Abstract
Rift Valley fever virus (RVFV) (genus Phlebovirus, family Bunyaviridae) causes mosquito-borne epidemic diseases in humans and livestock. The virus carries three RNA segments, L, M, and S, of negative or ambisense polarity. L protein, an RNA-dependent RNA polymerase, encoded in the L segment, and N protein, encoded in the S segment, exert viral RNA replication and transcription. Coexpression of N, hemagglutinin (HA)-tagged L, and viral minigenome resulted in minigenome replication and transcription, a finding that demonstrated HA-tagged L was biologically active. Likewise L tagged with green fluorescent protein (GFP) was biologically competent. Coimmunoprecipitation analysis using extracts from cells coexpressing HA-tagged L and GFP-tagged L showed the formation of an L oligomer. Bimolecular fluorescence complementation analysis and coimmunoprecipitation studies demonstrated the formation of an intermolecular L-L interaction through its N-terminal and C-terminal regions and also suggested an intramolecular association between the N-terminal and C-terminal regions of L protein. A biologically inactive L mutant, in which the conserved signature SDD motif was replaced by the amino acid residues GNN, exhibited a dominant negative phenotype when coexpressed with wild-type L in the minigenome assay system. Expression of this mutant L also inhibited viral gene expression in virus-infected cells. These data provided compelling evidence for the importance of oligomerization of RVFV L protein for its polymerase activity.
Rift Valley Fever virus (RVFV), which belongs to the genus Phlebovirus in the family Bunyaviridae, is endemic in sub-Saharan African countries and causes large outbreaks in endemic areas and countries outside of the endemic area, including Egypt, Saudi Arabia, and Yemen (2). RVFV is transmitted by mosquitoes, and the virus infection causes a high rate of abortions in pregnant ruminants and acute lethal hepatitis in newborn lambs (26). Most human patients show an acute febrile myalgic syndrome, with a small minority of patients, perhaps ∼1%, experiencing severe hemorrhagic fever or encephalitis. Also, some patients show retinal vasculitis, which results in partial blindness for an undefined period (22).
RVFV has three single-stranded genomic RNA segments, designated L, M, and S. The L and M RNA segments are of negative polarity. The L segment contains a 6,279-nucleotide-long open reading frame (ORF) encoding L protein and a viral RNA-dependent RNA polymerase (RdRp), while the M segment has a single ORF encoding NSm protein, a 78-kDa protein, and two major viral envelope proteins, Gn and Gc. The S segment is of ambisense polarity and encodes NSs protein and N protein; the former and the latter are encoded in anti-viral-sense RNA and in viral-sense RNA, respectively. In bunyaviruses, both L protein and N protein are needed for viral RNA replication and viral mRNA synthesis, the latter of which uses a host mRNA-derived cap structure as a primer (3, 16, 29). N protein encapsidates viral RNAs to form helical nucleocapsids, which serve as a template for viral RNA synthesis (21, 23).
RVFV L protein plays a central role in viral RNA synthesis, yet very little is known about how L protein exerts its functions; the properties of viral replication complexes, structures of viral RNA polymerases, and host factors needed for viral RNA synthesis have been characterized poorly for bunyaviruses. Past studies of other negative-stranded RNA viruses, including Sendai virus (36), parainfluenza virus 3 (37), and measles virus (4), all of which belong to the paramyxoviruses, and lymphocytic choriomeningitis virus (LCMV) (32), an arenavirus, suggested that their L proteins function as an oligomer to exert the RdRp function. In addition, the influenza A virus RdRp heterotrimer (18, 19) has been reported to form oligomers. Currently, it is unclear whether the L proteins of bunyaviruses also form an oligomer and exhibit RdRp activities in infected cells. We present here that RVFV L protein forms an oligomer. We further identified L protein regions that were involved in L oligomerization and explored the biological importance of the L protein oligomer for viral RNA synthesis.
MATERIALS AND METHODS
Cells and viruses.
BHK/T7-9 cells stably expressing T7 RNA polymerase (15) and Vero E6 cells were maintained as described previously (14). rMP12-rLuc (14), an RVFV MP-12 strain carrying the Renilla luciferase (rLuc) gene in place of the NSs gene in the S segment, was used.
Plasmids.
Plasmids encoding the mutant L protein were generated by modification of pT7-IRES-vL encoding the MP-12 L protein (14). pT7-IRES-HA-L and pT7-IRES-GFP-L were constructed by adding a hemagglutinin (HA) tag sequence (TAC CCC TAC GAC GTG CCC GAC TAC GCC) and by inserting a cassette of the green fluorescent protein (GFP) gene and 10-alanine linker sequence (5) between the encephalomyocarditis virus internal ribosome entry site (IRES) and the L gene ORF of pT7-IRES-vL, respectively. pT7-IRES-GFP was constructed by replacing the L gene of pT7-IRES-vL with a GFP gene. The nucleotide residues TCA GAT GAT encoding SDD at amino acids 1132 to 1134 in the L gene of pT7-IRES-GFP-L were replaced with GGA AAT AAT encoding GNN, resulting in pT7-IRES-GFP-L(GNN). Unique restriction enzyme sites (see Fig. 5A) were used to make pT7-IRES-vL-derived plasmids, each of which had a deletion at different sites of the L protein. A modified version of yellow fluorescent protein (YFP), Venus (25), was synthesized (Bio Basic) and inserted upstream of the L gene ORF, as described for pT7-IRES-GFP-L. A series of plasmids were constructed by adding the sequences encoding amino acid residues 1 to 172 of Venus (12) (VN) or 155 to 239 of Venus (VC) with a linker (GGGGS)3 sequence (34) to the N and/or C terminus of the L gene of pT7-IRES-vL. We also constructed a plasmid encoding VN-tubulin, a fusion protein consisting of N-terminal VN, a linker (GGGGS)3 sequence, and an alpha-tubulin gene.
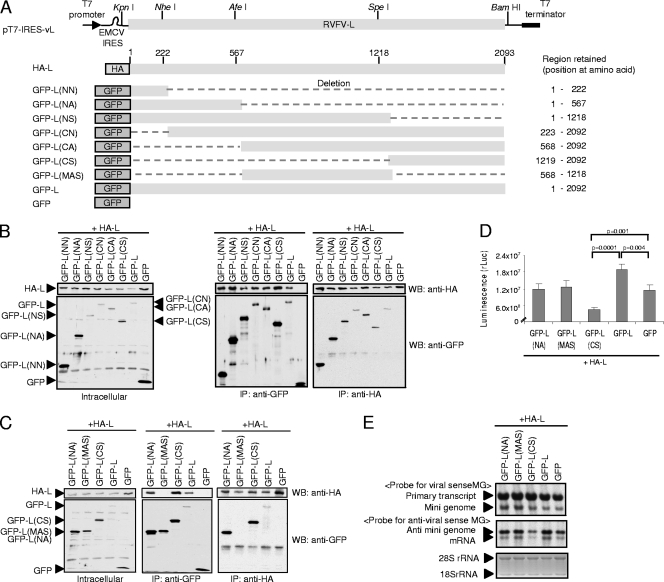
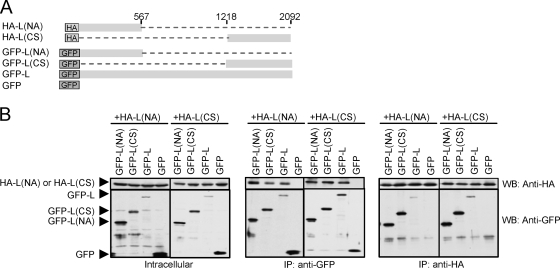
FIG. 5.
Interaction between L protein fragments and full-length L protein in expressed cells. (A) A schematic diagram of HA-L-, GFP-L-, GFP-, and GFP-L-derived deletion mutants. The structure of pT7-IRES-vL is shown at the top of the diagram; unique restriction enzyme sites in pT7-IRES-vL are also shown. Deletions are shown as dashed lines. The amino acid regions of the L protein, which are retained in the deletion mutants, are shown on the right. (B) BHK/T7-9 cells in a six-well plate were cotransfected with 2.0 μg of pT7-IRES-HA-L and 2.0 μg of the indicated plasmid. Intracellular proteins were extracted at 24 h posttransfection. Western blot analysis (left) of cell extracts using anti-HA antibody (top) and anti-GFP antibody (bottom) is shown. The same cell extracts were subjected to coimmunoprecipitation analysis using anti-GFP antibody (middle) or anti-HA antibody (right). Anti-HA antibody and anti-GFP antibody were used to detect immunoprecipitated proteins, as shown in the top and bottom panels, respectively. (C) Experiments similar to those described in the legend for panel B were performed by using a plasmid different from those shown at the top of panel B. (D) BHK/T7-9 cells in a 12-well plate were cotransfected with 1 μg of pT7-IRES-vN, 1 μg of pPro-T7-M-rLuc(−), 0.25 μg of pT7-IRES-HA-L, and 0.25 μg of plasmid encoding the protein shown in the diagram. Cell extracts were prepared at 36 h posttransfection, and rLuc activities were determined. The values represent the mean ± standard deviation of three independent experiments. The P values between the indicated samples were determined by Student's t test. (E) Plasmid transfection was performed as described in the legend for panel D, and intracellular RNAs were extracted at 36 h posttransfection. Northern blot analysis was performed as described in the legend for Fig. 1B.
Minigenome assay.
Subconfluent BHK/T7-9 cell monolayers in a 12-well plate were cotransfected with 0.5 μg of pT7-IRES-vL, 1 μg of pT7-IRES-vN encoding N protein (14), and 1 μg of pPro-T7-M-rLuc(−) encoding a viral-sense RVFV minigenome carrying an rLuc gene (13). The total amount of plasmid was adjusted to 2.5 μg by using empty-vector pT7-IRES (13). In some experiments, pT7-IRES-vL was substituted with another plasmid encoding a mutant L gene. Different amounts of plasmids encoding the L protein or those encoding mutant L proteins were also used as indicated below. Cell extracts were prepared for measuring rLuc activities and Northern blot analysis at 36 h posttransfection. For rLuc activities, cells were lysed with 1 ml of lysis buffer supplied by the manufacturer, and luminescences were measured according to the manufacturer's protocol (Promega). Statistical analysis was performed for three independent experiments to obtain mean and standard deviation. For Northern blot analysis, total intracellular RNA was extracted by using Trizol (Invitrogen) according to the manufacturer's instruction.
Coimmunoprecipitation analysis.
BHK/T7-9 cells in a six-well plate were cotransfected with 2 μg of plasmid pT7-IRES-HA-L and 2 μg of plasmid pT7-IRES-GFP-L. After washing the cells with cold phosphate-buffered saline twice at 24 h posttransfection, cell extracts were prepared by lysing cells on ice for 20 min in 300 μl of TN buffer (50 mM Tris-HCl, pH 8.0, and 150 mM NaCl) containing 1% NP-40 and protease inhibitor (complete mini EDTA free; Roche). Lysates were centrifuged at 10,000 × g for 10 min at 4°C to remove nuclei and large debris. Then the lysates were incubated with 25 μl of anti-GFP microbeads (Miltenyi Biotec) for 2 h on ice with occasional mixing, and immunoprecipitation was performed according to the manufacturer's protocol, with additional extensive washing. For immunoprecipitation with anti-HA antibody, precleared lysate was incubated with 40 μl (bed volume of 20 μl) of mouse anti-HA monoclonal antibody-conjugated agarose (Sigma) overnight with rotation. The agarose was washed five times with 1 ml of cold TN buffer containing 1% NP-40 and protease inhibitor and boiled with sodium dodecyl sulfate (SDS) sample buffer for 5 min. The supernatant of the agarose was subjected to SDS-polyacrylamide gel electrophoresis (PAGE) and subsequent Western blot analysis.
Western blot analysis.
Proteins that were separated by 7.5% or 12% SDS-PAGE were blotted on a polyvinylidene difluoride membrane (Immobilon-P; Millipore) with CAPS [3-(cyclohexylamino)-1-propanesulfonic acid] buffer (10 mM CAPS, pH 11.0; 10% methanol). Membranes were blocked with TBST buffer (10 mM Tris-HCl, pH 7.6; 130 mM NaCl; and 0.1% Tween 20) containing 1% bovine serum albumin (Santa Cruz) at room temperature for more than 1 h. Anti-GFP rabbit polyclonal antibody (Santa Cruz); anti-HA rabbit polyclonal antibody (Santa Cruz); anti-L 434 rabbit polyclonal antibody, which was produced by immunizing rabbits with a bacterially expressed fragment of the L protein corresponding to the amino acid positions 1 to 434; or anti-actin goat polyclonal antibody (Santa Cruz) was added in TBST buffer containing 1% bovine serum albumin and incubated with membrane at 4°C overnight. The membrane was washed with TBST buffer and incubated with horseradish peroxidase-conjugated, anti-rabbit antibody (Santa Cruz) for 45 min at room temperature. After the membranes were washed, they were developed with an ECL detection kit (GE Healthcare), according to the manufacturer's instructions. Light emission was detected by exposure to Blue autoradiography film (ISC Bioexpress).
Northern blot analysis.
Intracellular RNAs were dissolved in RNase-free distilled water, and 4 μg RNA was incubated in denaturing buffer, which included RBS (20 mM MOPS [morpholinepropanesulfonic acid], 1 mM EDTA, and 5 mM sodium acetate), 50% formamide, and 20% formaldehyde, for 10 min at 68°C. RNAs were separated on 1.5% agarose gels containing RBS and 18% formaldehyde and transferred onto a nylon membrane (Roche). Northern blot analysis was performed with strand-specific RNA probes for a sense and antisense rLuc gene (13) by using a DIG system (Roche) according to the manufacturer's protocol. rRNAs (28S and 18S) were detected by staining agarose gels with ethidium bromide.
Virus infection.
BHK/T7-9 cells in a 12-well plate were cotransfected with 1.0 μg of pT7-IRES-vN and 0.5 μg of one of the following plasmids: pT7-IRES-L, pT7-IRES-L(GNN), or pT7-IRES. At 24 h posttransfection, cells were infected with rMP12-rLuc (14) at a multiplicity of infection (MOI) of 5. Samples were prepared for an rLuc assay at the indicated hours postinfection (p.i.).
Bimolecular fluorescence complementation (BiFC) analysis.
BHK/T7-9 cells in a two-well chambered cover glass were transfected with plasmid encoding a fusion protein of VN and L proteins, a fusion protein of VC and L proteins, or an L protein carrying the N-terminal Venus protein (Venus-L) as controls. In another negative control, cells were cotransfected with plasmid encoding VN-tubulin and plasmid encoding the L protein carrying the N-terminal VC (VC-L). In the experimental group, cells were transfected with 1.0 μg of plasmid encoding a fusion protein and 1.0 μg of an empty vector, pT7-IRES, or cotransfected with 1.0 μg of two plasmids as indicated in the legend to Fig. 6. In all cases, the total amount of plasmids was adjusted to 2.0 μg. At 12 h posttransfection, live cells were visualized by the use of a Zeiss LSM 510 UV META laser-scanning confocal microscope with a YFP-specific filter. In a separate experiment, cells were harvested with SDS sample buffer and then subjected to SDS-PAGE and Western blot analysis.
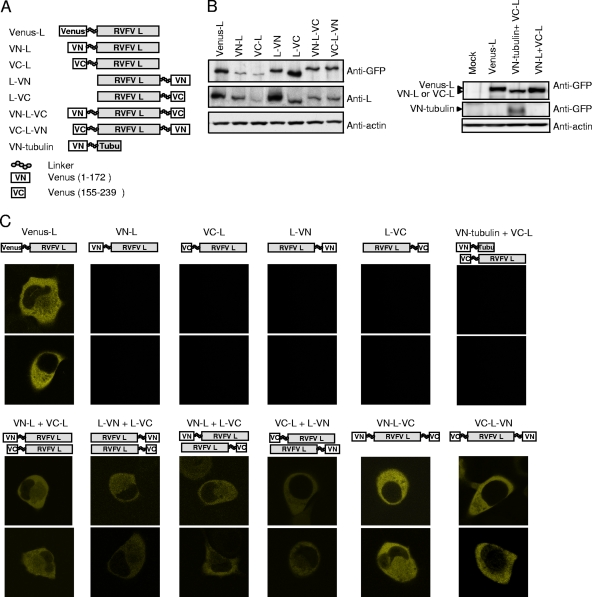
FIG. 6.
BiFC analysis of various full-length L proteins. (A) A schematic diagram of L protein carrying Venus protein at the N terminus (Venus-L) or L proteins carrying an N-terminal fragment of Venus (VN) or a C-terminal fragment of Venus (VC) at their N and/or C terminus with a linker. A schematic diagram of VN-tubulin is also shown. (B) BHK/T7-9 cells in a 12-well plate were independently transfected with 1.0 μg of plasmid expressing the indicated protein. At 12 h posttransfection, intracellular proteins were analyzed by Western blotting by using anti-GFP, anti-L 434, and anti-actin antibodies. (C) BHK/T7-9 cells in a two-well slide were transfected with 1.0 μg of a plasmid or cotransfected with 1.0 μg each of two plasmids as indicated. At 12 h posttransfection, fluorescence in the live cells was observed by confocal microscopy by the use of a YFP filter. For each group, two representative samples are shown.
RESULTS
RVFV L proteins carrying an N-terminal GFP tag or HA tag are biologically active.
We generated two L protein expression plasmids, pT7-IRES-HA-L encoding the L protein tagged with an HA epitope at the N terminus and pT7-IRES-GFP-L encoding the L protein tagged with GFP at the N terminus. BHK/T7-9 cells stably expressing T7 polymerase (15) were independently transfected with pT7-IRES-HA-L, pT7-IRES-GFP-L, and the parental plasmid pT7-IRES-vL encoding the wild-type (wt) L protein (14) or mock transfected. Western blot analysis of cell extracts obtained at 24 h posttransfection showed an efficient expression of GFP-L, HA-L, and wt L proteins; due to an addition of a 27-kDa GFP to the L protein, GFP-L showed a slower migration than HA-L and wt L in the gels (Fig. 1A). No signal was detected in mock-transfected cells.
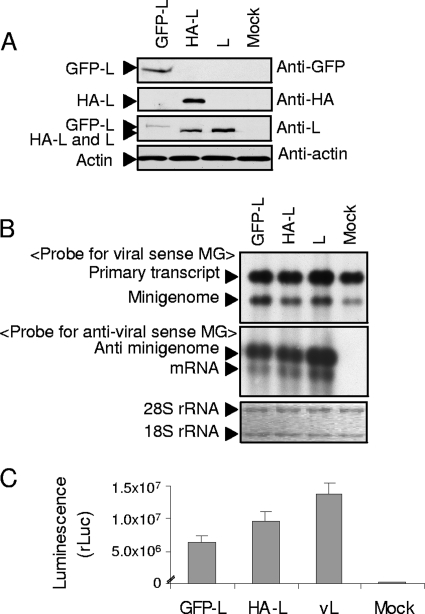
FIG. 1.
Expression and biological activities of HA-L and GFP-L. (A) BHK/T7-9 cells in a 12-well plate were independently transfected with 2.0 μg of pT7-IRES-GFP-L, pT7-IRES-HA-L, and pT7-IRES-vL encoding wt L protein or mock transfected. Cell extracts were prepared at 24 h posttransfection and subjected to Western blot analysis using anti-GFP antibody (top), anti-HA antibody (second from top), anti-L 434 rabbit polyclonal antibody (second from bottom), and anti-actin antibody (bottom). (B and C) BHK/T7-9 cells in a 12-well plate were cotransfected with 0.5 μg of pT7-IRES-vL (lane L), 1.0 μg of pT7-IRES-vN encoding N protein, and 1.0 μg of pPro-T7-M-rLuc(−) encoding a viral-sense RVFV minigenome carrying an rLuc gene. pT7-IRES-GFP-L, pT7-IRES-HA-L, and an empty vector, pT7-IRES, were used in place of pT7-IRES-vL in the lanes labeled GFP-L, HA-L, and Mock, respectively. At 36 h posttransfection, cell extracts were prepared for Northern blot analysis and measurement of rLuc activities. Northern blot analysis (panel B) was performed using the total intracellular RNAs prepared from the plasmid-transfected cells. Expressed primary viral-sense minigenome RNA transcripts (primary transcript) and full-length minigenome RNA (Minigenome) were detected by using a riboprobe (<Probe for viral sense MG>) that specifically binds to the rLuc gene of the expressed minigenome RNA (viral sense polarity) (top). The anti-viral sense of the minigenome (anti-minigenome) and mRNA encoding the rLuc gene (mRNA) were detected by a riboprobe (<Probe for antiviral sense MG>) that specifically binds to the rLuc gene of the antiviral-sense polarity (middle). The bottom panel shows 28S and 18S rRNAs in these RNA samples. The rLuc activities (panel C) from three independent experiments with standard deviation bars are shown.
To test whether HA-L and GFP-L are biologically active, we performed a minigenome assay, in which expressed L and N proteins drove RNA replication and transcription of the expressed viral-sense minigenome RNA carrying a reporter gene in BHK/T7-9 cells; all L genes, the N gene, and the minigenome were cloned in T7 polymerase-driven expression vectors (13, 14). BHK/T7-9 cells were cotransfected with pPro-T7-M-rLuc(−) encoding viral-sense M RNA carrying the rLuc gene in the place of the M gene ORF (13), pT7-IRES-vN encoding the N protein (14), and one of the L expression plasmids, pT7-IRES-GFP-L, pT7-IRES-HA-L, or pT7-IRES-vL. As a negative control, cells were cotransfected with pPro-T7-M-rLuc(−), pT7-IRES-vN, and an empty plasmid, pT7-IRES. At 36 h posttransfection, intracellular RNAs were extracted and subjected to Northern blot analysis. Consistent with our previous study (13), a strand-specific RNA probe that was designed to bind to the rLuc gene of the expressed viral-sense minigenome detected two RNA signals; one was primary transcripts, which did not undergo hepatitis delta virus ribozyme-mediated cleavage of nascent T7 RNA transcripts from pPro-T7-M-rLuc(−), and the other was the expected size of the minigenome RNA (Fig. 1B, top). Northern blot analysis with another strand-specific probe that binds to antiviral sense minigenome RNA showed that wt L, GFP-L, and HA-L were all biologically active to support minigenome RNA replication and transcription (Fig. 1B, middle). The production of comparable levels of rLuc protein, as determined by measuring rLuc activities in extracts from cells expressing wt L, GFP-L, and HA-L further confirmed that all three L proteins were biologically competent (Fig. 1C).
Interaction of GFP-L and HA-L in expressed cells.
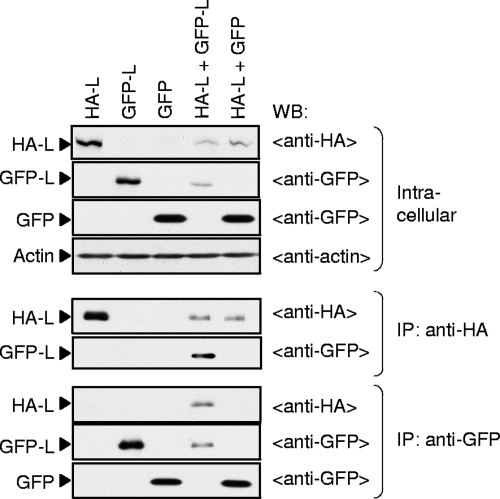
We tested whether HA-L interacted with GFP-L in coexpressed cells. BHK/T7-9 cells were independently transfected with pT7-IRES-HA-L, pT7-IRES-GFP-L, and pT7-IRES-GFP expressing GFP or cotransfected with pT7-IRES-GFP-L and pT7-IRES-HA-L or pT7-IRES-HA-L and pT7-IRES-GFP; 4 μg of plasmid was used for expression of a single protein, while 2 μg of each plasmid was used for coexpression. Western blot analysis of the cell extracts using anti-HA and anti-GFP antibodies showed an efficient accumulation of HA-L, GFP-L, and GFP in the cells expressing a single protein, while the amounts of HA-L and GFP-L in the cells coexpressing both proteins and that of HA-L in the cells coexpressing HA-L and GFP were less prominent, probably partly due to the use of reduced amounts of each plasmid in coexpressing cells (Fig. 2, top). Anti-HA antibody coimmunoprecipitated GFP-L, but not GFP, along with HA-L (Fig. 2, middle). A reciprocal experiment, in which cell extracts were immunoprecipitated with anti-GFP antibody, also showed coprecipitation of HA-L with GFP-L but not with GFP (Fig. 2, bottom). These data demonstrated that HA-L interacted with the L protein region of the GFP-L protein in coexpressed cells.
FIG. 2.
Coimmunoprecipitation analysis of HA-L and GFP-L. BHK/T7-9 cells in a six-well plate were independently transfected with 4.0 μg of pT7-IRES-HA-L (HA-L), pT7-IRES-GFP-L (GFP-L), or pT7-IRES-GFP (GFP) or cotransfected with 2.0 μg of pT7-IRES-HA-L and 2.0 μg of pT7-IRES-GFP-L (HA-L + GFP-L) or 2.0 μg of pT7-IRES-HA-L and 2.0 μg of pT7-IRES-GFP (HA-L + GFP). Cell extracts were prepared at 24 h posttransfection and directly subjected to Western blot analysis using anti-HA antibody, anti-GFP antibody, or anti-actin antibody (top). The same cell extracts were subjected to coimmunoprecipitation analysis using anti-HA antibody (middle) or anti-GFP antibody (bottom). The immunoprecipitated samples were examined by Western blot analysis by using anti-HA or anti-GFP antibodies.
Biological significance of L oligomerization.
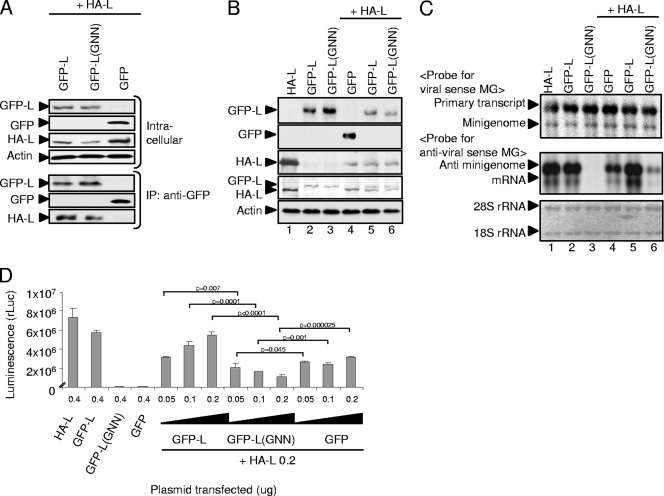
To assess the biological significance of L oligomerization, we generated a functionally inactive L protein mutant and tested whether the interaction between mutant L and wt L inhibited the RdRp activity of the wt L protein; if the L protein functions as an oligomer for viral RNA synthesis, then the physical interaction of an inactive L mutant with an active L protein(s) in cells coexpressing both proteins would result in the formation of a biologically inactive L oligomer, causing the inhibition of the RdRp activity of the active L protein. Because the amino acid sequence SDD in the characteristic motif C of the RVFV L protein is highly conserved in all segmented negative-strand RNA viruses (1, 17, 20, 30) and is essential for RdRp functions (9, 17, 32), pT7-IRES-GFP-L(GNN) was constructed by mutating the amino acids SDD, at the positions 1132 to 1134 of the L protein region in pT7-IRES-GFP-L, to GNN. To know whether GFP-L(GNN) interacted with a biologically active L protein, coimmunoprecipitation analysis was performed using extracts of cells coexpressing GFP-L(GNN) and HA-L (Fig. 3A). Expressed GFP-L(GNN) bound to coexpressed HA-L (Fig. 3A, bottom), an observation demonstrating that the introduced mutation did not prevent L oligomerization. The coexpression of GFP-L(GNN) with N protein and minigenome RNA did not induce minigenome RNA replication and transcription (Fig. 3C, lane 3), regardless of efficient GFP-L(GNN) expression (Fig. 3B, lane 3), demonstrating that GFP-L(GNN) was biologically inactive for RNA synthesis. To determine whether GFP-L(GNN) exerted a dominant negative effect on the RdRp activity of a biologically active L, the minigenome assays were performed in cells coexpressing HA-L and GFP-L(GNN). As controls for the assay, GFP-L and GFP were coexpressed with HA-L. The abundance of minigenome RNA and mRNA was higher in the cells coexpressing GFP-L and HA-L than in those coexpressing HA-L and GFP (Fig. 3C, lanes 3 and 4), which suggested to us that an increase in the amounts of the L protein augmented minigenome RNA synthesis. In contrast, reduced levels of minigenome RNA replication and transcription occurred in the cells coexpressing HA-L and GFP-L(GNN) (Fig. 3C, lane 6), strongly suggesting a dominant negative phenotype of GFP-L(GNN). Consistent with Northern blot analysis, background levels of rLuc activities were detected in the cells expressing GFP-L(GNN) in a minigenome assay, whereas expression of HA-L or GFP-L resulted in high levels of rLuc activities (Fig. 3D). Increasing the amounts of pT7-IRES-HA-L and pT7-IRES-GFP-L to 0.5 μg, however, barely caused a rise in rLuc activities (data not shown). Cotransfection of a fixed amount of pT7-IRES-HA-L with increasing amounts of pT7-IRES-GFP-L, but not of pT7-IRES-GFP, resulted in an increase in rLuc activities (Fig. 3D), whereas cotransfection of a fixed amount of pT7-IRES-HA-L, with increasing amounts of pT7-IRES-GFP-L(GNN), resulted in a greater reduction in rLuc activities. The data that an increase in the amounts of pT7-IRES-GFP-L resulted in an increase in rLuc activities implied that minigenome RNA templates and all factors that are required for viral RNA synthesis were not limiting factors for minigenome RNA synthesis. Accordingly, it was less likely that a reduction of rLuc activities in the cells coexpressing GFP-L(GNN) was simply due to competition for the use of necessary sources for the minigenome RNA synthesis between GFP-L(GNN) and HA-L. Rather, our finding of the decreased levels of rLuc activities in the cells that were transfected with the increased amounts of the plasmid encoding GFP-L(GNN) implied that an oligomer comprising HA-L and GFP-L(GNN) was inactive for minigenome RNA synthesis. These data suggested that an L oligomer, rather than an L monomer, had biological activities for RVFV RNA synthesis.
FIG. 3.
Characterization of a dominant negative mutant of L protein, GFP-L(GNN). (A) BHK/T7-9 cells in a six-well plate were cotransfected with 2.0 μg of pT7-IRES-HA-L and 2.0 μg of one of the following plasmids: pT7-IRES-GFP-L (GFP-L), pT7-IRES-GFP-L(GNN) encoding GFP-L(GNN), or pT7-IRES-GFP (GFP). Cell extracts were prepared at 24 h posttransfection and directly subjected to Western blot analysis (Intracellular) by using anti-GFP antibody, anti-HA antibody, or anti-actin antibody. The same cell extracts were subjected to coimmunoprecipitation analysis using anti-GFP antibody (IP: anti-GFP), and the immunoprecipitated proteins were detected by anti-GFP antibody or anti-HA antibody by Western blot analysis. (B) BHK/T7-9 cells in a 12-well plate were cotransfected with 1.0 μg of pT7-IRES-vN, 1.0 μg of pPro-T7-M-rLuc(−), and 0.5 μg of one of the following plasmids: pT7-IRES-HA-L (lane 1); pT7-IRES-GFP-L (lane 2); pT7-IRES-GFP-L(GNN) (lane 3); a mixture of 0.25 μg of pT7-IRES-HA-L and 0.25 μg of pT7-IRES-GFP (lane 4); a mixture of 0.25 μg of pT7-IRES-HA-L and 0.25 μg of pT7-IRES-GFP-L (lane 5); or a mixture of 0.25 μg of pT7-IRES-HA-L and 0.25 μg of pT7-IRES-GFP-L(GNN) (lane 6). Cell extracts were prepared for Western blot analysis at 36 h posttransfection. Anti-GFP antibody (top two rows), anti-HA antibody (third row from top), anti-L 434 rabbit polyclonal antibody (fourth row from top), and anti-actin antibody (bottom) were used for Western blot analysis. (C) Cells were transfected and prepared as described in the legend for panel B, and intracellular RNAs were subjected to Northern blot analysis as described in the legend for Fig. 1B. (D) BHK/T7-9 cells in a 12-well plate were cotransfected with 1 μg of pT7-IRES-vN, 1 μg of pPro-T7-M-rLuc(−), and one plasmid or two plasmids shown in the diagram at indicated amounts. Cell extracts were prepared at 36 h posttransfection, and rLuc activities were measured. The values represent the mean ± standard deviation of three independent experiments. The P values between the indicated samples were determined by Student's t test.
Effect of dominant negative L mutant on viral gene expression in RVFV-infected cells.
Next, we examined whether the expression of L(GNN), an untagged version of the L mutant carrying the same SDD-to-GNN amino acid mutations as those in GFP-L(GNN), in cells infected with rMP12-rLuc also inhibited viral gene expression. BHK/T7-9 cells were cotransfected with pT7-IRES-L(GNN) encoding the L protein mutant and pT7-IRES-vN. Either pT7-IRES-vL or pT7-IRES was used in place of pT7-IRES-L(GNN) in control groups. At 24 h posttransfection, cells were infected with rMP12-rLuc at an MOI of 5. Cell extracts were prepared at 4 h, 6 h, and 8 h p.i., and rLuc activities were measured (Fig. 4A). Expression of wt L slightly augmented rLuc activities at 4 and 6 h p.i., whereas expression of L(GNN) inhibited rLuc activities at 4, 6, and 8 h p.i, which led us to suggest that expressed L(GNN) formed a biologically inactive oligomer with virally encoded L protein and inhibited the viral RNA synthesis in rMP12-rLuc-infected cells. Consistent with this notion, anti-GFP antibody coimmunoprecipitated the virally encoded L protein along with GFP-L and GFP-L(GNN) from the cell extracts prepared from rMP12-rLuc-infected cells expressing GFP-L and GFP-L(GNN), respectively (Fig. 4B). These data revealed that the expressed GFP-L or GFP-L(GNN) formed a complex with the viral L protein in infected cells.
FIG. 4.
Effects of L(GNN) expression on virus replication. (A) BHK/T7-9 cells in a 12-well plate were cotransfected with 1.0 μg of pT7-IRES-vN and 0.5 μg of one of the following plasmids: pT7-IRES-L (L), pT7-IRES-L(GNN) [L(GNN)], or pT7-IRES (EV). Cells were infected with rMP12-rLuc at an MOI of 5 at 24 h posttransfection. rLuc activities were measured by using cell extracts prepared at 4, 6, and 8 h p.i. The values represent the mean ± standard deviation of three independent experiments. The P values between the indicated samples were determined by Student's t test. (B) BHK/T7-9 cells in a six-well plate were cotransfected with 2.0 μg of pT7-IRES-vN and 1.0 μg of one of the following plasmids: pT7-IRES-vL (L), pT7-IRES-GFP-L (GFP-L), pT7-IRES-GFP-L(GNN) [GFP-L(GNN)], or pT7-IRES-GFP (GFP). Then, cells were infected with rMP12-rLuc at an MOI of 5 at 24 h posttransfection. Cell extracts were prepared at 8 h p.i. and subjected to Western blot analysis by using anti-L 434 antibody (top) and anti-actin antibody (middle). Immunoprecipitation was performed by using anti-GFP antibody and subjected to Western blot analysis by using anti L-434 antibody (bottom).
Analysis of the L protein region(s) important for oligomerization.
To identify the region(s) of the L protein, which are important for L oligomerization, two series of L protein deletion mutants were generated and used for coimmunoprecipitation analysis. The deletion mutants were constructed by taking advantage of the several unique restriction enzyme sites within the L gene (Fig. 5A). One series of the mutants, including GFP-L(NN), GFP-L(NA), and GFP-L(NS), had deletions of different lengths from the C terminus of the L protein, while the deletions of different lengths from the N terminus of the L protein generated mutants GFP-L(CN), GFP-L(CA), and GFP-L(CS); all mutants had GFP fused to the N terminus of L. BHK/T7-9 cells were cotransfected with pT7-IRES-HA-L and a plasmid encoding one of these L deletion mutants. As a control, GFP-L or GFP was coexpressed with HA-L. Western blot analysis using anti-GFP antibody and anti-HA antibody confirmed expression of these L deletion mutants, GFP-L, GFP, and HA-L proteins in transfected cells (Fig. 5B, left). Following coimmunoprecipitation analyses of the cell extracts by the use of anti-HA antibody or anti-GFP antibody, all L deletion mutants were found to have interacted with coexpressed HA-L protein (Fig. 5B, middle and right). These data demonstrated that both the N-terminal and C-terminal regions of the L protein bound to the full-length L protein.
To know whether the middle region of the L protein was involved in L protein oligomerization, we constructed plasmid encoding GFP-L(MAS), a fusion protein consisting of N-terminal GFP and a C-terminal L protein fragment corresponding to amino acids from positions 568 to 1218 (Fig. 5A), and coexpressed it with HA-L; the size of GFP-L(MAS) was comparable to that of GFP-L(NA) or GFP-L(CS) (Fig. 5C, left). Anti-GFP antibody failed to coimmunoprecipitate HA-L along with GFP-L(MAS) from the cell extracts coexpressing HA-L and GFP-L(MAS) (Fig. 5C, middle). We also did not detect an interaction between GFP-L(MAS) and HA-L in a reciprocal experiment, in which anti-HA antibody was used for coimmunoprecipitation (Fig. 5C, right). These data demonstrated that the N- and C-terminal regions, but not the middle region, of the L protein interacted with the full-length L protein.
We next tested effects of the expression of these L fragments, including GFP-L(NA), GFP-L(MAS), and GFL-L(CS), in the minigenome assay, wherein expression of GFP-L or GFP served as a control. Consistent with the data shown in Fig. 3, the coexpression of HA-L with GFP-L resulted in higher rLuc activities and rLuc mRNA abundance than did the coexpression of HA-L and GFP (Fig. 5D and E). We did not see reductions in rLuc activities and rLuc mRNA abundance in the cells coexpressing GFP-L(NA) and HA-L and in those coexpressing GFP-L(MAS) and HA-L. In contrast, GFP-L(CS) expression inhibited HA-L-mediated minigenome RNA synthesis and rLuc activities (Fig. 5D and E). These data suggest that the binding of the C-terminal fragment of the L protein, but not the N-terminal fragment of L protein, to HA-L inhibited HA-L protein-mediated minigenome RNA synthesis.
BiFC analysis of L oligomerization.
To further understand the status of L oligomerization, we performed BiFC analysis. This assay is based on the ability of two split proteins of fluorescence protein to interact with each other and reconstitute fluorophore (7, 10, 12, 27, 31, 35). Amino acid residues 1 to 172 (VN) and 155 to 239 (VC) of the Venus protein, a modified YFP (25), were fused to the N terminus or the C terminus of the full-length L protein (Fig. 6A). We reasoned that if the expressed VN-tagged L protein and the VC-tagged L protein interact, they may bring together the nonfluorescing VN fragment and VC fragment in such a way as to permit refolding of the fluorophore and the subsequent restoration of fluorescence. We demonstrated the accumulation of the expressed proteins by Western blot analysis (Fig. 6B) and determined the fluorescent signal intensities by fluorescence microscopy (Fig. 6C). As a positive control, we used a plasmid encoding a fusion protein (Venus-L) that consisted of the N-terminal Venus protein and the C-terminal full-length L protein. Venus-L was efficiently expressed in the transfected cells (Fig. 6B) and yielded strong fluorescent signals (Fig. 6C). As expected, no fluorescence signal was observed with cells expressing VN-L, L carrying VN in its N terminus; L-VN, L carrying the VN in its C terminus; L-VC, L carrying VC in its C terminus; or VC-L, L carrying VC in its N terminus (Fig. 6A and C). Likewise, coexpression of VC-L and VN-tubulin, a fusion protein consisting of the N-terminal VN and C-terminal tubulin (Fig. 6A), resulted in no fluorescent signal (Fig. 6C). In contrast, coexpression of VN-L and VC-L resulted in low fluorescent signals, demonstrating that there was an intermolecular interaction between the N termini of the L proteins (Fig. 6C). Coexpression of L-VN and L-VC also resulted in low levels of fluorescent signals (Fig. 6C), suggestive of an intermolecular interaction between the C termini of the expressed L proteins. To know the presence of an intermolecular interaction between the N terminus of L and the C terminus of L in an L oligomer, we subjected cells coexpressing VN-L and L-VC and those coexpressing VC-L and L-VN to BiFC analysis (Fig. 6C). Both samples showed similarly low levels of fluorescent signals that demonstrated to us that the N terminus of L and the C terminus of L in an L oligomer interacted intermolecularly. Independent expression of both VN-L-VC, L carrying the N-terminal VN and the C-terminal VC, and VC-L-VN, L carrying the N-terminal VC and the C-terminal VL, resulted in strong fluorescent signals, comparable to those detected with Venus-L-expressing cells (Fig. 6C). The fluorescent signal intensities detected in cells expressing VN-L-VC were substantially higher than those detected in cells coexpressing VN-L and L-VC. Likewise, VC-L-VN expression resulted in substantially higher fluorescence signals than did the coexpression of VC-L and L-VN (Fig. 6C). These data strongly implied that there was an intramolecular interaction between the N terminus and the C terminus of the expressed L protein.
In summary, BiFC analysis led us to conclude that intermolecular interactions occurred among the C termini of L proteins and the N termini of L proteins and also between the C-terminal region and the N-terminal region of L proteins in the L oligomer(s). In addition, the findings pointed to an intramolecular interaction between the N-terminal and the C-terminal regions of an L protein.
Analysis of L oligomerization using deletion mutants of L protein.
To further confirm the data obtained in the BiFC analysis, we expressed various combinations of the N- and C-terminal fragments of the L protein in cultured cells and tested their intermolecular interactions. BHK/T7-9 cells were cotransfected with plasmid encoding a fusion protein, HA-L(NA), consisting of the N-terminal HA tag, a 567-amino-acid-long N-terminal fragment of L protein, and one of the following plasmids: plasmids encoding GFP-L(NA), GFP-L(CS), GFP-L, and GFP (Fig. 7A). We also performed similar cotransfection experiments by using a plasmid encoding a fusion protein, HA-L(CS), consisting of the N-terminal HA tag and L protein fragment corresponding to the C-terminal amino acids at positions 1219 to 2092 in place of the plasmid encoding HA-L(NA) (Fig. 7B). As expected, coimmunoprecipitation analyses using anti-GFP or anti-HA antibodies resulted in binding of HA-L(NA) and HA-L(CS) to GFP-L, but not to GFP (Fig. 7). We found interactions between both HA-L(NA) and GFP-L(NA) and HA-L(NA) and GFP-L(CS). Likewise, HA-L(CS) interacted with GFP-L(NA) and with GFP-L(CS) (Fig. 7). These data revealed that the N-terminal fragment interacted with both N- and C-terminal fragments, while the C-terminal fragment also interacted with both N-terminal and C-terminal fragments. Hence, the data shown in Fig. 7 were in good agreement with those obtained in the BiFC analysis.
FIG. 7.
Interaction between L protein fragments. (A) A schematic diagram of L fragments. Deletions are shown as dashed lines. The number represents amino acids from the N terminus of the L protein. (B) BHK/T7-9 cells in a six-well plate were cotransfected with plasmid expressing HA-L(NA) (2.0 μg) and one of the indicated plasmids (2.0 μg) or a plasmid expressing HA-L(CS) and one of the indicated plasmids. Intracellular proteins were extracted at 24 h posttransfection. Western blot analysis of cell extracts (left, Intracellular) and coimmunoprecipitation analysis using anti-GFP antibody (middle, IP: anti-GFP), or anti-HA antibody (right, IP: anti-HA) are shown. Western blot analysis using anti-HA antibody and anti-GFP antibody is indicated as WB: anti-HA and WB: anti-GFP, respectively.
DISCUSSION
In the present study, we tested for the presence of an RVFV L protein oligomer in expressed cells and explored the biological significance of an L oligomer by expressing a biologically inactive mutant L protein in a minigenome assay and infected cells. Coimmunoprecipitation analyses (Fig. 2, 5, and 7) and BiFC analysis (Fig. 6) gave evidence of intermolecular interactions among the N termini of L proteins and the C termini of L proteins and also between the C-terminal and N-terminal regions of L proteins in the L oligomer. The evidence also pointed to an intramolecular interaction between the N and C termini of L. Expressed L(GNN), an L protein containing the GNN residues in place of the highly conserved SDD residues, exhibited a dominant negative effect on the RNA synthesis functions of wt L in both the minigenome assay (Fig. 3) and infected cells (Fig. 4), suggesting the importance of an L oligomer for viral RNA synthesis.
Expression of GFP-L(CS), the C-terminal fragment of L protein, but not GFP-L(NA), the N-terminal fragment of the L protein, suppressed rLuc activities in a minigenome assay (Fig. 5D and E). This finding led us to suggest that an intermolecular interaction between the C-terminal region of L proteins and/or between the C-terminal region of L protein and the N-terminal region of L protein was important for the L oligomer to exert the RdRp function. BiFC analysis, however, showed low fluorescence signals in the cells coexpressing L-VN and L-VC, those coexpressing VN-L and L-VC, and those coexpressing VC-L and L-VN. When we used a minigenome assay, we found that all of the L protein mutants carrying a C-terminal tag were unable to support viral RNA synthesis (data not shown), which suggested to us that the addition of a tag at the C terminus of L altered the structure of the L protein, rendered the L protein to be biologically inactive, and possibly inhibited efficient intermolecular interaction at the C termini of the L proteins and/or between the C terminus of L protein and the N terminus of the L protein (Fig. 6C). In contrast to L protein mutants carrying the C-terminal tag, GFP-L(CS) did not carry a C-terminal tag (Fig. 5A); it is quite possible that GFL-L(CS) efficiently interacted with coexpressed HA-L and suppressed the RdRp function of the HA-L in a minigenome assay (Fig. 5D and E).
In contrast to the suggested biological importance of an intermolecular interaction that was mediated by the C-terminal regions of L proteins and/or that was mediated between the C-terminal region of the L protein and the N-terminal region of the L protein for the RdRp functions (Fig. 5D and E), the biological significance of an intermolecular interaction between the N-terminal regions of L proteins was less obvious. The BiFC assay showed an interaction between VN-L and VC-L (Fig. 6C), and coimmunoprecipitation analysis also revealed an interaction between the expressed N-terminal fragments (Fig. 7B). Hence GFP-L(NA) most probably interacted with coexpressed L protein in a minigenome assay. However, binding of GFP-L(NA) to HA-L protein did not disrupt the RdRp function of HA-L, which was indicative that the formation of an L oligomer, mediated by the N-terminal regions of L, may not be crucial for viral RNA synthesis but may have another biological role. For example, this interaction may be important for the L protein to form a viral ribonucleoprotein complex with viral RNAs and N protein during viral assembly. It is tempting to speculate that interactions between the C-terminal regions of L and between the C-terminal region and the N-terminal region of L exert different biological activities. Others also hypothesized that the different regions of Sendai virus L interact with one another, with respect to the individual steps in viral RNA synthesis (36).
In addition to the intermolecular interactions of L protein, the BiFC analysis findings point to the presence of an intramolecular interaction between the N and C termini of the L protein (Fig. 6). The presence of a putative intramolecular interaction in RVFV L protein was not surprising, as intramolecular interactions have been observed with the RdRp of positive RNA viruses (6), and at least a portion of viral RdRps probably have similar structures (8, 24, 30, 39). Because intramolecular and intermolecular interactions of poliovirus RNA polymerase have been considered to be important for viral RNA synthesis (11, 28, 38), it is highly likely that the intramolecular L protein interaction is also essential for the biological activities of RVFV L protein. This putative intramolecular L protein interaction may occur cotranslationally or posttranslationally. Moreover, the formation of this intramolecular L protein interaction may facilitate the intermolecular L protein interactions.
Although the L protein mutants, GFP-L(GNN) (Fig. 3) and L(GNN) (Fig. 4), exhibited a dominant negative effect on wt L protein in the minigenome assay and in virus-infected cells, respectively, the inhibition of viral RNA synthesis by GFP-L(GNN) in the minigenome assay was not very strong; the rLuc activity in the cells coexpressing HA-L and GFP-L(GNN) was roughly one-fourth of the rLuc activity detected with the cells coexpressing HA-L and GFP-L, suggesting an approximately 75% reduction in the rLuc activity by the coexpression of GFP-L(GNN) (Fig. 3D). In contrast, the coexpression of the same level of wt LCMV L protein and its dominant negative mutant in a similar minigenome assay resulted in a 20- to 30-fold reduction in minigenome RNA synthesis (32). These data suggest that the status of a biologically active L oligomer probably differed between RVFV and LCMV. The finding that a dominant negative mutant of LCMV L protein efficiently suppressed minigenome RNA synthesis suggested to us that a biologically active LCMV L oligomer carries many L proteins and that inclusion of one molecule of a dominant negative mutant in an L oligomer renders the L oligomer biologically incompetent. In contrast, a modest inhibition of minigenome RNA synthesis by a dominant negative mutant of RVFV L protein implied to us that a biologically active L oligomer included a small number of L proteins. Further studies are required to determine the stoichiometry of the L protein oligomer that exerted the RdRp activity.
HA-L and GFP-L were biologically active to support minigenome RNA synthesis, demonstrating that the addition of the N-terminal tag did not suppress the RdRp function. However, we were unable to rescue an RVFV mutant virus carrying L RNA encoding HA-L (data not shown), which suggested to us that adding an HA tag sequence at the 5′ end of the L gene ORF was not tolerated at a certain step(s) in RVFV replication. In this regard, it is worth noting that Shi and Elliott reported the generation of recombinant Bunyamwera orthobunyaviruses expressing a V5 epitope tag within the L gene ORF but not at the termini of the L gene (33). Because various reagents that are suitable for purification of HA-tagged or GFP-tagged proteins are available, expressed HA-L or GFP-L may be useful for identifying host factors that associate with the RVFV L protein and characterizing viral replication/transcription complexes.
Acknowledgments
We thank Krishna Narayanan for discussion and critical reading of the manuscript and Thomas Albrecht for supporting our experiments via confocal microscopy.
This work was supported by grants from NIAID to S.M. and C.J.P. through the Western Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research, NIH grant number U54 AI057156, and by NIH-NIAID-DMID-02-24 Collaborative Grant on Emerging Viral Diseases. K.T. was supported by the James W. McLaughlin fellowship fund.
Footnotes
Published ahead of print on 7 October 2009.
REFERENCES
- 1.Aquino, V. H., M. L. Moreli, and L. T. Moraes Figueiredo. 2003. Analysis of oropouche virus L protein amino acid sequence showed the presence of an additional conserved region that could harbour an important role for the polymerase activity. Arch. Virol. 148:19-28. [DOI] [PubMed] [Google Scholar]
- 2.Bird, B. H., T. G. Ksiazek, S. T. Nichol, and N. J. Maclachlan. 2009. Rift Valley fever virus. J. Am. Vet. Med. Assoc. 234:883-893. [DOI] [PubMed] [Google Scholar]
- 3.Bishop, D. H., M. E. Gay, and Y. Matsuoko. 1983. Nonviral heterogeneous sequences are present at the 5′ ends of one species of snowshoe hare bunyavirus S complementary RNA. Nucleic Acids Res. 11:6409-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cevik, B., D. E. Holmes, E. Vrotsos, J. A. Feller, S. Smallwood, and S. A. Moyer. 2004. The phosphoprotein (P) and L binding sites reside in the N-terminus of the L subunit of the measles virus RNA polymerase. Virology 327:297-306. [DOI] [PubMed] [Google Scholar]
- 5.Doyle, T., and D. Botstein. 1996. Movement of yeast cortical actin cytoskeleton visualized in vivo. Proc. Natl. Acad. Sci. USA 93:3886-3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferrer-Orta, C., A. Arias, C. Escarmis, and N. Verdaguer. 2006. A comparison of viral RNA-dependent RNA polymerases. Curr. Opin. Struct. Biol. 16:27-34. [DOI] [PubMed] [Google Scholar]
- 7.Frieman, M., B. Yount, M. Heise, S. A. Kopecky-Bromberg, P. Palese, and R. S. Baric. 2007. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J. Virol. 81:9812-9824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hansen, J. L., A. M. Long, and S. C. Schultz. 1997. Structure of the RNA-dependent RNA polymerase of poliovirus. Structure 5:1109-1122. [DOI] [PubMed] [Google Scholar]
- 9.Hass, M., M. Lelke, C. Busch, B. Becker-Ziaja, and S. Gunther. 2008. Mutational evidence for a structural model of the Lassa virus RNA polymerase domain and identification of two residues, Gly1394 and Asp1395, that are critical for transcription but not replication of the genome. J. Virol. 82:10207-10217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hemerka, J. N., D. Wang, Y. Weng, W. Lu, R. S. Kaushik, J. Jin, A. F. Harmon, and F. Li. 2009. Detection and characterization of influenza A virus PA-PB2 interaction through a bimolecular fluorescence complementation assay. J. Virol. 83:3944-3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hobson, S. D., E. S. Rosenblum, O. C. Richards, K. Richmond, K. Kirkegaard, and S. C. Schultz. 2001. Oligomeric structures of poliovirus polymerase are important for function. EMBO J. 20:1153-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu, C. D., and T. K. Kerppola. 2003. Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat. Biotechnol. 21:539-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ikegami, T., C. J. Peters, and S. Makino. 2005. Rift Valley fever virus nonstructural protein NSs promotes viral RNA replication and transcription in a minigenome system. J. Virol. 79:5606-5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ikegami, T., S. Won, C. J. Peters, and S. Makino. 2006. Rescue of infectious Rift Valley fever virus entirely from cDNA, analysis of virus lacking the NSs gene, and expression of a foreign gene. J. Virol. 80:2933-2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ito, N., M. Takayama-Ito, K. Yamada, J. Hosokawa, M. Sugiyama, and N. Minamoto. 2003. Improved recovery of rabies virus from cloned cDNA using a vaccinia virus-free reverse genetics system. Microbiol. Immunol. 47:613-617. [DOI] [PubMed] [Google Scholar]
- 16.Jin, H., and R. M. Elliott. 1993. Characterization of Bunyamwera virus S RNA that is transcribed and replicated by the L protein expressed from recombinant vaccinia virus. J. Virol. 67:1396-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin, H., and R. M. Elliott. 1992. Mutagenesis of the L protein encoded by Bunyamwera virus and production of monospecific antibodies. J. Gen. Virol. 73:2235-2244. [DOI] [PubMed] [Google Scholar]
- 18.Jorba, N., E. Area, and J. Ortin. 2008. Oligomerization of the influenza virus polymerase complex in vivo. J. Gen. Virol. 89:520-524. [DOI] [PubMed] [Google Scholar]
- 19.Jorba, N., R. Coloma, and J. Ortin. 2009. Genetic trans-complementation establishes a new model for influenza virus RNA transcription and replication. PLoS Pathog. 5:e1000462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kinsella, E., S. G. Martin, A. Grolla, M. Czub, H. Feldmann, and R. Flick. 2004. Sequence determination of the Crimean-Congo hemorrhagic fever virus L segment. Virology 321:23-28. [DOI] [PubMed] [Google Scholar]
- 21.Le May, N., N. Gauliard, A. Billecocq, and M. Bouloy. 2005. The N terminus of Rift Valley fever virus nucleoprotein is essential for dimerization. J. Virol. 79:11974-11980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meegan, J. M., and C. J. Peters. 1989. Rift Valley fever. CRC Press, Boca Raton, FL.
- 23.Mohl, B. P., and J. N. Barr. 2009. Investigating the specificity and stoichiometry of RNA binding by the nucleocapsid protein of Bunyamwera virus. RNA 15:391-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muller, R., O. Poch, M. Delarue, D. H. Bishop, and M. Bouloy. 1994. Rift Valley fever virus L segment: correction of the sequence and possible functional role of newly identified regions conserved in RNA-dependent polymerases. J. Gen. Virol. 75:1345-1352. [DOI] [PubMed] [Google Scholar]
- 25.Nagai, T., K. Ibata, E. S. Park, M. Kubota, K. Mikoshiba, and A. Miyawaki. 2002. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 20:87-90. [DOI] [PubMed] [Google Scholar]
- 26.Nichol, S. T. 2001. Bunyaviruses, 4th ed. Lippincott-Raven, Philadelphia, PA.
- 27.Nyfeler, B., S. W. Michnick, and H. P. Hauri. 2005. Capturing protein interactions in the secretory pathway of living cells. Proc. Natl. Acad. Sci. USA 102:6350-6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pata, J. D., S. C. Schultz, and K. Kirkegaard. 1995. Functional oligomerization of poliovirus RNA-dependent RNA polymerase. RNA 1:466-477. [PMC free article] [PubMed] [Google Scholar]
- 29.Patterson, J. L., B. Holloway, and D. Kolakofsky. 1984. La Crosse virions contain a primer-stimulated RNA polymerase and a methylated cap-dependent endonuclease. J. Virol. 52:215-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poch, O., I. Sauvaget, M. Delarue, and N. Tordo. 1989. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 8:3867-3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Remy, I., A. Montmarquette, and S. W. Michnick. 2004. PKB/Akt modulates TGF-beta signalling through a direct interaction with Smad3. Nat. Cell Biol. 6:358-365. [DOI] [PubMed] [Google Scholar]
- 32.Sánchez, A. B., and J. C. de la Torre. 2005. Genetic and biochemical evidence for an oligomeric structure of the functional L polymerase of the prototypic arenavirus lymphocytic choriomeningitis virus. J. Virol. 79:7262-7268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi, X., and R. M. Elliott. 2009. Generation and analysis of recombinant Bunyamwera orthobunyaviruses expressing V5 epitope-tagged L proteins. J. Gen. Virol. 90:297-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shimozono, S., and A. Miyawaki. 2008. Engineering FRET constructs using CFP and YFP. Methods Cell Biol. 85:381-393. [DOI] [PubMed] [Google Scholar]
- 35.Shyu, Y. J., C. D. Suarez, and C. D. Hu. 2008. Visualization of AP-1 NF-κB ternary complexes in living cells by using a BiFC-based FRET. Proc. Natl. Acad. Sci. USA 105:151-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smallwood, S., B. Cevik, and S. A. Moyer. 2002. Intragenic complementation and oligomerization of the L subunit of the Sendai virus RNA polymerase. Virology 304:235-245. [DOI] [PubMed] [Google Scholar]
- 37.Smallwood, S., and S. A. Moyer. 2004. The L polymerase protein of parainfluenza virus 3 forms an oligomer and can interact with the heterologous Sendai virus L, P and C proteins. Virology 318:439-450. [DOI] [PubMed] [Google Scholar]
- 38.Thompson, A. A., R. A. Albertini, and O. B. Peersen. 2007. Stabilization of poliovirus polymerase by NTP binding and fingers-thumb interactions. J. Mol. Biol. 366:1459-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vieth, S., A. E. Torda, M. Asper, H. Schmitz, and S. Gunther. 2004. Sequence analysis of L RNA of Lassa virus. Virology 318:153-168. [DOI] [PubMed] [Google Scholar]