Abstract
Thirteen different glycoproteins are incorporated into mature herpes simplex virus type 1 (HSV-1) virions. Five of them play important roles during entry, while others intervene during egress of the virus. Although HSV-1 gM is not essential in cell culture, its deletion reduces viral yields and promotes syncytium formation. Furthermore, gM is conserved among herpesviruses, is essential for several of them, and can redirect the gD and gH/gL viral glycoproteins from the cell surface to the trans-Golgi network, where gM presumably modulates final capsid envelopment. Late in infection, gM reaches the nuclear envelope and decorates perinuclear virions. This process seemingly requires UL31 and UL34 and occurs when several markers of the trans-Golgi network have relocalized to the nucleus. However, the precise mechanism of gM nuclear targeting is unclear. We now report that gM is quickly and specifically targeted to nuclear membranes in a virus-dependent manner. This occurs prior to the HSV-1-induced reorganization of the trans-Golgi network and before gM enters the secretory pathway. The presence of a high-mannose glycosylation pattern on gM further corroborated these findings. While gM was targeted to the inner nuclear membrane early in infection, its partners gD, gH, gN, VP22, UL31, and UL34 did not colocalize with gM. These data suggest that nuclear gM fulfills an early nuclear function that is independent of its known interaction partners and its function in viral egress.
Herpes simplex virus type 1 (HSV-1) virions are composed of a DNA core, an icosahedral capsid, a tegument layer, and an envelope containing 13 different glycoproteins (52). These glycoproteins are involved in various aspects of the viral life cycle, including entry, egress, and acquisition of the final envelope. Hence, gB, gC, gD, and the gH/gL complex mediate attachment and fusion of the virus at the cell surface (70). Following new protein expression, these proteins, along with gM, are eventually localized in the nuclear envelope (7, 15, 24, 35, 46, 79, 84). While it has been suggested that some of the viral glycoproteins mediate the passage of the viral capsids through the nuclear membranes, others have argued that this process is independent of these proteins (24, 39, 48, 93). Late during the viral life cycle, gB, gD, gK, UL20, and the gE/gI, gH/gL, and gM/gN complexes anchor the progeny capsids to the trans-Golgi network (TGN) via partially redundant interactions with components of the tegument (29, 47, 56, 57). The presence of the viral glycoproteins at the TGN, the presumed site of final envelopment (30-33, 41, 68, 69, 85, 89-91), ensures their incorporation into mature virions and proper viral egress. The viral glycoproteins are thus important players in the biology of herpesviruses.
The UL10 gene codes for a glycoprotein of 473 amino acids referred to as gM. This N-glycosylated viral glycoprotein, which is predicted to contain eight transmembrane domains (18), is expressed as a precursor of 47 kDa, modified with high- mannose-type oligosaccharides to yield a 50-kDa molecule, and finally processed into a 53- to 63-kDa mature protein (6). Though formally classified as a late protein, its transcript can be detected as early as 2 h postinfection (p.i.) and before DNA replication takes place (72, 80). HSV-1 gM is not essential in cell culture but is conserved throughout the herpesvirus family (58), arguing for a significant role during the viral life cycle. Consistently, its deletion reduces viral titers by 10- to 100-fold in various herpesviruses, including HSV-1 (5, 21, 27, 42, 48). Interestingly, a synergistic reduction of viral yields occurs when gM is depleted in combination with UL11 or gE/gI in the related pseudorabies virus (PRV), a swine herpesvirus, but surprisingly not for HSV-1 (9, 10, 43, 48). The accumulation of unenveloped, tegumented capsids in the cytosol in these mutants indicates that gM may play a role during secondary envelopment of the virus. Furthermore, gM prevents syncytium formation mediated by gB, gD, and gH/gL (40, 44, 86) and has also been reported to have a role during PRV cell entry (21). Thus, gM seems to have multiple roles during the viral life cycle.
The HSV-1 gM glycoprotein functionally interacts with multiple viral proteins. For instance, gM redirects gD and gH/gL to the TGN in transfected cells (17). This is due to the presence of a classical endocytic signal at its carboxyl terminus, which recycles gM from the plasma membrane and promotes its accumulation at the TGN late in infection as well as in transfected cells (17). Another interacting partner is the tegument protein VP22, encoded by UL49, itself interacting with the gE/gI viral glycoproteins (26, 81). The gM proteins of several herpesviruses also associate with gN (2, 36, 44, 45, 50, 54, 55, 73, 94), but this has not yet been experimentally demonstrated for HSV-1 gM. Finally, the nuclear localization of gM late during infection is partly dependent on UL31 and UL34 (92). These various interactions most likely contribute to and modulate gM functions.
HSV-1 gM is localized at the plasma membrane, at the TGN, and on mature extracellular virions (6, 17, 52). It has also recently been reported at nuclear envelopes and on perinuclear virions (7), suggesting gM may regulate the nuclear egress of capsids, though one study suggests gM is not involved in this process (48). The concomitant retrieval to the nucleus of TGN and Golgi markers, along with gM, suggests a mechanism whereby gM might be passively relocalized to the nucleus (7) upon viral disruption of the TGN (3, 4, 13, 20, 85). However, the modulation of the nuclear localization of gM by UL31 and UL34 (92) suggests a more active mechanism. Unfortunately, the functional relevance and mechanism of targeting gM to the nucleus have remained elusive so far.
To further characterize HSV-1 gM, we probed its nuclear targeting. We found that the protein was detected on nuclear membrane invaginations and speckles early in infection at a time when other HSV-1 glycoproteins (i.e., gB, gD, and gH) were not detectable in that compartment. Experiments to evaluate the mechanism of gM targeting indicated it was not passively recycled from the TGN but rather actively and specifically targeted to the nucleus. Thus, the vast majority of gM was localized to the nucleus at 4 to 6 h p.i., at a time when gB, gD, and gH had accumulated at an intact TGN and were undetectable on nuclear membranes. Analysis of gM nuclear targeting revealed that it did not enter the secretory pathway early during infection and therefore could not be retrieved to the nucleus from the TGN, the Golgi apparatus, or the endoplasmic reticulum (ER) Golgi intermediate compartment (ERGIC). The presence of a high-mannose, immature glycosylation pattern on gM at these early infection times was consistent with its localization on nuclear membranes. Altogether, it suggests gM most likely did not make it past the ER during the early phase of the infection. This phenotype requires the expression of at least one other viral protein, as transfection of gM alone does not result in gM localization to the nuclear compartment. However, none of its known interacting partners (gD, gH, gN, VP22, UL31, or UL34) colocalized with gM at 4 h p.i. Furthermore, the previously reported interaction between gM, gD, and gH/gL (17) did not occur on nuclear membranes at early time points. These findings highlight an early, active, and specific gM nuclear targeting that relies on a novel targeting mechanism that does not depend on the reorganization of the TGN late in infection or on the gM functional partners characterized so far.
MATERIALS AND METHODS
Cells and viruses.
BHK, Vero, and 143B tk− cells were cultured in Dulbecco's modified Eagle's medium (Sigma-Aldrich) supplemented with 10% fetal bovine serum (Medicorp), 2 mM l-glutamine (Invitrogen), and antibiotics (100 U/ml penicillin and 100 μg/ml streptomycin) at 37°C in 5% CO2. For 143B tk− cells, 15 μg/ml 5-bromo-2 deoxyuridine (Sigma-Aldrich) was added to the medium, except prior to infection. Wild-type HSV-1 strain 17+ and mutant viruses (see below) were all propagated on BHK cells, and their titers were determined by plaque assay on Vero cells.
Constructs.
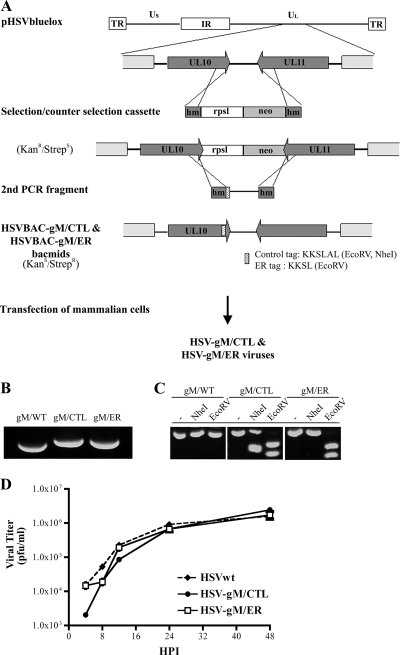
The ER retention (KKSL) and control (KKSLAL) trafficking signals were introduced into the gM of the previously described HSV-1 bacterial artificial chromosome (BAC) clone pHSV-1(17+)bluelox, which contains almost the entire genome of HSV-1 strain 17+, lacZ, and a eukaryotic Cre recombinase expression cassette but lacks the gene UL23, which codes for the viral thymidine kinase, and OriL (61). The BAC genes and the eukaryotic Cre recombinase expression cassette are flanked by LoxP sites (61). pHSV-1(17+)bluelox was maintained in the Escherichia coli strain DH10B under chloramphenicol selection. For mutagenesis, a selection and counterselection cassette was generated by PCR amplification of pRpsLneo (Gene Bridges; Germany) using the primers acgaccccgagcccgccgaggaccccgtgtacagcaccgtccgccgttggggcctggtgatgatggcgggatcg (forward primer) and ccaaaacaatgttctgttacggtcgcacgcgtgtcgtttttaaaaaacctcagaagaactcgtcaagaaggcg (reverse primer), which included sequences homologous to adjacent regions of the HSV-1 UL10 stop codon (underlined), as well as to the pRpsLneo plasmid (Fig. 1A). This RpsLneo cassette was electroporated into DH10B bacteria containing pHSV-1(17+)bluelox and pKD46, a plasmid encoding the Red recombinase (60, 97). Red expression was induced with 1% (wt/vol) l-arabinose at 37°C for 1 h, and recombinant BACs were selected for kanamycin resistance. In the second step, the RpsLneo cassette was replaced with a second PCR product that amplified the same end region of gM and that contained a KKSL or KKSLAL tag (indicated in uppercase boldface letters), as well as diagnostic restriction sites (EcoRV in both constructions and NheI for the KKSLAL construct; the restriction sites are underlined). The primers used for this second PCR were acgaccccgagcccgccgaggaccccgtgtacagcaccgtccgccgttggtacaccgatatcgatatcgaaatgaaccgtctgggtAAGAAGTCCCTGtagctgtttggttccgttttaataaac (KKSL forward primer), ccaaaacaatgttctgttacggtcgcacgcgtgtcgtttttaaaaaacc (KKSL reverse primer), acgaccccgagcccgccgaggaccccgtgtacagcaccgtccgccgttggtacaccgatatcgaaatgaaccgtctgggtAAGAAGTCGCTAGCACTAtagctgtttggttccgttttaataaac (KKSLAL forward primer), and ccaaaacaatgttctgttacggtcgcacgcgtgtcgtttttaaaaaacc (KKSLAL reverse primer). After selection for the loss of streptomycin sensitivity (i.e., removing the RpsL gene), the pHSV-1(17+)bluelox clones were analyzed for the correct insertion by PCR, restriction analysis, and sequencing. The BACs containing tagged versions of gM were named HSVBAC-gM/ER and HSVBAC-gM/CTL. BAC DNA was prepared from 500-ml overnight E. coli cultures using the NucleoBond BAC100 kit (Macherey & Nagel, Düren, Germany) and were used to transfect 143B cells (MBS mammalian transfection kit). Once significant cytopathic effects had developed, the cells were collected and freeze-thawed three times, and the resulting lysates were used to generate viral stocks. These mutant viruses were named HSV-gM/ER and HSV-gM/CTL, respectively. Note that these viruses also contained one loxP site and lacZ for the expression of beta-galactosidase.
FIG. 1.
Construction and generation of recombinant virus. (A) Schematic diagram of the selection and counterselection approach to introduce gM/KKSL (ER retention signal) or gM/KKSLAL (inactive signal) into the viral genome of HSV-1 strain 17+ present in the pHSV1(17+)bluelox bacmid. Shown at the top is the viral genome map with the unique long (Ul) and unique short (Us) regions, the internal repeats (IR), and the terminal repeats (TR) and the enlargement of the UL10 (gM) and UL11 gene regions. The mutagenesis involved the synthesis by PCR of a selection and counterselection cassette encoding a selectable (neo; light-gray box) and a counterselectable (RpsL; white box) marker flanked by HSV-1 sequences (dark-gray box). Following the insertion of this cassette into the pHSV1(17+)bluelox backbone by homologous recombination, a second PCR fragment covering the end region of the tagged gM and containing the control or ER retention tag was amplified from pHSV1(17+)bluelox and used to subsequently remove the selection cassette, once again by homologous recombination. The stippled box represents the tag sequences into which restriction sites were introduced for diagnostic purposes. hm, homologous sequences; RpsL, streptomycin sensitivity gene; neo, kanamycin resistance gene. (B and C) The recombinant BACs (HSVBAC-gM/CTL and HSVBAC-gM/ER) were finally transfected into 143B cells to reconstitute recombinant viruses. Analyses of these viruses by PCR (B) and restriction enzyme digestion (C) were performed to verify the mutagenesis (see the text for details). (D) A single-step growth kinetics assay of wild-type HSV (HSVwt), HSV-gM/CTL, and HSV-gM/ER was performed. 143B cells were infected at an MOI of 5, and the supernatant was harvested at the indicated time and concentrated by centrifugation. The yields of released infectious virus were then determined by plaque assays on Vero cells. All values are the means of three independent experiments, and the small error bars depict the standard deviations of the means.
To evaluate the quality of the mutagenesis, the engineered viruses were analyzed by various means. First, the insert was amplified by PCR, using wild-type HSV-1, HSV-gM/ER, and HSV-gM/CTL viral DNAs as templates, and analyzed on an agarose gel. As expected, the wild-type virus produced a 334-nucleotide fragment, while the two mutants ran slightly more slowly, consistent with the additional presence of the ER and control tags (Fig. 1B). Second, the above-mentioned PCR products were digested with NheI or EcoRV. The control wild-type PCR sequence did not contain any NheI and EcoRV restriction sites and therefore yielded the same 334-nucleotide fragment as when it was not digested (Fig. 1C, gM/WT). In contrast, the gM/CTL PCR fragment was cleaved once by NheI or once by EcoRV, as expected, to give two bands. Moreover, the gM/ER PCR fragment was predicted to contain an EcoRV restriction site but no NheI restriction site, as observed experimentally (Fig. 1C). Finally, sequencing of the gM region of the viruses confirmed their correct mutagenesis (data not shown). The presence of the ER and control targeting sequences had no impact on HSV-1 growth in noncomplementing cells and consequently had no effect on viral entry, replication, assembly, or egress (Fig. 1D).
For transient expression, we also constructed gM/ER and gM/CTL expression plasmids. To this end, the gM coding sequence was amplified from the above-mentioned HSVBAC-gM/ER or HSVBAC-gM/CTL template and subcloned into the pEGFP-C3 mammalian expression vector (Clontech). The resulting constructs, pEGFP-gM/ER and pEGFP-gM/CTL, were sequenced to confirm the presence of the insert and the desired tags.
Single-step growth curve.
143B tk− cells grown in six-well plates were infected at a multiplicity of infection (MOI) of 3 with wild-type HSV-1, HSV-gM/CTL, or HSV-gM/ER and overlaid with standard medium. At various times, culture supernatants were harvested, and the released virions were concentrated for 1 h at 39,000 × g, resuspended in MNT buffer (30 mM MES [morpholineethanesulfonic acid], 100 mM NaCl, and 20 mM Tris-HCl, pH 7.4), and snap-frozen in liquid nitrogen. All samples were assayed for infectious virus by titration on Vero cells.
HSV-1 infection.
143B tk− cells were grown overnight on glass coverslips. The cells were mock treated or infected with wild-type HSV-1 at an MOI of 2 to 5 for 1 h at 37°C for absorption and then grown in standard medium at 37°C for the times indicated. Two hours later, the medium was supplemented with 20 μg/ml taxol (Sigma), which stabilizes microtubules and limits the dispersal of the Golgi apparatus/TGN stacks by the infection without any effect on viral egress or host protein secretion (4, 77, 88). When indicated, the cells were then either shifted to 15°C or 20°C in standard medium containing 20 μg/ml cycloheximide (Sigma-Aldrich) to ensure a single pulse of viral assembly (14, 85). All samples were treated for immunofluorescence microscopy as described below.
Dual transfection and HSV-1 infection.
143B tk− cells grown in 24-well plates were transfected with 0.8 μg/well pEGFP-gM/ER or pEGFP-gM/CTL using Lipofectamine 2000 (Invitrogen). Twenty-four hours later, the cells were either immediately fixed and permeabilized for immunofluorescence microscopy or further infected to analyze the subcellular localization of green fluorescent protein (GFP)-labeled gM expressed from the transfected plasmid in the context of an HSV-1 infection. For the latter, the transfected cells were either infected with wild-type HSV-1 or mock treated as described above. At various times following infection, the cells were fixed and permeabilized for immunolabeling as detailed below.
Immunofluorescence microscopy.
Cells were fixed in 3% paraformaldehyde in phosphate-buffered saline (PBS) and washed with PBS, and any remaining fixative was inactivated with 50 mM NH4Cl in PBS. The cells were permeabilized using 0.1% Triton X-100 for 4 min, and nonspecific protein binding sites were blocked with 10% fetal bovine serum. The specimens were labeled for 1 h at room temperature or overnight at 4°C with primary antibodies diluted in 10% fetal bovine serum, washed, and incubated with secondary antibodies. The samples were mounted on glass slides in Mowiol containing 0.1 μg/ml Hoechst 33342 (Sigma-Aldrich) to stain the nuclei. For confocal microscopy, the nuclei were stained with Topro-3 (Invitrogen). The following antibodies were used: PAS980 polyclonal anti-gM (courtesy of Lynn Enquist), TGN46 (Serotec), Calnexin (Stressgen), G1/93 anti-ERGIC53 (from Hans-Peter Hauri), Golgin97 (Molecular Probes), p230 (BD Transduction Laboratories), monoclonal antibody (MAb) 414 anti-nuclear pore complex (NPC) proteins (Covance Research Products), ICP5 anti-VP5 (Cedarlane), ID3 anti-gD (from Gary Cohen and Rosalyn Eisenberg), LP11 anti-gH (from Helena Brown), MAb 15betaB2 anti-gB (from David C. Johnson), anti-ICP4 and anti-ICP0 (Abcam), AGV031 anti-VP22 (from Gillian Elliott), UL31 (from Joel Baines), and UL34 (two different antibodies, one from Richard Roller and the second from Susanne Bailer [M. Ott and S. Bailer, unpublished data]). All secondary antibodies (Alexa 350, 488, and 568) were from Molecular Probes. Fluorescence microscopy was performed with an Axiophot wide-field fluorescence microscope (Zeiss) equipped with filters and a Retiga 1300 camera (Q Imaging). The images were acquired and analyzed with Northern Eclipse imaging software (Empix Imaging). They were processed and assembled with Photoshop 6.0 (Adobe). Confocal microscopy was performed with a DM IRBE inverted microscope (Leica) equipped with a Leica SP1 spectrometer and argon (488-nm), argon-krypton (568-nm), and helium-neon (647-nm) lasers. The confocal sections were acquired using 100× objectives and were reconstructed and processed with LCS Lite software.
Analysis of gM glycosylation.
HeLa cells grown in suspension were infected with wild-type HSV-1 strain 17+ or mock infected for 6 to 8 h at 37°C, pelleted, washed, resuspended in RSB buffer (10 mM NaCl, 10 mM Tris-Cl, pH 8.4, 5 mM MgCl2), and broken mechanically, and the nuclei were harvested as previously described (67). For cell lysates, total-membrane preparations, and extracellular virions, 143B or HeLa cells grown on 100-mm petri dishes were infected with wild-type HSV-1 strain 17+ for 16 to 24 h at 37°C. Virions from the supernatants were harvested and concentrated by centrifugation for 1 h at 77,000 × g and then resuspended in 100 μl lysis buffer (10 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% NP-40, 1 mM EDTA, 10 mM NaF, and a cocktail of protease inhibitors). Cell lysates were prepared from the same dishes by scraping the cells in 1 ml of ice-cold PBS containing protease inhibitors and breaking them with three quick cycles of freezing and thawing. The total-membrane fractions were derived by fractionation of the above-mentioned cell lysates by centrifugation at 112,000 × g for 1 h and resuspension of the membrane pellets in 200 μl lysis buffer. These samples were denatured in denaturing buffer (0.5% sodium dodecyl sulfate [SDS], 1% mercaptoethanol) and digested for 1 h at 37°C with 500 U of endoglycosidase H (endo H) (New England Biolabs) according to the manufacturer's recommendations. The samples were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE), transferred to a polyvinylidene difluoride membrane, and probed with the PAS980 polyclonal antibody. The detection was performed with a Super Signal West Pico chemiluminescence kit (Pierce) and Kodak BioMax light film. Quantitative analysis of the data was performed with Image J version 1.42q using the analyze tool to determine the integrated densities (i.e., the area of the band multiplied by its darkness). For each glycoprotein, 100% represented the total amount of protein. The amount of the endo H-resistant form was expressed relative to that total.
Quantification of the subcellular localization of viral glycoproteins.
Where indicated, the localization of various glycoproteins labeled by immunofluorescence was quantified in cells arrested at 15°C or kept at 37°C as a control. Such cells were also labeled with Hoechst stain to detect nuclei and with antibodies directed against the ERGIC53 protein as a marker for the ERGIC. Ten random microscopy fields were scored for the number of cells exhibiting glycoproteins in either the nuclear or the ERGIC compartment (n = 34 to 147 per condition). A glycoprotein was considered nuclear only when it was not detected in any subcellular compartment except the nucleus. The data are presented as the proportion of cells relative to the total number of cells, which was set to 100%.
RT-PCR.
The relative levels of the mRNA transcripts for HSV-1 gM and gN were independently determined by reverse transcription (RT)-PCR assay. Total RNA was extracted from HSV-1- or mock-infected cells at 0, 2, 4, 7, 10, and 12 h p.i. using an SV total-RNA isolation kit (Promega). gM and gN were specifically amplified with an AccessQuick RT-PCR kit (Promega), which generates reverse-transcribed cDNA with the following primers: gagccttgtgggcacttatg (gM forward primer), gtgatctgcagcaaccaaga (gM reverse primer), taatacacacgcccatcgag (gN forward primer), and ggcctgttgtttgtcttgct (gN reverse primer).
RESULTS
HSV-1 gM is localized on nuclear membranes and intranuclear invaginations.
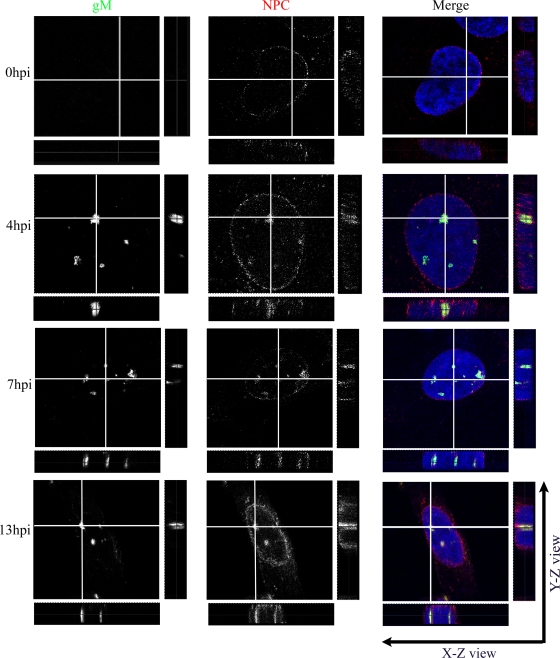
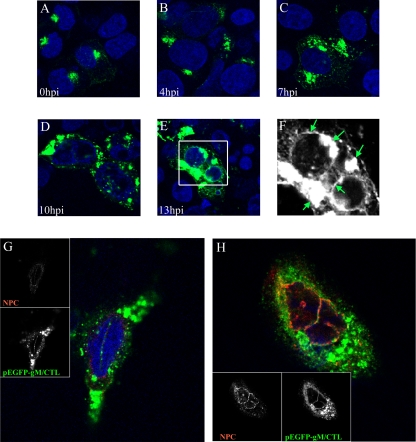
HSV-1 gM has previously been reported in nuclear membranes and on perinuclear virions late during infection (7). To determine when gM was targeted to the nucleus, we determined its subcellular localization at different times. For this purpose, 143B cells were chosen for their ability to resist quite well the cytopathic effects induced by HSV-1 (13, 85). The nuclear membranes were labeled with an antibody directed against several proteins of the NPC (19). In 143B cells infected at an MOI of 2, gM was expressed as early as 4 h p.i. and was distributed in a punctuated pattern throughout the entire nucleus (Fig. 2). At 13 h p.i., gM was also abundant on the nuclear rims. Given the numerous predicted transmembrane domains in gM (18), its localization in nuclear speckles was unexpected. The transmembrane nature of gM was confirmed, since it was found in a total-membrane fraction prepared from cells infected at 24 h p.i. (data not shown). Upon closer examination, it became evident that the speckles were often labeled with the NPC marker and hence delineated intranuclear tubules and invaginations (Fig. 2, X-Z and Y-Z). gM was thus predominantly, if not exclusively, on nuclear membranes as early as 4 h p.i. This phenomenon was not restricted to 143B cells and was also noted in the unrelated HeLa cell line (data not shown).
FIG. 2.
gM localizes primarily in punctate extensions and invaginations of nuclear membranes in infected cells. 143B cells were infected with wild-type HSV-1 strain 17+ at an MOI of 2. At the indicated time points, the cells were fixed and stained for gM (green), the NPC using MAb 414 (red), and the nucleus using Topro-3 (blue). The cells were then examined by laser scanning confocal microscopy. The individual color channels were scanned sequentially with only the fluorescence-stimulating laser powered on. Orthogonal slices in the X-Z and Y-Z planes were constructed in which the gray lines represent the cutting positions for the analysis.
gM reaches the nucleus well before the TGN is disrupted by the virus.
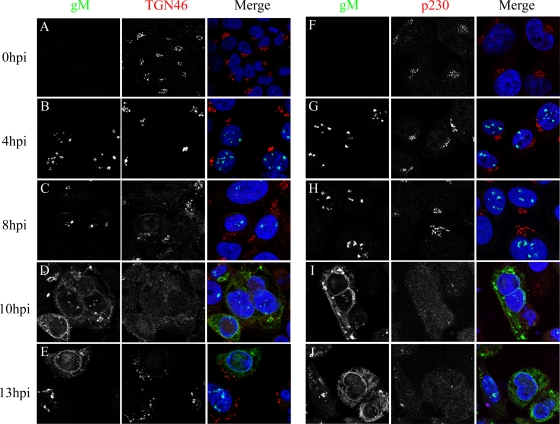
Several scenarios could be envisaged to explain the presence of gM on nuclear membranes. In the first model, gM would first be delivered to the TGN and then passively retrieved to the nucleus via a nonspecific redistribution of TGN proteins to the nucleus. This seemed a plausible idea, given that HSV-1 profoundly alters the intracellular architecture by remodeling the actin cytoskeleton, microtubules, Golgi apparatus, TGN, and mitochondria (1, 4, 13, 23, 74, 85). Furthermore, Baines and colleagues (7) showed that HSV-1 gM and various Golgi apparatus/TGN markers relocalize to nuclear membranes at 13 to 16 h p.i. (7). However, our data indicated that gM was already at the nucleus much earlier. We therefore examined the kinetics of these events. gM and two markers of the TGN (TGN46 and p230) were followed during the course of an infection. gM was again detected as early as 4 h p.i. in the nucleus, although its expression was maximal only later (Fig. 3), in agreement with a global HSV-1 microarray analysis (80). Interestingly, gM was exclusively nuclear at early time points and only started to escape from the nucleus around 9 to 10 h p.i. During the early stages of the infection, the TGN remained relatively intact and was perturbed only thereafter. Similar results were obtained upon analyzing the Golgi apparatus with the Golgin97 marker (data not shown). These experiments suggest that newly synthesized gM was actively retained at nuclear membranes and not passively redirected there upon TGN disruption.
FIG. 3.
Nuclear membrane localization of gM occurs prior to the dispersion of the TGN. 143B cells were infected with wild-type HSV-1 strain 17+ at an MOI of 2. The infected cells were fixed at various times during the infection and labeled for gM (green), TGN46 or p230 (red), and Topro-3 (blue) as indicated. The confocal images were processed as described in the legend to Fig. 2.
Contribution of the ER retrieval route to nuclear localization of gM.
Proteins synthesized in the ER and destined for that compartment occasionally escape and enter the secretory route toward the Golgi apparatus. To correct their mislocalization, the cell developed a retrieval apparatus to recycle mislocalized proteins back to the ER via retrograde transport pathways (34, 59, 62, 63). Though they achieve the same goal, two mechanisms operate in parallel. In the first, soluble ER proteins are retrieved via their carboxyl-terminal KDEL sequences (64). The second mechanism operates on integral proteins that contain a carboxyl-terminal KKXX motif. This motif interacts with COPI components that are packaged into vesicles destined for retrograde transport toward the ER (16, 49). Although such mechanisms are not known to direct proteins to the inner nuclear membrane, they do target proteins to the outer nuclear membrane, since it is continuous with the ER membrane. This is of particular interest given the presence of gM on the outer nuclear envelope (7, 92).
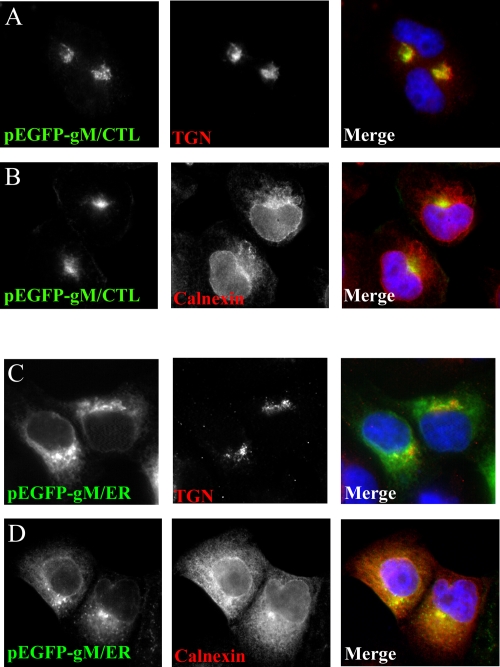
To evaluate a potential contribution of the COPI recycling pathway, gM was tagged with the ER retrieval signal KKSL. Such an approach has been used successfully with the herpesviral glycoproteins gD and gH (11, 79, 91). The ER tag was added to the very carboxyl-terminal end of GFP-gM encoded by a plasmid (pEGFP-gM/ER). As a control, an inactive form of such a tag, KKSLAL, was also generated (pEGFP-gM/CTL). Transfected gM/CTL colocalized very well with the TGN of transfected 143B cells (Fig. 4) and occasionally with the plasma membrane. This was consistent with an untagged gM construct (6, 17), indicating that the GFP tag did not perturb the normal localization of gM. In contrast, the localization of gM tagged with the KKSL signal was different. It colocalized to a significant extent with the ER and nuclear membranes, as indicated by its partial colocalization with the ER marker protein calnexin. As described for other KKSL-tagged HSV-1 glycoproteins (11, 79, 91), some gM was also found at the TGN, particularly in highly expressing cells, presumably because the ER retrieval apparatus had been overloaded. Once again, the GFP moiety did not alter the ability of the KKSL retrieval signal to function properly, since the molecules behaved exactly as for similar non-GFP-labeled constructions. The ER motif could therefore retrieve some of the transfected gM that had reached the Golgi apparatus back to the ER. Moreover, labeling with a gM-specific antibody gave the same results as the GFP signal, confirming the localization of gM (data not shown). Recycling of gM from the Golgi apparatus was therefore possible, but despite the presence of GFP-gM/ER at the nuclear periphery, its subcellular localization differed considerably from that of gM in HSV-1-infected cells (Fig. 2 and 3). Most prominently, gM expressed upon transient transfections was completely absent from nuclear speckles and invaginations (Fig. 4). Hence, these experiments were open to other interpretations. For instance, gM could be targeted to nuclear membranes by a different mechanism, or some component to transfer gM from the outer to the inner nuclear membrane might have been missing. Furthermore, viral proteins often exhibit a different subcellular localization when expressed in isolation or in the context of an infection. Another option would be that virus-expressed gM never gained access to the ER retrieval machinery in the first place.
FIG. 4.
ER tag is functional. 143B cells were transfected with enhanced GFP plasmids expressing the targeted forms of gM pEGFP-gM/CTL (A and B) or pEGFP-gM/ER (C and D). Twenty-four hours later, the cells were fixed, permeabilized, and stained in red for either TGN46 (TGN marker) or calnexin (ER marker).
gM accumulates in the nucleus upon infection with HSVBAC-gM/ER.
Given a potential discrepancy between transfection and infection and the efficacy of the KKSL targeting signal to redirect gM and other glycoproteins to the ER compartment, the KKSL or control KKSLAL sequence was introduced into the HSV-1 genome using the BAC clone pHSV-1(17+)bluelox (61). As detailed in Materials and Methods, the bacmids were as expected and, given the clonal nature of our approach, devoid of any remaining traces of wild-type gM. Tagging gM had no impact on HSV-1 growth in noncomplementing cells and consequently had no effect on viral entry, replication, assembly, or egress (Fig. 1D).
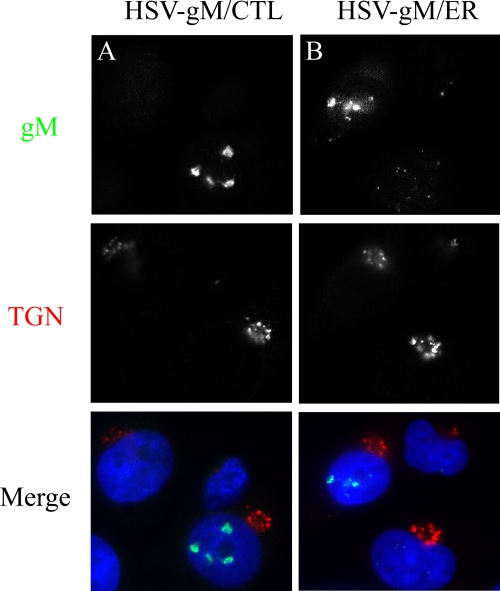
Next, we infected 143B cells with HSV-gM/ER or gM/CTL and analyzed the localization of gM using immunofluorescence microscopy. When infected at the physiological temperature of 37°C for 4 h, both gM/ER and gM/CTL proteins predominantly accumulated at the nuclear periphery and in nuclear speckles with limited colocalization with the TGN (Fig. 5). Thus, unless the ER tag did not work properly, the virus seemingly relocated gM from either the TGN (gM/CTL) or the ER (gM/ER) to the nucleus. These results were consistent with a potential retrieval of gM from the TGN and with a potential contribution of another viral factor in the targeting of gM to the nuclear membranes. However, there remained the formal possibility that gM had never reached the Golgi apparatus and in fact had never left the nucleus in the first place.
FIG. 5.
gM/CTL and gM/ER accumulate in the nucleus in infected cells. 143B cells were infected with HSV-gM/CTL (A) or HSV-gM/ER (B) for 4 h at 37°C. The cells were fixed, permeabilized, and double stained for gM (green) and TGN46 (red).
Early nuclear gM does not transit through the secretory pathway.
To probe whether gM expressed early during an infection enters the secretory pathway at all and is subsequently retrieved to the nuclear membranes from the Golgi/TGN, we inhibited the transport between the ER and the Golgi/TGN and asked whether this impedes the presence of gM at the nucleus. This approach relied on the ability of a 15°C incubation to trap all cargo at the ERGIC (38, 53, 75). Thus, if gM transits through the TGN en route to the nucleus, it should accumulate at the ERGIC at 15°C and colocalize with the ERGIC53 marker protein (78). Cells were infected with HSV-1 or mock treated, incubated for 4 h at 37°C to allow viral entry and replication, and then kept for an additional 4 h at 15°C. As a control, cells were incubated in parallel for 8 h at 37°C. Note that during this time, gM was already expressed (Fig. 2 and 3) but viral egress was still in its early stages. A MAb directed against ERGIC53 labeled a typical perinuclear compartment (Fig. 6A to F), which was particularly tight at 15°C, as reported before (38, 78, 82). In this experiment, gM remained exclusively associated with nuclear membranes, suggesting that newly synthesized gM had never gained access to the secretory pathway (Fig. 6C and D). In contrast, the HSV-1 glycoproteins gB, gD, and gH clearly colocalized with ERGIC53 after prolonged incubation at 15°C (Fig. 6E and F and data not shown). Quantification of these results revealed that gM was exclusively nuclear in 90% of the cells, while few gB (4%), gD (0%), and gH (11%) molecules were nuclear but rather localized at the ERGIC (Fig. 6G). The data strongly supported a model whereby gM was actively retained in nuclear membranes and did not enter the secretory pathway. Furthermore, this nuclear localization appeared to a specific feature of gM.
FIG. 6.
The early gM nuclear pool does not transit through the TGN. 143B cells were infected with wild-type HSV-1 strain 17+ (HSVwt) at an MOI of 2 for 4 h at 37°C, followed by an additional 4 h at 37°C or 15°C. The cells were fixed and stained against the ERGIC (red) and gM (A to D) or gB (E and F) (green). gD and gH were also analyzed, with results similar to those for gB (data not shown). (G) Quantification of the distribution of the glycoproteins in either the nucleus (NM [nuclear membrane]) or ERGIC in panels A to F was done for 10 random microscopy fields. The total cell number was between 34 and 147 for each condition and was normalized to 100%. (H) HeLa cells were mock infected or infected for 8 h with wild-type HSV-1 strain 17+. At that point, the nuclei were isolated as described in Materials and Methods and treated with endo H (+) or incubated in the absence of the enzyme (−) and loaded on an SDS-PAGE gel. The separated proteins were transferred onto a polyvinylidene difluoride membrane and probed for gM or gD by Western blotting. Note the various forms of the glycoproteins (nonglycosylated precursor, immature high-mannose form, and mature protein). (I) Quantification of endo H resistance by Image J. gM and gD on nuclei (H), in 24-h p.i. cell lysates, or in extracellular virions were resolved by SDS-PAGE, digested with endo H, probed by Western blotting, and quantified (see Materials and Methods). The error bars depict the standard deviations of the means.
Based on our results thus far, newly synthesized gM never seemed to reach the TGN but was specifically retained in the nucleus shortly after its synthesis. If this were the case, gM should mostly have an immature, high-mannose, N-linked glycosylation pattern. We therefore evaluated the sensitivity of gM to endo H, an enzyme that cleaves only high-mannose immature N-linked sugar chains, but not mature chains from glycoproteins (83). We proceeded to analyze gM from total 143B cell lysates. Unfortunately, although gM was detected by immunofluorescence as early as 4 h p.i., it was not detected by Western blotting at that time even upon loading up to 150 μg of total proteins per lane (data not shown). Given that gM is exclusively nuclear at 4 h p.i. (Fig. 2, 3, and 5), we reasoned that gM was perhaps marginal in the sea of host and viral proteins. The obvious alternative was to enrich the preparation for nuclei. Whereas the isolation of HSV-1-infected nuclei requires lengthy optimization steps, our laboratory had already reported the purification of infected nuclei from HeLa cells (67). Since HeLa cells behave the same way as 143B cells with respect to the presence of gM on nuclear membranes early during infection (data not shown), it was possible to use them for this purpose. Consequently, to specifically ask whether the nuclear pool of gM had a mature glycosylation profile, we used the previously described assay to purify nuclei from HSV-1-infected HeLa cells at 6 to 8 h p.i. (67). These relatively pure nuclei were lysed, treated with endo H, and analyzed for gM by immunoblotting. The vast majority of gM was sensitive to this enzyme (Fig. 6H), indicating that gM contained immature glycosylation chains and therefore never reached the Golgi apparatus or the TGN, where the N-linked sugar chains are further processed into the more mature forms. Quantification of the results with Image J software indicated that 91.0% ± 11.3% of gM was endo H sensitive and hence immature. This high rate of immaturity was specific to gM, as reprobing of the same blot indicated that gD was 37.6% ± 7% mature (Fig. 6I).
To show that gM did eventually mature, we performed two additional experiments. First, we isolated total cell lysates at 24 h p.i. from infected 143B and HeLa cells, digested the samples with endo H, and analyzed them as described above. As expected, a mixture of mature and immature gM molecules was detected (54.1% ± 4.5% mature) in 143B cells. Similar results were obtained for gD (50.4% ± 6.2% mature) in 143B cells or in HeLa cells (Fig. 6I). Second, we probed the maturity of the N-linked sugars of gM and gD in extracellular virions. Whereas gM was exclusively mature in extracellular virions (we did not detect any endo H-sensitive band), nearly all gD displayed mature sugars in 143B cells (92.8% ± 7%) (Fig. 6I). Once again, the data were similar in HeLa cells (Fig. 6I). Surprisingly, mature gM was also found in HSV-gM/ER virions (data not shown). Together, these data corroborated all previous results and indicated that gM was not simply targeted to nuclear membranes as a result of TGN reorganization or through its retrieval from the secretory pathway. Instead, gM was actively targeted to the nucleus shortly after its synthesis during the early stages of the infection.
gM is specifically targeted to the nucleus.
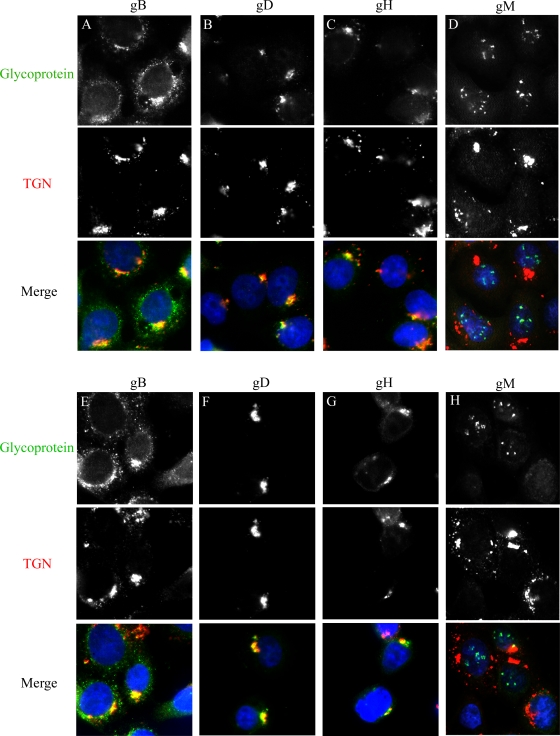
Our results suggested that the nuclear localization of gM was specific to this HSV-1 glycoprotein. This was unexpected, given that gM interacts with the HSV-1 glycoproteins gD and gH but not with gB and redirects gD and gH from the cell surface to the TGN upon cotransfection (17). To address whether gM interacted with gB, gD, or gH, their subcellular localization was analyzed in infected 143B cells. Since gB, gD, and gH are typically located at the TGN (17, 85), the infected cells were arrested at 20°C. Incubation at that temperature favors the detection of proteins even transiently residing in the TGN, since they cannot escape from that compartment (85). In cells infected with either HSVBAC-gM/ER or -gM/CTL at an MOI of 2, gM was again located on nuclear membranes (Fig. 7), while gB was in a perinuclear compartment partially overlapping with the TGN (85). In contrast, gD and gH were almost exclusively located at the TGN, with no indication of a presence in the nucleus. Thus, HSVBAC-gM/ER failed to target the other HSV-1 glycoproteins to the nuclear membranes and gM seemed to interact with these glycoproteins only later during infection and/or somewhere downstream in the secretory pathway. Alternatively, this interaction may only occur after their transient coexpression upon transfection. Most importantly, our data suggested that the early nuclear targeting of gM is highly specific.
FIG. 7.
gM/ER does not alter the targeting of other viral glycoproteins. 143B cells were infected with HSV-gM/CTL (A to D) or HSV-gM/ER (E to H) at an MOI of 2 for 7 h at 37°C and then for 6 h at 20°C. The 20°C block, which specifically stops newly made proteins in the TGN, has been shown to arrest several HSV-1 envelope glycoproteins in the TGN. The cells were fixed, permeabilized, and double stained with antibodies against viral glycoprotein gB, gD, gH, or gM (green) and TGN46 (red).
Nuclear targeting of gM depends on other viral proteins.
Upon transfection, gM/CTL was localized to the TGN and the cell surface (Fig. 4), in agreement with the results of others (17). In contrast, virus-expressed gM was predominantly located at nuclear membranes early during the HSV-1 infection (Fig. 2, 3, 5, and 6). Furthermore, the subcellular distribution of transfected gM/ER differed significantly from gM expressed during an infection (Fig. 4). Thus, gM localization to nuclear speckles and invaginations may require other viral proteins. To test this directly, cells were transfected with pEGFP-gM/CTL, subsequently infected with wild-type HSV-1, and examined at various times. Transfected gM was indeed relocated from the TGN to the nuclear membranes during the course of infection (Fig. 8A through F), albeit less efficiently than virus-expressed gM (Fig. 2 and 3). The nuclear localization of gM was particularly evident upon three-dimensional reconstruction of the data generated by confocal microscopy (Fig. 8G and H). Note that some gM was found at the TGN, but this was expected in the experimental design used, since the cells were transfected for 24 h prior to their infection with wild-type HSV-1 to allow the expression of the transgene. This allowed many gM molecules to reach the TGN before inoculation of the cells with the virus. Importantly, we never saw gM nuclear speckles in cells transfected with gM but not infected. Thus, HSV-1 actively redirected both transfected and virus-expressed gM to nuclear membranes in a process that required at least one other viral protein.
FIG. 8.
HSV-1 redistributes transfected gM to the nucleus. (A to F) 143B cells were transfected with the pEGFP-gM/CTL plasmid. Twenty-four hours posttransfection, the cells were infected with wild-type HSV-1 strain 17+ at an MOI of 2. Shown are confocal images of cells expressing enhanced GFP (EGFP)-tagged gM/CTL at the indicated times after infection. For the last time point (13 h p.i.), a zoom view (F) of the boxed area in panel E is provided to better show the presence of gM on nuclear membranes and the TGN (highlighted by arrows). (G and H) Enlarged images of cells expressing EGFP-gM/CTL at 13 h p.i., which were also probed for the nuclear membrane marker NPC (red) and with the nuclear DNA dye Topro-3 (blue). Confocal microscopy and image processing were performed as described in the legend to Fig. 2.
Early nuclear gM does not colocalize with its partners.
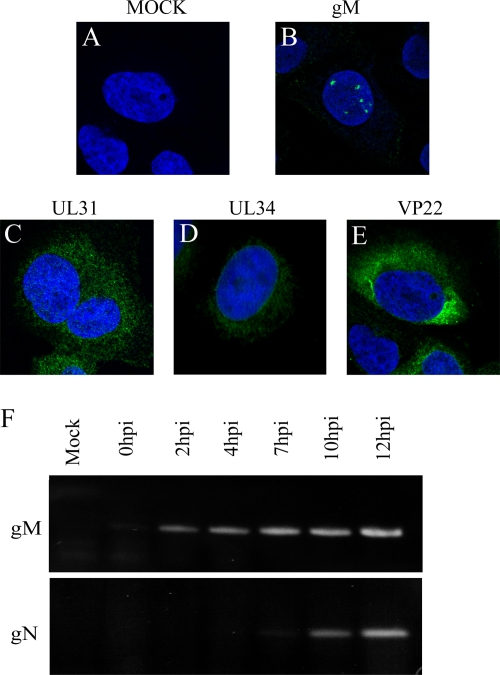
gM nuclear localization may be mediated via any one of its binding partners. Wills and colleagues recently reported the participation of UL31 and UL34 in this process late in infection (92). Unfortunately, it was not known whether this also is the case at earlier time points. We therefore sought to verify this. Cells were consequently mock treated or infected with wild-type virus, and UL31 and UL34 localization was monitored at 4 h p.i. The data showed that neither protein colocalized with gM at 4 h p.i. (Fig. 9). We therefore asked whether other gM partners might contribute to the nuclear targeting of gM. VP22 (Fig. 9), which directly binds to gM during a PRV infection (26, 81), or gD and gH (Fig. 7), which both functionally interact with gM, were not detected in the nucleus by 4 h p.i.
FIG. 9.
gM partners are not involved in gM nuclear localization. (A to E) 143B cells were infected with wild-type HSV-1 strain 17+ at an MOI of 2 for 4 h at 37°C and then fixed and stained against the viral proteins gM, VP22, UL31, and UL34 (green) and with the nuclear DNA dye Topro-3 (blue). Confocal microscopy and image processing were performed as described in the legend to Fig. 2. (F) Total RNA was extracted from HSV-1-infected cells and reverse transcribed, and gM and gN were PCR amplified as described in Materials and Methods. The upper blot shows gM RNA expression from the control mock-infected 143B cells or cells infected for 0, 2, 4, 7, 10, or 12 h. The lower blot shows the same conditions for gN.
Although not yet proven for HSV-1, gM from many herpesviruses forms a complex with the integral protein gN, encoded by the UL49.5 or UL49A gene (8, 37, 94). Interestingly, proper transport and processing of the gM/gN complex often depends on their mutual interaction (28, 44, 51, 55, 73). Unfortunately, it was not possible to determine the subcellular localization of gN due to the lack of antibodies, but we monitored gN expression by RT-PCR (Fig. 9F). gN was expressed starting at 7 to 10 h p.i. and could not be detected at 4 h p.i., a time when gM was already in nuclear membranes. gN thus could not be responsible for the nuclear retention of gM. In conclusion, none of the described gM interaction partners could explain the early nuclear targeting of gM.
DISCUSSION
We initiated this study to elucidate the specificity and the mechanism of HSV-1 gM targeting to nuclear membranes. Our time course experiments indicated that gM was rapidly targeted to the nucleus early during HSV-1 infection in at least two different cell lines (Fig. 2 and 3). This is in line with the results of Baines and colleagues, who focused on late events from 13 to 16 h p.i. and convincingly demonstrated that gM is localized at both nuclear membranes and on perinuclear virions (7). While that initial study suggested that the nuclear localization of gM may be due to a bulk retrieval of the TGN/Golgi components to the nucleus at those times (7), a subsequent study by the same group indicated that UL31 and UL34 may actively participate in this process (92).
The present study adds an additional layer of complexity, as gM was targeted to nuclear membranes well before the TGN was disrupted (Fig. 3). In contrast, gB, gD, and gH, as well as two classical TGN markers and one Golgi marker, were all absent from the nucleus during the first 8 to 10 h p.i., while gM was already detected in the nucleus as early as 4 h p.i. (Fig. 3), in agreement with published microarray data (80). Furthermore, gM displayed immature sugars and never transited through the Golgi apparatus and the TGN at early time points but acquired mature sugars later on (Fig. 6). HSV-1 gM is thus actively and specifically recruited to nuclear membranes early in the viral life cycle and never enters the secretory pathway. Consequently, the early gM nuclear localization cannot be explained on the basis of the HSV-1-induced TGN reorganization.
The absence of a prominent phenotype in HSV-gM/ER may at first seem unexpected (Fig. 1D). However, it may merely reflect the nonessential status of gM during an HSV-1 infection (5, 21) and/or reiterate the redundant nature of herpesvirus glycoproteins to ensure the propagation of the virus. Another explanation may be the sheer abundance of viral proteins, in comparison to host proteins, which could therefore overwhelm the ER retrieval machinery. However, in view of the nuclear retention of gM, the results made sense, since the KKSL retention signal can only function if the tagged protein escapes the ER and reaches the Golgi apparatus/TGN. As this occurred only late during infection, the ER tag could not operate until then and would not play any role early during infection. Altogether, the data confirmed that gM never transited through the ERGIC, the Golgi apparatus, or the TGN and therefore never encountered the ER retrieval machinery early during infection.
One surprising result was the incorporation of gM/ER into extracellular virions, since the protein should have been retained in the ER once it left the nucleus. This interpretation is consistent with reports regarding ER-tagged gD or gH (11, 79, 91). At this point, we cannot explain this discrepancy. It may be that enough gM/ER leaks out and makes it to the secondary site of reenvelopment later in the infection. This is supported by our findings that overexpressed gM/ER ends up in the TGN (Fig. 4), just like overexpressed gD/ER and gH/ER do (11, 79, 91). Though we sequenced the virions, found the anticipated ER retention signal, and used only low-passage viral stocks for our experiments, we cannot rule out the possibility that the ER tag did not work properly in the context of the infection. However, this caveat does not alter the overall conclusion that gM is found at the nuclear membranes early on.
The early presence of gM on nuclear membranes and its involvement in gD and gH recycling from the cell surface to the TGN later on (17) provided an experimental window to evaluate when such interactions occurred. Since gD and gH were absent from the nucleus at early time points, the results suggest that the interactions between them and gM occur later and/or somewhere downstream of the ER and nuclear membranes, once gM would have acquired a mature glycosylation (Fig. 6 and 7). Alternatively, these interactions may be measurable only under cotransfection conditions. Either way, the results underscored the unique property of gM to localize to nuclear membranes early in infection.
The targeting of integral proteins to nuclear membranes is the subject of intense scrutiny. To our knowledge, no specific trafficking signal has yet been indentified in these proteins. Furthermore, gM does not contain any of the characterized nuclear localization signals (PredictNLS software; data not shown). However, this was not surprising, as nuclear localization signal motifs are normally used to target soluble proteins into the nucleus and may not be relevant for gM. Clearly, gM was directed to nuclear invaginations and speckles only in the presence of other viral proteins (Fig. 8). The targeting of gM to nuclear membranes may depend on its folding status, posttranslational modifications, and/or interactions with other proteins. Possible viral candidates for gM nuclear retention included gD, gH, VP22, UL31, and UL34. However, none of them colocalized with gM in the nucleus at 4 h p.i. (Fig. 7 and 9). These observations are consistent with published studies, which have reported on the largely cytoplasmic localization of VP22 and UL34 at early time points (22, 66, 71, 95, 96) or on the appearance of UL31 at the nuclear membranes only from 8 h p.i. onward (71). Note that at 12 h p.i., both UL31 and UL34 stainings gave a characteristic nuclear ring pattern (data not shown). Furthermore, the UL31/UL34 complex does not physically interact with gM in coimmunoprecipitation assays (92). Baines and colleagues also reported few effects of the HSV-1 US3 kinase, which modulates UL31/UL34 localization, on the nuclear localization of gM (7). However, the UL31/UL34 complex seems to be involved late in infection, since at that time it does affect gM nuclear recruitment, as well as its relative distribution between the inner and outer nuclear membranes (92). Another potential gM-interacting partner, gN, was not detected at early times of infection when gM was retained at the nucleus (Fig. 9). Since gN seems to favor gM processing and transport, at least in some herpesviruses (28, 44, 51, 55, 73), it may facilitate gM exit from the nucleus later on, as the onset of gN transcription coincides with the nuclear egress of gM (Fig. 9). However, HSV-1 does not require gN to reach the TGN in transfected cells (17). In conclusion, none of the known interaction partners of gM seemed to be involved in its nuclear presence early in infection. Clearly, further experiments are needed to define how gM is targeted to nuclear membranes.
One stimulating issue is the potential role of HSV-1 gM at the nuclear membranes. Although work by Leege and colleagues suggests that deletion of gM does not impact the nuclear egress of both PRV and HSV-1 capsids (48), a compensatory mechanism cannot be ruled out in light of the built-in functional redundancy commonly observed in herpesviruses. In addition, the presence of gM on nuclear membranes is unlikely to be due merely to chance. This is substantiated by the numerous interactions between the related varicella-zoster virus and Kaposi's sarcoma-associated herpesvirus gM and viral proteins found in the nucleus, including ICP0, ICP4, the packaging UL33 protein, and the UL30 polymerase, as well as the UL31 and UL34 molecules (87, 92). Should gM be involved in HSV-1 nuclear capsid egress, various hypothetical scenarios may be considered. One possibility is that gM marks some kind of nuclear exit site where gM would assume a scaffolding role. However, this would be surprising, as nuclear gM does not overlap with the major capsid protein at these early times (data not shown). Second, gM may induce or maintain nuclear invaginations in an effort to reduce the overall distance between any intranuclear assembly sites and the nuclear periphery. Interestingly, this possibility has previously been proposed for at least one cytomegalovirus and another herpesvirus (12, 65). In addition, the biological significance of the HSV-1-stimulated nuclear invaginations that were seen in this study is highlighted by the fact they are readily detectable in the unrelated 143B, HeLa, and NRK cells (this study and data not shown). Furthermore, they are not unique to infected cells but have also been reported in normal uninfected cells (25, 76). Third, since gM modulates the syncytial activities of gB, gD, and gH (40, 44, 86), which are ultimately also found on nuclear membranes (24, 35, 79, 84), gM may prevent uncontrolled fusion between the inner and the outer nuclear membranes. The recruitment of gM to the nucleus prior to the other glycoproteins is consistent with this idea. Alternatively, gM may fine tune and positively modulate the fusion of HSV-1 perinuclear virions with the outer nuclear membrane. Of course, gM could also have other unsuspected functions on nuclear membranes.
Based on the present and past findings, a scenario emerges in which gM is targeted to nuclear membranes very early after its synthesis. This nuclear retention is achieved by one or several viral proteins, but not by gB, gD, gH, gN, VP22, UL31, or UL34 at that moment. At the nucleus, gM mediates some aspect of the viral life cycle and later reaches the TGN, a process possibly facilitated by gN. At the TGN, gM participates in secondary and final capsid envelopment and is incorporated into mature virions. Late in the infection, gM may be passively redistributed to the nuclear envelopes as a consequence of TGN rearrangement by the virus.
Acknowledgments
We are indebted to numerous people for reagents, including Hans-Peter Hauri (ERGIC-53 antibody), David Johnson (gB antibody), Gary Cohen and Rosalyn Eisenberg (gD antibody), Helena Brown (gH antibody), Lynn Enquist (gM antibody), Gillian Elliott (VP22 antibody), Joel Baines (UL31 antibody), Richard Roller and Susanne Bailer (UL34 antibodies), and Pascal Raymond and Gaudeline Rémillard-Labrosse (nucleus preparations). We also thank Sandra Loret, Camille Stegen, Yordanka Grudeva, and Gaudeline Rémillard-Labrosse for critical reading of the manuscript.
This work was funded by grants from the Canadian Institutes for Health Research (to R.L.; MOP 49547 and MOP 82921), the Canadian Foundation for Innovation (to R.L.; CFI 6908), and the German Research Council (to B.S.; DFG So403/2 and So403/3).
Footnotes
Published ahead of print on 7 October 2009.
REFERENCES
- 1.Ackermann, W. W., and H. Kurtz. 1952. The relation of herpes virus to host cell mitochondria. J. Exp. Med. 96:151-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams, R., C. Cunningham, M. D. Davison, C. A. MacLean, and A. J. Davison. 1998. Characterization of the protein encoded by gene UL49A of herpes simplex virus type 1. J. Gen. Virol. 79:813-823. [DOI] [PubMed] [Google Scholar]
- 3.Alconada, A., U. Bauer, B. Sodeik, and B. Hoflack. 1999. Intracellular traffic of herpes simplex virus glycoprotein gE: characterization of the sorting signals required for its trans-Golgi network localization. J. Virol. 73:377-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avitabile, E., S. Di Gaeta, M. R. Torrisi, P. L. Ward, B. Roizman, and G. Campadelli-Fiume. 1995. Redistribution of microtubules and Golgi apparatus in herpes simplex virus-infected cells and their role in viral exocytosis. J. Virol. 69:7472-7482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baines, J. D., and B. Roizman. 1991. The open reading frames UL3, UL4, UL10, and UL16 are dispensable for the replication of herpes simplex virus 1 in cell culture. J. Virol. 65:938-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baines, J. D., and B. Roizman. 1993. The UL10 gene of herpes simplex virus 1 encodes a novel viral glycoprotein, gM, which is present in the virion and in the plasma membrane of infected cells. J. Virol. 67:1441-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baines, J. D., E. Wills, R. J. Jacob, J. Pennington, and B. Roizman. 2007. Glycoprotein M of herpes simplex virus 1 is incorporated into virions during budding at the inner nuclear membrane. J. Virol. 81:800-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barnett, B. C., A. Dolan, E. A. Telford, A. J. Davison, and D. J. McGeoch. 1992. A novel herpes simplex virus gene (UL49A) encodes a putative membrane protein with counterparts in other herpesviruses. J. Gen. Virol. 73:2167-2171. [DOI] [PubMed] [Google Scholar]
- 9.Brack, A. R., J. M. Dijkstra, H. Granzow, B. G. Klupp, and T. C. Mettenleiter. 1999. Inhibition of virion maturation by simultaneous deletion of glycoproteins E, I, and M of pseudorabies virus. J. Virol. 73:5364-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Browne, H., S. Bell, and T. Minson. 2004. Analysis of the requirement for glycoprotein M in herpes simplex virus type 1 morphogenesis. J. Virol. 78:1039-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Browne, H., S. Bell, T. Minson, and D. W. Wilson. 1996. An endoplasmic reticulum-retained herpes simplex virus glycoprotein H is absent from secreted virions: evidence for reenvelopment during egress. J. Virol. 70:4311-4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buser, C., P. Walther, T. Mertens, and D. Michel. 2007. Cytomegalovirus primary envelopment occurs at large infoldings of the inner nuclear membrane. J. Virol. 81:3042-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campadelli-Fiume, G., R. Brandimarti, C. Di Lazzaro, P. L. Ward, B. Roizman, and M. R. Torrisi. 1993. Fragmentation and dispersal of Golgi proteins and redistribution of glycoproteins and glycolipids processed through the Golgi apparatus after infection with herpes simplex virus 1. Proc. Natl. Acad. Sci. USA 90:2798-2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Church, G. A., and D. W. Wilson. 1997. Study of herpes simplex virus maturation during a synchronous wave of assembly. J. Virol. 71:3603-3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Compton, T., and R. J. Courtney. 1984. Virus-specific glycoproteins associated with the nuclear fraction of herpes simplex virus type 1-infected cells. J. Virol. 49:594-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cosson, P., and F. Letourneur. 1994. Coatomer interaction with di-lysine endoplasmic reticulum retention motifs. Science 263:1629-1631. [DOI] [PubMed] [Google Scholar]
- 17.Crump, C. M., B. Bruun, S. Bell, L. E. Pomeranz, T. Minson, and H. M. Browne. 2004. Alphaherpesvirus glycoprotein M causes the relocalization of plasma membrane proteins. J. Gen. Virol. 85:3517-3527. [DOI] [PubMed] [Google Scholar]
- 18.Crump, C. M., C. H. Hung, L. Thomas, L. Wan, and G. Thomas. 2003. Role of PACS-1 in trafficking of human cytomegalovirus glycoprotein B and virus production. J. Virol. 77:11105-11113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davis, L. I., and G. Blobel. 1987. Nuclear pore complex contains a family of glycoproteins that includes p62: glycosylation through a previously unidentified cellular pathway. Proc. Natl. Acad. Sci. USA 84:7552-7556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Desai, P., G. L. Sexton, E. Huang, and S. Person. 2008. Localization of herpes simplex virus type 1 UL37 in the Golgi complex requires UL36 but not capsid structures. J. Virol. 82:11354-11361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dijkstra, J. M., N. Visser, T. C. Mettenleiter, and B. G. Klupp. 1996. Identification and characterization of pseudorabies virus glycoprotein gM as a nonessential virion component. J. Virol. 70:5684-5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elliott, G., and P. O'Hare. 1999. Live-cell analysis of a green fluorescent protein-tagged herpes simplex virus infection. J. Virol. 73:4110-4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Falke, D. 1997. Herpes simplex virus and the cytoskeleton. Trends Microbiol. 5:306. [DOI] [PubMed] [Google Scholar]
- 24.Farnsworth, A., T. W. Wisner, M. Webb, R. Roller, G. Cohen, R. Eisenberg, and D. C. Johnson. 2007. Herpes simplex virus glycoproteins gB and gH function in fusion between the virion envelope and the outer nuclear membrane. Proc. Natl. Acad. Sci. USA 104:10187-10192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fricker, M., M. Hollinshead, N. White, and D. Vaux. 1997. Interphase nuclei of many mammalian cell types contain deep, dynamic, tubular membrane-bound invaginations of the nuclear envelope. J. Cell Biol. 136:531-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fuchs, W., B. G. Klupp, H. Granzow, C. Hengartner, A. Brack, A. Mundt, L. W. Enquist, and T. C. Mettenleiter. 2002. Physical interaction between envelope glycoproteins E and M of pseudorabies virus and the major tegument protein UL49. J. Virol. 76:8208-8217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fuchs, W., and T. C. Mettenleiter. 1999. DNA sequence of the UL6 to UL20 genes of infectious laryngotracheitis virus and characterization of the UL10 gene product as a nonglycosylated and nonessential virion protein. J. Gen. Virol. 80:2173-2182. [DOI] [PubMed] [Google Scholar]
- 28.Fuchs, W., and T. C. Mettenleiter. 2005. The nonessential UL49.5 gene of infectious laryngotracheitis virus encodes an O-glycosylated protein which forms a complex with the non-glycosylated UL10 gene product. Virus Res. 112:108-114. [DOI] [PubMed] [Google Scholar]
- 29.Fulmer, P. A., J. M. Melancon, J. D. Baines, and K. G. Kousoulas. 2007. UL20 protein functions precede and are required for the UL11 functions of herpes simplex virus type 1 cytoplasmic virion envelopment. J. Virol. 81:3097-3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gershon, A. A., D. L. Sherman, Z. Zhu, C. A. Gabel, R. T. Ambron, and M. D. Gershon. 1994. Intracellular transport of newly synthesized varicella-zoster virus: final envelopment in the trans-Golgi network. J. Virol. 68:6372-6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Granzow, H., C. Birghan, T. C. Mettenleiter, J. Beyer, B. Kollner, and E. Mundt. 1997. A second form of infectious bursal disease virus-associated tubule contains VP4. J. Virol. 71:8879-8885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Granzow, H., B. G. Klupp, W. Fuchs, J. Veits, N. Osterrieder, and T. C. Mettenleiter. 2001. Egress of alphaherpesviruses: comparative ultrastructural study. J. Virol. 75:3675-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harley, C. A., A. Dasgupta, and D. W. Wilson. 2001. Characterization of herpes simplex virus-containing organelles by subcellular fractionation: role for organelle acidification in assembly of infectious particles. J. Virol. 75:1236-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jackson, M. R., T. Nilsson, and P. A. Peterson. 1990. Identification of a consensus motif for retention of transmembrane proteins in the endoplasmic reticulum. EMBO J. 9:3153-3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jensen, H. L., and B. Norrild. 1998. Herpes simplex virus type 1-infected human embryonic lung cells studied by optimized immunogold cryosection electron microscopy. J. Histochem. Cytochem. 46:487-496. [DOI] [PubMed] [Google Scholar]
- 36.Jons, A., J. M. Dijkstra, and T. C. Mettenleiter. 1998. Glycoproteins M and N of pseudorabies virus form a disulfide-linked complex. J. Virol. 72:550-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jons, A., H. Granzow, R. Kuchling, and T. C. Mettenleiter. 1996. The UL49.5 gene of pseudorabies virus codes for an O-glycosylated structural protein of the viral envelope. J. Virol. 70:1237-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klumperman, J., A. Schweizer, H. Clausen, B. L. Tang, W. Hong, V. Oorschot, and H. P. Hauri. 1998. The recycling pathway of protein ERGIC-53 and dynamics of the ER-Golgi intermediate compartment. J. Cell Sci. 111:3411-3425. [DOI] [PubMed] [Google Scholar]
- 39.Klupp, B., J. Altenschmidt, H. Granzow, W. Fuchs, and T. C. Mettenleiter. 2008. Glycoproteins required for entry are not necessary for egress of pseudorabies virus. J. Virol. 82:6299-6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klupp, B. G., R. Nixdorf, and T. C. Mettenleiter. 2000. Pseudorabies virus glycoprotein M inhibits membrane fusion. J. Virol. 74:6760-6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Komuro, M., M. Tajima, and K. Kato. 1989. Transformation of Golgi membrane into the envelope of herpes simplex virus in rat anterior pituitary cells. Eur. J. Cell Biol. 50:398-406. [PubMed] [Google Scholar]
- 42.Konig, P., K. Giesow, and G. M. Keil. 2002. Glycoprotein M of bovine herpesvirus 1 (BHV-1) is nonessential for replication in cell culture and is involved in inhibition of bovine respiratory syncytial virus F protein induced syncytium formation in recombinant BHV-1 infected cells. Vet. Microbiol. 86:37-49. [DOI] [PubMed] [Google Scholar]
- 43.Kopp, M., H. Granzow, W. Fuchs, B. Klupp, and T. C. Mettenleiter. 2004. Simultaneous deletion of pseudorabies virus tegument protein UL11 and glycoprotein M severely impairs secondary envelopment. J. Virol. 78:3024-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koyano, S., E. C. Mar, F. R. Stamey, and N. Inoue. 2003. Glycoproteins M and N of human herpesvirus 8 form a complex and inhibit cell fusion. J. Gen. Virol. 84:1485-1491. [DOI] [PubMed] [Google Scholar]
- 45.Lake, C. M., S. J. Molesworth, and L. M. Hutt-Fletcher. 1998. The Epstein-Barr virus (EBV) gN homolog BLRF1 encodes a 15-kilodalton glycoprotein that cannot be authentically processed unless it is coexpressed with the EBV gM homolog BBRF3. J. Virol. 72:5559-5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee, H. C., V. N. Chouljenko, D. V. Chouljenko, M. J. Boudreaux, and K. G. Kousoulas. 2009. The herpes simplex virus type 1 glycoprotein D (gD) cytoplasmic terminus and full-length gE are not essential and do not function in a redundant manner for cytoplasmic virion envelopment and egress. J. Virol. 83:6115-6124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee, J. H., V. Vittone, E. Diefenbach, A. L. Cunningham, and R. J. Diefenbach. 2008. Identification of structural protein-protein interactions of herpes simplex virus type 1. Virology 378:347-354. [DOI] [PubMed] [Google Scholar]
- 48.Leege, T., W. Fuchs, H. Granzow, M. Kopp, B. G. Klupp, and T. C. Mettenleiter. 2009. Effects of simultaneous deletion of pUL11 and glycoprotein M on virion maturation of herpes simplex virus type 1. J. Virol. 83:896-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Letourneur, F., E. C. Gaynor, S. Hennecke, C. Demolliere, R. Duden, S. D. Emr, H. Riezman, and P. Cosson. 1994. Coatomer is essential for retrieval of dilysine-tagged proteins to the endoplasmic reticulum. Cell 79:1199-1207. [DOI] [PubMed] [Google Scholar]
- 50.Liang, X., B. Chow, C. Raggo, and L. A. Babiuk. 1996. Bovine herpesvirus 1 UL49.5 homolog gene encodes a novel viral envelope protein that forms a disulfide-linked complex with a second virion structural protein. J. Virol. 70:1448-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lipinska, A. D., D. Koppers-Lalic, M. Rychlowski, P. Admiraal, F. A. Rijsewijk, K. Bienkowska-Szewczyk, and E. J. Wiertz. 2006. Bovine herpesvirus 1 UL49.5 protein inhibits the transporter associated with antigen processing despite complex formation with glycoprotein M. J. Virol. 80:5822-5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Loret, S., G. Guay, and R. Lippe. 2008. Comprehensive characterization of extracellular herpes simplex virus type 1 virions. J. Virol. 82:8605-8618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lotti, L. V., M. R. Torrisi, M. C. Pascale, and S. Bonatti. 1992. Immunocytochemical analysis of the transfer of vesicular stomatitis virus G glycoprotein from the intermediate compartment to the Golgi complex. J. Cell Biol. 118:43-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mach, M., B. Kropff, P. Dal Monte, and W. Britt. 2000. Complex formation by human cytomegalovirus glycoproteins M (gpUL100) and N (gpUL73). J. Virol. 74:11881-11892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mach, M., B. Kropff, M. Kryzaniak, and W. Britt. 2005. Complex formation by glycoproteins M and N of human cytomegalovirus: structural and functional aspects. J. Virol. 79:2160-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Melancon, J. M., R. E. Luna, T. P. Foster, and K. G. Kousoulas. 2005. Herpes simplex virus type 1 gK is required for gB-mediated virus-induced cell fusion, while neither gB and gK nor gB and UL20p function redundantly in virion de-envelopment. J. Virol. 79:299-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mettenleiter, T. C. 2006. Intriguing interplay between viral proteins during herpesvirus assembly or: the herpesvirus assembly puzzle. Vet. Microbiol. 113:163-169. [DOI] [PubMed] [Google Scholar]
- 58.Montague, M. G., and C. A. Hutchison III. 2000. Gene content phylogeny of herpesviruses. Proc. Natl. Acad. Sci. USA 97:5334-5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Munro, S., and H. R. Pelham. 1987. A C-terminal signal prevents secretion of luminal ER proteins. Cell 48:899-907. [DOI] [PubMed] [Google Scholar]
- 60.Murphy, K. C. 1998. Use of bacteriophage lambda recombination functions to promote gene replacement in Escherichia coli. J. Bacteriol. 180:2063-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nagel, C. H., K. Dohner, M. Fathollahy, T. Strive, E. M. Borst, M. Messerle, and B. Sodeik. 2008. Nuclear egress and envelopment of herpes simplex virus capsids analyzed with dual-color fluorescence HSV1(17+). J. Virol. 82:3109-3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nilsson, T., M. Jackson, and P. A. Peterson. 1989. Short cytoplasmic sequences serve as retention signals for transmembrane proteins in the endoplasmic reticulum. Cell 58:707-718. [DOI] [PubMed] [Google Scholar]
- 63.Pelham, H. R. 1988. Evidence that luminal ER proteins are sorted from secreted proteins in a post-ER compartment. EMBO J. 7:913-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pfeffer, S. R. 2007. Unsolved mysteries in membrane traffic. Annu. Rev. Biochem. 76:629-645. [DOI] [PubMed] [Google Scholar]
- 65.Pignatelli, S., P. Dal Monte, M. P. Landini, B. Severi, R. Nassiri, J. Gilloteaux, J. M. Papadimitriou, G. R. Shellam, T. Mertens, C. Buser, D. Michel, and P. Walther. 2007. Cytomegalovirus primary envelopment at large nuclear membrane infoldings: what's new? J. Virol. 81:7320-7321. (Author's reply, 81:7321-7322.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pomeranz, L. E., and J. A. Blaho. 1999. Modified VP22 localizes to the cell nucleus during synchronized herpes simplex virus type 1 infection. J. Virol. 73:6769-6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Remillard-Labrosse, G., G. Guay, and R. Lippe. 2006. Reconstitution of herpes simplex virus type 1 nuclear capsid egress in vitro. J. Virol. 80:9741-9753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Remillard-Labrosse, G., and R. Lippe. 2009. Meeting of conventional and unconventional pathways at the TGN. Commun. Integr. Biol. 2:1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Remillard-Labrosse, G., C. Mihai, J. Duron, G. Guay, and R. Lippe. 2009. Protein kinase D-dependent trafficking of the large herpes simplex virus type 1 capsids from the TGN to plasma membrane. Traffic 10:1074-1083. [DOI] [PubMed] [Google Scholar]
- 70.Reske, A., G. Pollara, C. Krummenacher, B. M. Chain, and D. R. Katz. 2007. Understanding HSV-1 entry glycoproteins. Rev. Med. Virol. 17:205-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Reynolds, A. E., B. J. Ryckman, J. D. Baines, Y. Zhou, L. Liang, and R. J. Roller. 2001. UL31 and UL34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J. Virol. 75:8803-8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Roizman, B., and A. E. Sears. 1993. Herpes simplex viruses and their replication, p. 11-68. In B. Roizman, R. J. Whitley, and C. Lopez (ed.), The human herpesviruses. Raven Press, New York, NY.
- 73.Rudolph, J., C. Seyboldt, H. Granzow, and N. Osterrieder. 2002. The gene 10 (UL49.5) product of equine herpesvirus 1 is necessary and sufficient for functional processing of glycoprotein M. J. Virol. 76:2952-2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Saffran, H. A., J. M. Pare, J. A. Corcoran, S. K. Weller, and J. R. Smiley. 2007. Herpes simplex virus eliminates host mitochondrial DNA. EMBO Rep. 8:188-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Saraste, J., and E. Kuismanen. 1984. Pre- and post-Golgi vacuoles operate in the transport of Semliki Forest virus membrane glycoproteins to the cell surface. Cell 38:535-549. [DOI] [PubMed] [Google Scholar]
- 76.Schermelleh, L., P. M. Carlton, S. Haase, L. Shao, L. Winoto, P. Kner, B. Burke, M. C. Cardoso, D. A. Agard, M. G. Gustafsson, H. Leonhardt, and J. W. Sedat. 2008. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science 320:1332-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schiff, P. B., and S. B. Horwitz. 1980. Taxol stabilizes microtubules in mouse fibroblast cells. Proc. Natl. Acad. Sci. USA 77:1561-1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schweizer, A., J. A. Fransen, T. Bachi, L. Ginsel, and H. P. Hauri. 1988. Identification, by a monoclonal antibody, of a 53-kD protein associated with a tubulo-vesicular compartment at the cis-side of the Golgi apparatus. J. Cell Biol. 107:1643-1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Skepper, J. N., A. Whiteley, H. Browne, and A. Minson. 2001. Herpes simplex virus nucleocapsids mature to progeny virions by an envelopment → deenvelopment → reenvelopment pathway. J. Virol. 75:5697-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stingley, S. W., J. J. Ramirez, S. A. Aguilar, K. Simmen, R. M. Sandri-Goldin, P. Ghazal, and E. K. Wagner. 2000. Global analysis of herpes simplex virus type 1 transcription using an oligonucleotide-based DNA microarray. J. Virol. 74:9916-9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stylianou, J., K. Maringer, R. Cook, E. Bernard, and G. Elliott. 2009. Virion incorporation of the herpes simplex virus type 1 tegument protein VP22 occurs via glycoprotein E-specific recruitment to the late secretory pathway. J. Virol. 83:5204-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tang, B. L., S. H. Low, H. P. Hauri, and W. Hong. 1995. Segregation of ERGIC53 and the mammalian KDEL receptor upon exit from the 15 degrees C compartment. Eur. J. Cell Biol. 68:398-410. [PubMed] [Google Scholar]
- 83.Tarentino, A. L., T. H. Plummer, Jr., and F. Maley. 1973. A beta-mannosidic linkage in the unit A oligosaccharide of bovine thyroglobulin. J. Biol. Chem. 248:5547-5548. [PubMed] [Google Scholar]
- 84.Torrisi, M. R., C. Di Lazzaro, A. Pavan, L. Pereira, and G. Campadelli-Fiume. 1992. Herpes simplex virus envelopment and maturation studied by fracture label. J. Virol. 66:554-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Turcotte, S., J. Letellier, and R. Lippe. 2005. Herpes simplex virus type 1 capsids transit by the trans-Golgi network, where viral glycoproteins accumulate independently of capsid egress. J. Virol. 79:8847-8860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Turner, A., B. Bruun, T. Minson, and H. Browne. 1998. Glycoproteins gB, gD, and gHgL of herpes simplex virus type 1 are necessary and sufficient to mediate membrane fusion in a Cos cell transfection system. J. Virol. 72:873-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Uetz, P., Y. A. Dong, C. Zeretzke, C. Atzler, A. Baiker, B. Berger, S. V. Rajagopala, M. Roupelieva, D. Rose, E. Fossum, and J. Haas. 2006. Herpesviral protein networks and their interaction with the human proteome. Science 311:239-242. [DOI] [PubMed] [Google Scholar]
- 88.Van De Moortele, S., R. Picart, A. Tixier-Vidal, and C. Tougard. 1993. Nocodazole and taxol affect subcellular compartments but not secretory activity of GH3B6 prolactin cells. Eur. J. Cell Biol. 60:217-227. [PubMed] [Google Scholar]
- 89.van Genderen, I. L., R. Brandimarti, M. R. Torrisi, G. Campadelli, and G. van Meer. 1994. The phospholipid composition of extracellular herpes simplex virions differs from that of host cell nuclei. Virology 200:831-836. [DOI] [PubMed] [Google Scholar]
- 90.Whealy, M. E., J. P. Card, R. P. Meade, A. K. Robbins, and L. W. Enquist. 1991. Effect of brefeldin A on alphaherpesvirus membrane protein glycosylation and virus egress. J. Virol. 65:1066-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Whiteley, A., B. Bruun, T. Minson, and H. Browne. 1999. Effects of targeting herpes simplex virus type 1 gD to the endoplasmic reticulum and trans-Golgi network. J. Virol. 73:9515-9520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wills, E., F. Mou, and J. D. Baines. 2009. The UL31 and UL34 gene products of herpes simplex virus 1 are required for optimal localization of viral glycoproteins D and M to the inner nuclear membranes of infected cells. J. Virol. 83:4800-4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wisner, T. W., C. C. Wright, A. Kato, Y. Kawaguchi, F. Mou, J. D. Baines, R. J. Roller, and D. C. Johnson. 2009. Herpesvirus gB-induced fusion between the virion envelope and outer nuclear membrane during virus egress is regulated by the viral US3 kinase. J. Virol. 83:3115-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wu, S. X., X. P. Zhu, and G. J. Letchworth. 1998. Bovine herpesvirus 1 glycoprotein M forms a disulfide-linked heterodimer with the UL49.5 protein. J. Virol. 72:3029-3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ye, G. J., K. T. Vaughan, R. B. Vallee, and B. Roizman. 2000. The herpes simplex virus 1 UL34 protein interacts with a cytoplasmic dynein intermediate chain and targets nuclear membrane. J. Virol. 74:1355-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yedowitz, J. C., A. Kotsakis, E. F. Schlegel, and J. A. Blaho. 2005. Nuclear localizations of the herpes simplex virus type 1 tegument proteins VP13/14, VHS, and VP16 precede VP22-dependent microtubule reorganization and VP22 nuclear import. J. Virol. 79:4730-4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang, Y., F. Buchholz, J. P. Muyrers, and A. F. Stewart. 1998. A new logic for DNA engineering using recombination in Escherichia coli. Nat. Genet. 20:123-128. [DOI] [PubMed] [Google Scholar]