Abstract
The flavivirus genome comprises a single strand of positive-sense RNA, which is translated into a polyprotein and cleaved by a combination of viral and host proteases to yield functional proteins. One of these, nonstructural protein 3 (NS3), is an enzyme with both serine protease and NTPase/helicase activities. NS3 plays a central role in the flavivirus life cycle: the NS3 N-terminal serine protease together with its essential cofactor NS2B is involved in the processing of the polyprotein, whereas the NS3 C-terminal NTPase/helicase is responsible for ATP-dependent RNA strand separation during replication. An unresolved question remains regarding why NS3 appears to encode two apparently disconnected functionalities within one protein. Here we report the 2.75-Å-resolution crystal structure of full-length Murray Valley encephalitis virus NS3 fused with the protease activation peptide of NS2B. The biochemical characterization of this construct suggests that the protease has little influence on the helicase activity and vice versa. This finding is in agreement with the structural data, revealing a single protein with two essentially segregated globular domains. Comparison of the structure with that of dengue virus type 4 NS2B-NS3 reveals a relative orientation of the two domains that is radically different between the two structures. Our analysis suggests that the relative domain-domain orientation in NS3 is highly variable and dictated by a flexible interdomain linker. The possible implications of this conformational flexibility for the function of NS3 are discussed.
Flaviviruses such as dengue virus (DENV), yellow fever virus (YFV), West Nile virus (WNV), and Japanese encephalitis virus (JEV) belong to the family Flaviviridae and are the causative agents of a range of serious human diseases including hemorrhagic fever, meningitis, and encephalitis (37). They remain a global health priority, as many viruses are endemic in large parts of the Americas, Africa, Australia, and Asia, and vaccines remain unavailable for most members (31, 46, 57).
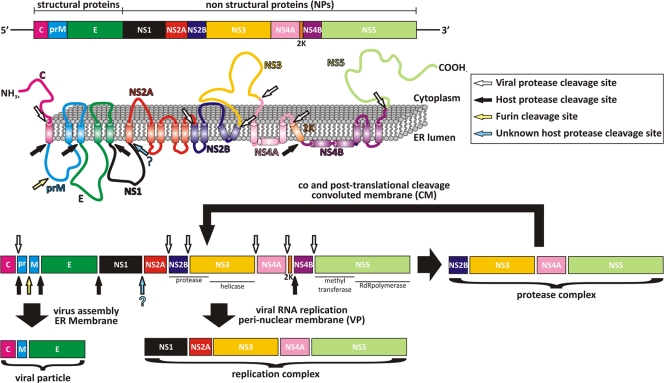
Flaviviruses have a positive-sense single-stranded RNA (ssRNA) genome (approximately 11 kb) that encodes one large open reading frame containing a 5′ type 1 cap and conserved RNA structures at both the 5′ and 3′ untranslated regions that are important for viral genome translation and replication. The genomic RNA is translated into a single polyprotein precursor (11) consisting of three structural (C [capsid], prM [membrane], and E [envelope]) and seven nonstructural (NS1, NS2a, NS2b, NS3, NS4a, NS4b, and NS5) proteins arranged in the order C-prM-E-NS1-NS2a-NS2b-NS3-NS4a-NS4b-NS5 (reviewed in reference 33) (Fig. 1). Only the structural proteins become part of the mature, infectious virion, whereas the nonstructural proteins are involved in polyprotein processing, viral RNA synthesis, and virus morphogenesis (33, 43). The precursor protein is directed by signal sequences into the host endoplasmic reticulum (ER), where NS1 and the exogenous domains of prM and E face the lumen, while C, NS3, and NS5 are cytoplasmic. NS2A, NS2B, NS4A, and NS4B are largely hydrophobic transmembrane proteins with small hydrophilic segments (Fig. 1). The post- and cotranslational cleavage of the polyprotein is performed by NS3 in the cytoplasm and by host proteases in the ER lumen to yield the mature proteins (Fig. 1) (33, 43). Of the nonstructural proteins, NS3 and NS5 are the best characterized, and both are essential for viral replication (23, 27, 41). Both proteins are multifunctional. The N-terminal one-third of NS3 contains the viral protease (NS3pro), which requires a portion of NS2B for its activity, while the remaining portion codes for the RNA helicase/NTPase/RTPase domain (NS3hel) (21, 22, 32, 55). NS5, however, contains both an N-terminal methyltransferase and a C-terminal RNA-dependent RNA polymerase (16, 51). The functions of NS1, NS2A, NS4A, and NS4B are not well understood, but they appear to play important roles in replication and virus assembly/maturation and have been found to bind to NS3 and NS5, possibly modulating their activity (33, 36).
FIG. 1.
Schematic diagram of flavivirus polyprotein organization and processing. (Top) Linear organization of the structural and nonstructural proteins within the polyprotein. (Middle) Putative membrane topology of the polyprotein predicted from biochemical and cellular analyses, which is then processed by cellular and viral proteases (indicated by arrows). (Bottom) Different complexes that are thought to arise in different cellular compartments during and following polyprotein processing.
Because of its enzymatic activities and its critical role in viral replication and polyprotein processing, NS3 constitutes a promising drug target for antiviral therapy (31). NS3pro (residues 1 to 169) is a trypsin-like serine protease with the characteristic catalytic triad (Asp-His-Ser) and a highly specific substrate recognition sequence, conserved in all flaviviruses, consisting of two basic residues in P2 and P1 followed by a small unbranched amino acid in P1′ (11). NS3pro has an aberrant fold compared to the canonical trypsin structure, and its folding and protease activity are dependent on a noncovalent association with a central 47-amino-acid hydrophilic domain of NS2B (19, 21). The remainder of NS2B contains three transmembrane helices involved in membrane associations. NS3 mediates cleavages at the C-terminal side of the highly conserved dibasic residue located at the coding junctions NS2A/NS2B, NS2B/NS3, NS3/NS4A, and NS4B/NS5 and also between the C terminus of C and NS4A (11, 33) (Fig. 1).
The C-terminal portion of NS3 (NS3hel, residues 170 to 619) performs several catalytically related activities, namely, RNA strand separation and (poly)nucleotide hydrolysis (5, 22, 32, 55) at a common, RecA-like NTPase catalytic center that couples the energy released from the hydrolysis of the triphosphate moieties of nucleotides to RNA unwinding. Although the precise role of NS3 in replication has not been established, its helicase activity is thought to separate nascent RNA strands from the template strands and to assist replication initiation by unwinding RNA secondary structure in the 3′ untranslated region (11, 13, 15, 33). NS3 is a member of the DEAH/D box family within helicase superfamily 2 (SF2) and is characterized by seven conserved sequence motifs involved in nucleic acid binding and hydrolysis (45). In addition, its RNA triphosphatase activity is thought to be involved in the capping of the viral RNA. In the process of replication, NS3 interacts, most likely via its C-terminal domain, with NS5 (13, 15, 24, 26, 58, 62). The NS3 5′ triphosphatase and NS5 methyltransferase activities probably cooperate in cap formation by removing the terminal γ-phosphate and performing sequential N7 and 2′ O methylations, respectively (16, 28, 46, 56). The guanylyltransferase activity required for cap formation remains elusive at present, although recent evidence suggests that it may be present in NS5 (8, 17). In addition, the interaction between NS3 and NS5 can stimulate NS3 helicase/NTPase activity (15, 62).
The atomic structures of NS3pro in the presence and absence of ligands and/or the NS2B activating domain (2, 19, 47) and NS3hel (35, 38, 39, 49, 58-60) are known, and recently, the structure of full-length DENV4 (one of four dengue virus serotypes) NS3 fused to an 18-residue NS2B cofactor (NS2B18NS3) was reported (34). This structure revealed an elongated conformation, with the protease domain interfacing with the NTP binding pocket and being separated from NS3hel by a relatively flexible linker, which suggested that the protease domain may have a positive effect on the activity of the NTPase/helicase domain. However, other reports suggested that NS3pro has no or a very limited effect on the activity of NS3hel (32, 62). In addition, since current evidence suggests that NS2B is not part of the replication complex (Fig. 1) (36), and it is known that in the absence of the NS2B cofactor, NS3pro is unfolded and inactive, it becomes hard to envisage what effect the NS3 protease domain may have on the helicase domain in a biologically relevant context. Equally, it is still not clear what role the helicase domain plays during polyprotein processing by NS3pro and, in general, why these two apparently distinct and unrelated catalytic activities are harbored within a single polypeptide.
In order to gain further insights into these questions, we report the biochemical analysis and crystallographic structure at a 2.75-Å resolution of full-length NS3 from Murray Valley encephalitis virus (MVEV), a member of the JEV group of flaviviruses, fused to the entire protease activation peptide of the NS2B cofactor (NS2B45NS3). The structure reveals the protease and helicase domains to be structurally independent and differs dramatically from the structure observed for DENV4 NS2B18NS3. We discuss the implications of this unexpectedly different configuration of the NS3 protein and argue that the structural flexibility observed is likely to be crucial for its multifunctional nature.
MATERIALS AND METHODS
Construction of the NS2B-NS3 protein.
Primers used were NS2Bfwd (5′-AAGTTCTGTTTCAGGGCCCGGCCACAGACATGTGGCTTGAAAGAGCAGCAGATGTGTCATGG-3′), NS2Brev (5′-GACTTTCATCTGCTCAATGACCCTGGAGTTGGTGGCGGAGGCAGTGGAGGAGGGGGT-3′), NS3fwd (5′-GGTGGCGGAGGCAGTGGAGGAGGGGGTGGGGGTGTTTTTTGGGATACGCCTTCC-3′), and NS3rev (5′-ATGGTCTAGAAAGCTTTACCGCTTCCCAGCTGCAAAGTCCTTG-3′).
PCR was performed by using KOD HotStart polymerase (Merck Biosciences) according to the manufacturer's recommendations. The MVEV NS2B fragment (amino acids 49 to 93) was amplified using NS2Bfwd and NS2Brev (10 pmol each), and the MVEV NS3 open reading frame was amplified with NS3fwd and NS3rev (10 pmol each). Primers NS2Brev and NS3fwd added an overlapping sequence to allow seamless gene fusion through PCR. Following PCR and PCR clean-up (Qiagen PCR clean-up kit), 1-ng aliquots of each fragment were combined in a fresh PCR mixture with primers NS2Bfwd and NS3rev (10 pmol each). This produced the NS2B-NS3 PCR fusion product with the NS2B fragment separated from NS3 by a sequence encoding a 9-amino-acid linker (GGGGSGGGG). The PCR product contained terminal 15-bp tags that allowed in-fusion cloning into pOPINF (7), adding an N-terminal His6-3C protease site tag to the construct. Cloning of the spin-column-purified PCR product was performed as detailed previously (7).
Expression and purification.
Methods used for expression and purification were essentially identical to a method for the purification of the MVEV NS5 methyltransferase domain (4) except that for size exclusion chromatography, a buffer containing 25 mM HEPES (pH 8.0), 300 mM NaCl, 0.5% glycerol, and 0.5 mM Tris(2-carboxyethyl)phosphine was used. The peak fractions were pooled, and 200 μg of 3C protease was added to remove the His tag, followed by gentle rocking at room temperature for 2 h. Imidazole was added to 40 mM, followed by the addition of 2 ml of Ni-Sepharose (GE Healthcare), and the suspension was incubated for 30 min at room temperature before passage over a disposable column. The flowthrough was collected, concentrated, and applied onto a HiLoad Superdex 200 column (GE Healthcare) equilibrated at 4°C in gel filtration buffer as described above. The peak fractions were pooled and concentrated to 5.5 mg/ml. The protein was characterized by liquid chromatography-electrospray ionization mass spectrometry and sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis analysis.
Crystallization, data collection, and processing.
Crystallization trials of the concentrated protein were set using the automated pipeline in the Oxford Protein Production Facility as described elsewhere previously (53). Thin, blade-like crystals formed within 2 days in a solution containing 100 mM MES (morpholineethanesulfonic acid) (pH 6.0) and 20% (wt/vol) polyethylene glycol 6000 with either 200 mM magnesium chloride or 200 mM calcium chloride (PACTpremier screen; Molecular Dimensions) or in a solution containing 100 mM MES (pH 6.0), 20% (wt/vol) polyethylene glycol 8000, and 200 mM calcium chloride (Emerald Wizard II screen; Emerald Biosystems). The crystals were often stacked and formed tight clusters, but isolated single crystals were obtained by using microseeding as described previously for NS3hel (38, 54). For data collection, crystals were briefly immersed in mother liquor containing 30% glycerol and flash-frozen in liquid nitrogen. A total of 360° of data were collected at the European Synchrotron Radiation Facility beam-line ID29 (Grenoble, France) as a series of 1.0° oscillations. Data were acquired by use of a Q315R charge-coupled-device detector (ADSC) with an exposure time of 2 s per frame and processed using XDS (25) and SCALA (20) to a resolution of 2.75 Å. The crystal parameters and data collection statistics are summarized in Table 1.
TABLE 1.
X-ray data collection and refinement statistics
| Statistic | Valuea |
|---|---|
| Data collection | |
| Space group | P21 |
| Unit cell dimensions a, b, c (Å) | a = 41.8, b = 105.2, c = 79.9 |
| β (°) | 97.4 |
| Resolution range (Å) | 39.60-2.75 (2.82-2.75) |
| Observations | 129,498 |
| Unique reflections | 17,578 |
| Completeness (%) | 98.6 (98.2) |
| I/σ(I) | 19.2 (3.2) |
| Rmergeb (%) | 8.7 (72.8) |
| Refinement | |
| Resolution range (Å) | 30.0-2.75 (2.82-2.75) |
| No. (%) of reflections in working set | 16,866 (1,226) |
| No. (%) of reflections in test set | 884 (66) |
| Wilson B (Å2) | 6.2 |
| Rfactorc working set (%) | 26.8 (32.4) |
| Rfreec (%) | 30.0 (31.1) |
| RMSD bond length (Å) | 0.006 |
| RMSD bond angle (°) | 0.899 |
| No. of atoms (protein/water) | 601 (91) |
| Average B factors (Å2) (main chain/water) | 51.3 (36.7) |
| Average B factors (Å2) NS3hel, NS3pro, NS2B | 52.2, 51.1, 50.0 |
The numbers in parentheses refer to the last (highest) resolution shell.
Rmerge = ΣhΣi|Ii(h) − <I(h)>|/ΣhΣi<Ii(h)>, where Ii(h) is the ith measurement and <I(h)> is the weighted mean of all measurements of Ii(h).
Rfactor and Rfree = Σh‖F(h)obs| − |F(h)calc‖/Σh|F(h)obs| for reflection in the working set and test set, respectively.
Structure solution and refinement.
The structure was solved by molecular replacement using the program PHASER (ccp4i suite [14]), with the MVEV NS3178-618 helicase (Protein Data Bank [PDB] accession number 2v8o) (38) and the WNV NS2B-NS3 protease (PDB accession number 2ijo) (2) as search models. One copy of each search model was located in the asymmetric unit with a solvent content of 46% (based on a Matthews coefficient Vm value of 2.26). The model was subjected to rigid body refinement and simulated annealing using CNS (10), manual model building with Coot (18), and refinement with PHENIX (1) and REFMAC (14). Buried surface areas and contacts between domains were calculated by using PISA (29). Superposition of structures was carried out by using the program SHP (50). Figures were prepared by using the program Pymol (http://www.pymol.org).
ATPase assay.
ATPase activity was measured bye using a malachite green-based ATPase assay according to the manufacturer's instructions (Innova Biosciences) with 5 nM of NS2B-NS3 or NS3hel and various amounts of ATP in a solution containing 25 mM HEPES (pH 7.5) and 5 mM MgCl2. The reaction was allowed to proceed for 10 min at 28°C, after which the reaction was terminated and the absorbance at 630 nm was recorded. Reaction velocities were determined by the inclusion of phosphate standards. Time course experiments showed that the absorbance increase was linear at the ATP concentrations tested (data not shown). Kinetic parameters were calculated with GraphPad (GraphPad Software Inc.).
Helicase activity assay.
The substrate of the partially double-stranded helicase assay was obtained by annealing RNA synthetic oligonucleotides to produce a 16-bp duplex with a 14-nucleotide 3′ overhang (Primm, Milan, Italy) (58). To form the partial double-stranded substrate, the 5′-CACCUCUCUAGAGUCGACCUGCAGGCAUCG-3′ strand was labeled with [γ-32P]ATP at its 5′ end by using T4 polynucleotide kinase and annealed with complementary primer 5′-CGACUCUAGAGAGGUG-3′. The annealed duplex was purified by high-performance liquid chromatography (Sephadex G25 columns; GE Healthcare).
The assay was performed using 20-μl reaction mixture volumes containing 25 mM HEPES (pH 7.5), 1 mM MgCl2, 2 mM dithiothreitol, 2 mM ATP, 5% glycerol, 5 U RNasin, and 10 fmol of RNA substrate. The reaction was initiated by adding the recombinant protein at various concentrations between 50 and 1,000 nM, or an equivalent volume of the buffer (negative control), and stopped after 30 min at 37°C by adding 6 μl of loading dye (50% EDTA, 0.5% SDS, 50% glycerol, 0.01% bromophenol blue) to the mixture. The helicase assay mixtures were resolved by electrophoresis using nondenaturing 17% polyacrylamide gels that were dried and analyzed by using a phosphorimager (Typhoon; GE Healthcare). The percentage of duplex unwinding was calculated by comparing the intensities of the two bands (ImageQuant software; Amersham Bioscience).
Protease activity assay.
The protease activity of NS2B-NS3 was measured by using an AMC (7-amino-4-methylcoumarin) fluorophore-based protease assay (SensoLyte AMC WNV protease assay kit; Anaspec). The peptide provided in the kit (Pyr-RTKR-AMC) generates free AMC upon NS3 protease cleavage, which can be detected by excitation at 354 nm and emission at 442 nm (SpectraFluorPlus; Tecan). For kinetic analysis, cleavage was allowed to proceed for 45 min at room temperature, after which fluorescence was measured. Time course experiments showed a linear increase in fluorescence emission over this period. For inhibitor studies, 150 pmol of inhibitor (undeca-d-Arg-NH2, included in the kit) was mixed for 5 min with 150 or 1,500 pmol of NS2B-NS3, and the increase in fluorescence was measured over time and compared to the fluorescence of reaction mixtures lacking an inhibitor.
Protein structure accession number.
Coordinates and structure factors have been deposited in the PDB under accession number 2WV9.
RESULTS AND DISCUSSION
Expression and crystallization of NS2B-NS3 and NS3hel.
The association of a central 47-amino-acid hydrophilic peptide of NS2B promotes the productive folding and activity of NS3pro. While the first 18 N-terminal residues of this peptide of NS2B are sufficient to express NS3 protease in a soluble form, this construct is catalytically inactive. To express soluble, catalytically competent full-length NS3, a fusion to NS2B residues 49 to 93 (NS2B45) was prepared by overlap PCR and in-fusion cloning to generate a construct encoding full-length NS3 coupled N terminally to NS2B45 via a nonapeptide linker (G4SG4) (see Materials and Methods and Fig. 3a). This construct is hereafter named NS2B45NS3. This method, analogous to that used previously to express and crystallize the NS2B-bound NS3pro of WNV and DENV (19), leads to high levels of soluble protein (∼10 mg/liter of culture) using Escherichia coli expression. The recombinant NS2B45NS3 possesses full protease and helicase activities (see below) and appears to be, at least within the time frame of the experiments, insensitive to proteolytic degradation (data not shown). Finally, we were able to show that full-length NS2B45NS3 was the molecular species contained in the crystals that led to structure determination (data not shown).
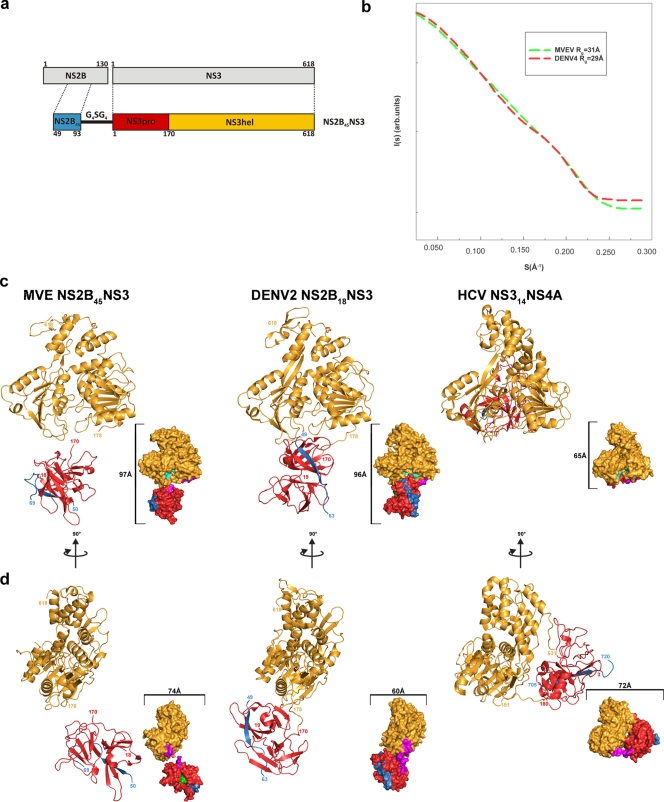
FIG. 3.
Overall structure of MVEV NS2B45NS. (a) Diagram of the MVEV NS2B and NS3 protein organization and of the NS2B45NS3 synthetic construct used for crystallization. (b) Superimposition of the scattering from an ab initio model of MVEV NS2B45NS3 and DENV4 NS2B18NS3 computed by use of the program CRYSOL (50). As a result of within-experiment errors, the two curves are indistinguishable. Rg is the radius of gyration. (c) Comparison of the structures of MVEV (MVE) NS2B45NS3, DENV4 NS2B18NS3, and HCVNS314NS4A. A cartoon diagram (main) and surface representation (inset) of equivalent views of the three synthetic constructs are shown. Color coding is the same as that in panel a: NS2B (and NS4A) is blue, the NS3pro domain is red, NS3hel is yellow, the NS3pro catalytic pocket is green, the NS3hel catalytic pocket is cyan, and the interdomain linker loop is magenta. (d) Orthogonal view of panel c.
The NS3hel (NS3 residues 178 to 619) construct was cloned and purified as described previously (38).
ATPase and RNA-unwinding activities.
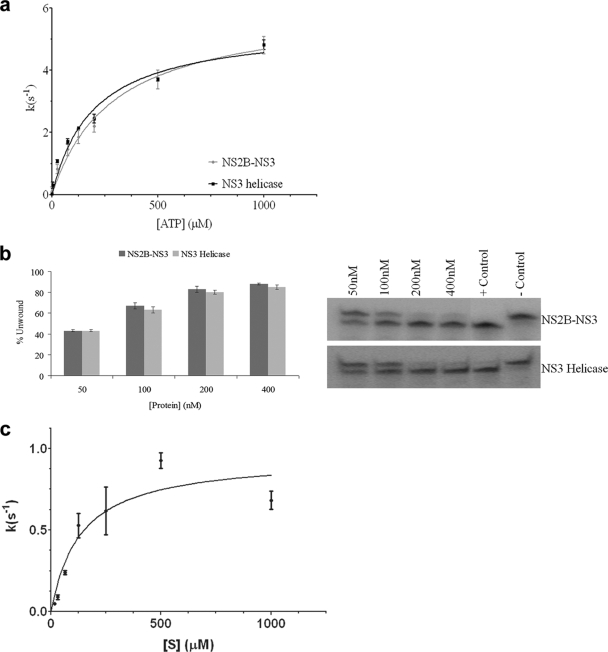
The ATPase activity of the NS2B45NS3 protein was compared with that of the NS3 helicase domain construct previously used for structural analysis (residues 178 to 619) (38). As observed previously, the reaction velocities followed Michaelis-Menten kinetics with respect to the ATP concentration. The results, shown in Fig. 2a, indicate that differences between the two protein constructs are small, although the helicase domain appears to exhibit a slightly higher affinity for ATP (kcat value of 6.0 s−1 and Km value of 0.29 mM ± 0.04 mM for NS2B45NS3, and kcat value of 5.5 s−1 and Km value of 0.19 mM ± 0.03 mM for NS3hel). The turnover value and affinity for ATP of NS2B45NS3 are remarkably similar to those observed previously for full-length NS3 of DENV2 (Km = 0.30 mM; kcat = 5.8 s−1) (59).
FIG. 2.
Catalytic activity studies of MVEV NS2B45NS3. (a) Comparison of the ATPase activities of MVEV NS2B45NS3 (light gray) and MVEV NS3171-618 (dark gray). The ATPase assay was carried out with 5 nM of enzyme in the presence of the indicated concentrations of ATP. The amount of inorganic phosphate released during catalysis was measured with malachite green. (b) Helicase activities of MVEV NS2B45NS3 (dark gray) and MVEV NS3171-618 (light gray). Unwinding activity was measured by using a radiolabeled double-stranded RNA substrate. Control lanes are included (positive control [heat denatured duplex] and negative control [in the absence of enzyme]). Enzyme concentrations are indicated. The values represent average data from three experiments. (c) Assay of the protease activity of NS2B45NS3 was carried out with 5 nM of enzyme in the presence of the indicated concentrations of peptide (see Materials and Methods). The amount of AMC released during proteolysis was detected by excitation at 354 nm and emission at 442 nm using a SpectraFluorPlus reader (Tecan).
RNA-unwinding activity was assessed, as described previously for the YFV NS3 protein (58), by using a partially double-stranded RNA molecule consisting of a 14-base 3′ single-stranded tail followed by a 16-bp double-stranded RNA region. A comparison of NS2B45NS3 and NS3hel, as shown in Fig. 2b, suggests that both are fully active helicases without an apparent difference between the two proteins under the conditions tested.
Taken together, the results suggest that although the cofactor-bound protease might have a small effect on ATP binding (1.5-fold), the overall effect of its presence on MVEV NS3 NTPase and helicase activities is small.
This is in apparent contrast to the observations described previously by Xu et al. (59) for DENV2, where full-length NS3 showed a 10-fold-lower ATP affinity than NS3hel (residues 171 to 618) while displaying a 30-fold-higher RNA-unwinding activity. A similar increase in the rate of RNA unwinding was found previously for Kunjin virus (KUNV) full-length NS3 (39). More recently, using fluorescence correlation spectroscopy, Luo et al. (34) showed that DENV4 NS2B18NS3 displays a 10-fold-higher ATP affinity than NS3hel (residues 177 to 618) (no attempt was made at measuring its ATPase or helicase activity). While these results appear to be irreconcilable, note that several studies have now shown that the choice of the N-terminal boundary of NS3hel can greatly affect the activity of this domain, indicating that comparisons between full-length and truncated NS3 molecules need to be regarded with caution. The helicase and the protease domains are connected via a flexible interdomain linker (residues 169 to 181). Generally, NS3hel proteins containing longer linker regions showed a similar ATPase activity compared to but a higher RNA helicase activity than those with short linkers (32, 39, 58), but the reason remains unclear; it may have a functional role (40) or cause structural artifacts, as observed previously for KUNV NS3hel (39). Remember, however, that there is no evidence to date for an in vivo role of the isolated NS3 helicase and that, therefore, a choice of boundaries for this domain is arbitrary.
Protease activity.
The protease activity of NS2B45NS3 was assessed by using the SensoLyte 440 WNV protease assay kit by using an AMC-coupled peptide: Pyr-RTKR-AMC. Kinetic analysis showed that the MVEV NS2B45NS3 protease had nearly identical activity compared to that of WNV NS2B-NS3pro (48), with a kcat of 1.0 s−1, and a slightly lower affinity, with a Km of 134 μM, compared to 43 μM for WNV (Fig. 2c). Furthermore, the MVEV NS2B45NS3 protease was inhibited by the undeca-d-Arg-NH2 peptide (data not shown), developed previously as an inhibitor for WNV NS2B-NS3pro (48).
Together, these results indicate that the protease domain in MVEV NS2B-NS3 was fully active and possessed properties comparable to those of the WNV NS2B-NS3 protease. The lack of an apparent effect due to the presence of the helicase domain, in line with recent observations for hepatitis C virus (HCV) NS3 (52) and WNV NS3 (12), suggests that the helicase domain has little influence on the protease activity in solution.
Structure determination.
Initial analyses suggested that the crystals belong to space group P21, with the following cell parameters: a equals 41.8 Å, b equals 105.2 Å, c equals 79.9 Å, and β equals 97.4°. Based on this assumption, the crystallographic asymmetric unit contains one monomer, and the structure could be solved and refined to a 2.75-Å resolution with a final R factor of 27% (Rfree = 30%), with good stereochemistry (more than 96% of residues in the most favored region of the Ramachandran plot [30]). However, careful inspection of the diffraction patterns reveals an alternative indexing solution, where the a axis is doubled but the crystal symmetry and other cell parameters remain the same. In this cell, there are two molecules of NS2B45NS3 related by the translation of half a unit cell along a. This results in half the reflections (h = 2n + 1) being systematically extremely weak, particularly at a low resolution. The additional data available by integrating the images in the larger cell are therefore limited, and a doubling of the degrees of freedom with refinement in the larger cell would result in an overfitting of the X-ray data. Rigid body refinement in the large-cell data of the two molecules in the small-cell positions, performed with the program XPLOR (10) without solvent correction between a 5- and 3.5-Å resolution, reduced the R factor for the h = 2n reflections only from 38.1 to 35.0%, while the R factor for the h = 2n + 1 reflections fell from 100% to 46.8%. Given the limitations of the current software, we therefore chose to refine the model for the smaller cell on the basis that the relationship between the two molecules in the larger cell is very close to being crystallographic (∼0.35 Å from the position required for a true “small cell”). This allowed a robust refinement of the model (although the resultant R factors are poor), and all the results below are based on this refinement (Table 1).
Regarding electron density, a total number of 580 out of 618 residues of full-length NS3 are well defined, together with 21 residues out of 45 of NS2B45. The 9 residues of the nonapeptide linker, 24 residues at the C terminus of the NS2B cofactor, 18 residues at the N terminus of the protease domain, and 7 residues in the linker region between protease and helicase are missing from the refined model.
Overall structure.
The structure of MVEV NS2B45NS3 comprises two separate globular folds, representing the protease and helicase/NTPase, respectively, coupled via a short, stretched, and partially disordered linker region (Fig. 3c and d). Despite the absence of visible density for residues 169 to 175 of the interdomain linker, there is an unequivocal choice for the protease/helicase pair forming the biologically relevant unit (18 Å separating residue 169 from residue 175 in the chosen pair as opposed to 38 Å for the next closest pair). The presence of the linker gives the molecule a beads-on-a-string appearance and a rather elongated shape (Zmax = 97 Å) that is dramatically different from that observed for HCV NS3-NS4A (Zmax = 66 Å), which is globular, with extensive interdomain interactions (61), but similar to the shape of DENV4 NS2B18NS3 (Zmax = 96 Å) (34) (Fig. 3c and d) and consistent with SAXS data obtained for both DENV4 NS2B18NS3 and full-length KUNV NS3 in solution (Dmax = 120 Å) (34, 39). Despite the similarity in overall shape and extensive structural homology of the individual domains (422/618 residues superimpose with a root mean square deviation [RMSD] of 1.5 Å for helicase domains, and 151/152 residues superimpose with an RMSD of 1.6 Å for protease domains), the relative orientations of the protease and helicase are dramatically different between MVEV NS2B45NS3 and DENV4 NS2B18NS3, as shown in Fig. 3b and c (a rotation of close to 180° and a translation of 17 Å are required to superimpose the protease domains after superimposition of the structures based on the helicase domains). This means that the relative positions of the protease and helicase catalytic centers are different in the two structures: in MVEV NS2B45NS3, they are both oriented away from the interdomain interface and exposed to the solvent, whereas in DENV4 NS2B18NS3, the helicase active site is at the interface between domains (Fig. 3c and d). Additionally, while the buried surface area between the protease and the helicase in DENV4 NS2B18NS3 is 568 Å2 (380 Å2 omitting the residues from the interdomain region), the area excluded in MVEV NS2B45NS3 is only 30 Å2, not consistent with a stable interface. Nevertheless, from simulated SAXS scattering curves from atomic coordinates, it is clear that it would be hard to distinguish between these different configurations in solution with a low-resolution technique such as SAXS (Fig. 3b). This means that previous SAXS data for DENV4 and KUNV NS3 are consistent with both the MVEV and DENV4 conformations.
Linker region and interdomain orientation.
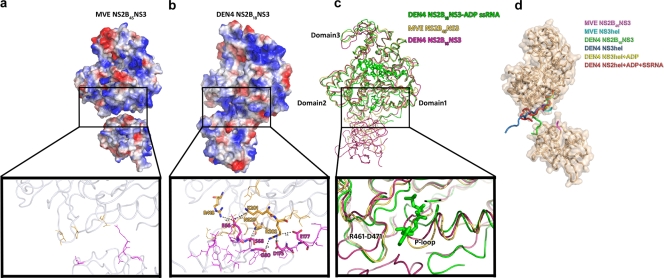
The protease and the helicase domains are connected by a short (13-residue), solvent-exposed, interdomain loop (“linker” loop, residues 169 to 181) that adopts a rather extended conformation spanning 24 Å, similar to the conformation seen for DENV4 NS2B18NS3, where the same peptide spans 25 Å. This linker region is rich in acidic amino acids and is partially disordered in our structure (residues 169 to 175 are not visible), although SDS-polyacrylamide gel electrophoresis analysis of the crystals shows the protein to be intact. In DENV4 NS2B18NS3, the interdomain linker is likely to contribute greatly to the charge distribution previously observed by Luo et al. (34), which is thought to optimize the diffusion of nucleotide substrates toward the ATP hydrolysis site, as shown in Fig. 4b. Such a charge distribution was not observed for MVEV NS2B45NS3, where the interdomain linker is partially disordered (Fig. 4a).
FIG. 4.
Structural analysis of protease-helicase interdomain interactions. (a) Electrostatic surface view of MVEV NS2B45NS3 and ribbon diagram of the protease (magenta)-helicase (yellow) interdomain region, with residues at the interacting interface represented as sticks (inset). Residues involved in hydrogen bonds and salt bridges are shown in boldface type, and interatomic distances are indicated. (b) DENV4 NS2B18NS3. (c) Structural superimposition of MVEV NS2B45NS3 (this study) (yellow), DENV4 NS2B18NS3 (PDB accession number 2vbc) (magenta), and DENV4NS3hel+ADP+ssRNA (PDB accession number 2jlz) (green). In DENV4, protease domain residue S68 contacts the catalytically relevant N329 in the helicase domain (which is blocked in a catalytically unfavorable conformation). E66 of the protease domain in particular is likely to block access to the ATP base, as its side chain sits directly in front of the binding pocket and appears to orientate R418 and K201 via long-range electrostatic interactions. In addition, D175 and E177 of the linker region contact R202 of the P loop, and D175 further contacts G80 of the protease domain. Indeed, superimposition of helicase domain 1 of DENV4 NS2B-NS3 and the ADP and ssRNA-bound helicase domain of DENV4 (PDB accession number 1jlv [34]), which represents the product-bound form, shows that domain 2 movement would cause a significant clash with the protease domain. (d) Analysis of the linker orientations in different flavivirus NS3 crystal structures. Shown is a ribbon diagram and transparent surface representation of the structure of MVEV NS2B45NS3. Shown is the superimposition of the interdomain linker region (residues 169 to 181) of MVEV NS2B45NS3 (magenta) (this study) with those of the MVEV NS3hel structure (cyan) (PDB accession number 1v80), DENV4 NS2B18NS3 (green) (PDB accession number 2vbc), and a number of DENV4 NS3hel structures solved in the presence and absence of cofactors (DENV4 NS3hel [blue] [PDB accession number 2jlq], DENV4 NS3hel+ADP [yellow] [PDB accession number 2jls], and DENV4NS3hel+ADP+ssRNA [red] [PDB accession number 2jlz]).
The flexibility of the linker loop is supported by several other lines of evidence. First, despite the high overall sequence identity (average of 72%), the linker residues are less conserved (average of 50%), suggesting limited structural constraints on this region. Furthermore, this region adopted distinct conformations in two different structures of NS3pro from WNV (PDB accession numbers 2ggv for WNV and 2ijo for WNV in complex with aprotinin [2]) and was disordered in the structures of DENV2 NS3pro apo and WNV NS3pro in complex with an inhibitor (PDB accession numbers 2fom and 2fp7, respectively [19]). The superimposition of the published DENV helicase structures that include the linker region further shows that even within a flavivirus species, the linker can adopt a range of conformations (Fig. 4d). Interestingly, the equivalent region in HCV NS3 is seen in different conformations in independent copies of HCV NS3-NS4A (NS4A is the protease activating domain of NS3 in HCV instead of NS2B) (61). Unsurprisingly, the three lowest-frequency modes identified by normal-mode analysis of our model (elNemo server) (data not shown) correspond to interdomain movements between the protease and helicase domains. In short, the evidence suggests that this loop is highly flexible and able to adopt several distinct conformations; indeed, it seems likely that the DENV4 NS2B18NS3 and MVEV NS2B45NS3 configurations interconvert in solution. Analysis of the structure of HCV NS3-NS4A suggests that it too, in principle, could adopt a conformation very similar to the DENV4 or MVEV conformations via simple rotations of the linker region. Conversely, it is less likely that flavivirus NS3 adopts an HCV-like conformation because in flaviviruses, the NS3-NS4A junction is cleaved in trans, whereas HCV NS3 cleaves this junction in cis. This could explain the lack of evidence for the existence of an HCV-like state for flavivirus NS3. The postulated flexibility of the linker region can now also explain the slow autolytic cis cleavage that was previously shown to occur within NS3hel (at a site corresponding to R459-G460) in cells infected with DENV2 (3) and that was more recently confirmed in vitro for WNV (6). This cis cleavage obviously requires R459-G460 in domain 2 of NS3hel to be accessible to the protease active site, which would require a high degree of flexibility between the two domains.
The protease domain.
The structure of the protease domain of MVEV NS2B45NS3 is very similar to that of the protease domain of WNV (NS2B40NS3pro) in both the inactive (apo) and active (inhibitor bound) conformations, with RMSDs of 1.6 Å for 153 atoms and 1.3 Å for 146 atoms, respectively (Fig. 5) (19). This, combined with the protease kinetics reported above, suggests that MVEV NS2B45NS3 is a fully active protease, with its activity and structure comparable to that of the isolated WNV NS2B40NS3pro.
FIG. 5.
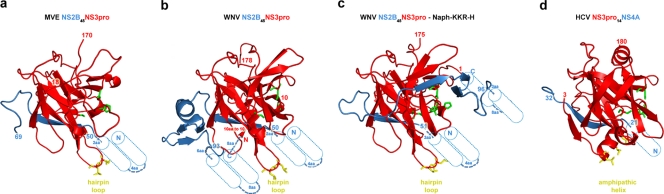
Structural comparison of cofactor-bound Flaviviridae NS3 protease domains. (a) Cartoon representation of MVEV NS2B45NS3pro (this study). (b) WNV NS2B45NS3pro (PDB accession number 2ggv). (c) WNV NS2B45NS3pro bound to Naph-KKR-H (PDB accession number 3e90). (d) HCV NS3pro18NS4A (PDB accession number 1cu1). Blue, NS2B and NS4A cofactors; red, NS3pro; green, stick representation of the NS3pro catalytic triad (His-Asp-Ser); yellow, hydrophobic residues in its membrane-inserted region. Putative transmembrane helices of the cofactors NS2B and NS4A are shown as cylinders, together with their distance to the nearest visualized residue in the atomic structure.
The NS3 N-terminal protease has been solved in the presence and absence of the cofactor domain of NS2B for DENV and WNV (2, 19, 47). The protease has a trypsin-like fold, containing two β-barrels with the Asp-His-Ser catalytic triad located in a cleft situated between the two barrels. In the absence of the NS2B cofactor, the protease is virtually inactive and less stable. Stabilization by NS2B is achieved through the insertion of a β-strand of NS2B into the N-terminal β-barrel of NS3pro, with minor additional interactions with the C-terminal β-barrel. As observed for the DENV NS2B-NS3pro structure, in the absence of a substrate, the MVEV NS2B region beyond residue 69 is disordered. Upon the addition of a substrate, the C-terminal part of NS2B undergoes a dramatic conformational change, wrapping around the C-terminal barrel and forming a stabilizing β hairpin that inserts into the active site (2, 19, 47).
These features of the flavivirus NS3 protease are similar to those observed for the more distantly related HCV NS3 protease (Fig. 5). The overall fold of the HCV protease is similar to that of the flavivirus NS3 proteases (Fig. 5). Like flaviviruses, HCV requires a cofactor for activity, but this is found in NS4A rather than NS2B. Crystallographic and nuclear magnetic resonance analyses showed a more subtle involvement of NS4A: the binding of NS4A induces stabilizing conformational changes in the NS3 protease without actually engaging with the active site (42, 61). Moreover, the activating region within NS4A was found to be much shorter (comprising a single β sheet) (Fig. 5).
Based on the recently reported crystal structure of WNV NS2B-NS3pro bound to the inhibitor Naph-KKR-H (47) and the predicted membrane domains of NS2B, the distances of the termini of the NS2B activating region to the ER membrane are rather short, 3 residues from the cofactor N terminus and 1 residue from the C terminus, placing the activated NS3 protease domain in a very tight membrane-anchored NS2B sling (Fig. 5b and c). As recently suggested (9), the NS3 membrane association is likely to be further stabilized by the membrane insertion of a β hairpin with conserved hydrophobic residues (MVEV NS3pro residues 27 to 34), analogous to the proposed membrane inserted amphipathic helix α0 in HCV NS3 (Fig. 5).
The NTPase/helicase domain.
The flavivirus NS3 C-terminal NTPase/helicase has been solved for YFV (58), DENV (35, 59), MVEV (38), KUNV (39), JEV (60), and Kokobera virus (49). It is a DEAH/D box helicase belonging to the SF2 superfamily of helicases containing the characteristic Walker A and B motifs responsible for NTP and Mg2+ binding and consisting of three domains of approximately equal size, forming a large positively charged cleft at their juncture, which is the RNA binding site. The structure is closely related to HCV NS3 helicase with respect to domains 1 and 2, but domain 3 shows a unique conformation typical of flaviviruses. Differences between the YFV, DENV, KUNV, Kokobera virus, JEV, and MVEV structures were found to be confined primarily to the orientation of domain 2 relative to domains 1 and 3, suggesting that the movement of domain 2 affects nucleic acid translocation in an ATP-dependent manner according to the inchworm model, which was recently confirmed by a detailed study of the catalytic cycle of the ATPase activity of a DENV4 helicase (35).
In the DENV4 NS2B-NS3 structure (34), the protease and linker regions directly contact the helicase NTP binding pocket, forming a network of hydrogen bonds and salt bridges (Fig. 4b). These interactions hold the NS3hel catalytic loop (P loop, or Walker A motif), critical for driving the conformational changes that occur upon NTP hydrolysis, in a conformation very similar to that of the apo form observed previously for the DENV2 helicase domain structure (35), limiting access to the nucleotide binding pocket (Fig. 4c). Another striking feature of the DENV4 conformation is that the loop between R461 and D471 (in domain 2) is moved outward compared to those of MVEV NS2B-NS3 and all other published helicase domain structures, where the loop is structurally much more conserved (Fig. 4c). The conformation of this loop in DENV4 NS2B18NS3 is likely to restrict the conformational freedom of helicase domain 2 and may therefore adversely affect NS3 helicase activity. In contrast, for the MVEV structure, these interactions are not present, leading to a more exposed NTP binding site and a lack of constraints on the R461-D471 loop (Fig. 4a).
Possible biological significance of the MVEV and DENV4 NS2B-NS3 conformations.
Our study has revealed a novel conformational state of a flavivirus NS3 protein where the protease and NTPase/helicase domains are spatially separated. In line with this, biochemical analyses of the helicase, ATPase, and protease activities suggest that the domains do not exert a significant effect on each other's activities. The individual MVEV helicase and protease domains are structurally similar to other flavivirus NS3 helicase and protease domains and to those of DENV4 NS2B-NS3. The crucial difference between the DENV4 and MVEV NS2B-NS3 structures is a large change in the relative orientations of the two domains. It seems likely that the flexible nature of the linker region and the small protease/helicase interfaces in both configurations allow their interconversion in solution. In other words, our analysis strongly suggests that the flavivirus NS3 protease and helicase domains are loosely tethered.
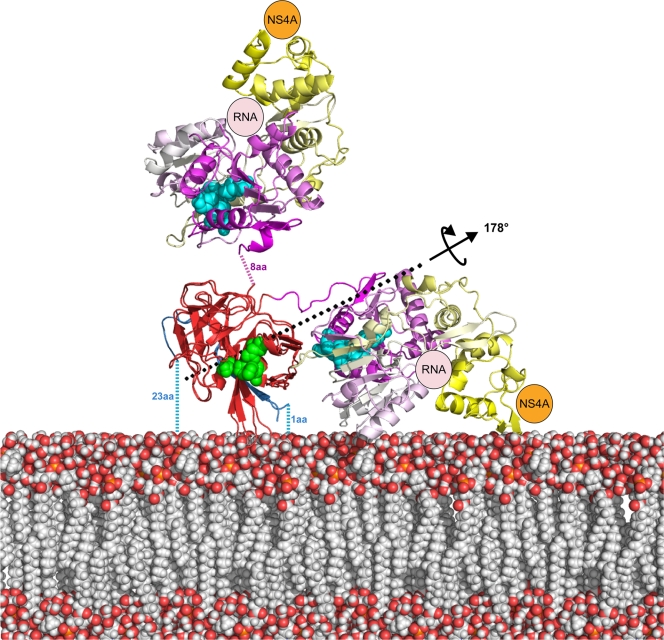
This does not resolve the question of what is the functional relevance, if any, of the yoking of two uncoupled activities within a single polypeptide chain. NS3 plays a major role in polyprotein processing, genome replication, and viral packing (44). Polyprotein processing and replication are thought to occur in adjacent but distinct compartments: convoluted membranes and vesicle packets, respectively (for a review, see reference 36). In both these processes, it is likely that the activities are performed in the vicinity of membranes. As outlined above, NS3 and NS2B associate with membranes yielding a rather rigidly membrane-bound NS3pro domain, especially in the fully activated state of NS3pro (Fig. 5). Figure 6 shows a cartoon representation of the MVEV and DENV4 NS2B-NS3 crystal structures with their protease domains superimposed and bound to the membrane, where the membrane positioning is based on the anchoring illustrated in Fig. 5c. This model for a possible structural organization of the NS2B-NS3 molecule at the cellular membrane suggests that the DENV4 NS2B18NS3 helicase is positioned very near the membrane, with the active site oriented toward the membrane and with very little space to accommodate RNA, while in the MVEV structure, the helicase domain is positioned away from the membrane, with the active site facing the cytoplasm. In both cases the protease active site is freely accessible. This might argue that the DENV4 conformation is not physiologically relevant (because the conformational flexibility of the NS3 linker may prevent its adoption in vivo); however, it is important that NS3 is also membrane anchored at its C terminus via its coupling to the membrane-bound NS4A (prior to the cleavage of the NS3-NS4A junction). This implies that additional in vivo constraints that probably restrict the conformational freedom of the NS3hel domain are present. Since the conformation of NS4A is not known, we cannot exclude the possibility that the DENV4 conformation may occur in vivo. Indeed, NS3 might adopt a DENV4-like conformation during polyprotein processing, where the activity of the C-terminal helicase is not needed. Indirect support for this possibility comes from the previously reported observation that NS2B is probably not part of the replication complex (36), which is instead formed by NS1, NS2A, NS3, NS4A, and NS5 (36) (Fig. 1), implying that the formation of the replication complex results in the inactivation of the NS3pro domain. Maintaining the helicase in an inactive conformation until it is required for replication could control the switch from polyprotein processing to replication, explaining why NS3 harbors two distinct activities within a single polypeptide chain. However, the significant gaps in our understanding of the role of the nonstructural proteins means that this remains a speculation, and an additional possibility is that linker flexibility establishes the appropriate interactions between NS3 and other nonstructural proteins as well as RNA. In particular, helicase domains 2 and 3 of NS3, constituting the most-C-terminal part of NS3 (Fig. 4c), have been implicated in interactions with other nonstructural proteins, and establishing these may require a flexible NS3hel arrangement.
FIG. 6.
Model for the structural organization of MVEV NS2B45NS3 and DENV4 NS2B18NS3 at the cellular membrane. Positioning on the membrane is based upon the tight anchoring of NS2B-NS3 at three points, at the NS2B N- and C-terminal transmembrane helices and at the NS3pro hydrophobic helix, as predicted by the structure of WNV NS2B45NS3pro-Naph-KKR-H and as shown in Fig. 5c. After positioning of WNV NS2B45NS3pro-Naph-KKR-H on the membrane, MVEV NS2B45NS3 and DENV4 NS2B18NS3 were superimposed based on the protease domain alone. A model for the membrane is shown as van der Waals balls and, atomic structures are shown in a cartoon representation and color coded according to the following convention: NS2B (blue), NS3pro (red), and NS3hel by subdomains (domain 1, magenta; domain 2, white; domain 3, yellow). NS4A (shown schematically in orange) was positioned at the NS3 C terminus (domain 3), and the RNA (shown schematically in pink) is positioned in the ssRNA binding groove (shown to be bound in DENV4NS3hel+ADP+ssRNA [PDB accession number 2jlz]). The NS3pro (green) and NS3hel catalytic pockets are shown in a ball representation. The position of hinge axis is shown. aa, amino acids.
Our data offer insights into the puzzle of the structural organization of the flavivirus NS3 protein. We propose that NS3 can assume a range of dramatically distinct conformations defined by the relative positioning of the protease and helicase via a flexible interdomain linker. Different configurations in NS3 may be important for modulating its catalytic activities in distinct locations during the virus life cycle, specifically for its membrane-anchored cis- and trans-protease activity and the switch to helicase and triphosphatase activities during replication, and possibly other functions.
Acknowledgments
We thank the staff at ID29 European Synchrotron Radiation Facility Grenoble for support with data collection, Joanne Nettleship for help with the mass spectrometry analysis, and R. Hurrelbrink and S. Fuller for useful discussions. We gratefully acknowledge Roman Tuma's help with Fig. 3b.
E.J.M. is supported by the Royal Society, and D.I.S., J.M.G., and the Oxford Protein Production Facility are supported by the UK Medical Research Council and European Commission grants LSHGCT-2006-031220 (SPINE2 COMPLEXES) and LSHG-CT-2004-511960 (VIZIER).
Footnotes
Published ahead of print on 30 September 2009.
REFERENCES
- 1.Adams, P. D., R. W. Grosse-Kunstleve, L. W. Hung, T. R. Ioerger, A. J. McCoy, N. W. Moriarty, R. J. Read, J. C. Sacchettini, N. K. Sauter, and T. C. Terwilliger. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58:1948-1954. [DOI] [PubMed] [Google Scholar]
- 2.Aleshin, A. E., S. A. Shiryaev, A. Y. Strongin, and R. C. Liddington. 2007. Structural evidence for regulation and specificity of flaviviral proteases and evolution of the Flaviviridae fold. Protein Sci. 16:795-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arias, C. F., F. Preugschat, and J. H. Strauss. 1993. Dengue 2 virus NS2B and NS3 form a stable complex that can cleave NS3 within the helicase domain. Virology 193:888-899. [DOI] [PubMed] [Google Scholar]
- 4.Assenberg, R., J. Ren, A. Verma, T. S. Walter, D. Alderton, R. J. Hurrelbrink, S. D. Fuller, S. Bressanelli, R. J. Owens, D. I. Stuart, and J. M. Grimes. 2007. Crystal structure of the Murray Valley encephalitis virus NS5 methyltransferase domain in complex with cap analogues. J. Gen. Virol. 88:2228-2236. [DOI] [PubMed] [Google Scholar]
- 5.Benarroch, D., B. Selisko, G. A. Locatelli, G. Maga, J. L. Romette, and B. Canard. 2004. The RNA helicase, nucleotide 5′-triphosphatase, and RNA 5′-triphosphatase activities of dengue virus protein NS3 are Mg2+-dependent and require a functional Walker B motif in the helicase catalytic core. Virology 328:208-218. [DOI] [PubMed] [Google Scholar]
- 6.Bera, A. K., R. J. Kuhn, and J. L. Smith. 2007. Functional characterization of cis and trans activity of the flavivirus NS2B-NS3 protease. J. Biol. Chem. 282:12883-12892. [DOI] [PubMed] [Google Scholar]
- 7.Berrow, N. S., D. Alderton, S. Sainsbury, J. Nettleship, R. Assenberg, N. Rahman, D. I. Stuart, and R. J. Owens. 2007. A versatile ligation-independent cloning method suitable for high-throughput expression screening applications. Nucleic Acids Res. 35:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bollati, M., M. Milani, E. Mastrangelo, S. Ricagno, G. Tedeschi, S. Nonnis, E. Decroly, B. Selisko, X. de Lamballerie, B. Coutard, B. Canard, and M. Bolognesi. 2009. Recognition of RNA cap in the Wesselsbron virus NS5 methyltransferase domain: implications for RNA-capping mechanisms in flavivirus. J. Mol. Biol. 385:140-152. [DOI] [PubMed] [Google Scholar]
- 9.Brass, V., J. M. Berke, R. Montserret, H. E. Blum, F. Penin, and D. Moradpour. 2008. Structural determinants for membrane association and dynamic organization of the hepatitis C virus NS3-4A complex. Proc. Natl. Acad. Sci. USA 105:14545-14550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brünger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54:905-921. [DOI] [PubMed] [Google Scholar]
- 11.Chambers, T. J., C. S. Hahn, R. Galler, and C. M. Rice. 1990. Flavivirus genome organization, expression, and replication. Annu. Rev. Microbiol. 44:649-688. [DOI] [PubMed] [Google Scholar]
- 12.Chappell, K. J., M. J. Stoermer, D. P. Fairlie, and P. R. Young. 2007. Generation and characterization of proteolytically active and highly stable truncated and full-length recombinant West Nile virus NS3. Protein Expr. Purif. 53:87-96. [DOI] [PubMed] [Google Scholar]
- 13.Chen, C. J., M. D. Kuo, L. J. Chien, S. L. Hsu, Y. M. Wang, and J. H. Lin. 1997. RNA-protein interactions: involvement of NS3, NS5, and 3′ noncoding regions of Japanese encephalitis virus genomic RNA. J. Virol. 71:3466-3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collaborative Computational Project, Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760-763. [DOI] [PubMed] [Google Scholar]
- 15.Cui, T., R. J. Sugrue, Q. Xu, A. K. Lee, Y. C. Chan, and J. Fu. 1998. Recombinant dengue virus type 1 NS3 protein exhibits specific viral RNA binding and NTPase activity regulated by the NS5 protein. Virology 246:409-417. [DOI] [PubMed] [Google Scholar]
- 16.Egloff, M. P., D. Benarroch, B. Selisko, J. L. Romette, and B. Canard. 2002. An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. EMBO J. 21:2757-2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Egloff, M. P., E. Decroly, H. Malet, B. Selisko, D. Benarroch, F. Ferron, and B. Canard. 2007. Structural and functional analysis of methylation and 5′-RNA sequence requirements of short capped RNAs by the methyltransferase domain of dengue virus NS5. J. Mol. Biol. 372:723-736. [DOI] [PubMed] [Google Scholar]
- 18.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126-2132. [DOI] [PubMed] [Google Scholar]
- 19.Erbel, P., N. Schiering, A. D'Arcy, M. Renatus, M. Kroemer, S. P. Lim, Z. Yin, T. H. Keller, S. G. Vasudevan, and U. Hommel. 2006. Structural basis for the activation of flaviviral NS3 proteases from dengue and West Nile virus. Nat. Struct. Mol. Biol. 13:372-373. [DOI] [PubMed] [Google Scholar]
- 20.Evans, P. R. 1997. SCALA. Joint CCP4+ESF-EAMCB Newslett. Protein Crystallogr. 1997:33. [Google Scholar]
- 21.Falgout, B., M. Pethel, Y. M. Zhang, and C. J. Lai. 1991. Both nonstructural proteins NS2B and NS3 are required for the proteolytic processing of dengue virus nonstructural proteins. J. Virol. 65:2467-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorbalenya, A. E., E. V. Koonin, A. P. Donchenko, and V. M. Blinov. 1989. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 17:4713-4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanley, K. A., J. J. Lee, J. E. Blaney, Jr., B. R. Murphy, and S. S. Whitehead. 2002. Paired charge-to-alanine mutagenesis of dengue virus type 4 NS5 generates mutants with temperature-sensitive, host range, and mouse attenuation phenotypes. J. Virol. 76:525-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johansson, M., A. J. Brooks, D. A. Jans, and S. G. Vasudevan. 2001. A small region of the dengue virus-encoded RNA-dependent RNA polymerase, NS5, confers interaction with both the nuclear transport receptor importin-beta and the viral helicase, NS3. J. Gen. Virol. 82:735-745. [DOI] [PubMed] [Google Scholar]
- 25.Kabsch, W. 1993. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26:795-800. [Google Scholar]
- 26.Kapoor, M., L. Zhang, M. Ramachandra, J. Kusukawa, K. E. Ebner, and R. Padmanabhan. 1995. Association between NS3 and NS5 proteins of dengue virus type 2 in the putative RNA replicase is linked to differential phosphorylation of NS5. J. Biol. Chem. 270:19100-19106. [DOI] [PubMed] [Google Scholar]
- 27.Khromykh, A. A., M. T. Kenney, and E. G. Westaway. 1998. trans-Complementation of flavivirus RNA polymerase gene NS5 by using Kunjin virus replicon-expressing BHK cells. J. Virol. 72:7270-7279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koonin, E. V. 1993. Computer-assisted identification of a putative methyltransferase domain in NS5 protein of flaviviruses and lambda 2 protein of reovirus. J. Gen. Virol. 74(Pt. 4):733-740. [DOI] [PubMed] [Google Scholar]
- 29.Krissinel, E., and K. Henrick. 2007. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372:774-797. [DOI] [PubMed] [Google Scholar]
- 30.Laskowski, R. A., D. S. Moss, and J. M. Thornton. 1993. Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol. 231:1049-1067. [DOI] [PubMed] [Google Scholar]
- 31.Lescar, J., D. Luo, T. Xu, A. Sampath, S. P. Lim, B. Canard, and S. G. Vasudevan. 2008. Towards the design of antiviral inhibitors against flaviviruses: the case for the multifunctional NS3 protein from dengue virus as a target. Antivir. Res. 80:94-101. [DOI] [PubMed] [Google Scholar]
- 32.Li, H., S. Clum, S. You, K. E. Ebner, and R. Padmanabhan. 1999. The serine protease and RNA-stimulated nucleoside triphosphatase and RNA helicase functional domains of dengue virus type 2 NS3 converge within a region of 20 amino acids. J. Virol. 73:3108-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lindenbach, B., H. Thiel, and C. Rice. 2007. Flaviviridae: the viruses and their replication, p. 1101-1152. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 34.Luo, D., T. Xu, C. Hunke, G. Gruber, S. G. Vasudevan, and J. Lescar. 2008. Crystal structure of the NS3 protease-helicase from dengue virus. J. Virol. 82:173-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luo, D., T. Xu, R. P. Watson, D. Scherer-Becker, A. Sampath, W. Jahnke, S. S. Yeong, C. H. Wang, S. P. Lim, A. Strongin, S. G. Vasudevan, and J. Lescar. 2008. Insights into RNA unwinding and ATP hydrolysis by the flavivirus NS3 protein. EMBO J. 27:3209-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mackenzie, J. 2005. Wrapping things up about virus RNA replication. Traffic 6:967-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mackenzie, J. S., D. J. Gubler, and L. R. Petersen. 2004. Emerging flaviviruses: the spread and resurgence of Japanese encephalitis, West Nile and dengue viruses. Nat. Med. 10:S98-S109. [DOI] [PubMed] [Google Scholar]
- 38.Mancini, E. J., R. Assenberg, A. Verma, T. S. Walter, R. Tuma, J. M. Grimes, R. J. Owens, and D. I. Stuart. 2007. Structure of the Murray Valley encephalitis virus RNA helicase at 1.9 Angstrom resolution. Protein Sci. 16:2294-2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mastrangelo, E., M. Milani, M. Bollati, B. Selisko, F. Peyrane, V. Pandini, G. Sorrentino, B. Canard, P. V. Konarev, D. I. Svergun, X. de Lamballerie, B. Coutard, A. A. Khromykh, and M. Bolognesi. 2007. Crystal structure and activity of Kunjin virus NS3 helicase; protease and helicase domain assembly in the full length NS3 protein. J. Mol. Biol. 372:444-455. [DOI] [PubMed] [Google Scholar]
- 40.Matusan, A. E., P. G. Kelley, M. J. Pryor, J. C. Whisstock, A. D. Davidson, and P. J. Wright. 2001. Mutagenesis of the dengue virus type 2 NS3 proteinase and the production of growth-restricted virus. J. Gen. Virol. 82:1647-1656. [DOI] [PubMed] [Google Scholar]
- 41.Matusan, A. E., M. J. Pryor, A. D. Davidson, and P. J. Wright. 2001. Mutagenesis of the dengue virus type 2 NS3 protein within and outside helicase motifs: effects on enzyme activity and virus replication. J. Virol. 75:9633-9643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCoy, M. A., M. M. Senior, J. J. Gesell, L. Ramanathan, and D. F. Wyss. 2001. Solution structure and dynamics of the single-chain hepatitis C virus NS3 protease NS4A cofactor complex. J. Mol. Biol. 305:1099-1110. [DOI] [PubMed] [Google Scholar]
- 43.Murray, C. L., C. T. Jones, and C. M. Rice. 2008. Architects of assembly: roles of Flaviviridae non-structural proteins in virion morphogenesis. Nat. Rev. Microbiol. 6:699-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patkar, C. G., and R. J. Kuhn. 2008. Yellow fever virus NS3 plays an essential role in virus assembly independent of its known enzymatic functions. J. Virol. 82:3342-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pyle, A. M. 2008. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu. Rev. Biophys. 37:317-336. [DOI] [PubMed] [Google Scholar]
- 46.Ray, D., and P. Y. Shi. 2006. Recent advances in flavivirus antiviral drug discovery and vaccine development. Recent Pat. Anti-Infect. Drug Discov. 1:45-55. [DOI] [PubMed] [Google Scholar]
- 47.Robin, G., K. Chappell, M. J. Stoermer, S. H. Hu, P. R. Young, D. P. Fairlie, and J. L. Martin. 2009. Structure of West Nile virus NS3 protease: ligand stabilization of the catalytic conformation. J. Mol. Biol. 385:1568-1577. [DOI] [PubMed] [Google Scholar]
- 48.Shiryaev, S. A., B. I. Ratnikov, A. V. Chekanov, S. Sikora, D. V. Rozanov, A. Godzik, J. Wang, J. W. Smith, Z. Huang, I. Lindberg, M. A. Samuel, M. S. Diamond, and A. Y. Strongin. 2006. Cleavage targets and the D-arginine-based inhibitors of the West Nile virus NS3 processing proteinase. Biochem. J. 393:503-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Speroni, S., L. De Colibus, E. Mastrangelo, E. Gould, B. Coutard, N. L. Forrester, S. Blanc, B. Canard, and A. Mattevi. 2008. Structure and biochemical analysis of Kokobera virus helicase. Proteins 70:1120-1123. [DOI] [PubMed] [Google Scholar]
- 50.Stuart, D. I., M. Levine, H. Muirhead, and D. K. Stammers. 1979. Crystal structure of cat muscle pyruvate kinase at a resolution of 2.6 A. J. Mol. Biol. 134:109-142. [DOI] [PubMed] [Google Scholar]
- 51.Tan, B. H., J. Fu, R. J. Sugrue, E. H. Yap, Y. C. Chan, and Y. H. Tan. 1996. Recombinant dengue type 1 virus NS5 protein expressed in Escherichia coli exhibits RNA-dependent RNA polymerase activity. Virology 216:317-325. [DOI] [PubMed] [Google Scholar]
- 52.Thibeault, D., M. J. Massariol, S. Zhao, E. Welchner, N. Goudreau, R. Gingras, M. Llinas-Brunet, and P. W. White. 2009. Use of the fused NS4A peptide-NS3 protease domain to study the importance of the helicase domain for protease inhibitor binding to hepatitis C virus NS3-NS4A. Biochemistry 48:744-753. [DOI] [PubMed] [Google Scholar]
- 53.Walter, T. S., J. M. Diprose, C. J. Mayo, C. Siebold, M. G. Pickford, L. Carter, G. C. Sutton, N. S. Berrow, J. Brown, I. M. Berry, G. B. Stewart-Jones, J. M. Grimes, D. K. Stammers, R. M. Esnouf, E. Y. Jones, R. J. Owens, D. I. Stuart, and K. Harlos. 2005. A procedure for setting up high-throughput nanolitre crystallization experiments. Crystallization workflow for initial screening, automated storage, imaging and optimization. Acta Crystallogr. D Biol. Crystallogr. 61:651-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Walter, T. S., E. J. Mancini, J. Kadlec, S. C. Graham, R. Assenberg, J. Ren, S. Sainsbury, R. J. Owens, D. I. Stuart, J. M. Grimes, and K. Harlos. 2008. Semi-automated microseeding of nanolitre crystallization experiments. Acta Crystallogr. F Struct. Biol. Cryst. Commun. 64:14-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wengler, G. 1991. The carboxy-terminal part of the NS 3 protein of the West Nile flavivirus can be isolated as a soluble protein after proteolytic cleavage and represents an RNA-stimulated NTPase. Virology 184:707-715. [DOI] [PubMed] [Google Scholar]
- 56.Wengler, G. 1993. The NS 3 nonstructural protein of flaviviruses contains an RNA triphosphatase activity. Virology 197:265-273. [DOI] [PubMed] [Google Scholar]
- 57.Whitehead, S. S., J. E. Blaney, A. P. Durbin, and B. R. Murphy. 2007. Prospects for a dengue virus vaccine. Nat. Rev. Microbiol. 5:518-528. [DOI] [PubMed] [Google Scholar]
- 58.Wu, J., A. K. Bera, R. J. Kuhn, and J. L. Smith. 2005. Structure of the flavivirus helicase: implications for catalytic activity, protein interactions, and proteolytic processing. J. Virol. 79:10268-10277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu, T., A. Sampath, A. Chao, D. Wen, M. Nanao, P. Chene, S. G. Vasudevan, and J. Lescar. 2005. Structure of the dengue virus helicase/nucleoside triphosphatase catalytic domain at a resolution of 2.4 Å. J. Virol. 79:10278-10288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamashita, T., H. Unno, Y. Mori, H. Tani, K. Moriishi, A. Takamizawa, M. Agoh, T. Tsukihara, and Y. Matsuura. 2008. Crystal structure of the catalytic domain of Japanese encephalitis virus NS3 helicase/nucleoside triphosphatase at a resolution of 1.8 A. Virology 373:426-436. [DOI] [PubMed] [Google Scholar]
- 61.Yao, N., P. Reichert, S. S. Taremi, W. W. Prosise, and P. C. Weber. 1999. Molecular views of viral polyprotein processing revealed by the crystal structure of the hepatitis C virus bifunctional protease-helicase. Structure 7:1353-1363. [DOI] [PubMed] [Google Scholar]
- 62.Yon, C., T. Teramoto, N. Mueller, J. Phelan, V. K. Ganesh, K. H. Murthy, and R. Padmanabhan. 2005. Modulation of the nucleoside triphosphatase/RNA helicase and 5′-RNA triphosphatase activities of dengue virus type 2 nonstructural protein 3 (NS3) by interaction with NS5, the RNA-dependent RNA polymerase. J. Biol. Chem. 280:27412-27419. [DOI] [PubMed] [Google Scholar]