasing concentrations of RNA were analyzed by using the nonlinear curve fitting method of GraphPad Prism-5, which uses the equation Y = (Ymax × Xh)/(Xh + Kh), where Y is the luciferase activity, X is the RNA concentration, K is the dissociation constant, and h is the Hill coefficient.
RESULTS
The 5′ leader of the pp14-encoding mRNA is intronic.
We have identified two viral transcripts from an MDV-transformed CD4+ T-cell line (MSB-1) derived from a chicken spleen lymphoma. Extensive screening of the MSB-1 cDNA library allowed us to identify two full-length cDNA clones that differed in their 5′ leader regions (Fig. 1B and C) and which corresponded to different pp14 protein sequences. The pp14 protein can be produced as two variants that differ in the amino acid compositions of their N termini; the common C terminus is encoded by exon 1c. One pp14 isoform has an N terminus with 13 amino acids that are encoded by exon 1a; this isoform is encoded by a bicistronic mRNA with a 613-nucleotide 5′ leader, designated 5L (Fig. 1B). The other pp14 isoform contains 20 amino acids in its N terminus encoded by exon 1b; this isoform is encoded by a bicistronic mRNA with a very short 5′ leader sequence of 14 nucleotides (Fig. 1C).
Sequence alignment mapped the 5L sequence within the second half of the intron between exons 1a and 1b in the MDV genome (Fig. 1A and B). The finding that the 5L sequence was part of the intron of another mRNA suggested that these mRNAs might arise from alternative splicing. Indeed, a donor splice site was found to be present upstream of the 5′ leader sequence (23). However, alternative promoter usage could represent a different mechanism to explain the existence of these transcript variants. We have explored this possibility and confirmed the findings of Shigekane et al. (40) with regard to a bidirectional promoter. In addition, we found a second promoter that was located downstream of exon 1a, in the region between nucleotides 128525 and 129142 (data not shown). This second promoter seems to explain the transcription of mRNAs containing the 5′ leader. We have used real-time RT-qPCR to quantify the relative expression of the two bicistronic viral mRNAs. The results from RT-qPCR indicated that in MDV-infected and transformed cells, the mRNA variant with the 5L sequence (mRNA 1b-1c) was two- to threefold more abundant than the mRNA variant lacking the 5L sequence (mRNA 1a-1c) (Fig. 1D).
The 5′ leader of the pp14-encoding mRNA contains a potential IRES.
Inspection of the 613-nucleotide 5L sequence revealed that it has features that were expected to be unfavorable for efficient cap-dependent translation. These features include multiple AUG codons (11 in total), several small upstream ORFs, and multiple predicted stem-loop structures (data not shown). However, these features, as well as a pyrimidine-rich tract that is contained in this 5′ leader, are sometimes found in IRESs (39). To investigate this possibility, we tested the ability of the 5L sequence to support translation in bicistronic reporter mRNAs.
The 5L sequence functions as an IRES in an ICR context.
To test the IRES activity of the 5L sequence in an ICR context, we used pR/MCS-F, a plasmid that encodes a bicistronic mRNA under control of the SV40 promoter with the R-Luc ORF, followed by a multicloning site and then the F-Luc ORF (Fig. 2A). The translation of the first cistron (i.e., R-Luc) is cap dependent, whereas translation of the second cistron (i.e., F-Luc) would require the presence of an IRES. We have previously shown that the 60-nucleotide sequence of the multicloning site has no IRES activity (41). The results showed that the 5L sequence mediated a 25-fold increase in relative F-Luc activity compared to the pR/MCS-F control (Fig. 2B), suggesting that the 5L sequence may function as an IRES in this context. This level was about 40% of that of the ECMV IRES (Fig. 2B). Northern blotting analysis of total RNA extracts from transfected cells showed that only the full-length bicistronic mRNAs were detected (Fig. 2C), consistent with the absence of a cryptic promoter or aberrant splicing.
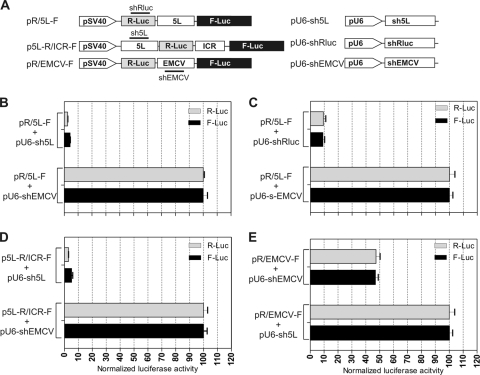
FIG. 2.
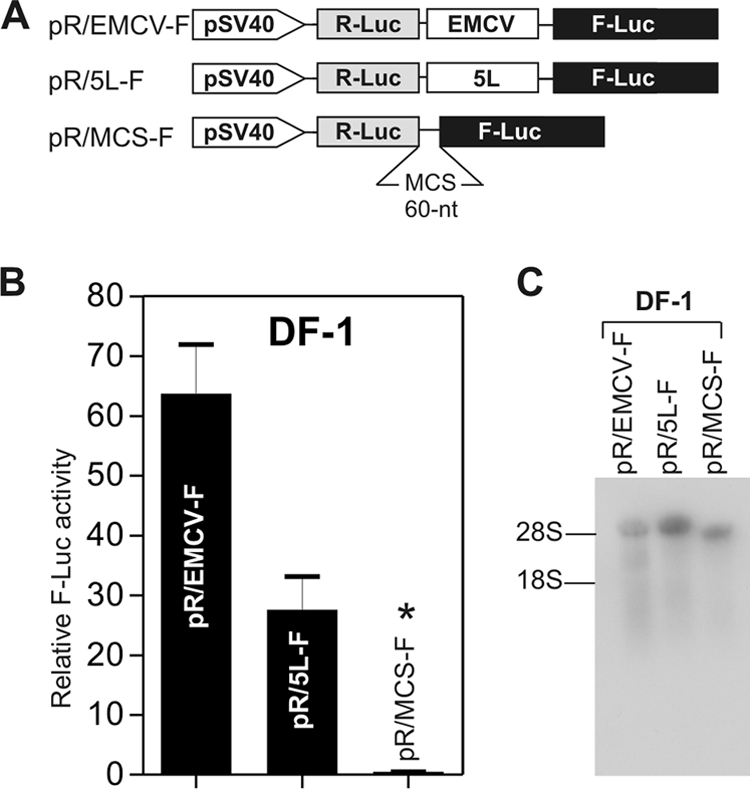
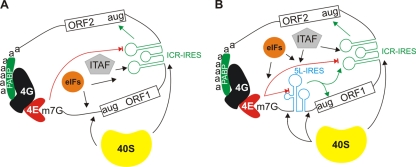
The 5′ leader sequence of the pp14 lytic protein has IRES activity. (A) Bicistronic luciferase constructs used for transfection. The 5L sequence was inserted between the R-Luc and F-Luc genes in the multicloning site (MCS) spacer of pR/MCS-F vector to give the DNA construct pR/5L-F. The EMCV IRES was cloned similarly to give the construct pR/EMCV-F and was used as a positive control. (B) Results of luciferase assay using DNA transfection of DF-1 cells. The relative F-Luc activity from each DNA construct was normalized to that from the pR/MCS-F empty vector, whose ratio was set to 1, as indicated by an asterisk above the bar graph. Error bars indicate standard errors of the means. (C) Northern blot analysis of total RNAs from DF-1 cells transfected with the DNA constructs depicted in panel A. The blots were hybridized with a 32P-labeled DNA probe corresponding to the 5′ end of the F-Luc gene. The positions of 18S and 28 rRNAs are shown to indicate the sizes of the detected RNA transcripts. The results indicate that only the full bicistronic RNAs are detected.
To determine whether these results could be explained by a cryptic promoter or splicing activity, we used an shRNA knockdown approach to determine whether both cistrons were translated from the same mRNA. We reasoned that an equivalent knockdown of both R-Luc and F-Luc activities by an appropriate shRNA would occur only if the two cistrons were translated from the same bicistronic mRNA. We tested the effect of shRNA on the 5L sequence using two different constructs. The first construct contained the 5L sequence in a configuration that would mimic the naturally occurring viral bicistronic configuration by including the MDV-1 ICR IRES in the ICR region, as illustrated by the construct p5L-R/ICR-F (Fig. 3A). The second construct contained the 5L sequence in the ICR region of the pR/MCS-F vector, which was identical to the one depicted in Fig. 2A and was named pR/5L-F. A construct that contained the EMCV IRES, pR/EMCV-F, was used as a technical validation control. We cotransfected DF-1 cells with the bicistronic vectors, along with a plasmid expressing shRNAs that targeted either the 5L IRES, the R-Luc cistron, or the EMCV IRES. The results showed an equivalent knockdown of more than 90% of both R-Luc and F-Luc activities with shRNAs that targeted either the 5L sequence (Fig. 3B) or the R-Luc gene (Fig. 3C) when the 5L IRES was located in the ICR region. Similar knockdown of both luciferase activities was achieved with the combination p5L-R/ICR-F and pU6-sh5L (Fig. 3D). This result indicated that both cistrons were part of the same bicistronic transcript generated from the constructs pR/5L-F and p5L-R/ICR-F and ruled out the possibility of cryptic splicing or cryptic promoter activity within the 5L sequence. This conclusion was validated by the results from the shRNA knockdown of the prototypic EMCV IRES (Fig. 3E).
FIG. 3.
siRNA knockdown using shRNA. (A) Constructs used for cotransfection experiments. p5L-R/ICR-F is a bicistronic DNA that mimics the naturally occurring viral transcript and in which the 5L sequence is cloned upstream from the R-Luc gene, whereas the ICR IRES is cloned upstream from the F-Luc gene. pR/5L-F and pR/EMCV-F are the same as shown in Fig. 2A. pU6-sh5L is the plasmid that expresses the shRNA against the 5L sequence. pU6-shRluc is the plasmid that expresses shRNA against R-Luc, and similarly, pU6-shEMCV is the plasmid that expresses the shRNA against EMCV IRES. (B to E) Results of cotransfection experiments with DF-1 cells using the bicistronic DNA constructs and their corresponding shRNAs. F-Luc and R-Luc activities were determined and are expressed as percentages of that of the nonsilencing shRNA control. We used pU6-shEMCV as a nonsilencing shRNA control for p5L-R/ICR-F and pR/5L-F and, conversely, pU6-sh5L as a nonsilencing shRNA control for pR/EMCV-F. Error bars indicate standard errors of the means.
The 5L sequence functions in a monocistronic context.
We next tested the 5L sequence in monocistronic mRNA. We cloned the 5L sequence in both sense and antisense orientations upstream of the R-Luc gene in the monocistronic vector pR (Fig. 4A). In this configuration the R-Luc gene with the 5L sequence was under the control of the SV40 promoter, whereas the transcription of the F-Luc gene, used as intraplasmid normalization control, was controlled by the herpes simplex virus thymidine kinase promoter. Following transfection of avian cells, normalized luciferase activity in cellular extracts was three to six times higher in the sense orientation than in the antisense orientation (Fig. 4B), which is used as a length-matched control. The reporter data also showed that the efficiency of translation initiation of R-Luc under the control of 5L (p5L-R construct) was comparable to that of R-Luc without the 5L sequence (pR construct). The luciferase data from the three constructs lacking an SV40 promoter were similar, providing additional evidence that the activity observed from the 5L sequence was not due to cryptic promoter activity (Fig. 4B). In addition, RT-PCR identified cDNA fragments that corresponded in predicted size to monocistronic RNAs that contained the 5L sequence and that were generated from the SV40 promoter (data not shown). This result suggests that the activity observed from the 5L sequence was not due to splicing activity.
FIG. 4.
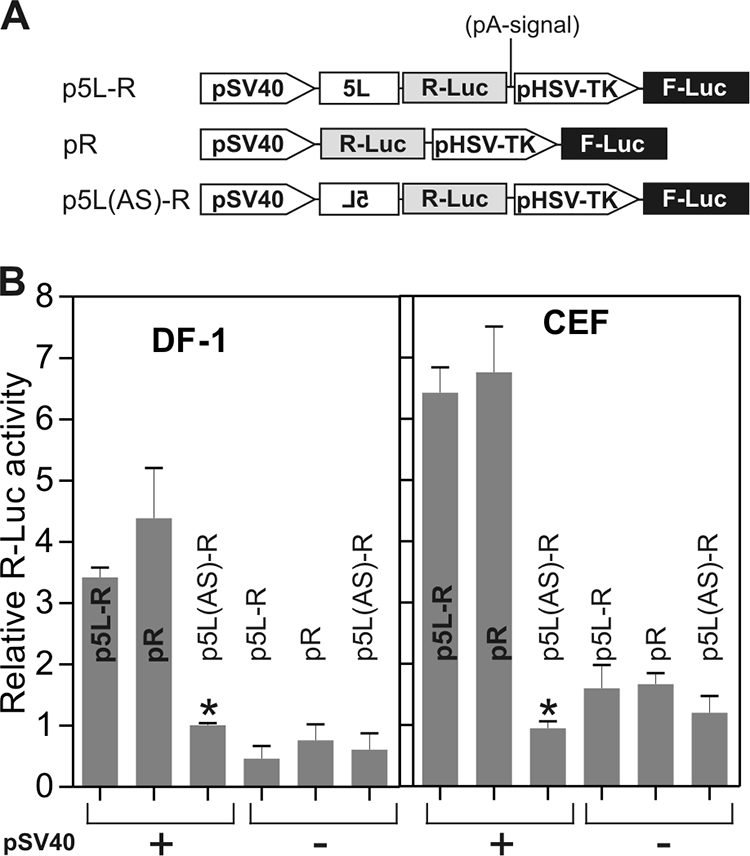
The 5′ leader sequence of the pp14 lytic protein enhances protein translation in an orientation-dependent manner and has no apparent cryptic promoter activity. (A) Monocistronic luciferase constructs used for transfection. The 5L sequence was cloned upstream from R-Luc in sense and antisense orientations to give p5L-R and p5L(AS)-R, respectively. A control vector without the 5L sequence was also used and was designated pR. Transcription of R-Luc in the three vectors is driven by the SV40 promoter. In order to control for the absence of cryptic promoter activity within the 5L sequence, we used three equivalent constructs from which the SV40 promoter was deleted. (B) Relative R-Luc activity from each construct was normalized with F-Luc activity, which is produced from a thymidine kinase promoter in the same plasmid encoding R-Luc. Each normalized R-Luc activity is presented as activity relative to that from SV40 promoter-driven 5L(AS)-R, as indicated by an asterisk above the bar graph, which is shown as 1. The means from three independent assays, each in triplicate, with standard errors of the means are shown.
The presence of multiple AUG codons within the 5L sequence prompted us to investigate their possible usage. If used during translation initiation, their mutation is expected to increase the translation of the reporter protein. In contrast, if the upstream AUG codons are not used, their mutation should not affect the translation of the reporter protein (29). Compared to the wild type, none of the mutated upstream AUG codons within the 5L sequence affected expression of the downstream reporter protein R-Luc in CEFs (data not shown). Importantly, even the mutations of those AUG codons that were in an excellent nucleotide context (24) had no significant effect on the ability of the 5L sequence to efficiently initiate translation of the reporter gene.
We concluded from these results that the 5L sequence of the mRNA encoding the MDV-1 pp14 protein contained an IRES that was capable of supporting protein translation of the downstream ORF independently of its location (whether it is located in the 5′ leader or ICR region) and that appeared to have no cryptic promoter activity.
The 5L IRES activity is enhanced when cap-dependent translation is reduced.
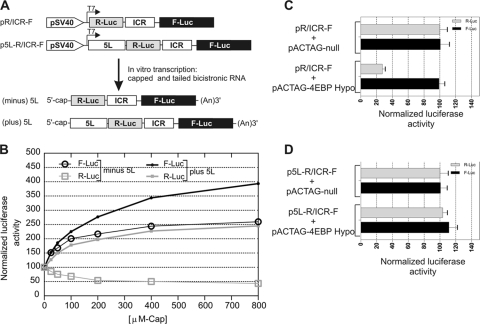
An important and well-documented property of IRESs is their ability to initiate translation in a cap-independent manner (30). This property is believed to confer an advantage in translating a subset of mRNAs under conditions where cap-dependent translation initiation is reduced, such as under virus-induced stress (16, 38). We used a reporter construct that mimicked the bicistronic viral mRNA with both the 5L IRES and the ICR IRES (Fig. 5A). The construct was used as a template to generate in vitro bicistronic mRNAs that were capped and polyadenylated (Fig. 5A). The resulting mRNAs were translated in vitro using rabbit reticulocyte lysate in the presence of increasing concentrations of the free m7GpppG cap analogue. The rationale behind this experiment was that the free cap analogue would bind to the cap binding pocket of eIF4E (1), block cap-dependent ribosome recruitment, and prevent the competitive influence of capped transcripts. Blocking cap-dependent translation should result in redirecting the translational resources to the IRES (1). The results showed that increasing the concentration of the free cap analogue in the translation reaction mixtures resulted in an increase in the IRES activity of the 5L sequence and, importantly, that there was a concomitant increase in the activity of the ICR IRES (Fig. 5B). As control, we used a construct that lacked the 5L sequence but still contained the ICR IRES (Fig. 5A). In the absence of the 5L sequence, the R-Luc gene was translated by a cap-dependent mechanism whereas the F-Luc gene was translated via the ICR IRES. In the presence of increasing concentrations of cap analogue and in the absence of the 5L sequence, the cap-dependent translation of the R-Luc decreased to about 50%; however, and as previously demonstrated (41), there was a concomitant increase in the cap-independent translation initiation of F-Luc. Importantly, we observed consistently higher relative activity of the ICR IRES with the construct containing the 5L IRES than of that without 5L. The observation that the cap-dependent translation of R-Luc in the absence of the 5L IRES was not completely abolished with the highest concentration of the free cap analogue (800 μM) was likely due to the fact that there was a level of background translation from uncapped transcripts that were produced during in vitro transcription of capped RNA, as previously reported (1, 41). The results of these studies support the notion that the 5L sequence mediates cap-independent translation initiation.
FIG. 5.
The 5′ leader sequence is capable of initiating translation when cap-dependent translation is inhibited. (A) DNA constructs used as templates to generate capped RNA transcripts in vitro that were subsequently poly(A) tailed. The bicistronic transcripts mimic the naturally occurring viral transcripts depicted in Fig. 1 with or without the 5L sequence. The ICR IRES is present in both transcripts. (B) Capped bicistronic transcripts were translated in vitro in the presence of increasing concentration of free cap analogue. F-Luc and R-Luc activities were determined and expressed as percentages of that in the control reaction in the absence of the cap analogue, whose value was set to 100%. (C and D) Results of cotransfection of mouse neuroblastoma N2a cells with the DNA constructs shown in panel A and with a construct encoding a hypophosphorylated 4E-BP gene (pACTAG-4E-BP Hypo) or null vector (pACTAG-null). The levels of F-Luc and R-Luc activities from each bicistronic construct were normalized to 100 when cotransfected with the pACTAG null vector. The F-Luc and R-Luc activities obtained in cells cotransfected with pACTAG-4E-BP-Hypo are represented as percentages of the activity obtained in cells cotransfected with control pACTAG null vector. Error bars indicate standard errors of the means.
To further test the ability of the 5L sequence to initiate cap-independent translation in vivo we cotransfected mouse neuroblastoma N2a cells with the DNA constructs depicted in Fig. 5A with either a hypophosphorylated form of 4E-BP (14) or a null vector. When expressed, the hypophosphorylated 4E-BP sequesters eIF4E, resulting in inhibition of cap-dependent translation (36). The percentage of luciferase activity remaining in the presence of the hypophosphorylated form of 4E-BP was calculated and compared to that obtained in the presence of the null vector. We used the R-Luc from the bicistronic mRNA lacking the 5L IRES as the internal control to monitor cap-dependent translation. Similarly, we used the F-Luc under the control of ICR IRES as a control for cap-independent translation (41). In the presence of hypophosphorylated 4E-BP, the R-Luc activity from the bicistronic mRNA without the 5L IRES decreased to about 30% of that from the control null vector (Fig. 5C), indicating that cap-dependent translation was being inhibited. In contrast, in the presence of the hypophosphorylated form of 4E-BP, both R-Luc and F-Luc activities from the bicistronic mRNA with the 5L IRES were not affected, and there was even a slight increase in the reporter gene expression (Fig. 5D). These results indicate that the inhibition of cap-dependent translation did not affect the activity of the 5L IRES; on the contrary, it seemed to enhance the cap-independent translation initiation mediated by both the 5L IRES and the ICR IRES.
The 5L IRES and the ICR IRES display allosteric behavior.
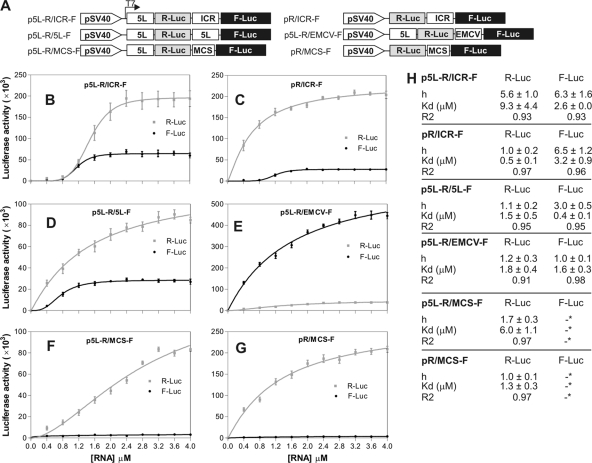
The results described above seemed to indicate that the 5L IRES had a stimulatory effect on the activity of the ICR IRES. To further investigate this finding, we produced additional constructs to assess the extent and specificity of the functional interaction between the 5L IRES and the ICR IRES from MDV-1 (Fig. 6A). These included a construct that harbored the 5L IRES both in the 5′ untranslated region (UTR) of the R-Luc and at the ICR region (i.e., 5′ UTR of the F-Luc), named p5L-R/5L-F. This construct should reveal whether a homologous interaction could occur between the same IRESs. We also included a construct that contained the EMCV IRES in the ICR region of the bicistronic construct in addition to the 5L IRES in the 5′ UTR of the R-Luc, named p5L-R/EMCV-F. This construct served as a control to assess the possibility of interactions between heterologous IRESs from two different viruses. Finally, we used a construct, named p5L-R/MCS-F, that contained only the 5L IRES in the 5′ UTR of the R-Luc without any IRES sequence in the ICR region but instead contained the multicloning site that we have previously shown to lack any detectable IRES activity (41). The pR/MCS-F construct was the control vector without any IRES. These constructs were used as DNA templates to produce capped mRNA transcripts by in vitro transcription. The resulting transcripts were then translated in vitro using rabbit reticulocyte lysate to produce dose-response curves that represented the amount of the reporter protein produced (luciferase activity) as a function of increased concentration of the mRNA substrate. This analysis is based on the assumption that there are three major binding or recruitment sites for the initiation of translation, depending on the mRNA used. For example, in the case of the p5L-R/ICR-F RNA, the 5L IRES and the ICR IRES form two recruitment sites and the third recruitment site is the cap structure. Similarly, in the case of the pR/ICR-F mRNA, there are two recruitment sites, the ICR IRES and the cap. The apparent dissociation constant is an approximate measure of the strength of the interaction between the recruitment sites and the translation complex. The apparent Hill coefficient is used as a measure of the cooperativity between the recruitment sites on the bicistronic mRNA template. With positive cooperativity, the Hill coefficient will have a value greater than 1.0. Analysis of the dose-response curve of the p5L-R/ICR-F mRNA revealed a sigmoidal shape, reflecting a substantial Hill coefficient, for both R-Luc and F-Luc (Fig. 6B and 6H), which is indicative of cooperative behavior between the 5L IRES and the ICR IRES within the bicistronic mRNA. It is unlikely that the cap structure has any contribution in this cooperativity, as we have shown before that inhibition of the cap resulted in enhanced IRES activity from both the 5L and ICR IRESs (Fig. 5B). In the absence of the 5L IRES (Fig. 6C and G), the dose-response curve of the R-Luc showed a hyperbola shape and a Hill coefficient value of 1.0, indicating the absence of cooperativity (Fig. 6H), whereas the F-Luc activity, under the control of the ICR IRES, maintained a sigmoidal shape, with the notable difference that the maximum activity was more than twofold less than that obtained in the presence of the 5L IRES (Fig. 6C). This result seems to indicate that the 5L IRES is required to induce maximum activity of the ICR IRES. Importantly, in the absence of the ICR IRES (Fig. 6D and F), the sigmoidal shape of the R-Luc activity (controlled by the 5L IRES) was mostly lost and the values of the apparent Hill coefficient were close to 1.0, indicating the loss of cooperativity. These data seem to indicate that the allosteric behavior of the 5L IRES in the 5′ UTR is dependent on the nature of the IRES in the ICR region (Fig. 6B versus D). Importantly, the optimal allosteric behavior of both IRESs was obtained in the configuration that mimicked their natural locations (Fig. 6B), which suggests that the 5L IRES and the ICR IRES from the MDV-1 immediate-early transcript have coevolved to initiate cap-independent translation, following an apparent allosteric model.
FIG. 6.
Allosteric interaction between the 5L IRES and the ICR IRES. (A) Bicistronic DNA constructs used as templates for in vitro transcription to generate capped and polyadenylated RNAs. The RNA transcripts were then used for in vitro translation using rabbit reticulocyte lysate. (B to G) Luciferase reporter results showing R-Luc and F-Luc activities as a function of increasing concentrations of RNA. The data were fitted to an allosteric sigmoidal model by the nonlinear curve fitting method of GraphPad Prism-5. The result of the fitting is represented by a solid line. (H) Summary of the results of the fitting data for each construct, including the Hill coefficient (h), the dissociation constant (Kd), and the goodness of the fit (r2). An asterisk indicates that the results are ambiguous. Error bars indicate standard errors of the means.
DISCUSSION
In this work we report the discovery of an intronic 5′ leader IRES in a major avian herpesvirus, MDV. The 5′ leader IRES is part of a the mature bicistronic transcript that is known to be the product of an immediate-early gene (18). The mature bicistronic transcript contains two ORFs, encoding the pp14 protein and a 12-kDa protein (RLORF9). In addition to the bidirectional promoter that was previously identified by Shigekane et al. (40), we have identified a second promoter sequence within the first intron that may explain the existence of bicistronic mRNA variants that encode two variants of the pp14 protein. The two variants of pp14 protein share the same C terminus, which is encoded by exon 1c (Fig. 1), while alternative promoter usage or alternative splicing can dictate the incorporation of either exon 1a or exon 1b at the N terminus. Antisera against the C terminus of the pp14 protein have revealed that the protein was expressed in cells which were lytically infected with oncogenic and attenuated MDV-1 strains as well as in latently MDV-infected and transformed cells (18). However, the antisera failed to give an indication of the relative abundance of each protein variant because they were raised against the C terminus, which is shared by both of the pp14 variants (19). Nothing is known about the biological function of the pp14 protein during the viral life cycle. However, it was found that antisense oligonucleotides against the 1.8-kb mRNA that encoded the pp14 protein could inhibit the proliferation of MD cells (23).
We have found that one mature bicistronic transcript variant had a long 5′ leader sequence that functioned as an IRES and that mapped to the intron between exon 1a and exon 1b. To our knowledge, this is the first report of a functional 5′ intronic IRES in a nuclear DNA virus. A proinsulin transcript isoform generated by the retention of an intron in the 5′ UTR was previously identified in chick embryos (27). However, this intron showed no IRES activity but rather inhibited the translation of the corresponding mRNA. In another situation, however, a retained intron containing a constitutive transport element enhanced the cytoplasmic export of mRNA in the presence of Tap and NXT proteins (22). Our preliminary search did not reveal the existence of a constitutive transport element within the 5L IRES. It would be of clear interest to see whether the Tap and NXT proteins have any effect on the IRES activity of the 5L sequence from the pp14 mRNA. It has been estimated that 10 to 18% of genes express alternative 5′ UTRs by using multiple promoters (44), and transcripts from about 12% of genes are alternatively spliced within their 5′ UTRs (31). The potential significance of these mRNA variants should become clear as our understanding of the influence that UTRs can have on gene expression increases (20). Our finding that the retained intronic 5′ leader sequence of the pp14 MDV-1 protein functions as IRES adds support to the hypothesized evolutionary link between IRESs and introns that was recently proposed by Hernandez (17).
We have previously shown that this immediate-early bicistronic mRNA contained an ICR IRES that controlled the translation of the 12-kDa protein (RLORF9) (41). To our knowledge, this is the first report of naturally occurring bicistronic capped mRNA that contains IRESs in both the 5′ leader and ICR region. The only other example where IRES elements are found in the 5′ UTR and the intergenic regions is the genomes of a number of dicistroviruses (for a review, see reference 37), with the fundamental difference that there is no cap on the single-stranded, positive-sense RNA genomes of dicistroviruses. In contrast, the MDV-1 dual-IRES bicistronic mRNA is transcribed from a double-stranded nuclear DNA virus whose transcription is mediated by RNA polymerase II in the host cell nucleus. Thus, the viral transcripts are capped and polyadenylated, which make them indistinguishable from the host mRNAs.
What are the advantages for a DNA virus such as MDV-1 to have a capped transcript with a 5′ leader that functions as an IRES? The presence of two structures that are known to function as recruitment sites for the initiation of protein translation, i.e., the cap structure and the IRES, is likely to provide the virus with the functional flexibility to use one or the other mechanism of protein translation initiation depending on the viral needs and the cellular status of the host cell translational apparatus. A possible explanation is that the 5L IRES may have dual function, with both a role as a structural element that mediates cap-independent translation and a regulatory role in preventing inhibition of initiation by the dsRNA-dependent protein kinase PKR, as it has been reported for the hepatitis C virus IRES (42). There might be, however, negative consequences of the presence of the cap structure and the IRES within the 5′ leader of the bicistronic mRNA as a result of the competitive recruitment of the translation initiation components. Indeed, we have demonstrated that when the activity of the cap structure as a recruitment site for the eIF4E was reduced, there was a concomitant enhancement of the 5L IRES activity. This finding raises the question of the impact of MDV-1 infection on cap-dependent translation of the host cell. As a cell-associated DNA virus, it must be critical for MDV-1 to maintain cap-dependent translation despite the cellular stress caused by the viral infection, but at the same time the virus needs to translate a subset of transcripts that require cap-independent and IRES-mediated translation. We are tempted to speculate that IRES-mediated translation is the preferred mode of translation for a subset of immediate-early transcripts due to the high level of the mRNA variant with the 5L IRES in cells infected with the oncogenic MDV-1 strain. Little is known about how MDV-1 controls the translation of the host cell in order to successfully move through its life cycle. The successful replication of DNA viruses requires that they gain control of key cellular signaling pathways that affect broad aspects of cellular macromolecular synthesis, metabolism, growth, and survival. The phosphatidylinositol 3′-kinase-Akt-mammalian target of rapamycin (mTOR) pathway is one such pathway. DNA viruses have evolved various mechanisms to activate this pathway to obtain the benefits of Akt activation, including the maintenance of translation through the activation of mTOR. In addition, viruses must overcome the inhibition of this pathway that results from the activation of cellular stress responses during viral infection (for an excellent review, see reference 5). The availability of oncogenic and nononcogenic strains of MDV-1 makes it an excellent model system to gain insight into the effect of MDV-1 infection on the phosphatidylinositol 3′-kinase-Akt-mTOR signaling pathway, and we are currently investigating this.
There are increasing reports on the occurrence of IRESs in DNA viruses. Most of these IRESs are ICR, such as in the case of the murine gammaherpesvirus 68 MK3 ORF (13) and the vp38 IRES from the white spot syndrome virus of marine shrimp (15). In the case of Kaposi's sarcoma-associated herpesvirus, the vFLIP IRES was found within the vCyclin ORF (2). Similarly, the VP2 ORF of the 19S late mRNA of simian virus was found to contain an IRES that controlled the translation of the VP3 ORF. All these IRESs are therefore ICR or within an ORF, with the exception of an IRES that was found within the U leader exon, located within the 5′ UTR of EBNA1 in Epstein-Barr virus (21). Our finding that an immediate-early gene from MDV-1 contains an IRES at the 5′ UTR and another IRES at the ICR region, both on the same bicistronic transcript, represents a hybrid situation. It appears to contain a prototypic viral configuration and at the same time mimics a cellular transcript with 5′ leader IRES. In terms of translational control, this naturally occurring bicistronic transcript represents a unique model system to study the functional interaction between two IRESs. This model may prove highly relevant in dissecting the mechanisms of IRES-mediated translation initiation. The first indication that the two IRESs were functionally related came from the observation that the sequestration of eIF4E by excess free cap analogue during in vitro translation of capped dual-IRES bicistronic transcripts led to a concomitant increase in the activities of both IRESs. This result gave a strong indication of functional synergism between the two IRESs and suggested that the 5L IRES and the ICR IRES have coevolved to initiate cap-independent translation in MDV-1 immediate-early transcripts. Our data on functional synergism between the two IRESs are supported by the finding that a 9-nucleotide IRES module from the Gtx 5′ UTR had enhanced IRES activity when present in multiple copies (11). We propose an allosteric interaction, not in the canonical protein/enzymology sense but as an RNA allosteric model to account for the responsiveness of the activity of the ICR IRES to the presence and the activity of the 5L IRES on the same bicistronic RNA. The molecular mechanisms behind these allosteric positive interactions are yet to be elucidated. We speculate, however, that the colocalization of two IRESs within the same bicistronic RNA may increase the local concentration of IRES-trans-activating factors (ITAFs) as well as other initiation factors that might otherwise be limiting for translation (11). It is believed that colocalization is a major driving force in the evolution of allosteric models in proteins (25). Within the closed looping model of mRNA and as a result of colocalization of the two IRESs, the close proximity of the two IRESs may also facilitate quick dynamic exchanges of ITAFs and other initiation factors, leading to the observed functional synergism as depicted in our proposed model (Fig. 7). The functional synergism between the two IRESs may also act to counterbalance the strong competition that arises from the cap structure. Our model does not exclude direct interactions between the two IRESs that could be mediated by ITAFs and that may lead to changes in IRES conformation and enhanced recruitment and thus IRES activity.
FIG. 7.
Model of the interaction between the 5L IRES and the ICR IRES within the MDV-1 bicistronic transcript. The two RNA variants can be distinguished by the absence (A) or the presence (B) of the 5L IRES upstream of the first ORF. In the absence of the 5L IRES (A), ribosome recruitment is achieved through contact between the cap binding complex (partially shown to contain 4E and 4G) the small ribosomal subunit (40S), and other eukaryotic initiation factors (eIFs), which leads to cap-dependent translation of the first ORF, whereas the second ORF is translated by cap-independent IRES-mediated recruitment of the 40S ribosomal subunit, eIFs, and other ITAFs. Under these conditions, the translation of the second ORF will be minimal, as most of the translational machinery is diverted to cap-dependent initiation. In the presence of the 5L IRES, the number of recruitment sites for translation initiation is increased to three, i.e., the cap structure and the two IRESs. We speculate that the colocalization of two IRESs within the same bicistronic RNA may increase the local concentration of ITAFs as well as other initiation factors required for efficient translation initiation by both IRESs. Within the closed looping model of mRNA this may facilitate quick dynamic exchanges of ITAFs and others initiation factors, leading to the observed functional synergism. The functional synergism between the two IRESs may also act to counterbalance the strong competition that arises from the cap structure. Our model does not exclude direct interactions between the two IRESs that could be mediated by ITAFs and that may lead to changes in IRES conformation and enhanced recruitment.
Acknowledgments
Funding for this project was provided by the Biotechnology and Biology Sciences Research Council (BBSRC) and partly by the European Animal Disease Genomics Network of Excellence (EADGENE) for animal health and food safety.
We thank L. P. Smith and L. Kgosana for excellent technical help with cell culture. We thank Nahum Sonenberg for the generous gift of the 4E-BP1 and pACTAG-2 constructs.
Footnotes
Published ahead of print on 30 September 2009.
REFERENCES
- 1.Ali, I. K., L. McKendrick, S. J. Morley, and R. J. Jackson. 2001. Activity of the hepatitis A virus IRES requires association between the cap-binding translation initiation factor (eIF4E) and eIF4G. J. Virol. 75:7854-7863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bieleski, L., and S. J. Talbot. 2001. Kaposi's sarcoma-associated herpesvirus vCyclin open reading frame contains an internal ribosome entry site. J. Virol. 75:1864-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradley, G., M. Hayashi, G. Lancz, A. Tanaka, and M. Nonoyama. 1989. Structure of the Marek's disease virus BamHI-H gene family: genes of putative importance for tumor induction. J. Virol. 63:2534-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradley, G., G. Lancz, A. Tanaka, and M. Nonoyama. 1989. Loss of Marek's disease virus tumorigenicity is associated with truncation of RNAs transcribed within BamHI-H. J. Virol. 63:4129-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchkovich, N. J., Y. Yu, C. A. Zampieri, and J. C. Alwine. 2008. The TORrid affairs of viruses: effects of mammalian DNA viruses on the PI3K-Akt-mTOR signalling pathway. Nat. Rev. Microbiol. 6:266-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buckmaster, A. E., S. D. Scott, M. J. Sanderson, M. E. Boursnell, N. L. Ross, and M. M. Binns. 1988. Gene sequence and mapping data from Marek's disease virus and herpesvirus of turkeys: implications for herpesvirus classification. J. Gen. Virol. 69:2033-2042. [DOI] [PubMed] [Google Scholar]
- 7.Bulow, V. V., and P. M. Biggs. 1975. Differentiation between strains of Marek's disease virus and turkey herpesvirus by immunofluorescence assays. Avian Pathol. 4:133-146. [DOI] [PubMed] [Google Scholar]
- 8.Bulow, V. V., and P. M. Biggs. 1975. Precipitating antigens associated with Marek's disease viruses and a herpesvirus of turkeys. Avian Pathol. 4:147-162. [DOI] [PubMed] [Google Scholar]
- 9.Buza, J. J., and S. C. Burgess. 2007. Modeling the proteome of a Marek's disease transformed cell line: a natural animal model for CD30 overexpressing lymphomas. Proteomics 7:1316-1326. [DOI] [PubMed] [Google Scholar]
- 10.Cebrian, J., C. Kaschka-Dierich, N. Berthelot, and P. Sheldrick. 1982. Inverted repeat nucleotide sequences in the genomes of Marek disease virus and the herpesvirus of the turkey. Proc. Natl. Acad. Sci. USA 79:555-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chappell, S. A., G. M. Edelman, and V. P. Mauro. 2000. A 9-nt segment of a cellular mRNA can function as an internal ribosome entry site (IRES) and when present in linked multiple copies greatly enhances IRES activity. Proc. Natl. Acad. Sci. USA 97:1536-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Churchill, A. E., and P. M. Biggs. 1967. Agent of Marek's disease in tissue culture. Nature 215:528-530. [DOI] [PubMed] [Google Scholar]
- 13.Coleman, H. M., I. Brierley, and P. G. Stevenson. 2003. An internal ribosome entry site directs translation of the murine gammaherpesvirus 68 MK3 open reading frame. J. Virol. 77:13093-13105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haghighat, A., S. Mader, A. Pause, and N. Sonenberg. 1995. Repression of cap-dependent translation by 4E-binding protein 1: competition with p220 for binding to eukaryotic initiation factor-4E. EMBO J. 14:5701-5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han, F., and X. Zhang. 2006. Internal initiation of mRNA translation in insect cell mediated by an internal ribosome entry site (IRES) from shrimp white spot syndrome virus (WSSV). Biochem. Biophys. Res. Commun. 344:893-899. [DOI] [PubMed] [Google Scholar]
- 16.Hellen, C. U., and P. Sarnow. 2001. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 15:1593-1612. [DOI] [PubMed] [Google Scholar]
- 17.Hernandez, G. 2008. Was the initiation of translation in early eukaryotes IRES-driven? Trends Biochem. Sci. 33:58-64. [DOI] [PubMed] [Google Scholar]
- 18.Hong, Y., and P. M. Coussens. 1994. Identification of an immediate-early gene in the Marek's disease virus long internal repeat region which encodes a unique 14-kilodalton polypeptide. J. Virol. 68:3593-3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong, Y., M. Frame, and P. M. Coussens. 1995. A 14-kDa immediate-early phosphoprotein is specifically expressed in cells infected with oncogenic Marek's disease virus strains and their attenuated derivatives. Virology 206:695-700. [DOI] [PubMed] [Google Scholar]
- 20.Hughes, T. A. 2006. Regulation of gene expression by alternative untranslated regions. Trends Genet. 22:119-122. [DOI] [PubMed] [Google Scholar]
- 21.Isaksson, A., M. Berggren, and A. Ricksten. 2003. Epstein-Barr virus U leader exon contains an internal ribosome entry site. Oncogene 22:572-581. [DOI] [PubMed] [Google Scholar]
- 22.Jin, L., B. W. Guzik, Y. C. Bor, D. Rekosh, and M. L. Hammarskjold. 2003. Tap and NXT promote translation of unspliced mRNA. Genes Dev. 17:3075-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawamura, M., M. Hayashi, T. Furuichi, M. Nonoyama, E. Isogai, and S. Namioka. 1991. The inhibitory effects of oligonucleotides, complementary to Marek's disease virus mRNA transcribed from the BamHI-H region, on the proliferation of transformed lymphoblastoid cells, MDCC-MSB1. J. Gen. Virol. 72:1105-1111. [DOI] [PubMed] [Google Scholar]
- 24.Kozak, M. 1986. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 44:283-292. [DOI] [PubMed] [Google Scholar]
- 25.Kuriyan, J., and D. Eisenberg. 2007. The origin of protein interactions and allostery in colocalization. Nature 450:983-990. [DOI] [PubMed] [Google Scholar]
- 26.Lambeth, L. S., Y. Zhao, L. P. Smith, L. Kgosana, and V. Nair. 2009. Targeting Marek's disease virus by RNA interference delivered from a herpesvirus vaccine. Vaccine 27:298-306. [DOI] [PubMed] [Google Scholar]
- 27.Mansilla, A., C. Lopez-Sanchez, E. J. de la Rosa, V. Garcia-Martinez, E. Martinez-Salas, F. de Pablo, and C. Hernandez-Sanchez. 2005. Developmental regulation of a proinsulin messenger RNA generated by intron retention. EMBO Rep. 6:1182-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martinez-Salas, E., R. Ramos, E. Lafuente, and S. Lopez de Quinto. 2001. Functional interactions in internal translation initiation directed by viral and cellular IRES elements. J. Gen. Virol. 82:973-984. [DOI] [PubMed] [Google Scholar]
- 29.Mauro, V. P., S. A. Chappell, and J. Dresios. 2007. Analysis of ribosomal shunting during translation initiation in eukaryotic mRNAs. Methods Enzymol. 429:323-354. [DOI] [PubMed] [Google Scholar]
- 30.Merrick, W. C. 2004. Cap-dependent and cap-independent translation in eukaryotic systems. Gene 332:1-11. [DOI] [PubMed] [Google Scholar]
- 31.Nagasaki, H., M. Arita, T. Nishizawa, M. Suwa, and O. Gotoh. 2005. Species-specific variation of alternative splicing and transcriptional initiation in six eukaryotes. Gene 364:53-62. [DOI] [PubMed] [Google Scholar]
- 32.Osterrieder, N., J. P. Kamil, D. Schumacher, B. K. Tischer, and S. Trapp. 2006. Marek's disease virus: from miasma to model. Nat. Rev. Microbiol. 4:283-294. [DOI] [PubMed] [Google Scholar]
- 33.Peng, F., G. Bradley, A. Tanaka, G. Lancz, and M. Nonoyama. 1992. Isolation and characterization of cDNAs from BamHI-H gene family RNAs associated with the tumorigenicity of Marek's disease virus. J. Virol. 66:7389-7396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petherbridge, L., A. C. Brown, S. J. Baigent, K. Howes, M. A. Sacco, N. Osterrieder, and V. K. Nair. 2004. Oncogenicity of virulent Marek's disease virus cloned as bacterial artificial chromosomes. J. Virol. 78:13376-13380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reddy, S. M., B. Lupiani, I. M. Gimeno, R. F. Silva, L. F. Lee, and R. L. Witter. 2002. Rescue of a pathogenic Marek's disease virus with overlapping cosmid DNAs: use of a pp38 mutant to validate the technology for the study of gene function. Proc. Natl. Acad. Sci. USA 99:7054-7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richter, J. D., and N. Sonenberg. 2005. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature 433:477-480. [DOI] [PubMed] [Google Scholar]
- 37.Roberts, L. O., and E. Groppelli. 2009. An atypical IRES within the 5′ UTR of a dicistrovirus genome. Virus Res. 139:157-165. [DOI] [PubMed] [Google Scholar]
- 38.Sarnow, P. 2003. Viral internal ribosome entry site elements: novel ribosome-RNA complexes and roles in viral pathogenesis. J. Virol. 77:2801-2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schneider, R. J., and I. Mohr. 2003. Translation initiation and viral tricks. Trends Biochem. Sci. 28:130-136. [DOI] [PubMed] [Google Scholar]
- 40.Shigekane, H., Y. Kawaguchi, M. Shirakata, M. Sakaguchi, and K. Hirai. 1999. The bi-directional transcriptional promoters for the latency-relating transcripts of the pp38/pp24 mRNAs and the 1.8 kb-mRNA in the long inverted repeats of Marek's disease virus serotype 1 DNA are regulated by common promoter-specific enhancers. Arch. Virol. 144:1893-1907. [DOI] [PubMed] [Google Scholar]
- 41.Tahiri-Alaoui, A., L. P. Smith, S. Baigent, L. Kgosana, L. J. Petherbridge, L. S. Lambeth, W. James, and V. Nair. 2009. Identification of an intercistronic internal ribosome entry site in a Marek's disease virus immediate-early gene. J. Virol. 83:5846-5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vyas, J., A. Elia, and M. J. Clemens. 2003. Inhibition of the protein kinase PKR by the internal ribosome entry site of hepatitis C virus genomic RNA. RNA 9:858-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu, H., Y. Yao, Y. Zhao, L. P. Smith, S. J. Baigent, and V. Nair. 2008. Analysis of the expression profiles of Marek's disease virus-encoded microRNAs by real-time quantitative PCR. J. Virol. Methods 149:201-208. [DOI] [PubMed] [Google Scholar]
- 44.Zhang, T., P. Haws, and Q. Wu. 2004. Multiple variable first exons: a mechanism for cell- and tissue-specific gene regulation. Genome Res. 14:79-89. [DOI] [PMC free article] [PubMed] [Google Scholar]