Abstract
The nuclear magnetic resonance (NMR) structure of a globular domain of residues 1071 to 1178 within the previously annotated nucleic acid-binding region (NAB) of severe acute respiratory syndrome coronavirus nonstructural protein 3 (nsp3) has been determined, and N- and C-terminally adjoining polypeptide segments of 37 and 25 residues, respectively, have been shown to form flexibly extended linkers to the preceding globular domain and to the following, as yet uncharacterized domain. This extension of the structural coverage of nsp3 was obtained from NMR studies with an nsp3 construct comprising residues 1066 to 1181 [nsp3(1066-1181)] and the constructs nsp3(1066-1203) and nsp3(1035-1181). A search of the protein structure database indicates that the globular domain of the NAB represents a new fold, with a parallel four-strand β-sheet holding two α-helices of three and four turns that are oriented antiparallel to the β-strands. Two antiparallel two-strand β-sheets and two 310-helices are anchored against the surface of this barrel-like molecular core. Chemical shift changes upon the addition of single-stranded RNAs (ssRNAs) identified a group of residues that form a positively charged patch on the protein surface as the binding site responsible for the previously reported affinity for nucleic acids. This binding site is similar to the ssRNA-binding site of the sterile alpha motif domain of the Saccharomyces cerevisiae Vts1p protein, although the two proteins do not share a common globular fold.
The coronavirus replication cycle begins with the translation of the 29-kb positive-strand genomic RNA to produce two large polyprotein species (pp1a and pp1ab), which are subsequently cleaved to produce 15 or possibly 16 nonstructural proteins (nsp's) (11). Among these, nsp3 is the largest nsp and also the largest coronavirus protein. nsp3 is a glycosylated (16, 22), multidomain (36, 51), integral membrane protein (38). All known coronaviruses encode a homologue of severe acute respiratory syndrome coronavirus (SARS-CoV) nsp3, and sequence analysis suggests that at least some functions of nsp3 may be found in all members of the order Nidovirales (11). Hallmarks of the coronavirus nsp3 proteins include one or two papain-like proteinase domains (3, 12, 16, 31, 56, 62), one to three histone H2A-like macrodomains which may bind RNA or RNA-like substrates (5, 9, 48, 54, 55), and a carboxyl-terminal Y domain of unknown function (13). An extensive bioinformatics analysis of the coronavirus replicase proteins by Snijder et al. (51) provided detailed annotations of the then-recently sequenced SARS-CoV genome (35, 47), including the identification of a domain unique to SARS-CoV and the prediction of the ADP-ribose-1″-phosphatase (ADRP) activity of the X domain (since shown to be one of the macrodomains).
Only limited information is so far available regarding the ways in which the functions of nsp3 are involved in the coronavirus replication cycle. Some functions of nsp3 appear to be directed toward protein; e.g., the nsp3 proteinase domain cleaves the amino-terminal two or three nsp's from the polyprotein and has deubiquitinating activity (4, 6, 14, 30, 53, 60). Most homologues of the most conserved macrodomain of nsp3 appear to possess ADRP activity (9, 34, 41-43, 48, 59) and may act on protein-conjugated poly(ADP-ribose); however, this function appears to be dispensable for replication (10, 42) and may not be conserved in all coronaviruses (41). The potential involvement of nsp3 in RNA replication is suggested by the presence of several RNA-binding domains (5, 36, 49, 54, 55). nsp3 has been identified in convoluted membrane structures that are also associated with other replicase proteins and that have been shown to be involved in viral RNA synthesis (16, 24, 52), and nsp3 papain-like proteinase activity is essential for replication (14, 62). Other conserved structural features of nsp3 include two ubiquitin-like domains (UB1 and UB2) (45, 49). We have also recently reported that nsp3 is a structural protein, since it was identified as a minor component of purified SARS-CoV preparations, although it is not known whether nsp3 is directly involved in virogenesis or is incidentally incorporated due to protein-protein or protein-RNA interactions (36).
A nucleic acid-binding region (NAB) is located within the polypeptide segment of residues 1035 to 1203 of nsp3. The NAB is expected to be located in the cytoplasm, along with the papain-like protease, ADRP, a region unique to SARS-CoV (the SARS-CoV unique domain [SUD]), and nsp3a, since both the N and C termini of nsp3 were shown previously to be cytoplasmic (38). Two hydrophobic segments are membrane spanning (38), and the NAB is located roughly 200 residues in the N-terminal direction from the first membrane-spanning segment. This paper presents the next step in the structural coverage of nsp3, with the determination of the NAB structure. The structural studies included nuclear magnetic resonance (NMR) characterization of two constructs, an nsp3 construct comprising residues 1035 to 1181 [nsp3(1035-1181)] and nsp3(1066-1203), and complete NMR structure determination for the construct nsp3(1066-1181) (see Fig. 8). The structural data were then used as a platform from which to investigate the nature of the previously reported single-stranded RNA (ssRNA)-binding activity of the NAB (36). Since no three-dimensional (3D) structures for the corresponding domains in other group II coronaviruses are known and since the SARS-CoV NAB has only very-low-level sequence identity to other proteins, such data could not readily be derived from comparisons with structurally and functionally characterized homologues.
FIG. 8.
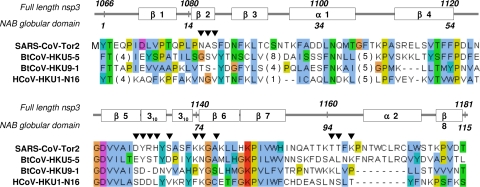
Sequence alignment of the polypeptide segment nsp3(1066-1181) that forms the globular domain of the SARS-CoV NAB with homologues from other group II coronaviruses. Protein multiple-sequence alignment was performed using ClustalW2 and included sequences from SARS-CoV Tor2 (accession no. AAP41036) and representatives of three protein clusters corresponding to three group II coronavirus lineages identified by a BLAST search: bat coronavirus HKU5-5 (BtCoV-HKU5-5; accession no. ABN10901), BtCoV-HKU9-1 (accession no. P0C6T6), and human coronavirus HKU1-N16 (HCoV-HKU1-N16; accession no. ABD75496). Above the sequences, the positions in full-length SARS-CoV nsp3, the locations of the regular secondary structures in the presently solved NMR structure of the SARS-CoV NAB globular domain, and the residue numbering in this domain are indicated. Amino acids are colored according to conservation and biochemical properties, following ClustalW conventions. Residues implicated in interactions with ssRNA are marked with inverted black triangles. In the present context, the key features are that there is only one position with conservation of K or R (red) and that there are extended sequences with conservation of hydrophobic residues (blue) (see the text).
MATERIALS AND METHODS
Production of nsp3(1066-1203), nsp3(1066-1181), and nsp3(1035-1181).
Initially, the protein production core of the Consortium for Functional and Structural Proteomics of the SARS Coronavirus cloned and expressed a construct encoding nsp3 residues 1066 to 1225 by using expression vector pMH1F with a six-His tag and a pBAD derivative in Escherichia coli DL41 cells. The 1D 1H NMR spectrum of the protein indicated the presence of a globular domain, as well as of flexibly disordered polypeptide segments (data not shown). Next, two new constructs, nsp3(1066-1203) and nsp3(1066-1181), were designed based on the disordered-region prediction by GlobPlot (29). Sequences encoding the two constructs were subcloned into pET-28b and pET-25b (Novagen), respectively, and the resulting plasmids were used to transform E. coli strain BL21-CodonPlus (DE3)-RIL (Stratagene). The expression of the uniformly 13C,15N-labeled proteins was carried out by growing freshly transformed cells in M9 minimal medium containing 1 g/liter 15NH4Cl and 4 g/liter d-[13C6]glucose as the sole nitrogen and carbon sources. Cell cultures were grown at 37°C with vigorous shaking to an optical density at 600 nm of 0.8 to 0.9. The temperature was then lowered to 18°C, and after induction with 1 mM isopropyl-β-d-thiogalactopyranoside, the cell cultures were grown for 18 h.
The cells producing nsp3(1066-1203) from the pET-28b vector were harvested by centrifugation, resuspended in extraction buffer (50 mM sodium phosphate at pH 7.5, 150 mM NaCl, 5 mM imidazole, 0.1% Triton X-100, and Complete protease inhibitor tablets [Roche]), and lysed by sonication. The cell debris was removed by centrifugation (20,000 × g for 20 min). For the first purification step, the soluble protein was loaded onto an Ni2+ affinity column (HisTrap; Amersham) equilibrated with 50 mM sodium phosphate buffer, pH 7.5, containing 150 mM NaCl and 5 mM imidazole. The bound proteins were eluted with a 5 to 500 mM imidazole gradient. Fractions containing nsp3(1066-1203) were pooled and concentrated to a volume of 2 ml using centrifugal ultrafiltration devices (Millipore). The buffer was then exchanged by dilution with 8 ml of 50 mM sodium phosphate buffer, pH 7.5, containing 150 mM NaCl and subsequent concentration to 2 ml. After three cycles, 20 μl of thrombin (Enzyme Research Laboratories) was added and the reaction was monitored by gel electrophoresis. After 5 h at room temperature, the sample was loaded onto a size exclusion column (Superdex 75; Amersham) equilibrated with 50 mM sodium phosphate buffer, pH 7.5, containing 150 mM NaCl and then eluted with the same buffer. The fractions containing nsp3(1066-1203) were again pooled and concentrated to a final volume of 550 μl for a final protein concentration of 1.4 mM.
For the production of nsp3(1066-1181) from the pET-25b vector, cells were lysed as described for nsp3(1066-1203) except that the extraction buffer was 50 mM sodium phosphate, pH 6.5, with 50 mM NaCl, 0.1% Triton X-100, and Complete protease inhibitor tablets. For the first purification step, the soluble protein was loaded onto an anion-exchange column (HiTrap Q FF; Amersham) equilibrated with 50 mM sodium phosphate buffer, pH 6.5, containing 50 mM NaCl. The proteins were eluted with a 50 to 1,000 mM NaCl gradient. Fractions containing the protein were pooled and concentrated to a volume of 10 ml by using centrifugal ultrafiltration devices (Millipore). The sample was loaded onto a size exclusion column (Superdex 75; Amersham) equilibrated with 50 mM sodium phosphate buffer, pH 6.5, containing 50 mM NaCl and then eluted with the same buffer. The fractions containing nsp3(1066-1181) were again pooled and concentrated to a final volume of 500 μl. The solution was then supplemented with 50 μl of D2O and 5.5 μl of 200 mM NaN3 for a protein concentration in the NMR sample of 1.4 mM.
Uniformly 15N-labeled nsp3(1035-1181) was produced using the same protocol used for nsp3(1066-1181) to obtain an NMR sample containing 1.1 mM protein.
NMR spectroscopy.
NMR measurements were performed at 298 K with Avance 600, DRX 700, and Avance 800 spectrometers equipped with TXI HCN z- or xyz-gradient probe heads (Bruker BioSpin, Billerica, MA). Proton chemical shifts were referenced to internal 3-(trimethylsilyl)-1-propanesulfonic acid sodium salt (DSS). The 13C and 15N chemical shifts were referenced indirectly to DSS by using the absolute frequency ratios (58). Sequence-specific resonance assignments for nsp3(1066-1181) were obtained as reported previously (50), and the same approach was used to assign the residues 1182 to 1203 in nsp3(1066-1203). In nsp3(1035-1181), the 15N and 1HN resonances of the residues 1036 to 1066 were assigned as a group by comparison with nsp3(1066-1181). Steady-state 15N{1H}-nuclear Overhauser effects (NOEs) were measured using transverse relaxation optimized spectroscopy (TROSY)-based experiments (46, 61) on a Bruker Avance 600 spectrometer, with a saturation period of 3.0 s and a total interscan delay of 5.0 s.
Determination of amide proton Pf.
Amide proton protection factors (Pf) for nsp3(1066-1181) were determined using a 1.2 mM 15N-labeled protein sample that was lyophilized from H2O solution and then redissolved in 99% D2O. The decay of the signal intensity of the 15N-1H correlation peaks due to the amide proton chemical exchange with D2O was monitored by acquiring a series of 2D [15N,1H]-heteronuclear single-quantum coherence ([15N,1H]-HSQC) spectra at different times after preparation of the D2O solution, for a total period of 2 weeks. These data were analyzed using the software CARA (23), and for each peak, the volume was plotted versus the reaction time. The exponential decay constants yielded preliminary values for the Pf, which were then corrected for primary structure effects as described by Bai et al. (2).
Structure determination.
The input for the structure calculation was obtained from a 3D 15N-resolved [1H,1H]-NOE spectroscopy ([1H,1H]-NOESY) spectrum and from two 3D 13C-resolved [1H,1H]-NOESY spectra recorded with the carrier frequency in the aliphatic and the aromatic regions, respectively. All three data sets were recorded with a mixing time of 60 ms. The software ATNOS/CANDID (17, 18) was used in combination with the torsion angle dynamics algorithm of CYANA (15). Seven cycles of automated NOE cross-peak identification with ATNOS (18), automated NOE assignment with CANDID (17), and structure calculation with CYANA were performed. In the second and subsequent cycles, the intermediate protein structure was used as an additional guide for the interpretation of the NOESY spectra. During the first six cycles, ambiguous distance restraints were used, and in the final cycle, only distance restraints that could be attributed to a single pair of hydrogen atoms were retained. The 20 conformers with the lowest residual CYANA target function values obtained from the seventh ATNOS/CANDID/CYANA cycle were subjected to energy minimization in a water shell with the program OPALp (25, 33), using the AMBER force field (8). The program MOLMOL (26) was used to analyze the ensemble of 20 energy-minimized conformers.
Structure validation.
Analyses of the stereochemical qualities of the molecular models were accomplished using the Joint Center for Structural Genomics Validation Central Suite (http://www.jcsg.org).
Study of the interaction of nsp3(1066-1181) with ssRNA.
The interaction of nsp3(1066-1181) with unlabeled ssRNA1 (5′-AAAUACCUCUCAAAAAUAACACCACACCAUAUACCACAU-3′) was evaluated by comparison of the 2D [15N,1H]-HSQC spectra of uniformly 15N-labeled nsp3(1066-1181) recorded at four protein/ssRNA1 ratios, 3:1, 1:1, 1:2, and 1:3. As a control, a 2D [15N,1H]-HSQC spectrum was obtained after addition of single-stranded 5′-CUUGUUCAUU-3′ in fourfold excess with respect to the protein concentration under otherwise identical conditions.
Electrophoretic mobility shift assays (EMSAs).
nsp3(1066-1181) was mixed with an ssRNA or single-stranded DNA substrate in an assay buffer containing 50 mM NaCl and 50 mM sodium phosphate at pH 6.5. The following custom-synthesized RNA oligomers (Integrated DNA Technologies, Inc., San Diego, CA) were tested: randomized 20-mer DNA and RNA; the homopolymers A10, C10, and U10; 5′-CCCGAUACCC-3′, which contains the core GAUA sequence that was shown previously to bind to nsp3a (49); 5′-CUAAACGAAC-3′, which is the leader transcription-regulatory sequence (TRS) from the SARS-CoV genome [TRS(+)]; 5′-GUUCGUUUAG-3′, which is the leader TRS from the SARS-CoV antigenome [TRS(−)]; the decamers 5′-GAGAGAGAGA-3′, 5′-GGAGGAGGAG-3′, and 5′-GGGAGGGAGG-3′; the GGGA repeat oligomers 5′-GGGAGGGA-3′ [(GGGA)2] and 5′-GGGAGGGAGGGAGGGAGGGA-3′ [(GGGA)5]; and the G-positional decamers 5′-GGGAAAAAAA-3′, 5′-AAAGGGAAAA-3′, and 5′-AAAAAAAGGG-3′. Each reaction mixture contained between 0 and 495 μM protein and 10 μg of the RNA or DNA substrate, equivalent to 80 μM 20-mer, 160 μM decamer, or 190 μM octamer nucleic acids. Protein-nucleic acid mixtures were incubated for 1 h at 37°C and then analyzed by native electrophoresis on precast 6% acrylamide DNA retardation gels (Invitrogen). Nucleic acid was detected using SYBR gold poststain (Invitrogen) and photographed using a UV light source equipped with a digital camera. SYBR gold was rinsed out, and protein was subsequently detected using SYPRO ruby poststain (Invitrogen).
Protein structure accession number.
The atomic coordinates of the bundle of 20 conformers used to represent the solution structure of nsp3(1066-1181) have been deposited in the Protein Data Bank (PDB; http://www.rcsb.org/pdb/) with the accession code 2k87.
RESULTS AND DISCUSSION
At the outset of this project, 1D 1H NMR spectroscopy analysis of the 160-residue construct nsp3(1066-1225) indicated the presence of both a globular domain and flexibly disordered polypeptide segments. Based on the prediction of disordered segments by GlobPlot (29), we prepared two new constructs comprising the residues 1066 to 1203 and 1066 to 1181. Initial NMR data then showed that while nsp3(1066-1181) contained the entire globular domain, nsp3(1066-1203) also included a flexibly disordered C-terminal tail. nsp3(1066-1181), which also provided higher-quality NMR data, was therefore selected for complete NMR structure determination. To further investigate the linker region in the N-terminal direction from the globular domain, we also expressed and purified uniformly 15N-labeled nsp3(1035-1181).
NMR structure of nsp3(1066-1181).
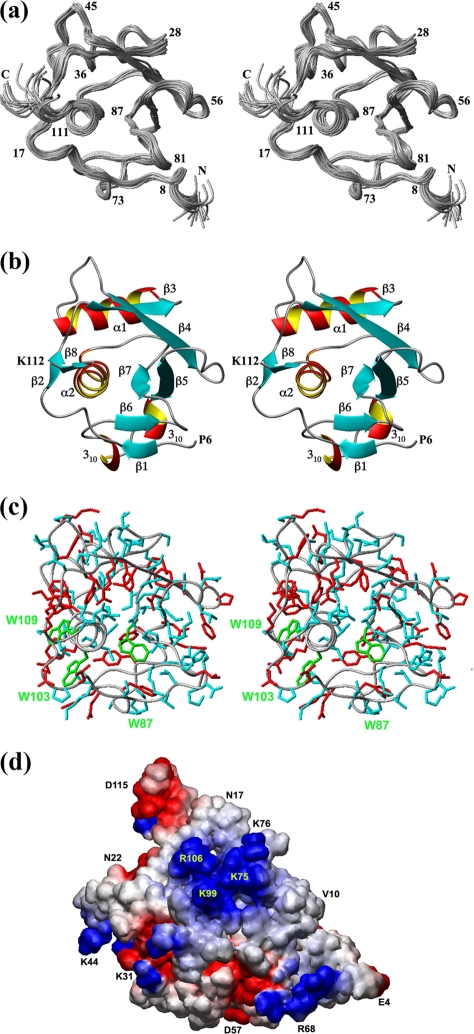
The NMR structure determination was based on the previously reported resonance assignments (50). Three 3D heteronuclear resolved [1H,1H]-NOESY spectra were recorded with a mixing time of 60 ms. NOESY peak picking, NOE assignment, and the structure calculation were carried out with the programs ATNOS, CANDID, and CYANA (see Materials and Methods for details). The seventh cycle of the ATNOS/CANDID/CYANA calculation yielded 2,368 meaningful NOE upper distance limits. In the resulting structure, the average global root mean square deviation relative to the mean coordinates calculated for the backbone atoms of residues 1071 to 1178 in the energy-refined bundle of 20 conformers (Fig. 1a) was 0.44 ± 0.10 Å. Combined with the residual CYANA target function value of 0.65 ± 0.23 Å2 (Table 1), this is indicative of high-quality NMR structure determination.
FIG. 1.
(a to c) Stereo views of the NMR structure of nsp3(1066-1181). (a) Bundle of 20 energy-minimized CYANA conformers. The polypeptide backbone is shown as a gray spline function through the Cα positions. Selected sequence positions in the globular domain are indicated by numerals, where the numbers 1 to 116 correspond to nsp3 residues 1066 to 1181. (b) Ribbon representation of the conformer in panel a that is closest to the mean coordinates. The regular secondary structure elements and the two chain ends at positions 6 and 112 are indicated. (c) All-heavy-atom presentation of the conformer in panel b. The backbone is represented by a gray spline function through the Cα atoms, amino acid side chains with local displacements of ≤0.6 Å are colored blue, those with local displacements of >0.6 Å are red, and the three Trp residues are highlighted in green (see the text). (d) Representation of the electrostatic potential surface showing the area that was found, by chemical shift perturbation experiments, to contain the residues involved in ssRNA binding. The locations of selected residues are identified.
TABLE 1.
Input for the structure calculation and characterization of the bundle of 20 energy-minimized CYANA conformers that represent the NMR structure of nsp3(1066-1181)
| Quantity | Valuea |
|---|---|
| No. of NOE upper distance limits | 2,368* |
| Intraresidual | 544* |
| Short range | 670* |
| Medium range | 397* |
| Long range | 757* |
| Dihedral angle constraint | 118* |
| Residual target function (Å2) | 0.65 ± 0.23* |
| Residual NOE violations | |
| No. ≥ 0.1 Å | 3 ± 2* |
| Maximum (Å) | 0.35 ± 0.01* |
| Residual dihedral angle violations | |
| No. ≥ 2.5° | 3 ± 1* |
| Maximum (°) | 71.3 ± 2.74* |
| AMBER energies (kcal/mol) | |
| Total | −4,054.95 ± 86.30 |
| van der Waals | −342.05 ± 11.19 |
| Electrostatic | −4,259.94 ± 85.38 |
| rmsd from ideal geometry | |
| Bond length (Å) | 0.0075 ± 0.0001 |
| Bond angle (°) | 1.941 ± 0.36 |
| rmsd relative to mean coordinates (Å)b | |
| bb (1071-1178) | 0.44 ± 0.10 |
| ha (1071-1178) | 0.85 ± 0.15 |
| Ramachandran plot—residuesc in: | |
| Most favored regions (%) | 78 |
| Additional allowed regions (%) | 19 |
| Generously allowed regions (%) | 3 |
| Disallowed regions (%) | 0 |
Entries marked with asterisks refer to the 20 CYANA conformers with the lowest residual target function values; the remaining entries refer to the same conformers after energy minimization with OPALp. The ranges indicate the minimum and maximum values. Where applicable, values are given as means ± standard deviations.
bb indicates the backbone atoms N, Cα, and C′; ha stands for all heavy atoms. The numbers in parentheses indicate the residues for which the rmsd was calculated.
As determined by PROCHECK (27).
Overall, the NMR structure of nsp3(1066-1181) includes eight β-strands, two α-helices, and two 310-helices (Fig. 1b), which are arranged in the sequential order β1-β2-β3-α1-β4-β5-310-310-β6-β7-α2-β8. The eight β-strands form two antiparallel β-sheets, the first containing β1 and β6 and the second containing β2 and β8, and a parallel half-barrel comprising β3, β4, β5, and β7. Helices α1 and α2 are oriented antiparallel to strands β3 and β7, respectively (Fig. 1b). In the 3D fold, the two 310-helices and the two short antiparallel β-sheets are anchored against the surface of the barrel-like molecular core formed by the four-strand β-sheet and the two α-helices.
Along the polypeptide chain, the first regular secondary structure, β1, comprises residues 9 and 10 (where residue numbers 1 to 116 correspond to nsp3 residues 1066 to 1181) and is connected via a well-defined 6-amino-acid linker to strand β2, containing residues 17 and 18. A 3-residue turn leads to β3, which spans residues 23 to 27 and is connected via a short turn to helix α1, with residues 30 to 40, and then a 7-residue loop affords the link to strand β4, with residues 48 to 54. Next, a type VIa turn (7) with cis-proline in position 56 connects to β5, with residues 62 to 66, which is oriented parallel to β4. Τwo 310-helices with resides 67 to 69 and 72 to 74 are part of the linker sequence to β6, which is formed by residues 78 and 79. A well-defined 6-residue loop leads to β7, with residues 85 to 88, a further 6-residue linker leads to α2, with residues 95 to 108, and the last regular secondary structure is β8, containing residues 110 to 112.
Overall, the electrostatic potential surface of nsp3(1066-1181) shows a homogeneous charge distribution, with the sole exception that there is a positive patch constituted by the residues K75, K76, K99, and R106, which are located in the loop preceding strand β6 and helix α2 (Fig. 1d).
Since a search of the PDB provided evidence that the NAB forms a new polypeptide fold, we performed additional NMR experiments to obtain independent support for this novel structure. We measured the exchange rates of the amide protons of the NAB with deuterons from the solvent by dissolving a sample of the lyophilized protein in D2O. The rates at which the amide proton signal intensities decrease provide information on the amount of protection of each proton by the protein secondary and tertiary structures, with greater protection and lower exchange rates reflecting involvement in the hydrogen bonds of regular secondary structures and/or sequestration from the solvent by the protein's tertiary structure. This method is distinct from the NMR experiments that had been used to provide conformational constraints for the structure determination and thus provides an independent check on the compatibility of the 3D structure with experimental data.
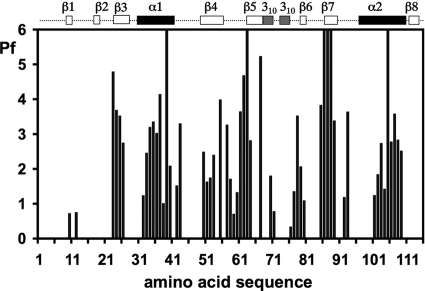
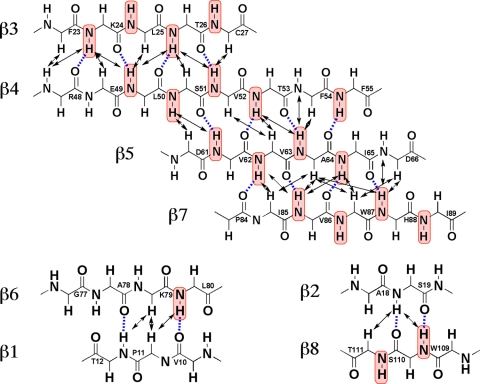
Amide proton Pf are defined as log(kin/kex), where kex is the measured hydrogen/deuterium exchange rate constant and kin is the intrinsic hydrogen/deuterium exchange rate constant for the same residue type when completely exposed to the solvent water. The reference value kin is determined by the nature of the residue and its sequential neighbors (2). High Pf values corrected for the primary structure effects prevail for nearly all the backbone amide groups in the regular secondary structures (Fig. 2), thus providing independent support for the NOE-derived novel fold of nsp3(1066-1181). The residues with the highest Pf values are located in strands β5 and β7, which exhibit a dense hydrogen bonding network (Fig. 3) and are buried in the core of the protein. The expected pattern of Pf values for amide protons in COi-NH(i+ 3) hydrogen bonds of α-helices (where i represents the position of the residue of interest in the protein sequence) is clearly seen for helices α1 and α2, with outstandingly high Pf values for the residues M39 and C107. The only apparent discrepancy from the NOE-based regular secondary structure determination was noted for the short β-sheet formed by strands β2 and β8 (Fig. 3), where no measurable exchange protection was seen. This β-sheet is solvent exposed near the protein surface (Fig. 1b), which is probably the reason for the implicated weak protection. The β-sheet topology in the NMR structure of nsp3(1066-1181) reveals that the residues with high Pf values are in most instances structurally constrained by larger numbers of NOEs than those with lower Pf values (Fig. 3). There are also a few residues in nonregular secondary structure regions that exhibit significant Pf. These are either hydrogen bonded or buried in the molecular core or both. An example is F42, with an amide group that interacts with the carbonyl group of N37 and the side chain hydroxyl group of T40.
FIG. 2.
Histogram of the amide proton Pf versus the amino acid sequence of nsp3(1066-1181); the numbers 1 to 116 along the horizontal axis correspond to nsp3 residues 1066 to 1181. At the top, the positions of the regular secondary structure elements are indicated. Pf is defined as log(kin/kex), where kex is the measured hydrogen/deuterium exchange rate constant and kin is the intrinsic hydrogen/deuterium exchange rate constant for the same residue type when completely exposed to the solvent water (2). Higher Pf values reflect lower amide proton exchange rates with the solvent, which result from involvement in hydrogen bonding networks and/or sequestration from the solvent by the protein's tertiary structure.
FIG. 3.
β-Sheet topology in nsp3(1066-1181), where blue lines represent hydrogen bonds that were identified by MOLMOL (26) in at least 10 conformers out of the ensemble of 20 conformers depicted in Fig. 1. Interstrand 1H-1H NOEs are indicated by double-headed black arrows. Amide groups of residues with Pf values of ≥2.0 are color coded in red.
Similar information was obtained for the tryptophan indole protons. The two indole protons of W103 and W109, with ω1(15N) values near 129 ppm and with 1H chemical shifts of 11.0 and 10.1 ppm, respectively (Fig. 4a and b), do not show measurable protection, whereas the indole proton of W87, at ω2(1H) of 10.6 ppm, is observable for more than 12 days after the dissolution of the protein in D2O. This behavior correlates with the locations of these residues in the 3D structure, where W87 is located in the core of the protein and W103 and W109 are solvent accessible near the protein surface (Fig. 1c).
FIG. 4.
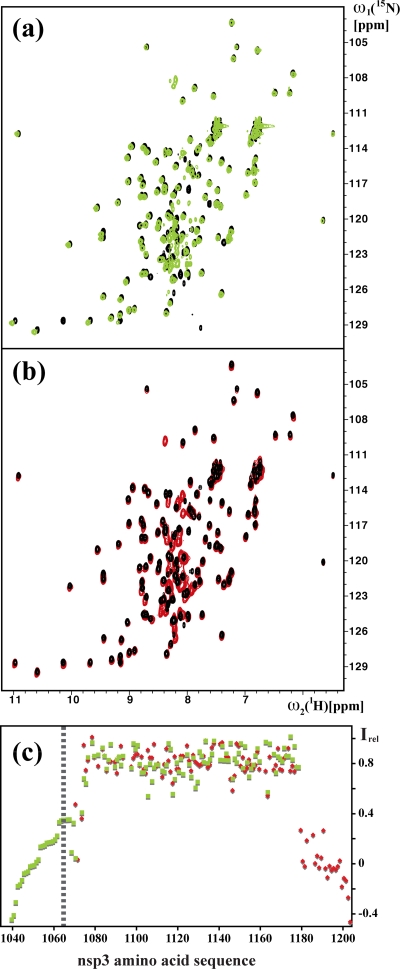
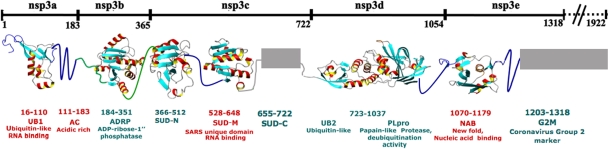
(a) Superposition of the 2D [15N,1H]-HSQC spectra of nsp3(1066-1181) (black) and nsp3(1035-1181) (green). (b) Superposition of the 2D [15N,1H]-HSQC spectra of nsp3(1066-1181) (black) and nsp3(1066-1203) (red). (c) Plot of relative 15N{1H}-NOE intensities, Irel, versus the amino acid sequence of the nsp3 fragment comprising residues 1035 to 1203. Red squares represent the experimental measurements for the backbone amide groups in the construct nsp3(1066-1203), and green squares represent those for nsp3(1035-1181). The broken vertical line is used to indicate that the NMR signals of the residues 1035 to 1065 were assigned as a group (see Materials and Methods), and the data points to the left of this line have arbitrarily been arranged in the order of decreasing Irel values. The 15N{1H}-NOEs were recorded at a 1H frequency of 600 MHz by using a saturation period of 3.0 s and a total interscan delay of 5.0 s (46, 61).
3D structure homology search using the PDB, SCOP, and CATH databases.
A 3D structure homology search was performed with the software DALI (19, 20), using the conformer closest to the mean coordinates of nsp3(1066-1181) (Fig. 1b) as the input. The result implicated more than 300 homologues, all with DALI Z scores below 3.3 Å. This outcome is due to a certain degree of similarity among parts of the polypeptide folds of the NAB globular domain (Fig. 1b) and proteins from the signal transduction protein CheY family, which has a large representation in the PDB. However, visual inspection showed that although there is some overlapping of regular secondary structure elements, the arrangements of the β-strands are characteristically different in each pairwise comparison with individual CheY proteins. Similar structure homology searches using the SCOP (32) and CATH (39) databases provided similar results, and no protein was identified in the three databases that would form a globular fold of the type seen for the SARS-CoV NAB. Although new folds were previously identified in other regions of the SARS-CoV proteome (see, for example, reference 1), we have here the first domain within nsp3 for which standard homology searches indicate that it exhibits a new fold. This is an intriguing finding in the context that previous observations indicate a trend for 3D structure redundancy of nsp3 domains, as exemplified by the three macrodomain-like folds of ADRP [nsp3(184-351)] and the N-terminal and middle regions of the SUD {SUD-N [nsp3(389-517)] and SUD-M [nsp3(527-651)]} and the ubiquitin-like folds of UB1 [nsp3(1-112)] and UB2 [nsp3(723-792)] (5, 45, 48, 49, 55).
Exploring the overall organization of the NAB domain within nsp3.
To characterize the linker segments that flank the nsp3(1066-1181) globular domain, we studied the two constructs nsp3(1035-1181) and nsp3(1066-1203). The additional 31 residues in nsp3(1035-1181) correspond to the segment linking the globular domains of the papain-like protease [nsp3(723-1037)] and the NAB (Fig. 5). For nsp3(1066-1203), the C-terminal extension by 22 residues was somewhat arbitrary, due to the lack of information on the location of the nearest following domain. In the 2D [15N,1H]-HSQC spectra of the two constructs (Fig. 4a and b), the peaks from residues 1071 to 1177 maintain the same chemical shifts that were observed for nsp3(1066-1181), indicating that all three constructs contain identical globular domains. Nearly all the peaks of the polypeptide segments of residues 1035 to 1070 and 1178 to 1203 are in the random-coil chemical shift region, with 1H chemical shifts between 7.5 and 8.5 ppm. The increased intensities of the resonances from the two tails, compared with the peaks from the globular domain, are indicative of flexibly disordered polypeptide segments. Increased mobility of both tail regions was confirmed by 15N{1H}-NOE experiments (Fig. 4c). The residues 1071 to 1178, for which the motion of the backbone 15N-1H moieties is essentially restricted to the overall tumbling of the molecule, have positive NOE values of about 0.8, whereas the residues 1035 to 1065 and 1180 to 1203 have values in the range of −0.4 to 0.4, indicating increased dynamics on the subnanosecond time scale for these polypeptide segments.
FIG. 5.
Structural organization of the nsp3 fragment containing residues 1 to 1318. The solid line at the top indicates the initial domain annotation based on bioinformatics and phylogenetic analyses. The dashed line to the right represents the C-terminal segment of residues 1318 to 1922 of nsp3. Below, the presently known structural coverage with globular domains and flexibly disordered linker segments is shown. Ribbon representations are used for the globular domains. Flexibly disordered regions revealed by NMR spectroscopy and disordered segments implicated by X-ray crystallography are shown as blue and green lines, respectively. Gray lines and rectangles represent polypeptide segments with so far unknown structures. SUD-C, C-terminal region of SUD.
Overall, nsp3 is characterized by the arrangement of small globular domains linked by flexibly disordered polypeptide segments (Fig. 5). This domain distribution may play a functional role by governing substrate accessibility and protein-protein interactions, which could then result in spatial proximity of multiple activities of proteases, deubiquitination factors, and nucleic acid-binding domains (45, 48, 49). Possible interactions between nsp3 and other SARS-CoV proteins have been investigated using binding studies with nsp3 fragments of various lengths (21, 40, 57). Considering those fragments that contain the NAB, Imbert et al. (21) reported that nsp3(1033-1418) interacts with multiple other SARS-CoV nonstructural proteins, including nsp5, nsp12, nsp13, nsp14, nsp15, nsp16, and other nsp3 domains; von Brunn et al. (57) reported interactions of nsp3(722-1921) with nsp2, ORF3a, and ORF9b; Pan et al. (40) reported interactions of nsp3(726-1438) with nsp4 and nsp12. These results indicate that nsp3 may have multiple functional partners within the SARS-CoV proteome. The now available structural coverage of nsp3 (Fig. 5) provides a basis for the design of additional interaction studies that could yield specific information about individual nsp3 domains.
The combination of flexibly disordered regions and structured binding motifs, such as the Staufen double-stranded RNA-binding domains and the RNA recognition domains of the polypyrimidine tract-binding protein, is a common feature in RNA interaction proteins. Modular organization can increase the specificity and affinity of binding compared with those of the individual domains, for example, by allowing simultaneous interactions with different segments of an RNA sequence. Within nsp3, potential partners of the NAB for such concerted action include UB1, which exhibits a ubiquitin-like fold with nucleic acid-binding activity (49), and SUD-N and SUD-M, which adopt macrodomain folds and bind RNA (5, 55).
Mapping of ssRNA-binding sites.
The NAB has been shown to interact with nucleic acids (36). Here, we used binding experiments with exogenous ssRNA to investigate the locations of nucleic acid-binding sites. Chemical shift perturbation studies were performed using uniformly 15N-labeled nsp3(1066-1181) and unlabeled ssRNA1, which has the sequence 5′-AAAUACCUCUCAAAAAUAACACCACACCAUAUACCACAU-3′. This RNA was chosen as a follow-up to our earlier study, in which EMSAs showed binding of the NAB to this sequence (36). The oligonucleotide was designed to contain the sequence AUA, which copurified with nsp3a during expression in E. coli (49), and to be single stranded with minimal secondary structure.
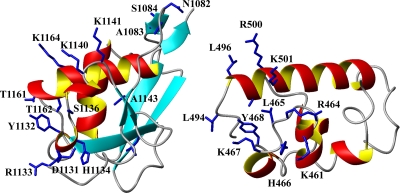
Upon the addition of a threefold excess of the ssRNA, a small number of the residues show highly selective chemical shift perturbations (Fig. 6). These residues are located in a positively charged protein surface patch defined by the residues K1140, K1141, K1164, and R1171 (K75, K76, K99, and R106 in the construct numeration in Fig. 1d; see also Fig. 8). The nearby residues N1082, A1083, S1084, D1131, H1134, and T1162 are also affected by the presence of ssRNA. It is worth mentioning that although the NAB exhibits a new fold with no apparent similarity to other RNA-binding proteins, a detailed comparison of the nucleic acid-binding site thus identified with those of other RNA-binding polypeptides revealed significant homologies. These RNA interaction sites typically consist of a surface patch of positively charged and aromatic residues, some of which are also neighbors in the sequence (28, 36, 37, 44). The NAB shows particularly close similarity to the RNA-binding site of the sterile alpha motif (SAM) of the Saccharomyces cerevisiae Vts1p protein, which exhibits high affinity for ssRNA with the sequence CNGGN, where N can be any of the four ribonucleotides. Vts1p is an α-helical protein in which the residues involved in binding to ssRNA are located in a loop between helices α2 and α3 (464RLHKY468) and in the first two turns of helix α5 (496LGARK501). In the NAB, most of the interacting residues are located in the linker segment comprising the two 310-helices and at the start of helix α2. In addition to the common arrangement of a patch of positively charged residues in the two proteins, conservation of other residues exists. For example, Y1132 and H1134 in the SARS-CoV NAB (Y67 and H69 in Fig. 1d) correspond to Y468 and H466 in Vts1p (Fig. 7).
FIG. 6.
Studies of RNA binding using chemical shift perturbation experiments. Panel a shows the 2D [15N,1H]-HSQC spectra of nsp3(1066-1181) in the absence (red) and presence (blue) of a threefold excess of ssRNA1 (see the text for the sequence). Residues with chemical shift changes are indicated, and some are also shown in panels b to d in expanded plots of the superimposed 2D [15N,1H]-HSQC spectra.
FIG. 7.
Ribbon representations of residues 1071 to 1177 in the NMR structure of nsp3(1066-1181) (left) and residues 445 to 517 in the SAM of Vst1p (right). In nsp3(1066-1181), the side chains of the residues with significant chemical shift perturbations (Fig. 6) are indicated. For the Vts1p SAM (PDB code 2ese), the residues located in the ssRNA interaction site described by Oberstrass et al. (37) are indicated.
Investigation of SARS-CoV NAB homologues with the use of BLAST searches revealed four relatively distant protein clusters corresponding to the four major group II coronavirus lineages (Fig. 8). Comparison of the species listed in Fig. 8 reveals significant sequence conservation in the segments corresponding to the β-strands of SARS-CoV nsp3(1066-1181), indicating that at least some features of this structure might be present also in the homologous proteins. One observes further that there is pronounced divergence in the polypeptide regions implicated in RNA binding, suggesting that conservation on the level of the 3D structure would not necessarily go along with conservation of the physiological function.
RNA-binding specificity.
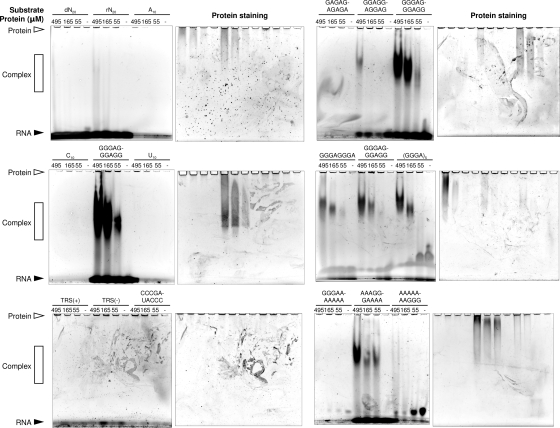
The results described above allowed us to delineate the RNA-binding site on nsp3e. We followed this up by carrying out additional EMSA experiments to test the range of RNA sequences to which nsp3e might bind. We selected a group of RNAs that had previously also been studied with other nsp3 domains (5, 36; Johnson, M. A., A. Chatterjee, et al., unpublished data); thus, in addition to characterizing binding to the NAB, the experiments enabled us to conduct comparisons of RNA binding to the NAB with that to the other nsp3 domains. Figure 9 shows that nsp3e binds strongly to A- and G-containing RNAs, especially (GGGA)5, (GGGA)2, GGGAGGGAGG, GGAGGAGGAG, AAAAAAAGGG, and AAAGGGAAAA. The protein has weak affinity for random RNA sequences and minimal affinity for random DNA sequences.
FIG. 9.
Results from EMSA experiments demonstrating nucleic acid binding to nsp3e. In each column, pairs of gel photographs are shown: on the left, the gels are stained for RNA, and on the right, the same gels are stained for protein. The nucleic acid sequence and the protein concentrations, ranging from 0 to 495 μM, are given above each gel. To the left, the position of the protein is indicated by an open triangle, that of the RNA is indicated by a closed triangle, and the positions of protein-RNA complexes are indicated by open rectangles. The nucleic acid concentrations used were 80 μM for 20-mers, 160 μM for decamers, and 190 μM for octamers. dN20 and rN20, randomized 20-mer DNA and RNA.
nsp3e appears to bind most strongly to the sequences containing repeats of three consecutive guanosines; for example, in Fig. 9, middle right panels, a majority of the (GGGA)2 and GGGAGGGAGG RNAs are bound to the protein even at an approximately 1:1 RNA/protein ratio. GGAGGAGGAG is also bound, but not quite as strongly. No evidence was seen for binding to A10, U10, or C10, to TRS(+) or its reverse complement, TRS(−), or to RNA oligomer 5′-CCCGAUACCC-3′, which contains the GAUA sequence that is recognized by the N-terminal domain of nsp3, nsp3a.
An earlier NMR study demonstrated that the macrodomain which forms SUD-M binds to A10 but has very little affinity for U10. EMSAs showed weak binding of SUD-M to A15, (ACUG)5, and TRS(−). No affinity for TRS(+) or for the nsp3a oligomer was observable (5). Recent work in our laboratory (Johnson et al., unpublished) showed that SUD-M and the peptide comprising SUD-M and the SUD C-terminal region are both purine RNA-binding proteins and also have affinities for a range of G- and A-containing RNA sequences. The peptide comprising SUD-N and SUD-M has been shown to bind guanosines (54, 55). Thus, overall, the binding behavior of nsp3e appears to be similar to that of at least two other regions of nsp3, namely, the N-terminal nsp3a domain and the SUD. This may indicate functional linkage among the domains of nsp3.
In conclusion, the present paper extends the structural and functional coverage of the SARS-CoV nsp3 to the N-terminal part of the initially annotated nsp3e domain. Figure 5 illustrates that this extension with flexible linkers and a globular domain is in line with the “string-of-pearls” appearance of the preceding nsp3 polypeptide region. In addition to performing the global structural characterization, we identified an ssRNA-binding site on the surface of the globular domain nsp3(1066-1181). The overall flexible arrangement of the globular domains along the nsp3 polypeptide chain (Fig. 5) indicates the possibility of concerted actions by multiple functionalities represented by different regions of the polypeptide chain, including ssRNA binding by the nsp3(1066-1181) globular domain.
Acknowledgments
This study was supported by NIAID/NIH contract no. HHSN266200400058C, “Functional and Structural Proteomics of the SARS-CoV,” and the Joint Center for Structural Genomics through NIH/NIGMS grant no. U54-GM074898 and SARS RO1 grant AI059799. P.S. was supported by a fellowship from the Spanish Ministry of Science and Education and by the Skaggs Institute for Chemical Biology. M.A.J. was supported by a fellowship from the Canadian Institutes of Health Research and by the Skaggs Institute for Chemical Biology. K.W. is the Cecil H. and Ida M. Green Professor of Structural Biology at The Scripps Research Institute.
Footnotes
Published ahead of print on 14 October 2009.
REFERENCES
- 1.Almeida, M. S., M. A. Johnson, T. Herrmann, M. Geralt, and K. Wuthrich. 2007. Novel beta-barrel fold in the nuclear magnetic resonance structure of the replicase nonstructural protein 1 from the severe acute respiratory syndrome coronavirus. J. Virol. 81:3151-3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bai, Y., J. S. Milne, L. Mayne, and S. W. Englander. 1993. Primary structure effects on peptide group hydrogen exchange. Proteins 17:75-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker, S. C., K. Yokomori, S. Dong, R. Carlisle, A. E. Gorbalenya, E. V. Koonin, and M. M. Lai. 1993. Identification of the catalytic sites of a papain-like cysteine proteinase of murine coronavirus. J. Virol. 67:6056-6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barretto, N., D. Jukneliene, K. Ratia, Z. Chen, A. D. Mesecar, and S. C. Baker. 2005. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 79:15189-15198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chatterjee, A., M. A. Johnson, P. Serrano, B. Pedrini, J. S. Joseph, B. W. Neuman, K. Saikatendu, M. J. Buchmeier, P. Kuhn, and K. Wuthrich. 2009. Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold. J. Virol. 83:1823-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, Z., Y. Wang, K. Ratia, A. D. Mesecar, K. D. Wilkinson, and S. C. Baker. 2007. Proteolytic processing and deubiquitinating activity of papain-like proteases of human coronavirus NL63. J. Virol. 81:6007-6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chou, P. Y., and G. D. Fasman. 1977. Beta-turns in proteins. J. Mol. Biol. 115:135-175. [DOI] [PubMed] [Google Scholar]
- 8.Cornell, W. D., P. Cieplak, C. I. Bayly, I. R. Gould, K. M. Merz, D. M. Ferguson, D. C. Spellmeyer, T. Fox, J. W. Caldwell, and P. A. Kollman. 1995. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 117:5179-5197. [Google Scholar]
- 9.Egloff, M. P., H. Malet, A. Putics, M. Heinonen, H. Dutartre, A. Frangeul, A. Gruez, V. Campanacci, C. Cambillau, J. Ziebuhr, T. Ahola, and B. Canard. 2006. Structural and functional basis for ADP-ribose and poly(ADP-ribose) binding by viral macro domains. J. Virol. 80:8493-8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eriksson, K. K., L. Cervantes-Barragan, B. Ludewig, and V. Thiel. 2008. Mouse hepatitis virus liver pathology is dependent on ADP-ribose-1″-phosphatase, a viral function conserved in the alpha-like supergroup. J. Virol. 82:12325-12334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gorbalenya, A. E., L. Enjuanes, J. Ziebuhr, and E. J. Snijder. 2006. Nidovirales: evolving the largest RNA virus genome. Virus Res. 117:17-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorbalenya, A. E., E. V. Koonin, A. P. Donchenko, and V. M. Blinov. 1989. Coronavirus genome: prediction of putative functional domains in the non-structural polyprotein by comparative amino acid sequence analysis. Nucleic Acids Res. 17:4847-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorbalenya, A. E., E. V. Koonin, and M. M. Lai. 1991. Putative papain-related thiol proteases of positive-strand RNA viruses. Identification of rubi- and aphthovirus proteases and delineation of a novel conserved domain associated with proteases of rubi-, alpha- and coronaviruses. FEBS Lett. 288:201-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graham, R. L., and M. R. Denison. 2006. Replication of murine hepatitis virus is regulated by papain-like proteinase 1 processing of nonstructural proteins 1, 2, and 3. J. Virol. 80:11610-11620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Güntert, P., C. Mumenthaler, and K. Wüthrich. 1997. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 273:283-298. [DOI] [PubMed] [Google Scholar]
- 16.Harcourt, B. H., D. Jukneliene, A. Kanjanahaluethai, J. Bechill, K. M. Severson, C. M. Smith, P. A. Rota, and S. C. Baker. 2004. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J. Virol. 78:13600-13612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herrmann, T., P. Güntert, and K. Wüthrich. 2002. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 319:209-227. [DOI] [PubMed] [Google Scholar]
- 18.Herrmann, T., P. Güntert, and K. Wüthrich. 2002. Protein NMR structure determination with automated NOE-identification in the NOESY spectra using the new software ATNOS. J. Biomol. NMR 24:171-189. [DOI] [PubMed] [Google Scholar]
- 19.Holm, L., S. Kaariainen, P. Rosenstrom, and A. Schenkel. 2008. Searching protein structure databases with DaliLite v. 3. Bioinformatics 24:2780-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holm, L., and C. Sander. 1995. Dali: a network tool for protein structure comparison. Trends Biochem. Sci. 20:478-480. [DOI] [PubMed] [Google Scholar]
- 21.Imbert, I., E. J. Snijder, M. Dimitrova, J.-C. Guillemot, P. Lecine, and B. Canard. 2008. The SARS-coronavirus Plnc domain of the nsp3 as a replication/transcription scaffolding protein. Virus Res. 133:136-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanjanahaluethai, A., Z. Chen, D. Jukneliene, and S. C. Baker. 2007. Membrane topology of murine coronavirus replicase nonstructural protein 3. Virology 361:391-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keller, R. L. J. 2005. Optimizing the process of NMR spectrum analysis and computer aided resonance assignment. Thesis. ETH Zürich no. 15947. Swiss Federal Institute of Technology Zürich, Zürich, Switzerland.
- 24.Knoops, K., M. Kikkert, S. H. Worm, J. C. Zevenhoven-Dobbe, Y. van der Meer, A. J. Koster, A. M. Mommaas, and E. J. Snijder. 2008. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 6:e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koradi, R., M. Billeter, and P. Güntert. 2000. Point-centered domain decomposition for parallel molecular dynamics simulation. Comp. Phys. Commun. 124:139-147. [Google Scholar]
- 26.Koradi, R., M. Billeter, and K. Wüthrich. 1996. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 14:51-55, 29-32. [DOI] [PubMed] [Google Scholar]
- 27.Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283-291. [Google Scholar]
- 28.Lewis, H. A., K. Musunuru, K. B. Jensen, C. Edo, H. Chen, R. B. Darnell, and S. K. Burley. 2000. Sequence-specific RNA binding by a Nova KH domain: implications for paraneoplastic disease and the fragile X syndrome. Cell 100:323-332. [DOI] [PubMed] [Google Scholar]
- 29.Linding, R., R. B. Russell, V. Neduva, and T. J. Gibson. 2003. GlobPlot: exploring protein sequences for globularity and disorder. Nucleic Acids Res. 31:3701-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindner, H. A., N. Fotouhi-Ardakani, V. Lytvyn, P. Lachance, T. Sulea, and R. Menard. 2005. The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J. Virol. 79:15199-15208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, D. X., K. W. Tibbles, D. Cavanagh, T. D. Brown, and I. Brierley. 1995. Identification, expression, and processing of an 87-kDa polypeptide encoded by ORF 1a of the coronavirus infectious bronchitis virus. Virology 208:48-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lo Conte, L., B. Ailey, T. J. Hubbard, S. E. Brenner, A. G. Murzin, and C. Chothia. 2000. SCOP: a structural classification of protein database. Nucleic Acids Res. 28:257-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luginbühl, P., P. Güntert, M. Billeter, and K. Wüthrich. 1997. The new program OPAL for molecular dynamics simulations and energy refinements of biological macromolecules. J. Biomol. NMR 8:136-146. [DOI] [PubMed] [Google Scholar]
- 34.Malet, H., B. Coutard, S. Jamal, H. Dutartre, N. Papageorgiou, M. Neuvonen, T. Ahola, N. Forrester, E. A. Gould, D. Lafitte, F. Ferron, J. Lescar, A. E. Gorbalenya, X. de Lamballerie, and B. Canard. 2009. The crystal structures of Chikungunya and Venezuelan equine encephalitis virus nsP3 macro domains define a conserved adenosine binding pocket. J. Virol. 83:6534-6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marra, M. A., S. J. Jones, C. R. Astell, R. A. Holt, A. Brooks-Wilson, Y. S. Butterfield, J. Khattra, J. K. Asano, S. A. Barber, S. Y. Chan, A. Cloutier, S. M. Coughlin, D. Freeman, N. Girn, O. L. Griffith, S. R. Leach, M. Mayo, H. McDonald, S. B. Montgomery, P. K. Pandoh, A. S. Petrescu, A. G. Robertson, J. E. Schein, A. Siddiqui, D. E. Smailus, J. M. Stott, G. S. Yang, F. Plummer, A. Andonov, H. Artsob, N. Bastien, K. Bernard, T. F. Booth, D. Bowness, M. Czub, M. Drebot, L. Fernando, R. Flick, M. Garbutt, M. Gray, A. Grolla, S. Jones, H. Feldmann, A. Meyers, A. Kabani, Y. Li, S. Normand, U. Stroher, G. A. Tipples, S. Tyler, R. Vogrig, D. Ward, B. Watson, R. C. Brunham, M. Krajden, M. Petric, D. M. Skowronski, C. Upton, and R. L. Roper. 2003. The genome sequence of the SARS-associated coronavirus. Science 300:1399-1404. [DOI] [PubMed] [Google Scholar]
- 36.Neuman, B. W., J. S. Joseph, K. S. Saikatendu, P. Serrano, A. Chatterjee, M. A. Johnson, L. Liao, J. P. Klaus, J. R. Yates III, K. Wuthrich, R. C. Stevens, M. J. Buchmeier, and P. Kuhn. 2008. Proteomics analysis unravels the functional repertoire of coronavirus nonstructural protein 3. J. Virol. 82:5279-5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oberstrass, F. C., A. Lee, R. Stefl, M. Janis, G. Chanfreau, and F. H.-T. Allain. 2006. Shape-specific recognition in the structure of the Vts1p SAM domain with RNA. Nat. Struct. Mol. Biol. 13:160-167. [DOI] [PubMed] [Google Scholar]
- 38.Oostra, M., M. C. Hagemeijer, M. van Gent, C. P. Bekker, E. G. te Lintelo, P. J. Rottier, and C. A. de Haan. 2008. Topology and membrane anchoring of the coronavirus replication complex: not all hydrophobic domains of nsp3 and nsp6 are membrane spanning. J. Virol. 82:12392-12405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Orengo, C. A., A. D. Michie, S. Jones, D. T. Jones, M. B. Swindells, and J. M. Thornton. 1997. CATH—a hierarchic classification of protein domain structures. Structure 5:1093-1108. [DOI] [PubMed] [Google Scholar]
- 40.Pan, J., X. Peng, Y. Gao, Z. Li, X. Lu, Y. Chen, M. Ishaq, D. Liu, M. DeDiego, L. Enjuanes, and D. Guo. 2008. Genome-wide analysis of protein-protein interactions and involvement of viral proteins in SARS-CoV replication. PLoS One 3:e3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Piotrowski, Y., G. Hansen, A. L. Boomaars-van der Zanden, E. J. Snijder, A. E. Gorbalenya, and R. Hilgenfeld. 2009. Crystal structures of the X-domains of a group-1 and a group-3 coronavirus reveal that ADP-ribose-binding may not be a conserved property. Protein Sci. 18:6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Putics, A., W. Filipowicz, J. Hall, A. E. Gorbalenya, and J. Ziebuhr. 2005. ADP-ribose-1″-monophosphatase: a conserved coronavirus enzyme that is dispensable for viral replication in tissue culture. J. Virol. 79:12721-12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Putics, A., A. E. Gorbalenya, and J. Ziebuhr. 2006. Identification of protease and ADP-ribose 1″-monophosphatase activities associated with transmissible gastroenteritis virus non-structural protein 3. J. Gen. Virol. 87:651-656. [DOI] [PubMed] [Google Scholar]
- 44.Ramos, A., S. Grünert, J. Adams, D. R. Micklem, M. R. Proctor, S. Freund, M. Bycroft, D. S. Johnson, and G. Varani. 2000. RNA recognition by a Staufen double-stranded RNA-binding domain. EMBO J. 19:997-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ratia, K., K. S. Saikatendu, B. D. Santarsiero, N. Barretto, S. C. Baker, R. C. Stevens, and A. D. Mesecar. 2006. Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. Proc. Natl. Acad. Sci. USA 103:5717-5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Renner, C., M. Schleicher, L. Moroder, and T. A. Holak. 2002. Practical aspects of the 2D 15N-{1H}-NOE experiment. J. Biomol. NMR 23:23-33. [DOI] [PubMed] [Google Scholar]
- 47.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M. H. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Gunther, A. D. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394-1399. [DOI] [PubMed] [Google Scholar]
- 48.Saikatendu, K. S., J. S. Joseph, V. Subramanian, T. Clayton, M. Griffith, K. Moy, J. Velasquez, B. W. Neuman, M. J. Buchmeier, R. C. Stevens, and P. Kuhn. 2005. Structural basis of severe acute respiratory syndrome coronavirus ADP-ribose-1″-phosphate dephosphorylation by a conserved domain of nsP3. Structure 13:1665-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Serrano, P., M. A. Johnson, M. S. Almeida, R. Horst, T. Herrmann, J. S. Joseph, B. W. Neuman, V. Subramanian, K. S. Saikatendu, M. J. Buchmeier, R. C. Stevens, P. Kuhn, and K. Wuthrich. 2007. Nuclear magnetic resonance structure of the N-terminal domain of nonstructural protein 3 from the severe acute respiratory syndrome coronavirus. J. Virol. 81:12049-12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Serrano, P., M. A. Johnson, A. Chatterjee, B. Pedrini, and K. Wüthrich. 2008. NMR assignment of the nonstructural protein nsp3(1066-1181) from SARS-CoV. Biomol. NMR Assign. 2:135-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Snijder, E. J., P. J. Bredenbeek, J. C. Dobbe, V. Thiel, J. Ziebuhr, L. L. Poon, Y. Guan, M. Rozanov, W. J. Spaan, and A. E. Gorbalenya. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331:991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stertz, S., M. Reichelt, M. Spiegel, T. Kuri, L. Martinez-Sobrido, A. Garcia-Sastre, F. Weber, and G. Kochs. 2007. The intracellular sites of early replication and budding of SARS-coronavirus. Virology 361:304-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sulea, T., H. A. Lindner, E. O. Purisima, and R. Menard. 2005. Deubiquitination, a new function of the severe acute respiratory syndrome coronavirus papain-like protease? J. Virol. 79:4550-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tan, J., Y. Kusov, D. Mutschall, S. Tech, K. Nagarajan, R. Hilgenfeld, and C. L. Schmidt. 2007. The “SARS-unique domain” (SUD) of SARS coronavirus is an oligo(G)-binding protein. Biochem. Biophys. Res. Commun. 364:877-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tan, J., C. Vonrhein, O. S. Smart, G. Bricogne, M. Bollati, Y. Kusov, G. Hansen, J. R. Mesters, C. L. Schmidt, and R. Hilgenfeld. 2009. The SARS-unique domain (SUD) of SARS coronavirus contains two macrodomains that bind G-quadruplexes. PLoS Pathog. 5:e1000428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thiel, V., K. A. Ivanov, A. Putics, T. Hertzig, B. Schelle, S. Bayer, B. Weissbrich, E. J. Snijder, H. Rabenau, H. W. Doerr, A. E. Gorbalenya, and J. Ziebuhr. 2003. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 84:2305-2315. [DOI] [PubMed] [Google Scholar]
- 57.von Brunn, A., C. Teepe, J. C. Simpson, R. Pepperkok, C. C. Friendel, R. Zimmer, R. Roberts, R. Baric, and J. Haas. 2007. Analysis of intraviral protein-protein interactions of the SARS coronavirus ORFeome. PLoS One 2:e459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wishart, D. S., C. G. Bigam, J. Yao, F. Abildgaard, H. J. Dyson, E. Oldfield, J. L. Markley, and B. D. Sykes. 1995. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J. Biomol. NMR 6:135-140. [DOI] [PubMed] [Google Scholar]
- 59.Xu, Y., L. Cong, C. Chen, L. Wei, Q. Zhao, X. Xu, Y. Ma, M. Bartlam, and Z. Rao. 2009. Crystal structures of two coronavirus ADP-ribose-1″-monophosphatases and their complexes with ADP-ribose: a systematic structural analysis of the viral ADRP domain. J. Virol. 83:1083-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zheng, D., G. Chen, B. Guo, G. Cheng, and H. Tang. 2008. PLP2, a potent deubiquitinase from murine hepatitis virus, strongly inhibits cellular type I interferon production. Cell Res. 18:1105-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu, G., Y. Xia, L. K. Nicholson, and K. H. Sze. 2000. Protein dynamics measurements by TROSY-based NMR experiments. J. Magn. Reson. 143:423-426. [DOI] [PubMed] [Google Scholar]
- 62.Ziebuhr, J., B. Schelle, N. Karl, E. Minskaia, S. Bayer, S. G. Siddell, A. E. Gorbalenya, and V. Thiel. 2007. Human coronavirus 229E papain-like proteases have overlapping specificities but distinct functions in viral replication. J. Virol. 81:3922-3932. [DOI] [PMC free article] [PubMed] [Google Scholar]