Abstract
Endogenous retroviruses present in the human genome provide a rich record of ancient infections. All presently recognized elements, including the youngest and most intact proviruses of the human endogenous retrovirus K(HML-2) [HERV-K(HML-2)] family, have suffered postinsertional mutations during their time of chromosomal residence, and genes encoding the envelope glycoprotein (Env) have not been spared these mutations. In this study, we have, for the first time, reconstituted an authentic Env of a HERV-K(HML-2) provirus by back mutation of putative postinsertional amino acid changes of the protein encoded by HERV-K113. Aided by codon-optimized expression, we demonstrate that the reconstituted Env regained its ability to be incorporated into retroviral particles and to mediate entry. The original ancient HERV-K113 Env was synthesized as a moderately glycosylated gp95 precursor protein cleaved into surface and transmembrane (TM) subunits. Of the nine N-linked oligosaccharides, four are part of the TM subunit, contributing 15 kDa to its apparent molecular mass of 41 kDa. The carbohydrates, as well as the cytoplasmic tail, are critical for efficient intracellular trafficking, processing, stability, and particle incorporation. Whereas deletions of the carboxy-terminal 6 residues completely abrogated cleavage and virion association, more extensive truncations slightly enhanced incorporation but dramatically increased the ability to mediate entry of pseudotyped lentiviruses. Although the first HERV-K(HML-2) elements infected human ancestors about 30 million years ago, our findings indicate that their glycoproteins are in most respects remarkably similar to those of classical contemporary retroviruses and can still mediate efficient entry into mammalian cells.
Retroviruses that infect germ line cells or their precursors can become vertically transmitted genetic elements and spread in a host population during subsequent generations (35). About 8% of the human genome consists of stably integrated endogenous retroviruses acquired during early and more recent evolution by our primate and hominid ancestors. These fossils are grouped into several distinct families (2, 42). Their sequences and genomic structures typically resemble those of one of the genera of current exogenous retroviruses, providing an exceptional archive for the study of many aspects of viral and host coevolution and its dynamics (25). In contrast to humans, several animals, including mice and sheep, contain replication-competent present-day exogenous and endogenized forms of the same retrovirus (1, 10). The most striking example is the koala, a species with a profound ongoing endogenization burst with a highly oncogenic gammaretrovirus (52).
The most recently integrated human elements belong to the betaretrovirus-like human endogenous retrovirus K(HML-2) [HERV-K(HML-2)] family (7, 25, 54). Infectious viruses of this family appear to have started invading the chromosomes before the evolutionary split of Old World monkeys and hominoids about 30 million years ago (41, 50). Several of its members are human specific, indicating continuing active replication in our ancestors following the deviation of the chimpanzee lineage 5 to 6 million years ago (4, 7, 12). The recent acquisition of several HERV-K(HML-2) elements is further substantiated by the lack of fixation (not present in all individuals) (6, 54). However, during their residency in the host genome, every one of the currently known proviruses has suffered from mutations, deletions, or recombination events. In many cases, homologous recombination has left only a single long terminal repeat at the integration site (31). None of the more complete proviruses appear to be replication competent, although some of them have retained the capacity to form particles (8, 11). Recently, infectious HERV-K(HML-2) viruses have been produced by generating consensus sequences based on human-specific elements and, in an alternative approach, by assembling the functional regions of three authentic proviruses into a single element (21, 39). These studies clearly demonstrate that (i) HERV-K(HML-2) is able to form viral particles infecting human cells and (ii) certain recombination events, such as template switching during reverse transcription or gene conversion, might reestablish fully functional chimeric HERV-K(HML-2) elements by combining conserved sequences from partially crippled proviruses.
One of the best preserved full-length HERV-K(HML-2) elements is HERV-K113 (5, 54). Due to its low prevalence of less than 20%, it escaped recognition by the Human Genome Project, and the provirus was identified in a human bacterial artificial chromosome library on chromosome 19p13.11 of an unknown DNA donor (54). As previously reported by us and by others, despite a functional long terminal repeat promoter and open reading frames for all proteins, a few substitutions in the reverse transcriptase and one critical substitution in the gag gene of the provirus contribute to its lack of replication (5, 29, 54). In addition, the envelope protein of the cloned HERV-K113 provirus is not incorporated into viral particles, suggesting further postinsertional damage by mutation(s) (5, 20). These changes are not necessarily present in all HERV-K113 variants in the human population, since some degree of polymorphism between carriers from different ethnic groups can be expected (44). Thus, functional HERV-K113 envelope genes might still exist in some humans, as has been demonstrated for the related HERV-K108 element (20).
Retroviral envelope proteins are synthesized as fusion-inactive precursor glycoproteins, which trimerize and are cleaved by furin-like endopeptidases in the Golgi compartment of the cell into surface (SU) and transmembrane (TM) subunits (32, 38). The mature N-glycosylated trimeric proteins eventually reach the cell membrane, where they become incorporated into particles (17). The cytoplasmic domain of the protein has been shown to be critical for the transport, incorporation, and fusogenicity of virions (9, 13, 15, 37). Some retroviruses release a short carboxy-terminal R peptide, a step necessary for full fusogenic activity of the glycoprotein (27). With the exception of the syncytin-1 and -2 glycoproteins, which are the fusogenic envelope proteins of HERV-W and HERV-FRD elements involved in the formation of the placental syncytiotrophoblast layer, the envelope proteins of other HERVs are poorly characterized (14).
In the present report, we identify inactivating mutations in the envelope gene of a present day HERV-K113 provirus. All or almost all of the changes most likely occurred gradually following integration in the genome more than a million years ago. In this case, the already endogenous virus was probably able to replicate for a certain time, although damaging preintegrational mutations introduced by the RNA polymerase or during reverse transcription by errors of the enzyme and by the action of APOBEC proteins cannot be completely ruled out. We reconstituted the original sequence presumed to exist at the time of integration. Aided by codon-optimized expression, we investigated various aspects of the glycoprotein and carboxy-terminal truncation mutants in terms of cleavage, N-glycosylation, incorporation into viral particles, and mediation of viral entry into target cells.
MATERIALS AND METHODS
Cell culture.
HEK 293T, Tera-1, HeLa, and Crandell feline kidney (CrFK) cells were cultured in complete Dulbecco's modified Eagle medium (DMEM) containing 10% fetal bovine serum, penicillin (50 U/ml), streptomycin (50 μg/ml), and l-glutamine (2 mM).
Codon optimization, mutagenesis, and cloning of Env constructs.
A synthetic version of the HERV-K113 (GenBank AY037928) Env protein, optimized for expression in mammalian cells, was designed and synthesized by Geneart (Regensburg, Germany). The codon-optimized sequence (coEnv) (see Fig. S1 in the supplemental material) was cloned into a C-terminal V5-tag-containing pcDNA3 expression vector (Invitrogen) using the EcoRI/NotI cleavage sites. The construct for the expression of the putative ancestral (original) oricoEnv-V5 (pcDNAoricoEnv-V5), as well as all derived mutants, was generated using the QuikChange Multi Site-Directed Mutagenesis Kit or the QuikChange Site-Directed Mutagenesis Kit (Stratagene) and the primer pairs listed in Fig. S2 in the supplemental material. These mutants include the N-glycosylation-deficient variant oricoEnvGly−; the cleavage site mutants oricoEnvCS− and oricoEnvCS_MMTV; the C-terminal truncation mutants oricoEnvΔ659-699, oricoEnvΔ680-699, and oricoEnvΔ693-699; and the envelope proteins without a V5 tag. The last four were generated by the introduction of stop codons at the respective positions in pcDNAoricoEnv-V5. All mutations were confirmed by DNA sequencing.
Envelope expression analysis.
For Env expression analysis, 4 × 106 cells grown in 100-mm dishes were transfected with the appropriate expression plasmid using the calcium phosphate method. The cells were lysed in 500 μl lysis buffer (1% Triton X-100, 50 mM Tris HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, and a protease inhibitor mixture [Roche]), and cell debris was removed by centrifugation. For immunoprecipitation experiments, 25 μl of αV5 Sepharose (Sigma) was added to the lysate and incubated on an overhead shaker for 4 h. The Sepharose beads were washed five times with washing buffer (1% Triton X-100, 50 mM Tris HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, 0.02% sodium dodecyl sulfate, and a protease inhibitor mixture), mixed with Laemmli sample buffer (Bio-Rad), briefly boiled, and pelleted. For simple immunoblotting, the cleared lysates were directly mixed with sample buffer and cleared by centrifugation before being subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The separated proteins were transferred to a polyvinylidene difluoride membrane (Roth) using the semidry transfer method. The membranes were blocked in blocking buffer (phosphate-buffered saline [PBS], 5% skim milk powder, 0.1% Tween) and finally incubated with specific antibodies. An αV5-horseradish peroxidase (HRP) antibody (Invitrogen) was used for quantification. It was detected by a secondary Alexa Fluor 680 donkey anti-mouse immunoglobulin G before measurement of the band fluorescence intensity using an Odyssey Scanning and Analysis (LI-COR) unit. In experiments without quantification, αV5-HRP or TM-specific (HERM-1811-5; Austral Biologicals) monoclonal antibodies were used in conjunction with an HRP-conjugated secondary anti-mouse antibody. Bands were visualized using enhanced chemiluminescence reagents (Pierce).
Immunofluorescence microscopy.
Cells (1 × 105) were grown in chamber slides (Nalge Nunc) and transfected with 0.2 μg of plasmid DNA using Effectene reagent (Qiagen). After 48 h, the cells were fixed with 2% paraformaldehyde in PBS for 30 min, rinsed briefly with PBS, permeabilized with PBS-buffered 0.5% Triton X-100, and again washed three times with PBS. After being blocked with Marvel (1% nonfat milk in PBS) for 20 min, the cells were incubated with primary antibodies diluted in blocking buffer for 60 min at 37°C. HERM-1811-5 (Austral Biologicals) or αV5 (Serotec) was used as a primary antibody. The slides were washed three times with PBS, and a Cy3-conjugated mouse-specific secondary antibody (Sigma) diluted 1:200 was added for 30 min. After extensive washing, the cells were mounted in Mowiol and examined using a Zeiss LSM 510 confocal laser scanning microscope.
Deglycosylation experiments.
Peptide-N-glycosidase F (PNGase F) deglycosylation experiments were performed with precipitated V5-tagged envelope proteins immunoprecipitated from lysates of transfected cells grown in six-well plates. The cells were lysed 48 h posttransfection in 200 μl lysis buffer. The precipitates were briefly boiled in 1× denaturation buffer at 98°C for 10 min, and 3 μl 10× G7 buffer (New England Biolabs), 3 μl 10% NP-40, and 1,500 units of PNGase F (New England Biolabs) were added for digestion. The tubes were incubated for various times at 37°C, and the deglycosylation reaction was stopped by adding Laemmli sample buffer (Bio-Rad). The proteins were immunoblotted using the αV5-HRP specific antibody (Invitrogen).
To inhibit intracellular N-glycosylation, cells were preincubated for 2.5 h with DMEM containing 30 or 50 μg/ml of tunicamycin. After the pretreatment, the cells were transfected using the Polyfect Transfection Reagent Kit (Qiagen) and incubated for an additional 24 h in medium containing tunicamycin before lysis and immunoblotting.
Production of pseudotyped SHIV reporter viruses.
The simian immunodeficiency virus (SIVmac)-based retroviral vectors pSIvec1ΔenvLuc and pSIvec1ΔenvGFP, the human immunodeficiency virus type 1 (HIV-1) Rev-expressing plasmid pCMV-Rev, and the plasmid pSVIII-ΔKS (expressing a nonfunctional HIV-1 envelope to allow nonspecific virus uptake to be determined) have been described elsewhere (3). To generate infectious simian-human immunodeficiency virus (SHIV) particles capable of a single round of infection, 4 × 106 HEK 293T cells were transfected with 15 μg of pSIvec1ΔenvLuci, 5 μg pCMV-Rev, and 0.5 μg of an envelope expression vector in 10-cm dishes using the calcium phosphate method. At 2 days posttransfection, the supernatants were harvested, centrifuged at 6,000 rpm for 10 min, and filtered through 0.45-μm-pore-size membranes to remove residual cells. Gag-specific enzyme-linked immunosorbent assays and reverse transcriptase activity (GE Healthcare) were used to assess and normalize virus preparations.
Ultracentrifugation of cell supernatants.
Viral particles were concentrated by ultracentrifugation of cell supernatants through a 20% sucrose cushion for 2 h at 4°C and 175,000 × g. The viral pellet was resuspended in 50 μl of 0.05 M HEPES, pH 7.2.
Infectivity assays.
For entry experiments, CrFK target cells were sown in six-well plates (600,000 cells/well). The next day, the cells were rinsed with PBS and incubated with 400 μl normalized virus supernatant and 200 μl DMEM for 2 h before the addition of 1.5 ml DMEM and culturing for 2 days. Infection was determined by measuring the luciferase activity in cell lysates using the Luciferase Assay System (Promega) according to the manufacturer's instructions.
RESULTS
Enhanced expression of the HERV-K113 envelope protein.
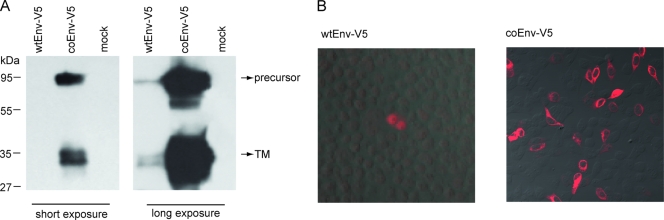
The Env protein of HERV-K(HML-2) is translated from a singly spliced transcript. A second splicing event removing large parts of the coding region resulted in the mRNA for the nucleocytoplasmic shuttle protein Rec at the expense of Env (Fig. 1). This internal splicing, the low GC content of 42%, and the use of rare codons are the most likely causes for the poor expression of the HERV-K(HML-2) envelope protein from cytomegalovirus promoter-driven constructs (5). To overcome this limitation of the Env of HERV-K113, which significantly hampers the functional characterization of the protein, we generated a synthetic version optimized for expression in mammalian cells by synonymous exchange of 71% of the codons (no change in amino acid sequence) and by raising the GC content of the sequence to 63%. The codon optimization and the concomitant removal of the internal splice sites resulted in a drastic increase in expression (over 50-fold), as demonstrated by immunoprecipitation (Fig. 2A) of the glycoproteins from transiently transfected HEK 293T cells and by immunofluorescence microscopy (Fig. 2B). For immunoprecipitation and detection purposes, a V5 tag that increases the molecular mass by 1.7 kDa was fused to the C terminus of wild-type Env (wtEnv) and coEnv. On immunoblots, specific bands with apparent molecular masses in the range of about 75 to 95 kDa and 32 to 38 kDa were visible (Fig. 2A), sizes that match those expected for the glycosylated precursor protein and the TM subunit (5).
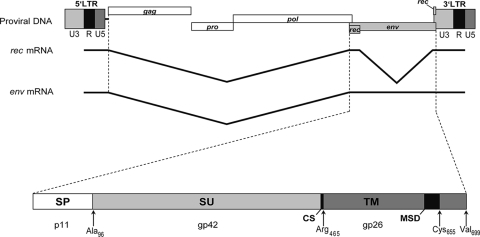
FIG. 1.
Genomic organization of the HERV-K113 provirus and its envelope protein. The Env precursor protein is translated from a singly spliced RNA and is composed of the N-terminal signal peptide (SP), the SU glycoprotein, and the TM subunit comprising the hydrophobic membrane-spanning domain (MSD). The cleavage site (CS) for cellular furin proteases is indicated. The theoretical molecular masses of each subunit (SP, TM, and SU), as well as the last amino acid of relevant domains, are shown.
FIG. 2.
Expression of wtEnv and coEnv. HEK 293T cells were transfected with pcDNAwtEnv-V5 and pcDNAcoEnv-V5. (A) Western blot analyses of immunoprecipitates using an αV5-HRP antiserum (1:5,000). Assumed precursor proteins at 75 to 90 kDa and TM subunits at 32 to 38 kDa are visible. Quantification of the band intensities indicated an increase in expression of 50-fold or more resulting from Env codon optimization. (B) Immune fluorescence analysis of wtEnv and coEnv using an αV5-Cy3 antibody. The signal intensity and the number of cells expressing Env were significantly higher in cells transfected with coEnv.
Reconstitution of the ancestral sequence of the HERV-K113 envelope protein.
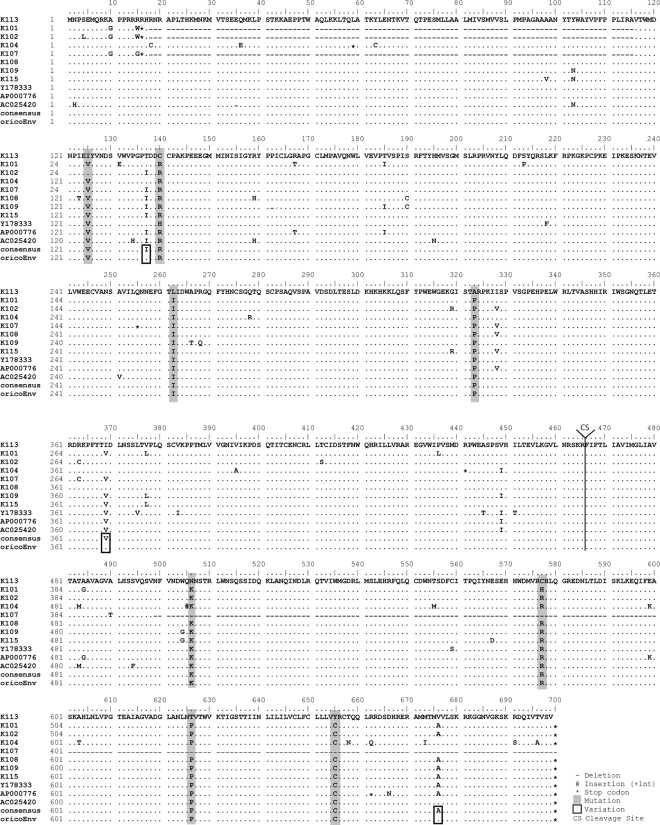
We and others have previously reported that the HERV-K113 envelope protein is not incorporated into viral particles (5, 20), and it was assumed that the cause lay in postinsertional mutations acquired during its time in the human genome. In an approach similar to that recently used to generate two related HERV-K(HML-2) consensus sequences (21, 39), we aligned the amino acid sequence of HERV-K113 Env with the corresponding sequences of 10 well-preserved human-specific elements belonging to this family to identify and repair postinsertional amino acid substitutions in the HERV-K113 coEnv protein (Fig. 3). To differentiate between authentic polymorphisms in the integrating ancient viruses and postinsertional changes, we assumed a nonsynonymous postinsertional mutation if none or only one of the aligned proviruses matched the amino acid of HERV-K113 and assumed a shared polymorphism if two or more of the elements had the same amino acid as HERV-K113, even if it was different from the consensus sequence. Three such variable positions (T137, I369, and V676) were identified and left unchanged (Fig. 3). In contrast, eight putative protein-relevant postinsertional substitutions were identified, four in the SU and four in the TM region (Fig. 3). Using site-directed mutagenesis, we reversed these 8 amino acids in the pcDNAcoEnv-V5 plasmid, taking the consensus amino acids as the originals. The resulting construct, pcDNAoricoEnv-V5, encodes a V5-tagged version of the putatively ancestral HERV-K113 envelope protein existing at the time of integration into a human ancestor.
FIG. 3.
Alignment of the amino acid sequences of 11 HERV-K(HML-2) envelope proteins. The sequences of HERV-K101 (AF164609), K102 (AF164610), K104 (AC116309), K107 (AF164613), K108 (AC072054), K109 (AC055116), K115 (AY037929), Y178333, AP000776, and AC025420 are compared to the K113 (AY037928) wtEnv sequence. The consensus sequence and the putative original sequence (oricoEnv) obtained by applying an algorithm to identify shared polymorphisms (see Results for details) are included. The consensus and oricoEnv sequences differ at positions 137, 369, and 676 due to shared polymorphisms (highlighted by rectangles). The eight sites in HERV-K113 wtEnv changed to the consensus sequence in order to reconstitute the ancestral protein sequence are highlighted in gray.
The original HERV-K113 envelope protein is moderately N-glycosylated.
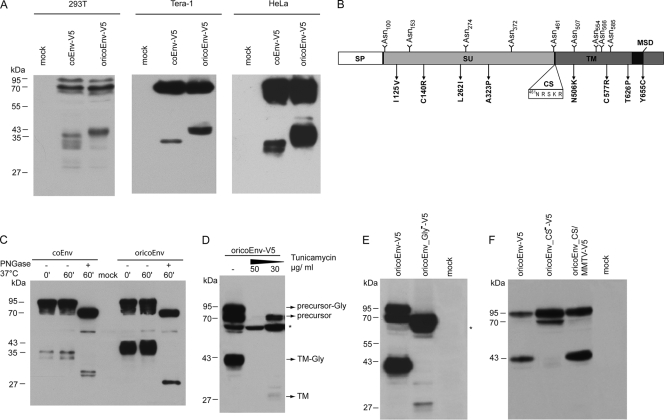
We first compared the expression of coEnv (present day) and oricoEnv (original) by immunoblotting of lysates from transfected HEK 293T, Tera-1, and HeLa cells. Although the two glycoproteins had similar levels of expression, considerable differences in the processing and migration patterns of the presumed TM subunits were seen. Although the ratio of precursor to TM varied somewhat from lysate to lysate, oricoEnv was more efficiently cleaved than coEnv in all experiments. The presumed TM subunit of oricoEnv-V5 migrated as a 43-kDa band in all cells tested. In contrast, coEnv-V5 was very inefficiently cleaved into a 35-kDa protein in Tera-1 cells, and in HEK 293T and HeLa cells, a fuzzy doublet of about 33 and 35 kDa could be seen (Fig. 4A).
FIG. 4.
Reconstitution of the ancient K113 oricoEnv sequence alters TM subunit processing. (A) Western blot analysis of coEnv-V5 and oricoEnv-V5 expressed in HEK 293T, Tera-1, and HeLa cells. (B) Positions of the potential glycosylation sites (NXS/T) within the Env molecule. The locations of the cleavage site (CS) and of amino acid changes resulting in oricoEnv are indicated. (C) PNGase F treatment of cell lysates from HeLa cells expressing coEnv-V5 and oricoEnv-V5. The lysates were incubated with and without PNGase F at 37°C for 1 h. The treated samples showed a shift in the precursor and TM bands. (D) Incubation of HEK 293T cells expressing oricoEnv-V5 with tunicamycin (30 and 50 μg/ml). The generation of a 64-kDa N-terminal Env precursor degradation product (marked by an asterisk) was enhanced by tunicamycin treatment. (E) Elimination of the predicted glycosylation sites by mutagenesis results in a decrease in the molecular masses of the precursor and TM subunit and inhibits processing. (F) Analysis of cleavage site mutants in HEK 293T cells. In oricoEnv_CS−-V5, the cleavage site sequence 461NRSKR was changed to 461NASAA. OricoEnv_CS/MMTV-V5 carries a mutation (461NRSKR to 461NRAKR) that prevents potential glycosylation at N461 and matches the MMTV cleavage site.
Differences in the carbohydrate compositions of the two proteins were considered to be a likely reason for the discrepancy in size. coEnv contains 10 consensus NXS/T motifs for N-linked glycosylation (23, 55), with two of the sites (N506 and N507) being adjacent (Fig. 3). The N506K back mutation altered this motif, giving nine potential sites for N-linked carbohydrates in oricoEnv: five in the SU subunit and four in the ectodomain of TM (Fig. 4B). Treatment of precipitated glycoproteins from transfected HeLa cells with PNGase F, which removes all N-linked glycans, decreased the apparent molecular mass of the presumed oricoEnv-V5 TM subunit to 28 kDa (Fig. 4C), the size calculated for the nonglycosylated TM-V5 subunits. However, the supposed TM doublet of coEnv-V5 was also shifted by PNGase F treatment, although the deglycosylated proteins clearly migrated above 28 kDa, indicating further N-terminal processing sites and a lower carbohydrate content of the TM subunit than that of oricoEnv. The precursor masses of approximately 95 and 70 kDa are consistent with the molecular masses of the glycosylated and nonglycosylated proteins (the theoretical mass of the unmodified protein without a signal peptide is 68 kDa).
To ascertain the nature of the detected protein bands, we incubated pcDNAoricoEnv-V5-transfected HEK 293T cells with tunicamycin, an agent that prevents the transfer of saccharidic units to the nascent peptide chain in the endoplasmic reticulum. Immunoblot analysis showed that tunicamycin treatment resulted in the disappearance of the glycosylated precursor and TM bands, and instead, a protein pattern similar to that observed after PNGase F treatment (a 70-kDa precursor and 28-kDa TM) was seen (Fig. 4D). The TM protein in tunicamycin-treated cells was barely visible, indicating an inefficient furin protease-mediated cleavage of the nonglycosylated protein. This assumption was further strengthened by the expression and immunoblot analysis of a mutant oricoEnv protein with all nine potential N-glycosylation sites mutated by changing the serine or threonine residue of the NXS/T motif into alanine. The oricoEnvGly− protein showed a band pattern similar to that already described for cells treated with moderate levels of tunicamycin (Fig. 4E). Together, these results illustrate that prevention of N-linked glycosylation results in a 70-kDa precursor protein whose processing is extremely inefficient. Furthermore, the strength of a specific 64-kDa band increases with treatment. This protein appears to represent an N-terminal truncation product of the unglycosylated precursor, suggesting a protective role for the N-terminal glycans against cellular proteases and degradation.
Interestingly, the position of one of the potentially glycosylated asparagines (N461) is only 5 amino acids upstream of the predicted furin cleavage site (Fig. 4B). An N-glycan at this location might influence the processing efficiency of the precursor protein. To test this by preventing glycosylation at this position, we replaced the serine in the 461NRSKR sequence with alanine (461NRAKR). This mutation obliterated the glycosylation signal and changed the supposed cleavage site motif of oricoEnv into a sequence matching that of the closely related betaretrovirus mouse mammary tumor virus (MMTV) (Fig. 4B). As shown in Fig. 4F, this substitution had no significant effect on the processing efficiency. In contrast, replacing the positively charged amino acids in the cleavage site with alanine (the 461NASAA mutant) completely abolished the release of the 43-kDa TM subunit. This result therefore implies either that N461 is not glycosylated or that the carbohydrate at this location neither inhibits nor enhances processing. Furthermore, the 461NASAA mutant provides further evidence that the 43-kDa TM protein is generated by processing at the assumed SU/TM cleavage site.
Incorporation of the reconstituted HERV-K113 Env into lentiviral particles is modulated by C-terminal sequences.
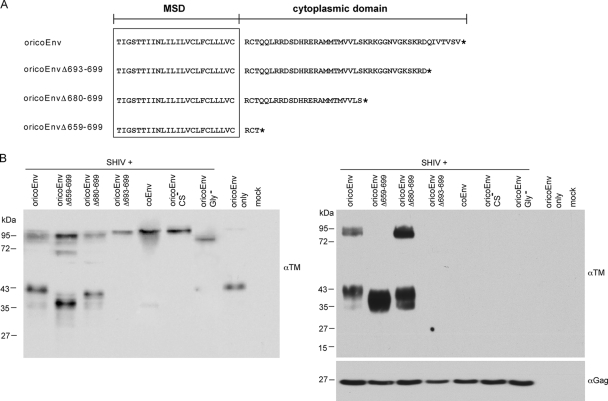
We next investigated whether the HERV-K113 oricoEnv protein is incorporated into lentiviral particles. Together with vectors for the expression of the lentiviral proteins and the packaged RNA containing a luciferase gene, 0.5 μg of an Env plasmid of choice was included to produce pseudotyped SHIV reporter particles in transfected HEK 293T cells, as described previously (3). Furthermore, for several retroviruses, a C-terminal truncation of the cytoplasmic tail has been shown to influence particle incorporation and/or fusogenic properties of the envelope protein (13-15). We therefore created three mutants with C-terminal truncations, namely, oricoEnvΔ659-699, oricoEnvΔ680-699, and oricoEnvΔ693-699 (Fig. 5A), and included them with the cleavage- and glycosylation-minus mutants oricoEnvGly− and oricoEnvCS− in the experiments. All of the proteins were expressed at approximately equivalent levels in HEK 293T cells (Fig. 5B). The Env proteins were detected using a commercially available monoclonal antibody that recognizes an epitope in the ectodomain of the TM subunit, and the changes in size of the truncated TM subunits were noticeable for oricoEnvΔ659-699 and oricoEnvΔ680-699 (Fig. 5). Interestingly, no processing occurred with the oricoEnvΔ693-699 mutant that lacks just 7 amino acids (Fig. 5B, left). To examine incorporation into virions, the supernatants were harvested, carefully clarified, and concentrated by ultracentrifugation through a 20% sucrose cushion. Immunoblot analysis of the pellets using the TM-specific antibody revealed various degrees of incorporation into SHIV particles (Fig. 5B, right). Whereas no incorporation was detectable for coEnv, oricoEnvΔ693-699, and the glycosylation- and cleavage-deficient oricoEnv mutants, substantial incorporation of oricoEnvΔ680-699 and oricoEnvΔ659-699 was observed at levels approximately two and five times higher, respectively, than those of the parental oricoEnv. It is of particular interest that the mutant almost completely lacking the cytoplasmic tail (oricoEnvΔ659-699) was exclusively incorporated as the cleaved glycoprotein.
FIG. 5.
Incorporation of oricoEnv and C-terminal truncation mutants into lentiviral particles. (A) The amino acid sequences of the membrane-spanning domain (MSD) (boxed) and the cytoplasmic region are shown. The Env truncation mutants are named according to the number of amino acids deleted by the introduction of a stop codon (*). (B) On the left is an immunoblot of HEK 293T cells transfected with vectors used to produce pseudotyped SHIV reporter viruses plus the respective Env expression plasmids, visualized by a TM-ectodomain-specific monoclonal antibody. Note the size shifts of the mutated TM subunits. The corresponding supernatants were collected, concentrated by ultracentrifugation through a sucrose cushion, and also subjected to immunoblotting with the TM-specific antibody (right). Finally, the blot was stripped and incubated with an SIV Gag-specific antibody to determine the particle load.
Together, these results demonstrate that progressive truncation of the HERV-K Env cytoplasmic tail influences processing and particle association.
The ancestral HERV-K113 envelope protein mediates entry of retroviral particles, and its infectivity can be augmented by an extensive C-terminal truncation.
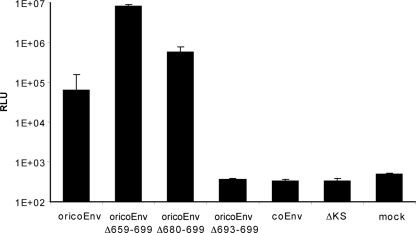
The infectivity of the SHIV-luciferase reporter particles carrying various envelope proteins was tested next. The single-round entry assays were performed using CrFK cells or dog Cf2Th cells as targets and virus suspensions normalized for reverse transcriptase activity. CrFK cells have been previously reported to be susceptible to infection with HERV-K(HML-2) (39). Although specific entry of virions carrying coEnv or oricoEnvΔ693-699 could not be detected (Fig. 6), significant entry was achieved by pseudotyping with the ancestral oricoEnv, giving levels of luciferase activity 2 orders of magnitude higher than the background determined with a nonfunctional HIV envelope protein (ΔKS) (3). Similar results were obtained in experiments with Cf2Th cells (data not shown). Unexpectedly, the efficiencies of entry with oricoEnvΔ680-699 and oricoEnvΔ659-699 pseudotypes were even 10- or 100-fold higher, respectively (Fig. 6). In parallel experiments using pseudotypes expressing green fluorescent protein instead of luciferase, flow cytometry revealed that up to 30% of the target cells were infected with oricoEnvΔ659-699 (data not shown). Therefore, in contrast to the modern proviral HERV-K113 envelope protein, the putative ancestral envelope is efficiently incorporated into retroviral particles and mediates entry. Removal of the last 7 amino acids of the glycoprotein completely blocked cleavage and particle incorporation, and although truncation of the cytoplasmic tail by 20 or 42 C-terminal amino acids resulted in only a slightly higher incorporation rate, there was a much stronger enhancement of virus entry.
FIG. 6.
Infectivity assay with pseudotyped SHIV reporter particles carrying a luciferase gene. The lentiviral particles were produced in HEK 293T cells, normalized, and used to infect CrFK cells. Forty-eight hours later, the cells were lysed and assayed for luciferase activity. The background level was determined by infection with a truncated nonfunctional HIV Env (ΔKS). Mock-treated cells were not infected. The means and standard deviations of six replicates are shown. Similar results were achieved in four independent experiments. RLU, relative light units.
Two arginine-to-cysteine mutations are the predominant cause of the loss of coEnv function.
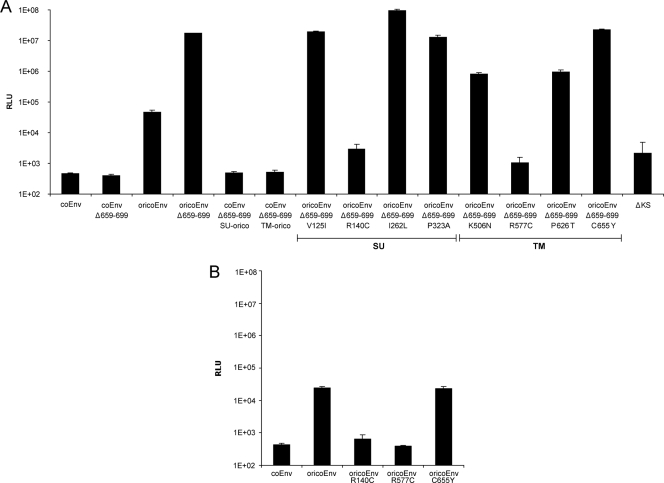
To further analyze the impacts of the postinsertional mutations in the env gene of HERV-K113, we generated coEnvΔ659-699 constructs containing the SU or TM sequence of oricoEnv. These two envelope proteins (coEnvΔ659-699 SU-orico and coEnvΔ659-699 TM-orico) did not facilitate entry of pseudotyped reporter viruses, indicating that mutations in both the SU and the TM subunits of coEnv contribute to the lack of entry (Fig. 7A). Eight oricoEnvΔ659-699 mutants, each carrying a single reversion, were then generated in order to identify the extent to which each of the eight postinsertional mutations contributed to the loss of coEnv function. Entry experiments using these pseudotyped reporter viruses showed that only the R140C mutation in the SU subunit and the R577C mutation in the TM subunit essentially completely abolished entry, whereas the K506N and P626T mutations each reduced entry by a factor of approximately 10 (Fig. 7A). Similar results were obtained with mutants in the context of the full-length oricoEnv protein (Fig. 7B).
FIG. 7.
Impacts of postinsertional mutations in the envelope protein of HERV-K113 on the entry efficiency of pseudotyped SHIV reporter viruses. (A) Mutations in the context of the C-terminal truncation mutant with the highest entry level. (B) The substitutions had similar effects on the function of the full-length oricoEnv. The means and standard deviations of six replicates are shown. Similar results were obtained in two independent experiments. RLU, relative light units.
DISCUSSION
Cell entry of retroviruses is triggered by a specific interaction of the viral envelope glycoprotein with one or more receptor proteins in the cellular membrane. The receptor expression profile largely defines the virus's cellular tropism. Despite being present in many healthy and tumorigenic human tissues and being capable of pseudotyping other retroviruses, the envelope protein of HERV-K(HML-2) has been only poorly characterized (19, 20). Two major obstacles have long hampered the study of the protein's fundamental characteristics: the inefficient protein expression in mammalian cells, even when under the control of strong promoters, and the uncertainty concerning the impacts of postinsertional mutations (53). Recently, however, recognition that the HERV-K108 glycoprotein has residual functional activity plus the in silico generation of consensus prototypic HERV-K(HML-2) sequences have added considerably to our basic understanding of the virus and its proteins (20, 21, 39).
The data presented here describe a strategy to overcome the poor expression and document efforts to reconstitute a HERV-K(HML-2) Env protein coded for by the HERV-K113 element that originally entered the human germ line as an infectious retrovirus more than 800,000 years ago (33, 54). This provirus is one of the most studied elements. It has preserved open reading frames for all viral proteins but has been inactivated by postinsertional mutations affecting, among other things, the expression and function of Env (5, 29).
The use of a synthetic DNA sequence optimized for mammalian cells elevated the expression of HERV-K113 Env by a factor of over 50 compared with that of the proviral sequence. Codon optimization has previously been successfully used to boost the expression of several other retroviral proteins, including envelope glycoproteins (18, 45). Full-length retroviral genomes require a balanced expression of their proteins for optimal replication, and their DNA, RNA, and protein sequences are under a multitude of constraints and selective pressures. The consequential use of rare codons, low GC content, unstable RNA, and internal splicing, (e.g., the generation of Rec transcripts from HERV-K env mRNA) result in submaximal levels of expression. A limited expression of toxic viral proteins can also minimize adverse effects on cell viability. Viral RNA is also a target for antiviral responses that result in specific transcript degradation and low protein expression. Recently, a large number of microRNAs that target exogenous and endogenous retroviruses, including HERV-K, have been identified (28). The roles played by such silencing small RNAs in the inefficient expression of HERV-K proteins deserve further investigation.
The efficient expression of the Env protein prompted us to identify and correct mutations that have altered the original protein sequence of the primordial infectious retrovirus and to perform a comparative study with the contemporary Env. Such sequence changes may have already been introduced before integration, e.g., by the RNA polymerase in the producer cell, during reverse transcription, as a result of the inherent low fidelity of the enzyme, or by the action of APOBEC deaminases. Alternatively, the mutations might have occurred gradually over evolutionary time since integration. Because it is unlikely that there are more than one or two reverse transcription errors in a single round and as the action of APOBEC deaminases was not shown to significantly affect HERV-K113 in a recent report (40), it is likely that most, if not all, of the mutations occurred after integration.
Although the method used to identify postinsertional changes was based, like those previously used to generate consensus sequences (21, 39), on sequence alignments, it also allows likely mutations and shared polymorphisms in a minority of elements to be discriminated (29). The young age of the 11 proviruses used for alignment makes it very unlikely that the shared polymorphisms are the result of matching postinsertional mutations in more than two individual proviruses or of ectopic recombination events (30) between the env genes of HERV-K113 and two or more of the other aligned elements, although these cannot be completely ruled out.
The assumed shared polymorphisms account for three amino acid differences between the reconstituted HERV-K113 Env and the deduced consensus sequence (Fig. 3). In contrast, among the aligned sequences, 8 amino acids are unique to the contemporary HERV-K113 Env or match only one element. The corresponding nonsynonymous mutations were therefore assumed to be postinsertional changes and were inverted to restore the original protein sequence. As shown by the immunoblot TM band, the reversions yielding oricoEnv altered the protein's N-glycan pattern. Although one consensus N-glycosylation motif in the TM subunit was lost by the N506K back mutation, an increase in the apparent molecular mass was seen. Aberrant folding and impaired transport through the endoplasmic reticulum-Golgi compartment are the likely reasons for the peculiar glycosylation, the migration characteristic in the Western blot, and the reduced stability of the modern HERV-K113 Env. This is consistent with the very low level of surface expression and lack of incorporation into viral particles (20). In contrast, the TM subunit of the reconstituted original protein (oricoEnv) displayed a more regular glycosylation pattern, reached the cell surface (data not shown), and was efficiently incorporated into lentiviral particles. In agreement with the known consensus sequences for HERV-K(HML-2), oricoEnv contains nine potential N-glycosylation sites, five in the SU subunit and four in the TM subunit. The numbers and the SU/TM ratio of 5/4 are similar to those of Jaagsiekte sheep retrovirus (ratio, 6/3). The MMTV ratio (3/2) and that of syncytin 1 (HERV-W; ratio, 6/1) are less glycosylated (14). Deglycosylation experiments shifted the V5-tagged oricoEnv TM by 15 kDa from 43 kDa to 28 kDa. Assuming a reduction of at least 2.8 kDa per oligosaccharide (14), it is likely that all four N-glycosylation sites predicted for TM are modified in HEK 293T cells and probably in most other human cells. This seems also to be true for the five sites in the oricoEnv SU subunit. Focusing on the HERV-K108 glycoprotein, which contains one additional N-glycosylation site in its SU subunit, Dewannieux and colleagues demonstrated a 20-kDa shift following PNGase F digestion (20). This result and the size difference between the glycosylated and nonglycosylated precursor proteins indicate that most, if not all, theoretical sites carry an N-linked carbohydrate. Of particular interest in this respect is N461, which lies only 5 amino acids upstream of the furin cleavage site (461NRSKR). Since cleavage occurs after initial glycosylation, an oligosaccharide at this position might influence the processing efficiency (55), and indeed, impaired cleavage has previously been shown for HERV-K(HML-2) Env proteins (8). To test this, we replaced the serine with alanine, eliminating the glycosylation motif and immediately creating the cleavage site of MMTV (26). The mutated protein was cleaved with the same efficiency as the oricoEnv protein, indicating that in HEK 293T cells, at least, the presence of an N-glycosylation site close to the cleavage site does not impede the furin protease or asparagine 461 is not glycosylated. Alternatively, a cleavage enhancement by N-glycan prevention in the 461NRAKR mutant might be exactly balanced by a negative effect of the serine-to-alanine mutation. As tunicamycin treatment or expression of nonglycosylated oricoEnv resulted in poor processing and susceptibility to intracellular proteolysis, it appears that efficient cleavage of Env depends on proper glycosylation. Several studies have described the importance of extensive SU and TM glycosylation for folding, transport, processing, fusogenicity, and immunological shielding of numerous retroviral envelope proteins (36, 47, 49). For HIV-1, which contains between 18 and 33 N-linked carbohydrates (4 in TM), mutation of glycosylation sites in gp41 has been shown to lead to considerable inhibition of Env transport, cleavage, and fusion activities (16, 23, 55). This impediment results from a substantial arrest of the protein in the Golgi apparatus (23). Furthermore, nonglycosylated HERV-W envelope proteins also remain trapped in the secretory compartment as misfolded proteins (14).
In agreement with these observations and with previously published data, it was not possible to demonstrate incorporation of the contemporary HERV-K Env (coEnv) into lentiviral particles or the production of infectious virus (5, 20). It is likely that one or both of the arginine-to-cysteine mutations that render the protein nonfunctional prevent the correct folding of the Env protein, which in turn results in aberrant glycosylation and a lack of particle incorporation. In contrast to coEnv, pseudotyping of SHIV particles by the ancient oricoEnv was very efficient. The protein was able to efficiently mediate entry into target cells, demonstrating a functional reconstitution. In contrast, abrogating N-glycosylation or preventing processing into SU and TM subunits by mutation of the cleavage site dramatically reduced oricoEnv particle incorporation.
HERV-K(HML-2) Env has a relatively short putative cytoplasmic tail of only 44 amino acids (20), and truncation of the cytoplasmic tail is known to modulate particle incorporation and the fusogenicity of retroviral glycoproteins (13, 15, 37). In the case of MLV and related gammaretroviruses, the R peptide, a short C-terminal peptide, is cleaved off by the viral protease during maturation, a step necessary for full fusogenicity. Although there is no evidence for the presence of an R peptide in HERV-K(HML-2), we addressed whether arbitrary progressive C-terminal truncations of oricoEnv by insertion of stop codons would affect Env incorporation and associated viral functions. Surprisingly, the mutant with the shortest deletion (7 amino acids) had a late maturation defect with no cleavage or particle incorporation. The intermediate truncation of 18 amino acids did not significantly affect processing and incorporation efficiency, but particle infectivity was increased by a factor of 10. A further truncation of 41 amino acids, leaving only 3 amino acids of the putative cytoplasmic domain, yielded particles bearing approximately five-fold more Env but having a capacity for fusion almost 100-fold higher than with the full-length oricoEnv. Although the underlying mechanism for this difference remains unclear, it is possible that the higher glycoprotein content with the short-tailed Env helps the budding lentiviral particle by reducing the interference between the matrix protein of the nascent Gag shell and the C-terminal tail (13, 24, 43). Indeed, envelopes with short cytoplasmic tails are known to be better incorporated into SIV particles (34, 48). Alternatively, the mutant might have improved kinetics of transport and maturation or a lower rate of endocytosis, resulting in a higher glycoprotein concentration at the budding site.
Truncation of the Env cytoplasmic tail has been reported to augment fusogenicity for a number of retroviruses (13, 46, 51). For the HERV-K Env, it is tempting to speculate that the predominant or almost exclusive incorporation of completely processed glycoproteins is one reason for the considerably higher infectivity using the short-tailed truncation mutants. Other possible explanations include increased mobility in the viral membrane (which could promote Env clustering during fusion) or an inherent structural difference in the TM ectodomain that favors fusogenicity (22, 56). Further studies are needed to elucidate the exact cause of this enhancement and to determine its relevance for integration into HERV-K particles.
Given the abundance and polymorphism of endogenous retrovirus K elements in humans and other primates, it is likely that many more functional HERV-K(HML-2) glycoproteins are to be found in several genomes, in addition to the known functional HERV-K108 Env. The reconstitution and efficient expression of the original ancient HERV-K113 Env protein facilitated rational studies of the N-glycosylation, processing, incorporation, and mediation of entry of this fossil retrovirus. The results show that the protein can pseudotype retroviruses and mediate efficient entry into cells. Its use as a functional model protein should assist in the search for the cellular receptor and in studies focusing on the pathophysiologic, as well as physiologic, implications of HERV-K(HML-2) Env expression.
Supplementary Material
Acknowledgments
We thank Sandra Klein for her excellent technical support and Kazimierz Madela for providing his expertise with the confocal microscope. We are grateful to Steve Norley for his critical reading of the manuscript and helpful discussions.
The work was supported in part by a donation from the Heinz Kuthe de Mouson Legacy to R.K.
Footnotes
Published ahead of print on 7 October 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Arnaud, F., M. Varela, T. E. Spencer, and M. Palmarini. 2008. Coevolution of endogenous betaretroviruses of sheep and their host. Cell Mol. Life Sci. 65:3422-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bannert, N., and R. Kurth. 2006. The evolutionary dynamics of human endogenous retroviral families. Annu. Rev. Genomics Hum. Genet. 7:149-173. [DOI] [PubMed] [Google Scholar]
- 3.Bannert, N., D. Schenten, S. Craig, and J. Sodroski. 2000. The level of CD4 expression limits infection of primary rhesus monkey macrophages by a T-tropic simian immunodeficiency virus and macrophagetropic human immunodeficiency viruses. J. Virol. 74:10984-10993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbulescu, M., G. Turner, M. I. Seaman, A. S. Deinard, K. K. Kidd, and J. Lenz. 1999. Many human endogenous retrovirus K (HERV-K) proviruses are unique to humans. Curr. Biol. 9:861-868. [DOI] [PubMed] [Google Scholar]
- 5.Beimforde, N., K. Hanke, I. Ammar, R. Kurth, and N. Bannert. 2008. Molecular cloning and functional characterization of the human endogenous retrovirus K113. Virology 371:216-225. [DOI] [PubMed] [Google Scholar]
- 6.Belshaw, R., A. L. Dawson, J. Woolven-Allen, J. Redding, A. Burt, and M. Tristem. 2005. Genomewide screening reveals high levels of insertional polymorphism in the human endogenous retrovirus family HERV-K(HML2): implications for present-day activity. J. Virol. 79:12507-12514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belshaw, R., V. Pereira, A. Katzourakis, G. Talbot, J. Paces, A. Burt, and M. Tristem. 2004. Long-term reinfection of the human genome by endogenous retroviruses. Proc. Natl. Acad. Sci. USA 101:4894-4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bieda, K., A. Hoffmann, and K. Boller. 2001. Phenotypic heterogeneity of human endogenous retrovirus particles produced by teratocarcinoma cell lines. J. Gen. Virol. 82:591-596. [DOI] [PubMed] [Google Scholar]
- 9.Blot, V., S. Lopez-Verges, M. Breton, C. Pique, C. Berlioz-Torrent, and M. P. Grange. 2006. The conserved dileucine- and tyrosine-based motifs in MLV and MPMV envelope glycoproteins are both important to regulate a common Env intracellular trafficking. Retrovirology 3:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boeke, M., and J. P. Stoye. 1997. Retrotransposons, endogenous retroviruses and the evolution of retroelements., p. 343-436. In S. H. H. J. M. Coffin, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, NY. [PubMed]
- 11.Boller, K., K. Schonfeld, S. Lischer, N. Fischer, A. Hoffmann, R. Kurth, and R. R. Tonjes. 2008. Human endogenous retrovirus HERV-K113 is capable of producing intact viral particles. J. Gen. Virol. 89:567-572. [DOI] [PubMed] [Google Scholar]
- 12.Buzdin, A., S. Ustyugova, K. Khodosevich, I. Mamedov, Y. Lebedev, G. Hunsmann, and E. Sverdlov. 2003. Human-specific subfamilies of HERV-K (HML-2) long terminal repeats: three master genes were active simultaneously during branching of hominoid lineages. Genomics 81:149-156. [DOI] [PubMed] [Google Scholar]
- 13.Celma, C. C., M. G. Paladino, S. A. Gonzalez, and J. L. Affranchino. 2007. Importance of the short cytoplasmic domain of the feline immunodeficiency virus transmembrane glycoprotein for fusion activity and envelope glycoprotein incorporation into virions. Virology 366:405-414. [DOI] [PubMed] [Google Scholar]
- 14.Cheynet, V., A. Ruggieri, G. Oriol, J. L. Blond, B. Boson, L. Vachot, B. Verrier, F. L. Cosset, and F. Mallet. 2005. Synthesis, assembly, and processing of the Env ERVWE1/syncytin human endogenous retroviral envelope. J. Virol. 79:5585-5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cote, M., Y. M. Zheng, L. M. Albritton, and S. L. Liu. 2008. Fusogenicity of Jaagsiekte sheep retrovirus envelope protein is dependent on low pH and is enhanced by cytoplasmic tail truncations. J. Virol. 82:2543-2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dash, B., A. McIntosh, W. Barrett, and R. Daniels. 1994. Deletion of a single N-linked glycosylation site from the transmembrane envelope protein of human immunodeficiency virus type 1 stops cleavage and transport of gp160 preventing env-mediated fusion. J. Gen. Virol. 75:1389-1397. [DOI] [PubMed] [Google Scholar]
- 17.Demirov, D. G., and E. O. Freed. 2004. Retrovirus budding. Virus Res. 106:87-102. [DOI] [PubMed] [Google Scholar]
- 18.Deml, L., A. Bojak, S. Steck, M. Graf, J. Wild, R. Schirmbeck, H. Wolf, and R. Wagner. 2001. Multiple effects of codon usage optimization on expression and immunogenicity of DNA candidate vaccines encoding the human immunodeficiency virus type 1 Gag protein. J. Virol. 75:10991-11001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Parseval, N., V. Lazar, J. F. Casella, L. Benit, and T. Heidmann. 2003. Survey of human genes of retroviral origin: identification and transcriptome of the genes with coding capacity for complete envelope proteins. J. Virol. 77:10414-10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dewannieux, M., S. Blaise, and T. Heidmann. 2005. Identification of a functional envelope protein from the HERV-K family of human endogenous retroviruses. J. Virol. 79:15573-15577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dewannieux, M., F. Harper, A. Richaud, C. Letzelter, D. Ribet, G. Pierron, and T. Heidmann. 2006. Identification of an infectious progenitor for the multiple-copy HERV-K human endogenous retroelements. Genome Res. 16:1548-1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edwards, T. G., S. Wyss, J. D. Reeves, S. Zolla-Pazner, J. A. Hoxie, R. W. Doms, and F. Baribaud. 2002. Truncation of the cytoplasmic domain induces exposure of conserved regions in the ectodomain of human immunodeficiency virus type 1 envelope protein. J. Virol. 76:2683-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fenouillet, E., J. C. Gluckman, and I. M. Jones. 1994. Functions of HIV envelope glycans. Trends Biochem. Sci. 19:65-70. [DOI] [PubMed] [Google Scholar]
- 24.Freed, E. O., and M. A. Martin. 1995. Virion incorporation of envelope glycoproteins with long but not short cytoplasmic tails is blocked by specific, single amino acid substitutions in the human immunodeficiency virus type 1 matrix. J. Virol. 69:1984-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gifford, R., P. Kabat, J. Martin, C. Lynch, and M. Tristem. 2005. Evolution and distribution of class II-related endogenous retroviruses. J. Virol. 79:6478-6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goodman, L. J., S. R. Kain, and G. L. Firestone. 1993. Trafficking of wild-type and an endoproteolytic-site mutant of the mouse mammary tumor virus glycoprotein. J. Biol. Chem. 268:2329-2336. [PubMed] [Google Scholar]
- 27.Green, N., T. M. Shinnick, O. Witte, A. Ponticelli, J. G. Sutcliffe, and R. A. Lerner. 1981. Sequence-specific antibodies show that maturation of Moloney leukemia virus envelope polyprotein involves removal of a COOH-terminal peptide. Proc. Natl. Acad. Sci. USA 78:6023-6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hakim, S. T., M. Alsayari, D. C. McLean, S. Saleem, K. C. Addanki, M. Aggarwal, K. Mahalingam, and O. Bagasra. 2008. A large number of the human microRNAs target lentiviruses, retroviruses, and endogenous retroviruses. Biochem. Biophys. Res. Commun. 369:357-362. [DOI] [PubMed] [Google Scholar]
- 29.Heslin, D. J., P. Murcia, F. Arnaud, K. Van Doorslaer, M. Palmarini, and J. Lenz. 2009. A single amino acid substitution in a segment of the CA protein within Gag that has similarity to human immunodeficiency virus type 1 blocks infectivity of a human endogenous retrovirus K provirus in the human genome. J. Virol. 83:1105-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hughes, J. F., and J. M. Coffin. 2005. Human endogenous retroviral elements as indicators of ectopic recombination events in the primate genome. Genetics 171:1183-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hughes, J. F., and J. M. Coffin. 2004. Human endogenous retrovirus K solo-LTR formation and insertional polymorphisms: implications for human and viral evolution. Proc. Natl. Acad. Sci. USA 101:1668-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hunter, E., and R. Swanstrom. 1990. Retrovirus envelope glycoproteins. Curr. Top. Microbiol. Immunol. 157:187-253. [DOI] [PubMed] [Google Scholar]
- 33.Jha, A. R., S. K. Pillai, V. A. York, E. R. Sharp, E. C. Storm, D. J. Wachter, J. N. Martin, S. G. Deeks, M. G. Rosenberg, D. F. Nixon, and K. E. Garrison. 2009. 371 cross sectional dating of novel haplotypes of HERV-K 113 and HERV-K 115 indicate these proviruses originated in Africa before Homo sapiens. Mol. Biol. Evol. doi: 10.1093/molbev/msp180. [DOI] [PMC free article] [PubMed]
- 34.Johnston, P. B., J. W. Dubay, and E. Hunter. 1993. Truncations of the simian immunodeficiency virus transmembrane protein confer expanded virus host range by removing a block to virus entry into cells. J. Virol. 67:3077-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Katzourakis, A., A. Rambaut, and O. G. Pybus. 2005. The evolutionary dynamics of endogenous retroviruses. Trends Microbiol. 13:463-468. [DOI] [PubMed] [Google Scholar]
- 36.Kayman, S. C., R. Kopelman, S. Projan, D. M. Kinney, and A. Pinter. 1991. Mutational analysis of N-linked glycosylation sites of Friend murine leukemia virus envelope protein. J. Virol. 65:5323-5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim, F. J., N. Manel, Y. Boublik, J. L. Battini, and M. Sitbon. 2003. Human T-cell leukemia virus type 1 envelope-mediated syncytium formation can be activated in resistant mammalian cell lines by a carboxy-terminal truncation of the envelope cytoplasmic domain. J. Virol. 77:963-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kowalski, M., J. Potz, L. Basiripour, T. Dorfman, W. C. Goh, E. Terwilliger, A. Dayton, C. Rosen, W. Haseltine, and J. Sodroski. 1987. Functional regions of the envelope glycoprotein of human immunodeficiency virus type 1. Science 237:1351-1355. [DOI] [PubMed] [Google Scholar]
- 39.Lee, Y. N., and P. D. Bieniasz. 2007. Reconstitution of an infectious human endogenous retrovirus. PLoS Pathog. 3:e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee, Y. N., M. H. Malim, and P. D. Bieniasz. 2008. Hypermutation of an ancient human retrovirus by APOBEC3G. J. Virol. 82:8762-8770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Macfarlane, C., and P. Simmonds. 2004. Allelic variation of HERV-K(HML-2) endogenous retroviral elements in human populations. J. Mol. Evol. 59:642-656. [DOI] [PubMed] [Google Scholar]
- 42.Mager, D. L., and P. Medstrand. 2003. Retroviral repeat sequences, p. 57-63. In D. N. Cooper (ed.), Nature encyclopedia of the human genome. Nature Pub. Group, London, United Kingdom.
- 43.Manrique, J. M., C. C. Celma, E. Hunter, J. L. Affranchino, and S. A. Gonzalez. 2003. Positive and negative modulation of virus infectivity and envelope glycoprotein incorporation into virions by amino acid substitutions at the N terminus of the simian immunodeficiency virus matrix protein. J. Virol. 77:10881-10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mayer, J., T. Stuhr, K. Reus, E. Maldener, M. Kitova, F. Asmus, and E. Meese. 2005. Haplotype analysis of the human endogenous retrovirus locus HERV-K(HML-2.HOM) and its evolutionary implications. J. Mol. Evol. 61:706-715. [DOI] [PubMed] [Google Scholar]
- 45.Megati, S., D. Garcia-Hand, S. Cappello, V. Roopchand, A. Masood, R. Xu, A. Luckay, S. Y. Chong, M. Rosati, S. Sackitey, D. B. Weiner, B. K. Felber, G. N. Pavlakis, Z. R. Israel, L. R. Smith, J. H. Eldridge, M. K. Sidhu, and M. A. Egan. 2008. Modifying the HIV-1 env gp160 gene to improve pDNA vaccine-elicited cell-mediated immune responses. Vaccine 26:5083-5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mulligan, M. J., G. V. Yamshchikov, G. D. Ritter, Jr., F. Gao, M. J. Jin, C. D. Nail, C. P. Spies, B. H. Hahn, and R. W. Compans. 1992. Cytoplasmic domain truncation enhances fusion activity by the exterior glycoprotein complex of human immunodeficiency virus type 2 in selected cell types. J. Virol. 66:3971-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pinter, A., W. J. Honnen, and J. S. Li. 1984. Studies with inhibitors of oligosaccharide processing indicate a functional role for complex sugars in the transport and proteolysis of Friend mink cell focus-inducing murine leukemia virus envelope proteins. Virology 136:196-210. [DOI] [PubMed] [Google Scholar]
- 48.Puffer, B. A., S. Pohlmann, A. L. Edinger, D. Carlin, M. D. Sanchez, J. Reitter, D. D. Watry, H. S. Fox, R. C. Desrosiers, and R. W. Doms. 2002. CD4 independence of simian immunodeficiency virus Envs is associated with macrophage tropism, neutralization sensitivity, and attenuated pathogenicity. J. Virol. 76:2595-2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quinones-Kochs, M. I., L. Buonocore, and J. K. Rose. 2002. Role of N-linked glycans in a human immunodeficiency virus envelope glycoprotein: effects on protein function and the neutralizing antibody response. J. Virol. 76:4199-4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reus, K., J. Mayer, M. Sauter, H. Zischler, N. Muller-Lantzsch, and E. Meese. 2001. HERV-K(OLD): ancestor sequences of the human endogenous retrovirus family HERV-K(HML-2). J. Virol. 75:8917-8926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ritter, G. D., Jr., M. J. Mulligan, S. L. Lydy, and R. W. Compans. 1993. Cell fusion activity of the simian immunodeficiency virus envelope protein is modulated by the intracytoplasmic domain. Virology 197:255-264. [DOI] [PubMed] [Google Scholar]
- 52.Tarlinton, R. E., J. Meers, and P. R. Young. 2006. Retroviral invasion of the koala genome. Nature 442:79-81. [DOI] [PubMed] [Google Scholar]
- 53.Tonjes, R. R., C. Limbach, R. Lower, and R. Kurth. 1997. Expression of human endogenous retrovirus type K envelope glycoprotein in insect and mammalian cells. J. Virol. 71:2747-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Turner, G., M. Barbulescu, M. Su, M. I. Jensen-Seaman, K. K. Kidd, and J. Lenz. 2001. Insertional polymorphisms of full-length endogenous retroviruses in humans. Curr. Biol. 11:1531-1535. [DOI] [PubMed] [Google Scholar]
- 55.Vigerust, D. J., and V. L. Shepherd. 2007. Virus glycosylation: role in virulence and immune interactions. Trends Microbiol. 15:211-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wyss, S., A. S. Dimitrov, F. Baribaud, T. G. Edwards, R. Blumenthal, and J. A. Hoxie. 2005. Regulation of human immunodeficiency virus type 1 envelope glycoprotein fusion by a membrane-interactive domain in the gp41 cytoplasmic tail. J. Virol. 79:12231-12241. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.