Abstract
The Epstein-Barr virus early protein EB2 (also called BMLF1, Mta, or SM), which allows the nuclear export of a subset of early and late viral mRNAs derived from intronless genes, is essential for the production of infectious virions. An important feature of mRNA export factors is their capacity to shuttle continuously between the nucleus and the cytoplasm. In a previous study, we identified a novel CRM1-independent transferable nuclear export signal (NES) at the N terminus of EB2, between amino acids 61 and 146. Here we show that this NES contains several small arginine-rich domains that cooperate to allow efficient interaction with TAP/NXF1. Recruitment of TAP/NXF1 correlates with this NES-mediated efficient nuclear export when it is fused to a heterologous protein. Moreover, the NES can export mRNAs bearing MS2 RNA-binding sites from the nucleus when tethered to the RNA via the MS2 phage coat protein RNA-binding domain.
In metazoans, nuclear export of most mRNAs generated from intronless and intron-containing genes is mediated by conserved proteins which coat the nascent RNA cotranscriptionally to form export-competent mRNPs that are capped at the 5′ end and spliced and cleaved/polyadenylated at the 3′ end (for a review, see reference 18). Translocation of mRNPs through the nuclear pore complex requires binding of a heterodimer composed of TAP (also called NXF1) and p15 (also called NXT1) (Mex67 and Mtr2, respectively, in Saccharomyces cerevisiae), which escorts competent mRNPs out of the nucleus via direct interactions with nucleoporins lining the nuclear pore. Because of its low affinity for binding of mRNAs, the heterodimer TAP-p15 requires adaptor proteins, among which the best characterized is REF (also called Aly) (30). It is now clear that splicing is an important step for the recruitment of the heterodimer TAP-p15 and for efficient mRNA nuclear export. Two multiprotein complexes are known to be recruited as a consequence of splicing: these are the exon junction complex, which is deposited 20 to 24 nucleotides upstream of each exon-exon junction during splicing and is associated with nonsense-mediated mRNA decay, and the human transcription/export complex (hTREX). hTREX contains the hTHO complex (composed of hHpr1, hTho2, fSAP79, fSAP35, and fSAP24) associated with the RNA helicase UAP56 and the adaptor protein REF. Recent studies strongly suggest that the cap-binding protein CBP80 and factors deposited at the first exon-exon junction by splicing cooperate to recruit hTREX to spliced mRNA (6). The presence of REF in this complex is thought to stimulate the recruitment of TAP onto the mRNP and to lead to its efficient export through the nuclear pore. For mRNAs generated from intronless genes, two export pathways have been described. One implies binding of SR proteins 9G8 and SRp20 to mRNAs (17). Similarly to REF, these two shuttling SR proteins have been found to serve as RNA-binding adaptors for TAP (16). The other pathway implies recruitment of REF at the 5′ end of the mRNA by the cap-binding protein CBP20 (27).
In contrast to the majority of mammalian genes, herpesvirus early and late genes are almost all intronless. The export and efficient expression of most of these intronless viral mRNAs appear to be strictly dependent on a viral gene product that shares properties with known mRNA export adaptors, for example, the herpes simplex virus type 1 protein ICP27 (5, 20), the cytomegalovirus (CMV) protein UL69 (22), the Kaposi's sarcoma-associated herpesvirus protein ORF57 (2, 23), the herpesvirus saimiri protein ORF57 (31), and the Epstein-Barr virus (EBV) protein EB2 (7, 29). The EB2 protein of EBV is absolutely required for production of infectious viral particles (9). It binds to RNA in vitro and in vivo, via an arginine-rich region (13), and induces the cytoplasmic accumulation of most early and late viral mRNAs (1, 9, 10), likely by interacting with cellular adaptors of the TAP receptor pathway, such as REF (14) and OTT1/RBM15 (14), a novel RNA-binding mRNA export factor that binds TAP (21).
An important feature of mRNA export factors is their capacity to shuttle continuously between the nucleus and the cytoplasm. This shuttling is mediated by specific factors interacting with peptide motifs called nuclear localization signals (NLS) and nuclear export signals (NES). EB2 shuttles between the nucleus and the cytoplasm (7, 29), and we previously identified two NLS and a CRM1-independent transferable NES, located at the N terminus of EB2, between amino acids (aa) 61 and 146 (14). This NES appears to be important for the function of the protein, since its deletion drastically affects the mRNA export function of the protein (14). In the present study, we show that this NES contains several small domains rich in arginine residues that participate in both interaction with TAP and nuclear export. Moreover, we show that tethering of a fusion protein consisting of the RNA-binding domain of the MS2 phage coat protein fused to EB2's TAP-binding domain is sufficient to induce efficient nuclear export of mRNAs bearing MS2 RNA-binding sites.
MATERIALS AND METHODS
Plasmids.
Eukaryotic expression vectors for EB2 and the various deletion mutants used were derived from CMV immediate-early promoter-based plasmid pCI (Promega). Each protein was tagged at the N terminus with the Flag epitope, and this is indicated by the letter F before the name of the protein. PCI.F.EB2 expresses the EB2 protein initiated at the BSLF2 AUG. pCI.F.EB2Nter expresses the first 184 N-terminal amino acids of EB2. pCI.F.EB2.Δ2 and pCI.FNLS.EB2.ΔB express EB2 proteins which have aa 39 to 63 and aa 61 to 140 deleted, respectively. The 50, 70, 90, 110, 130, or 184 N-terminal amino acids of EB2 have been deleted from EB2 in pCI.F.EB2M1, pCI.F.EB2M2, pCI.F.EB2M3, pCI.F.EB2M4, pCI.FNLS.EB2M5, and pCI.FNLS.EB2Cter, respectively. The simian virus 40 (SV40) T antigen NLS was introduced between the Flag epitope and the EB2-encoding sequence in plasmids pCI.FNLS.EB2M5, pCI.FNLS.EB2Cter, pCI.FNLS.EB2.ΔB, and pCI.FNLS.EB2NterΔ4. In pCI.Myc.EB2 and pCI.Myc.EB2Nter, the Flag epitope of pCI.Flag.EB2 was replaced with the Myc epitope. pCMV.NLS.βgal and pCMV.NLS.B.βgal have been described elsewhere (14). pCMV.NLS.B.βgal-Δb1, -Δb2, -Δb3, -Δb4, -Δb5, -Δb6, and -Δb7 were generated by site-directed mutagenesis of pCMV.NLS.B.βgal (Stratagene QuikChange XL site-directed mutagenesis kit), using the following double-stranded oligonucleotides: Δb1, 5′-CATCCTCAGAGGAGGAGAGCGTGGTCATAACC-3′; Δb2, 5′-CCGGCCCACGCCATATTTGTGATTCCCAGAAAG-3′; Δb3, 5′-CAGCGTGGTCATAACCCAGGACAAGACAGTCAC-3′; Δb4, 5′-CGTTTGTGATTCCCAGACCCCTGTGCAGGGAC-3′; Δb5, 5′-CAGGACAAGACAGTCGAGACTGGCAACTCC-3′; Δb6, 5′-CCCCTGTGCAGGGACCACAAAAGGCGACGC-3′; and Δb7, 5′-GAGACTGGCAACTCCTGCACCGATGAAAG-3′.
pNTAP.EB2Nter expresses the EB2 N-terminal 184 aa fused at the N terminus to a calmodulin-binding peptide and to a streptavidin-binding peptide (pNTAP vector from Stratagene) (25). pGEX-EB2Nter, pGEX-A, pGEX-B, pGEX-C, and pGEX-EB2Cter have been described previously (14, 25). pGEX-EB2Nter-Δ1, -Δ2, -Δ3, -Δ4, and -Δ5 and pCI.FlagEB2Nter-Δ1, -Δ2, -Δ3, -Δ4, and -Δ5 were generated by subcloning the EB2 N-terminal region, amplified by PCR from pAAC-FlagEB2-Δ1, -Δ2, -Δ3, -Δ4, and -Δ5 (3), into pGEX-3X and PCI.Flag, respectively. The Δ1, Δ2, Δ3, Δ4, and Δ5 mutants have aa 8 to 40, 38 to 63, 63 to 96, 95 to 131, and 148 to 184 deleted, respectively. pGEX-EB2Nter R/A was generated by site-directed mutagenesis (Stratagene QuikChange XL site-directed mutagenesis kit) using (successively) the following double-stranded oligonucleotides: 5′-CGGCATCGTTTGTGATTGCCGCAGCGGCGTGGGACCTACAGG-3′, 5′-CACCAGAGGCCACGCAGCGGCAGCCGGAGAGGTCCATGG-3′, 5′-CGGTCACGCTGCATGCCTCACCCCTGTGC-3′, and 5′-TCACCCCTGTGCGCGGACGAGGACGAG-3′.
PCI.FlagEB2Nter R/A was generated by subcloning a PCR-amplified EB2Nter R/A fragment from pGEX-EB2Nter R/A into a PCI.Flag gateway vector, using Gateway technology (Invitrogen). pBSK-TAP was a kind gift from E. Izaurralde. pGEX-TAP and pCI-FlagTAP were derived from pBSK-TAP. pCIF.NLS.9G8Nter was generated by subcloning a 9G8 PCR-amplified DNA fragment, obtained with the following oligonucleotides, into pCI-FlagNLS: 5′-GCCCCTCTAGAATGTCGCGTTACGGGCGGTACG-3′ and 5′-GCCCCTCTAGATCAGTAACGATGACAATCATAAGC-3′.
pDM128/PL, pDM128-4xMS2, and pCMV-MS2 were a kind gift from B. R. Cullen (32). pCMV-MS2 B has been published elsewhere (14), and pCMV-MS2 BC was generated by cloning a PCR-amplified fragment, obtained using the following oligonucleotides, into pCMV-MS2: 5′-CGGGATCCTCTTACACCAGAGGC-3′ and 5′-CGGCTCGAGCTGCCAGGCTCCAAT-3′. All MS2 constructs contained the hemagglutinin (HA) epitope in fusion with MS2.
Transfections and TAP-tagging experiments.
HeLa cells, HEK293 cells, and HEK293T cells were grown at 37°C in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal calf serum. Transfections were performed by the calcium phosphate precipitation method. HEK293T cells were transfected with 1 μg of pNTAP (Stratagene) or pNTAP.EB2Nter. Cells were harvested 48 h later, and a nuclear extract was prepared from 4 × 107 cells as previously described (25). To purify TAP-tagged EB2Nter and associated proteins, nuclear extracts were processed following the manufacturer's instructions (Interplay mammalian TAP system; Stratagene). Eluted proteins were analyzed by two-dimensional gel electrophoresis with silver staining and identified by matrix-assisted laser desorption ionization-time-of-flight mass spectrometry as previously described (25).
In vitro GST pull-down assays.
Glutathione S-transferase (GST) and GST fusion proteins were purified from Escherichia coli BL21 Codon Plus extracts, using glutathione-Sepharose 4B beads (Amersham Biosciences). Beads carrying GST or GST fusion proteins were equilibrated in TNTB binding buffer (10 mM Tris-HCl, pH 8, 250 mM NaCl, 0.1% NP-40, and 2 mg/ml bovine serum albumin) in the presence of protease inhibitors (Complete EDTA-free cocktail; Roche Molecular Biochemicals) and then incubated with in vitro radiolabeled proteins as follows. [35S]methionine-labeled TAP was synthesized in vitro from plasmid pBSK-TAP, using the TnT coupled transcription/translation system (Promega), and incubated with TNTB-equilibrated glutathione-Sepharose 4B beads loaded with GST or the GST fusion proteins for 4 h at 4°C. RNase treatment was performed by adding 1 μl of RNase A (10 mg/ml) and 1 μl of RNase T1 (10 U/μl) to the incubation mixture. Beads were washed five times in TNTB buffer without bovine serum albumin, and bound proteins were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and revealed by autoradiography.
Coimmunoprecipitation assays.
Transfected HeLa cells were harvested from 100-mm dishes at 48 h posttransfection and lysed in 300 μl IP buffer (10 mM Tris-HCl, pH 8, 100 mM NaCl, and 1 mM EDTA) plus protease inhibitors (Roche Molecular Biochemicals). For immunoprecipitation of the transiently expressed F.EB2Nter protein, cell extracts were incubated with 30 μl anti-Flag M2 affinity gel (Sigma) for 4 h at 4°C, and immunopurified proteins were analyzed by Western blotting, using either a rabbit polyclonal antibody generated against the GST-TAP protein produced in bacteria or an anti-Flag rabbit polyclonal antibody (Sigma). For immunoprecipitation of the endogenous TAP protein, cell extracts were incubated with the anti-TAP rabbit polyclonal antibody and the protein-antibody complexes were purified using 30 μl protein A-Sepharose beads (Amersham Biosciences). RNase treatment was performed by adding 1 μl of RNase A (10 mg/ml) and 1 μl of RNase T1 (10 U/μl) to the incubation mixture. Immunopurified proteins were analyzed by Western blotting, using either the anti-TAP rabbit polyclonal antibody or the M2 anti-Flag monoclonal antibody (MAb; Sigma). Western blots were revealed by enhanced chemiluminescence (Pierce).
Heterokaryon assay.
Twenty-four hours after transfection, 3.5 × 105 HeLa cells were seeded onto glass coverslips with an equal number of NIH 3T3 cells in six-well plates. The cells were allowed to grow overnight and then were treated for 2 h with 200 μg/ml of cycloheximide to inhibit de novo protein synthesis. The cells were then washed with phosphate-buffered saline (PBS), and heterokaryon formation was performed by incubating cells for 2 min in 50% polyethylene glycol 3000 to 3700 (Sigma) in PBS. Following polyethylene glycol-induced fusion, cells were washed extensively with PBS and returned to fresh medium containing 200 μg/ml of cycloheximide. After 2 h at 37°C, cells were fixed with 4% paraformaldehyde. For indirect immunofluorescence experiments, the M2 anti-Flag MAb (Sigma), the 9E10 anti-Myc MAb, an anti-hnRNP C MAb (kindly provided by G. Dreyfuss) (26), and an anti-Flag polyclonal antibody were used as primary antibodies. Alexa Fluor 488-conjugated goat anti-mouse immunoglobulin G (IgG) (heavy plus light chains [H+L]) or anti-rabbit antibody (Invitrogen) or a Fluorolink Cy3-labeled goat anti-rabbit or anti-mouse IgG (H+L) antibody (Amersham) was used as secondary antibody. Cell nuclei were stained by incubation with 5 μg/ml Hoescht 33258 (Sigma). The percentage of heterokaryons in which the proteins were found in both the HeLa and NIH 3T3 nuclei among the heterokaryons expressing the proteins were evaluated by counting the heterokaryons via conventional microscopy (Axioplan 2; Zeiss). The percentage was obtained by counting 20 to 60 heterokaryons in several independent experiments. The endogenous nonshuttling protein hnRNP C was used as a control and, as expected, was not transported from human to mouse nuclei (data not shown).
Quantification of CAT protein.
In order to evaluate chloramphenicol acetyltransferase (CAT) protein expression, we used a CAT enzyme-linked immunosorbent assay (ELISA) (Roche). After transfection, cells were collected in PBS. Half of the cells were use for CAT assays according to the manufacturer's instructions. The other half of the cells were used to monitor protein expression by Western blotting, using either the anti-Flag M2 MAb (Sigma), an anti-HA MAb (Boehringer), or an anti-TAP rabbit polyclonal antibody. Horseradish peroxidase-conjugated goat anti-mouse IgG (Amersham) or goat anti-rabbit IgG (Amersham) was used as the secondary antibody. For immunoblot detection, an ECL system (Amersham) was used.
Statistics.
Analyses were done by either Student's t test (for Fig. 8) or the Wilcoxon test (for Fig. 6) at the www.u707.jussieu.fr website.
FIG. 8.
Stimulation of pDM128 unspliced mRNA export by MS2-B and MS2-BC fusion proteins. (A) Schematic representation of pDM128-4xMS2 reporter plasmid. pDM128-4xMS2 is a modified version of pDM128/PL, depicted in Fig. 5, in which four RNA-binding sites for the MS2 coat protein have been introduced within the intron, immediately downstream of the CAT open reading frame. MS2 fusion proteins can thus be tethered to RNA via the MS2 binding sites, allowing the mRNA export properties of the MS2 fusions to be tested. (B) pDM128-4xMS2 was transfected into HeLa cells either alone or together with an expression plasmid for HA.MS2, HA.MS2-B (peptide B; aa 61 to 140), HA.MS2-BC (peptide BC; aa 61 to 200), or F.EB2Nter. The efficiency of unspliced mRNA export was evaluated by quantification of CAT expression by CAT ELISA (top), and the amounts of MS2 fusion proteins and F.EB2Nter expressed in each assay were evaluated by Western blotting (bottom) using either an anti-HA MAb (for the MS2 fusions) or the anti-Flag MAb M2. The data shown are derived from a single experiment but are representative of five experiments. Statistical tests were performed using data collected from these five experiments. Both HA.MS2-B and HA.MS2-BC were significantly different from the HA.MS2 control (P < 0.05), whereas F.EB2Nter was not (P > 0.1).
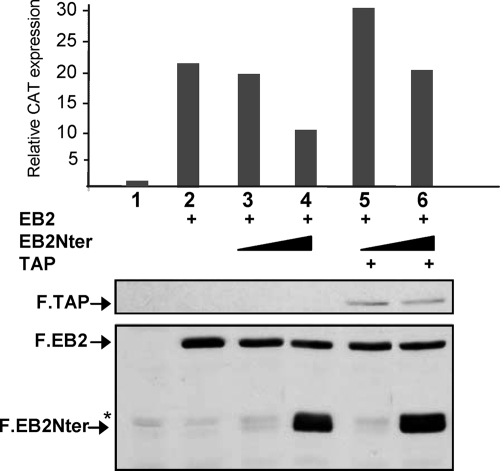
FIG. 6.
Overexpression of TAP rescues EB2-mediated export. pDM128/PL was transfected into HeLa cells either alone or together with expression plasmids for F.EB2 (0.5 μg), F.EB2Nter (0.1 and 1 μg), and F.TAP (1.5 μg), as indicated. The efficiency of unspliced mRNA export was evaluated by quantification of CAT expression by CAT ELISA (top), and the levels of expression of F.EB2, F.EB2Nter, and F.TAP proteins were visualized by Western blotting using the M2 anti-Flag MAb (bottom). The data shown are derived from a single experiment but are representative of four experiments. Statistical tests were performed using data collected from these four experiments. CAT expression was significantly increased when F.TAP was overexpressed at both EB2Nter concentrations used (P < 0.05). *, unspecific band.
RESULTS
Several regions in the N terminus of EB2 participate in the nuclear-cytoplasmic shuttling of the protein.
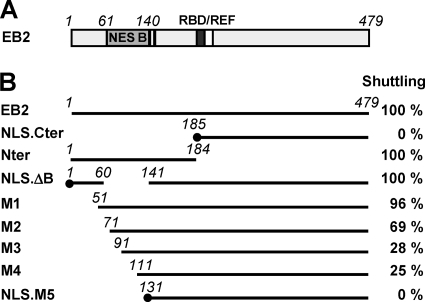
In a previous study, we identified the presence of a CRM1-independent transferable NES in the N terminus of EB2 (14) (NES B; aa 61 to 140) (Fig. 1A). This NES was characterized for its capacity to confer a shuttling property to a heterologous protein, such as β-galactosidase when fused to its N terminus. In an effort to characterize further the limits of this NES in the context of the EB2 protein, we generated a series of deletion mutants (Fig. 1B). We added the SV40 NLS sequence to the N termini of the EB2Cter, EB2.ΔB, and EB2.M5 proteins, since deletions removed one or two of the previously mapped NLS motifs (positions 126 to 130 and 143 to 145) (14) (Fig. 1B, mutants NLS.EB2Cter, NLS.EB2.ΔB, and NLS.EB2.M5). We then tested the capacity of these mutants to shuttle between the nucleus and the cytoplasm. For this purpose, EB2 and the various mutant proteins were expressed in HeLa cells, and their capacity to shuttle between the nucleus and the cytoplasm was evaluated in a human-mouse heterokaryon assay. Indirect immunofluorescence was used to visualize the proteins, and the number of heterokaryons in which the proteins shuttled was counted. The percentage calculated for each protein is indicated in Fig. 1B. As previously published, we found that wild-type EB2 shuttled in all of the heterokaryons observed, whereas NLS.EB2Cter remained strictly restricted to HeLa cell nuclei (14). Similar to EB2, EB2Nter also shuttled in all of the heterokaryons observed, which confirmed the presence of an NES in the N-terminal half of the protein. However, when we deleted NES B (aa 61 to 140), the resulting protein, NLS. EB2.ΔB, still shuttled in 100% of the heterokaryons observed, suggesting the presence of a second NES (NES A) outside the region of aa 61 to 140. We then tested a series of N-terminal deletion mutants. The first observation was that the shuttling ability of the protein was completely abolished after deletion of the first 130 N-terminal amino acids (mutant NLS.EB2.M5), which indicated that the second NES was located in the first 60 N-terminal amino acids of EB2. However, deletion of the first 50 N-terminal amino acids (mutant EB2.M1) had no effect on the shuttling of the protein, and deletion of the first 70 N-terminal amino acids (mutant EB2.M2) had only a limited effect. This suggests that the N terminus of the EB2 protein carries two independent NES, whose functions can complement each other for the shuttling of the protein. Further analysis of the shuttling efficiencies of the remaining deletion mutants (EB2.M3 and EB2.M4) revealed that several sequences within NES B cooperate for its full activity. Taken together, these results suggest that two independent NES contribute to EB2's capacity to shuttle and that several sequences within NES B cooperate for its full activity.
FIG. 1.
Localization of critical domains of EB2 involved in nucleocytoplasmic shuttling of the protein. (A) Schematic representation of EB2 protein. The light gray box represents the NES domain previously identified in the protein (NES B), the dark gray box represents the RNA-binding domain (RBD), the white box represents the REF interacting domain, and the two vertical bars represent the NLS. (B) Heterokaryon assay. EB2 and the various deletion mutants represented in the figure were expressed in HeLa cells from pCI-Flag expression vectors, and a heterokaryon assay was performed. Proteins were then visualized by indirect immunofluorescence, using the anti-Flag M2 MAb (for EB2 and its derivatives). An Alexa Fluor 488-conjugated goat anti-mouse IgG (H+L) antibody was used as the secondary antibody. Cell nuclei were stained with Hoescht 33258. The percentage of heterokaryons in which the proteins were found in both the HeLa and NIH 3T3 cell nuclei was evaluated by counting heterokaryons via conventional microscopy. The percentages are indicated in the right column. External boundaries of deletions are indicated in italics. The black dots at the N termini of mutants NLS.Cter, NLS.ΔB, and NLS.M5 indicate that the SV40 NLS was fused to the N terminus.
The N terminus of EB2 interacts directly with TAP.
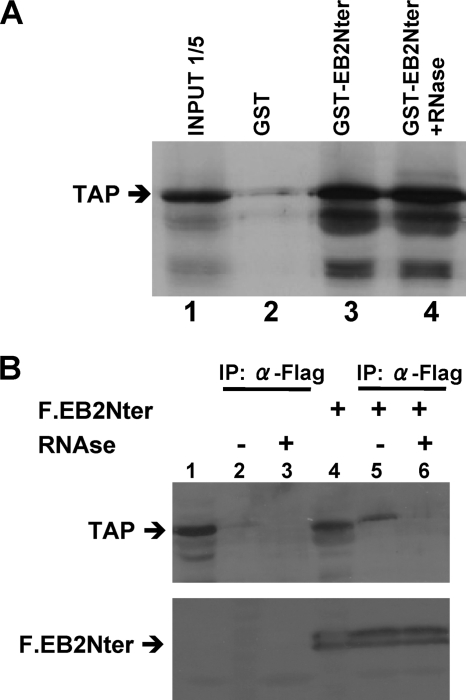
None of the regions identified in the N terminus of EB2 that participate in the nuclear export of the protein have homologies to previously known export sequences. In order to identify cellular factors that bind to the N-terminal region of EB2 (aa 1 to 184), we used a tandem affinity purification approach. HEK293T cells were transfected with pNTAP.EB2Nter, and the tagged EB2Nter protein was isolated from the cell lysate. EB2Nter-copurifying proteins were identified by matrix-assisted laser desorption ionization-time-of-flight mass spectrometry. Among the proteins we identified was the TAP nuclear export factor. To determine whether TAP binds directly to EB2Nter, we performed in vitro binding assays using the GST-EB2Nter fusion protein and [35S]methionine-labeled TAP, produced by coupled transcription-translation in vitro. As shown in Fig. 2A, TAP specifically bound to GST-EB2Nter (lane 3) but not to GST alone (lane 2), and this interaction was resistant to RNase treatment (lane 4), suggesting that TAP directly interacts with EB2Nter. The interaction between TAP and EB2Nter was then examined further in a coimmunoprecipitation assay. The EB2Nter protein, tagged at its N terminus with the Flag epitope (F.EB2Nter), was expressed in HeLa cells, and then immunoprecipitation was performed using anti-Flag MAb M2 affinity gel (Fig. 2B). As shown in Fig. 2B, immunoprecipitation of F.EB2Nter led to coimmunoprecipitation of significant amounts of endogenous TAP protein (lane 5). However, RNase treatment appeared to abolish the interaction (Fig. 2B, lane 6). Since F.EB2Nter does not contain the RNA-binding domain of the protein (13), it is unlikely that the interaction observed in our coimmunoprecipitation assays was mediated by RNA. However, interaction of TAP with RNA might be important to stabilize the interaction with EB2Nter in vivo.
FIG. 2.
The N terminus of EB2 interacts directly with the cellular protein TAP. (A) 35S-labeled TAP was incubated with purified GST or GST-EB2Nter bound to glutathione-Sepharose beads. The bound proteins were analyzed by SDS-PAGE and visualized by autoradiography. RNase treatment was performed where indicated. In lane 1, the equivalent of one-fifth of the TAP-expressing rabbit reticulocyte lysate used in each assay was loaded in the gel (input 1/5). (B) F.EB2Nter was expressed in HeLa cells as indicated. Cell lysates of either mock-transfected cells (lanes 1 to 3) or cells expressing F.EB2Nter (lanes 4 to 6) were immunoprecipitated with M2 anti-Flag MAb resin. The immunoprecipitates were immunoblotted with an anti-TAP rabbit polyclonal antibody (top) or with an anti-Flag polyclonal rabbit antibody (bottom). In lanes 1 and 4, the equivalent of 1/10 of the cell lysate used in each assay was loaded in the gel as an input control.
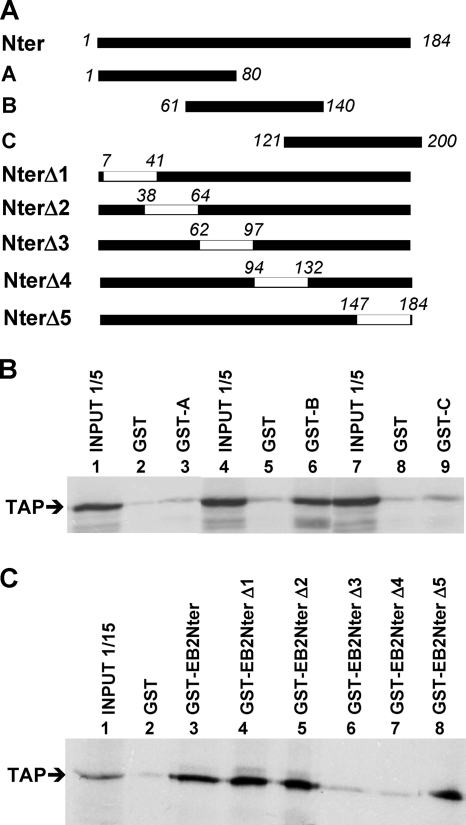
In order to localize more precisely the TAP interaction domain within EB2Nter, we performed in vitro binding assays using a battery of GST-EB2Nter mutant proteins (Fig. 3A) and [35S]methionine-labeled TAP, produced in vitro by coupled transcription-translation. In the first series of experiments, we used three overlapping peptides from the EB2Nter region (peptides A, B, and C) fused to GST. As shown in Fig. 3B, only GST-B (lane 6) bound TAP efficiently, whereas GST-A or GST-C did not (lanes 3 and 9). In a second series of experiments, we used GST-EB2Nter proteins in which overlapping deletions were introduced. As shown in Fig. 3C, GST-EB2Nter-Δ1, -Δ2, and -Δ5 bound TAP as efficiently as GST-EB2Nter (compare lanes 4, 5, and 8 to lane 3), whereas GST-EB2Nter-Δ3 and GST-EB2Nter-Δ4 did not (lanes 6 and 7). These results confirm the localization of the TAP interaction domain within peptide B. Taken together, these data indicate that one or several TAP interaction domains are localized between aa 61 and 140 of EB2.
FIG. 3.
TAP interacts with a region of EB2 between aa 61 and 140. (A) Schematic representation of EB2Nter and the various peptides and deletion mutants used as GST fusion proteins. External boundaries of deletions are indicated in italics. (B) 35S-labeled TAP was incubated with purified GST or GST fused with the overlapping peptide A, B, or C, depicted in panel A, bound to glutathione-Sepharose beads. The bound proteins were analyzed by SDS-PAGE and visualized by autoradiography. The equivalent of one-fifth of the TAP-expressing rabbit reticulocyte lysate used in each assay was loaded in the gel as an indication of the input (lanes 1, 4, and 7). (C) 35S-labeled TAP was incubated with purified GST, GST-EB2Nter, or the GST-EB2Nter deletion mutants depicted in panel A, bound to glutathione-Sepharose beads. The bound proteins were analyzed by SDS-PAGE and visualized by autoradiography. The equivalent of 1/15 of the TAP-expressing rabbit reticulocyte lysate used in each assay was loaded in the gel as an indication of the input (lane 1).
Several arginine residues within peptide B are important both for TAP binding and for EB2 shuttling.
Peptide B alone acts as an NES when fused to β-galactosidase, a heterologous nonshuttling protein whose size is incompatible with passive diffusion between the nucleus and the cytoplasm (NLS.B.βgal) (14). Peptide B also binds TAP. It was therefore important to know if TAP binding was required for the NES function of peptide B. We thus used the NLS.B.βgal fusion protein to map more precisely the domain(s) necessary for nuclear export within peptide B. The NLS.B.βgal protein used in the study is depicted in Fig. 4A. The SV40 NLS was added at the N terminus to ensure efficient nuclear import of the fusion protein, since only one of the two EB2 NLS is present in peptide B. We first generated a series of 15-aa deletion mutants in peptide B (Fig. 4A). These deletion mutants were expressed in HeLa cells, and their ability to shuttle was evaluated by a heterokaryon assay and compared to that of NLS.B.βgal (Fig. 4B). None of the NLS.B.βgal single-deletion mutants was drastically affected in its capacity to shuttle (data not shown) compared to NLS.B.βgal, which suggested that several regions within peptide B cooperate to confer nucleocytoplasmic shuttling properties to the fusion protein. We then generated mutants with various combinations of deletions made in the N-terminal half of peptide B (Δb1, Δb2, and Δb3) and in its C-terminal half (Δb4, Δb5, Δb6, and Δb7) and again tested their ability to shuttle in the heterokaryon assay. As shown in Fig. 4B, we observed a decrease of one-third to one-half of the shuttling activity compared to that of NLS.B.βgal with most of the double mutants (except for mutant NLS.B.βgal Δb1/b5, which shuttled more efficiently). However, it was only with a triple deletion mutant (NLS.B.βgal Δb3/b5/b7) that we observed a more drastic decrease in shuttling activity.
FIG. 4.
Several domains of peptide B cooperate for nuclear export. (A) Schematic representation of NLS.B.βgal fusion protein and the various deletion mutants derived from it. The light gray box represent the β-galactosidase open reading frame, the dark gray box represents the SV40 NLS, and the white box represents EB2 aa 61 to 140. The vertical black bar represent NLS1 of EB2. (B) Expression plasmids for the different fusion proteins listed in panel B (left column) were transfected into HeLa cells, and a heterokaryon assay was performed. The percentage of heterokaryons in which the transiently expressed proteins were found in both the HeLa and NIH 3T3 cell nuclei was evaluated by counting the heterokaryons via conventional microscopy. The percentages are indicated in the right column. (C) Amino acid sequence of EB2 peptide B (EBV strain Raji). Amino acids deleted from mutants NLS.B.βgal-Δb3, -Δb5, and -Δb7 are underlined in gray. The locations of the Δ3 and Δ4 deletions are indicated by brackets. Amino acids changed to alanines in the EB2Nter R/A mutant (see panel D) are indicated in bold. (D) 35S-labeled TAP was incubated with purified GST, GST-EB2Nter, or GST-EB2Nter R/A (in which the amino acids indicated in bold in panel C have been mutated to alanines) bound to glutathione-Sepharose beads. The bound proteins were analyzed by SDS-PAGE and visualized by autoradiography. In lane 1, the equivalent of 1/15 of the TAP-expressing rabbit reticulocyte lysate used in each assay was loaded in the gel (input 1/15).
Interestingly, regions b3, b5, and b7 contained all of the arginine residues found in peptide B (Fig. 4C), and TAP has been reported to bind arginine-rich motifs in both 9G8 and SRp20 SR proteins (11) as well as in REF (8), which suggests that these arginine residues might cooperate in the nuclear export of EB2 by recruiting the TAP protein. We then decided to introduce specific mutations (Fig. 4C; amino acids in bold were changed to alanines), including all of the arginines between aa 81 and 135, in the context of EB2Nter fused to GST (GST-EB2Nter R/A), and then we performed GST pull-down assays. EB2Nter R/A bound TAP with a much lower affinity than did EB2Nter (Fig. 4D). We also introduced the same mutations in the context of the NLS.B.βgal protein and performed a heterokaryon assay. The mutated protein shuttled in only 13% of the heterokaryons. Taken together, these results indicate that TAP interacts within a region of EB2 comprised of aa 81 to 135 and that the arginine residues present in this region are likely to be essential to the interaction. Furthermore, their mutation correlates with a strong decrease in the efficiency of NLS.B.βgal nucleocytoplasmic shuttling.
Overexpression of EB2Nter inhibits TAP-dependent export of unspliced pDM128/PL mRNA by EB2.
EB2 has previously been found to mediate export of unspliced mRNA generated from a pDM128/PL reporter plasmid (14). In the absence of EB2, the RNA transcript encoded by pDM128/PL is normally unable to exit the nucleus without being spliced, a process that deletes the cat open reading frame (15). The effect of EB2 on pDM128/PL unspliced mRNA has been linked to the recruitment of REF/TAP protein complexes via the C-terminal domain of EB2 (Fig. 5A) (14). Since EB2Nter lacks the RNA-binding domain and appeared to directly bind TAP, we hypothesized that EB2Nter could act as a dominant-negative mutant of EB2. We thus overexpressed increasing amounts of F.EB2Nter in HeLa cells and tested the effect on pDM128/PL unspliced mRNA export by F.EB2 (Fig. 5B). As shown previously, very little CAT protein was expressed when the pDM128/PL reporter plasmid was transfected into HeLa cells alone because of very poor export of unspliced mRNA generated from pDM128/PL (Fig. 5B, lane 1). However, as expected, we observed a strong increase in CAT expression in the presence of F.EB2 (Fig. 5B, lane 2), whereas F.EB2Nter, which does not bind RNA, had no stimulating effect (Fig. 5B, lane 3). But when we coexpressed increasing amounts of F.EB2Nter together with F.EB2, we observed a dose-dependent decrease in CAT expression (Fig. 5B, lanes 4 to 8). This decrease was not due to a decrease in EB2's expression levels (Fig. 5B, bottom panel).
FIG. 5.
EB2Nter behaves like a dominant-negative mutant of EB2. (A) Schematic representation of pDM128/PL reporter plasmid. The CAT coding sequence is inserted into intronic sequences. The positions of the 5′ and 3′ splice sites (SS) are indicated. In the absence of EB2, only the spliced mRNA generated from pDM128/PL is exported efficiently to the cytoplasm (no CAT expression). The presence of EB2 allows export of the unspliced mRNA (and consequently CAT expression) by a mechanism which involves binding of EB2 to the RNA and recruitment of the TAP-p15 complex via REF. Both the RNA-binding domain (indicated by the small black area) and the REF interaction domain have been localized to the C-terminal half of EB2. A direct interaction between EB2Nter and the TAP-p15 complex is also indicated. (B) Dose-dependent inhibition of EB2-mediated mRNA export by EB2Nter. pDM128/PL was transfected into HeLa cells either alone or together with 0.5 μg of expression plasmid for F.EB2 and increasing amounts (0.01 to 0.5 μg) of expression plasmid for F.EB2Nter, as indicated in the figure. The efficiency of unspliced mRNA export was evaluated by quantification of CAT expression by CAT ELISA (top), and the amounts of F.EB2 and F.EB2Nter proteins expressed in each assay were visualized by Western blotting using the M2 anti-Flag MAb (bottom). The data shown are derived from a single experiment but are representative of four experiments. (C) Relative activities of EB2Nter deletion mutants in EB2-mediated mRNA export inhibition assay. pDM128/PL was transfected into HeLa cells either alone or with expression plasmids for F.EB2 (0.5 μg), F.EB2Nter (0.5 μg), and F.EB2Nter internal deletion mutants (up to 3 μg), as indicated. The F.EB2Nter-Δ4 and -ΔB mutants are both tagged with the SV40 NLS. The efficiency of unspliced mRNA export was evaluated by quantification of CAT expression by CAT ELISA (top). The levels of expression of F.EB2, F.EB2Nter, and F.EB2Nter mutants were evaluated by Western blotting using the M2 anti-Flag MAb (bottom). The data shown are derived from a single experiment but are representative of four independent experiments. (D) Relative activities of EB2Nter and the EB2Nter R/A mutant in EB2-mediated mRNA export inhibition assay. pDM128/PL was transfected into HeLa cells either alone (lane 1) or with expression plasmids for F.EB2 (0.5 μg) (lanes 1 to 3), F.EB2Nter (0.5 μg) (lane 3), and F.EB2Nter R/A (2 μg) (lane 4). The efficiency of unspliced mRNA export was evaluated by quantification of CAT expression, using CAT ELISA (left). The levels of expression of F.EB2, F.EB2Nter, and F.EB2Nter R/A were evaluated by Western blotting using the M2 anti-Flag MAb (right).
In order to confirm that the competition effect observed above correlated with the presence of the TAP interaction domain within EB2Nter, we tested the effects of overexpression of deletion mutants F.EB2Nter-Δ1, -Δ2, -Δ3, -Δ4, -Δ5, and -ΔB, depicted in Fig. 1A and 3A. As shown in Fig. 5C, mutants F.EB2Nter-Δ1, -Δ2, and -Δ5, which were found to interact with TAP in vitro, were as efficient as F.EB2Nter at repressing the F.EB2-mediated mRNA export, whereas F.NLS.EB2NterΔ4 and F.NLS.EB2NterΔB, which did not interact with TAP in vitro, had no repressing effect. Surprisingly, however, F.EB2NterΔ3 did not bind TAP in vitro but still had a repressing effect, which suggested that the main EB2 interaction region with TAP in vivo is comprised between aa 95 and 131. Again, EB2's expression level remained constant in all assays (Fig. 5C, bottom panel). Furthermore, the EB2Nter R/A mutant, in which arginines of domain B were mutated and whose interaction with TAP was found to be strongly impaired (Fig. 4D), was accordingly much less efficient than EB2Nter in repressing F.EB2-mediated export (Fig. 5D). Lastly, we found that we could rescue EB2-induced pDML128/PL CAT expression by overexpressing TAP in our competition assays (Fig. 6), demonstrating that TAP is indeed the limiting factor.
Taken together, these results suggest that overexpressed F.EB2Nter sequesters a cellular factor, most likely TAP, which is necessary for EB2-dependent mRNA export of unspliced pDM128/PL mRNA.
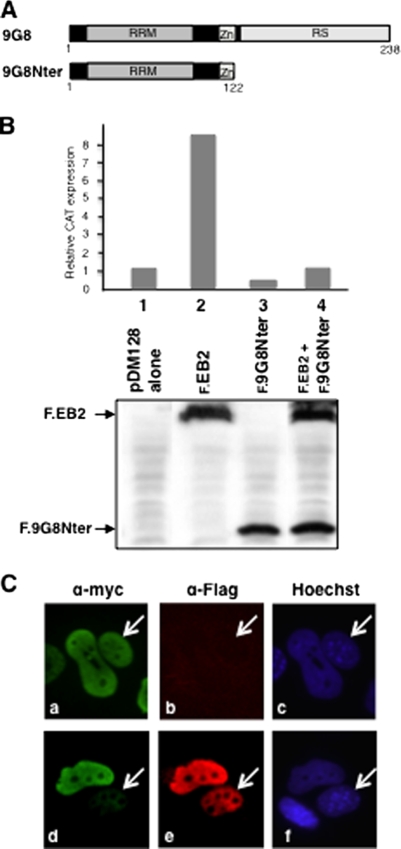
Titration of TAP by 9G8 inhibits EB2Nter's shuttling.
EB2Nter appears to interact with TAP via a region whose deletion is detrimental to its efficient export from the nucleus to the cytoplasm. To demonstrate a direct role for TAP in EB2Nter's shuttling, we titrated TAP. For this purpose, we used a mutant of the 9G8 SR protein which had its RS domain deleted but still interacted with TAP (11). This was tagged with the Flag epitope (F.9G8Nter) (Fig. 7A). We first tested whether F.9G8Nter could compete for EB2-mediated export of unspliced mRNA generated from pDM128/PL (Fig. 5A). Figure 7B shows that, as expected, and similar to the case for EB2Nter, as shown above, F.9G8Nter was very efficient in repressing unspliced pDM128/PL mRNA export by F.EB2 (compare lane 4 to lane 2). We then asked whether competition by F.9G8Nter for TAP binding would also directly affect EB2Nter's shuttling. To answer this question, we transfected HeLa cells with expression plasmids for Myc.EB2Nter and F.9G8Nter and tested for the ability of Myc.EB2Nter to shuttle in the interspecies heterokaryon assay. We compared the efficiencies of Myc.EB2Nter shuttling between heterokaryons which did not express detectable amounts of F.9G8Nter (Fig. 7C, panel b) and heterokaryons expressing significant amounts of F.9G8Nter (Fig. 7C, panel e). It is evident that when it is expressed, F.9G8Nter is found in both HeLa and NIH 3T3 cell nuclei, but this was expected, since 9G8 is known as a shuttling SR protein. As can be seen in Fig. 7C (panel a), equivalent amounts of Myc.EB2Nter were visualized in both HeLa and NIH 3T3 cell nuclei of the heterokaryon in the absence of F.9G8Nter. In contrast, the amount of Myc.EB2Nter detected in the NIH 3T3 cell nucleus was much lower than the amount detected in the HeLa cell nucleus when F.9G8Nter was expressed (Fig. 7C, panel d). These results strongly suggest that by interacting with EB2, TAP is indeed responsible for the nuclear export of the protein.
FIG. 7.
Competition for TAP's interaction with EB2Nter by 9G8 alters EB2's shuttling efficiency. (A) Schematic representation of the 9G8 SR protein and the C-terminal deletion mutant 9G8Nter. 9G8Nter retained both the TAP interaction domain and sequences necessary for 9G8 shuttling. (B) 9G8Nter inhibits EB2-dependent export of the unspliced mRNA generated from pDM128/PL (Fig. 6A). HeLa cells were transfected with pDM128/PL either alone or together with expression plasmids for F.EB2 (0.5 μg) and F.9G8Nter (0.5 μg), as indicated. The efficiency of unspliced mRNA export was evaluated by quantification of CAT expression by CAT ELISA (top). The levels of expression of F.EB2 and F.9G8Nter mutants were visualized by Western blotting using the M2 anti-Flag MAb (bottom). Data from a representative experiment out of four independent experiments are presented in the figure. (C) 9G8Nter competition for TAP binding interferes with EB2Nter nuclear export. HeLa cells were cotransfected with pCI.Myc.EB2Nter (0.5 μg) and pCI.F.9G8Nter (0.5 μg) expression plasmids, and a heterokaryon assay was performed. Proteins were visualized by indirect immunofluorescence, using the 9E10 anti-Myc antibody (for detection of Myc.EB2Nter) (a and d) and an anti-Flag polyclonal antibody (Sigma) (for detection of F.9G8Nter) (b and e). An Alexa Fluor 488-conjugated goat anti-mouse IgG (H+L) antibody (Invitrogen) (a and d) and a Fluorolink Cy3-labeled goat anti-rabbit antibody (Amersham) (b and e) were used as secondary antibodies. Cell nuclei were stained with Hoescht 33258 (Sigma) (c and f). NIH 3T3 cell nuclei are indicated by white arrows. A minimum of 20 heterokaryons of each sort (either expressing F.9G8Nter or not) were observed: only one example of each is presented in the figure. The phenotype presented in panel d was found in 75% of heterokaryons expressing 9G8Nter but in only 10 to 15% of heterokaryons that did not express 9G8Nter.
The domain comprising aa 61 to 140 of EB2 is sufficient to stimulate mRNA export when tethered to RNA by a heterologous RNA-binding domain.
We have shown above that EB2Nter interacts with TAP via several regions located between aa 61 and 140 and that this interaction appears to be at least partly responsible for mediating the export of EB2Nter from the nucleus to the cytoplasm. We next asked whether this TAP interaction domain was able to mediate mRNA nuclear export when tethered to RNA via a heterologous RNA-binding domain. For this experiment, we fused aa 61 to 140 and 61 to 200 to the MS2 phage coat protein RNA-binding domain (proteins MS2-B and MS2-BC, respectively) and used the pDM128/4xMS2 reporter construct, which contains the cat indicator gene and four copies of the MS2 operator RNA target, flanked by 5′ and 3′ splice sites, as indicated in Fig. 8A (24). As shown in Fig. 8B, the pDM128/4xMS2 indicator plasmid gave rise to very little CAT expression in transfected HeLa cells (Fig. 8B, lane 1) in the presence of MS2. However, expression of the MS2-B or MS2-BC fusion protein induced readily detectable expression of CAT from the pDM128/4xMS2 construct (Fig. 8B, lanes 3 and 4). These results suggest that tethering of the B domain of EB2 to the RNA is sufficient to stimulate nuclear export of the targeted mRNA.
DISCUSSION
Nuclear export of proteins from the nucleus to the cytoplasm is usually mediated by specific receptors, called β karyopherins, that belong to a large family of proteins conserved throughout evolution (28). Here we find that nuclear export of the viral EB2 protein is mediated by two contiguous but independent N-terminal NES, one of which (NES B) promotes export of a heterologous protein via a direct interaction with the mRNA export receptor TAP. Furthermore, we found that tethering of this TAP interaction domain to RNA by a heterologous RNA-binding domain is sufficient to stimulate translocation of an unspliced mRNA from the nucleus to the cytoplasm, whereas this mRNA would normally be exported very inefficiently.
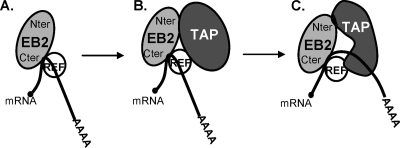
The interaction between EB2Nter and TAP was first revealed by a tandem affinity purification assay and then confirmed by GST pull-down experiments. In GST pull-down experiments, the interaction between the two proteins was resistant to RNase treatment, indicating that RNA was not making the link between the two proteins. Furthermore, EB2Nter lacks the EB2 RNA-binding domain (13), which also argues for a direct interaction between the two proteins. However, surprisingly, in vivo RNase treatment was highly detrimental to the interaction. This could be explained by a change in the conformation of TAP in association with RNA, increasing its affinity for EB2. Alternatively, in vivo, other cellular factors associated with TAP in an RNA-dependent manner could be important for TAP's association with EB2. Interestingly, we previously found that EB2 recruited TAP via REF through a C-terminal domain located between aa 213 and 236, adjacent to its RNA-binding domain. It is tempting to imagine a model in which TAP would first be recruited onto EB2's associated mRNPs by REF and then transferred to the RNA, as described by Hautbergue et al. (12), which would then stimulate a secondary interaction with the N-terminal domain of EB2 (Fig. 9). This model is supported by the fact that coimmunoprecipitation of TAP with full-length EB2 is more efficient than its coimmunoprecipitation with either EB2Nter or EB2Cter (aa 185 to 479) (data not shown) (14).
FIG. 9.
Model of recruitment of TAP to EB2-containing mRNPs. (A) EB2 first interacts with its target mRNA and recruits REF into the mRNP. (B) TAP recruited by REF onto the mRNP interacts with EB2 at a low affinity. (C) TAP is transferred to the mRNA following its interaction with REF and changes its conformation, thereby interacting with EB2 at a higher affinity.
We further dissected the N terminus of EB2 in order to precisely map the interaction domain with TAP. From our results, it appears that three small regions between aa 81 and 135 cooperate for optimum binding to TAP. A characteristic of these three regions is the presence of several arginine residues reminiscent of other TAP interaction domains characterized in the SR proteins 9G8 and SRp20 and in REF. TAP's binding motifs in 9G8 (LSTGMPRRSRFDRPPARR) and SRp20 (EKRSRNR) have little in common except for the presence of several arginine residues shown to be important for the interaction (11). Interaction with REF also involved a region rich in arginines (NRNQRRVNRGGGPRRNRPAIA) (12). By mutating most of the arginine residues present between aa 81 and 135, we significantly decreased the affinity of EB2Nter for TAP, which confirms an important role of arginine residues in the interaction between EB2Nter and TAP. The N terminus of EB2 is not predicted to be a very structured domain, and the various arginine residues, although distanced from each other in the primary structure, could participate in binding TAP.
It is noteworthy that the arginine cluster (KRRR) located at aa 128 to 130 was previously identified as a nuclear import signal for EB2 (NLS 1) (14). Interaction of TAP with this domain suggests that there could be competition between the import factor's binding to its NLS and to TAP. Interestingly, a second NLS (NLS 2), whose sequence (KRR) also contains arginines, has been localized at aa 143 to 144. This motif was not included in the region that we initially characterized as a nuclear export domain. However, in our experiments, we repeatedly observed that TAP interacted more efficiently with GST-EB2Nter than with GST-B (data not shown). Furthermore, the fusion protein MS2-BC, tethered to the RNA via MS2 binding sites, exported unspliced pDM128 mRNAs more efficiently than did MS2-B (Fig. 8). Taken together, these results suggest that a sequence outside peptide B, possibly the KRR motif of NLS 2, might also contribute to TAP's binding. Further study will be necessary to clarify this point, and in particular, it will be interesting to identify the importin(s) involved and to ask whether there is indeed competition with TAP for binding to EB2's N-terminal domain. This could be an efficient way to liberate TAP from EB2 in the cytoplasm.
The localization of TAP's interaction domain determined in vitro correlates well with the results obtained in our competition assays of TAP's interaction in vivo (Fig. 5). In effect, among the various internal deletion mutants tested (EB2Nter-Δ1, -Δ2, -Δ3, -Δ4, and -Δ5), only EB2Nter-Δ4, which has aa 85 to 131 deleted, did not compete for TAP interaction. There was, however, a discrepancy between the in vivo and in vitro results obtained with EB2Nter-Δ3, which competed for TAP's interaction in vivo as efficiently as EB2Nter but did not interact with TAP in vitro (GST pull-down assay). This lack of interaction in vitro could be explained by a problem of misconformation of GST-EB2Nter-Δ3 or a lack of accessibility of the TAP interaction domain because of the proximity of the GST domain induced by the deletion in the Δ3 mutant. Thus, the most critical region for interaction with TAP in EB2Nter is likely to be comprised between aa 94 to 132.
Following the demonstration of a direct interaction of TAP with an N-terminal domain of EB2, we found that tethering of this domain to pDM1284xMS2 unspliced mRNAs, via the MS2 phage coat RNA-binding protein, stimulated their nuclear export. Interestingly, we had previously shown that TAP also interacted with the C-terminal domain of EB2 (EB2Cter; aa 185 to 479), although indirectly, via REF (14). However, surprisingly, NLS.EB2Cter (or even a longer C-terminal construct, such as EB2.NLS.M5) did not exit the nucleus (Fig. 1), suggesting that this indirect recruitment of TAP by EB2 via REF might be only transitory and thus not sufficient to allow nuclear export of EB2Cter. This observation is again in favor of the model described above, in which TAP could be recruited primarily by EB2 via REF and then transferred to the RNA, and a direct interaction with EB2's N-terminal domain (aa 94 to 132) would then stabilize the complex.
EB2's functional homologue in herpes simplex virus, ICP27, has also been found to interact with REF via a domain located between aa 104 and 138 of the protein (4, 20). This interaction has been shown to be important for recruitment of REF to ICP4-containing transcription foci early in infection, away from SC35 speckles (5), but similar to what is seen with EB2, this interaction does not appear to be important for ICP27 export from the nucleus to the cytoplasm (5, 19). Interestingly, it was found that ICP27 also interacted with TAP without REF bridging. This interaction appeared to involve two distinct domains of the protein, one at the very N terminus of ICP27 and the second at its extreme C terminus (5). By using a dominant-negative TAP mutant (which has its C-terminal domain required for interaction with nucleoporins deleted but which still interacts with ICP27), Chen et al. (4) demonstrated that TAP was required for ICP27 export from the nucleus. Thus, EB2 and ICP27 both appear to use very similar mechanisms to recruit the mRNA export factor TAP.
In conclusion, our results clearly show that by directly binding to an arginine-rich domain of EB2, the general mRNA export factor TAP can mediate nuclear export. This direct interaction between EB2 and TAP is likely to be essential for efficient export of viral mRNAs.
Acknowledgments
This work was supported by the Institut National de la Santé et de la Recherche Médicale (INSERM), the Agence Nationale pour la Recherche (ANR) (ANR MIME grant RPV06120CSA to E.M.), and Lyon Biopole. We also thank the Réseau Herpesvirus and Cancer for its support. F.J. and E.H. are recipients of fellowships from the Ministère de la Recherche et de la Technologie (MRT). A.S. and E.M. are CNRS scientists.
We thank B. R. Cullen for providing plasmids pCMV-MS2, pDM128/PL, and pDM128/4xMS2. We also acknowledge the Platim microscope facilities of IFR128. Finally, we thank R. Buckland for reading the manuscript.
Footnotes
Published ahead of print on 30 September 2009.
REFERENCES
- 1.Batisse, J., E. Manet, J. Middeltorp, A. Sergeant, and H. Gruffat. 2005. The Epstein-Barr virus mRNA export factor EB2 is essential for intranuclear capsid assembly and production of gp350. J. Virol. 79:14102-14111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyne, J. R., K. J. Colgan, and A. Whitehouse. 2008. Recruitment of the complete hTREX complex is required for Kaposi's sarcoma-associated herpesvirus intronless mRNA nuclear export and virus replication. PLoS Pathog. 4:e1000194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buisson, M., F. Hans, I. Kusters, N. Duran, and A. Sergeant. 1999. The C-terminal region but not the Arg-X-Pro repeat of Epstein-Barr virus protein EB2 is required for its effect on RNA splicing and transport. J. Virol. 73:4090-4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, I.-H. B., K. S. Sciabica, and R. M. Sandri-Goldin. 2002. ICP27 interacts with the RNA export factor Aly/REF to direct herpes simplex virus type 1 intronless mRNAs to the TAP export pathway. J. Virol. 76:12877-12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, I. H., L. Li, L. Silva, and R. M. Sandri-Goldin. 2005. ICP27 recruits Aly/REF but not TAP/NXF1 to herpes simplex virus type 1 transcription sites although TAP/NXF1 is required for ICP27 export. J. Virol. 79:3949-3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng, H., K. Dufu, C. S. Lee, J. L. Hsu, A. Dias, and R. Reed. 2006. Human mRNA export machinery recruited to the 5′ end of mRNA. Cell 127:1389-1400. [DOI] [PubMed] [Google Scholar]
- 7.Farjot, G., M. Buisson, M. Duc Dodon, L. Gazzolo, A. Sergeant, and I. Mikaelian. 2000. Epstein-Barr virus EB2 protein exports unspliced RNA via a Crm-1-independent pathway. J. Virol. 74:6068-6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Golovanov, A. P., G. M. Hautbergue, A. M. Tintaru, L. Y. Lian, and S. A. Wilson. 2006. The solution structure of REF2-I reveals interdomain interactions and regions involved in binding mRNA export factors and RNA. RNA 12:1933-1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gruffat, H., J. Batisse, D. Pich, B. Neuhierl, E. Manet, W. Hammerschmidt, and A. Sergeant. 2002. Epstein-Barr virus mRNA export factor EB2 is essential for production of infectious virus. J. Virol. 76:9635-9644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han, Z., E. Marendy, Y. D. Wang, J. Yuan, J. T. Sample, and S. Swaminathan. 2007. Multiple roles of Epstein-Barr virus SM protein in lytic replication. J. Virol. 81:4058-4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hargous, Y., G. M. Hautbergue, A. M. Tintaru, L. Skrisovska, A. P. Golovanov, J. Stevenin, L. Y. Lian, S. A. Wilson, and F. H. Allain. 2006. Molecular basis of RNA recognition and TAP binding by the SR proteins SRp20 and 9G8. EMBO J. 25:5126-5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hautbergue, G. M., M. L. Hung, A. P. Golovanov, L. Y. Lian, and S. A. Wilson. 2008. Mutually exclusive interactions drive handover of mRNA from export adaptors to TAP. Proc. Natl. Acad. Sci. USA 105:5154-5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hiriart, E., L. Bardouillet, E. Manet, H. Gruffat, F. Penin, R. Montserret, G. Farjot, and A. Sergeant. 2003. A region of the Epstein-Barr virus (EBV) mRNA export factor EB2 containing an arginine-rich motif mediates direct binding to RNA. J. Biol. Chem. 278:37790-37798. [DOI] [PubMed] [Google Scholar]
- 14.Hiriart, E., G. Farjot, H. Gruffat, M. V. Nguyen, A. Sergeant, and E. Manet. 2003. A novel nuclear export signal and a REF interaction domain both promote mRNA export by the Epstein-Barr virus EB2 protein. J. Biol. Chem. 278:335-342. [DOI] [PubMed] [Google Scholar]
- 15.Hope, T. J., B. L. Bond, D. McDonald, N. P. Klein, and T. G. Parslow. 1991. Effector domains of human immunodeficiency virus type 1 Rev and human T-cell leukemia virus type I Rex are functionally interchangeable and share an essential peptide motif. J. Virol. 65:6001-6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang, Y., R. Gattoni, J. Stévenin, and J. A. Steitz. 2003. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol. Cell 11:837-843. [DOI] [PubMed] [Google Scholar]
- 17.Huang, Y., and J. A. Steitz. 2001. Splicing factors SRp20 and 9G8 promote the nucleocytoplasmic export of mRNA. Mol. Cell 7:899-905. [DOI] [PubMed] [Google Scholar]
- 18.Iglesias, N., and F. Stutz. 2008. Regulation of mRNP dynamics along the export pathway. FEBS Lett. 582:1987-1996. [DOI] [PubMed] [Google Scholar]
- 19.Johnson, L. A., L. Li, and R. M. Sandri-Goldin. 2009. The cellular RNA export receptor TAP/NXF1 is required for ICP27-mediated export of herpes simplex virus 1 RNA, but the TREX complex adaptor protein Aly/REF appears to be dispensable. J. Virol. 83:6335-6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koffa, M., J. Clements, E. Izaurralde, S. Wadd, S. Wilson, I. Mattaj, and S. Kuersten. 2001. Herpes simplex virus ICP27 protein provides viral mRNAs with access to the cellular mRNA export pathway. EMBO J. 20:5769-5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindtner, S., A. S. Zolotukhin, H. Uranishi, J. Bear, V. Kulkarni, S. Smulevitch, M. Samiotaki, G. Panayotou, B. K. Felber, and G. N. Pavlakis. 2006. RNA-binding motif protein 15 binds to the RNA transport element RTE and provides a direct link to the NXF1 export pathway. J. Biol. Chem. 281:36915-36928. [DOI] [PubMed] [Google Scholar]
- 22.Lischka, P., Z. Toth, M. Thomas, R. Mueller, and T. Stamminger. 2006. The UL69 transactivator protein of human cytomegalovirus interacts with DEXD/H-box RNA helicase UAP56 to promote cytoplasmic accumulation of unspliced RNA. Mol. Cell. Biol. 26:1631-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malik, P., D. J. Blackbourn, and B. Clements. 2004. The evolutionarily conserved Kaposi's sarcoma-associated herpesvirus ORF57 protein interacts with REF protein and acts as an mRNA export factor. J. Biol. Chem. 279:33001-33011. [DOI] [PubMed] [Google Scholar]
- 24.McDonald, D., T. J. Hope, and T. G. Parslow. 1992. Posttranscriptional regulation by the human immunodeficiency virus type 1 Rev and human T-cell leukemia virus type I Rex proteins through a heterologous RNA binding site. J. Virol. 66:7232-7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Medina-Palazon, C., H. Gruffat, F. Mure, O. Filhol, V. Vingtdeux-Didier, H. Drobecq, C. Cochet, N. Sergeant, A. Sergeant, and E. Manet. 2007. Protein kinase CK2 phosphorylation of EB2 regulates its function in the production of Epstein-Barr virus infectious viral particles. J. Virol. 81:11850-11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Michael, W. M., P. S. Eder, and G. Dreyfuss. 1997. The K nuclear shuttling domain: a novel signal for nuclear import and nuclear export in the hnRNP K protein. EMBO J. 16:3587-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nojima, T., T. Hirose, H. Kimura, and M. Hagiwara. 2007. The interaction between cap-binding complex and RNA export factor is required for intronless mRNA export. J. Biol. Chem. 282:15645-15651. [DOI] [PubMed] [Google Scholar]
- 28.Ossareh-Nazari, B., C. Gwizdek, and C. Dargemont. 2001. Protein export from the nucleus. Traffic 2:684-689. [DOI] [PubMed] [Google Scholar]
- 29.Semmes, O. J., L. Chen, R. T. Sarisky, Z. Gao, L. Zhong, and S. D. Hayward. 1998. Mta has properties of an RNA export protein and increases cytoplasmic accumulation of Epstein-Barr virus replication gene mRNA. J. Virol. 72:9526-9534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stutz, F., A. Bachi, T. Doerks, I. C. Braun, B. Séraphin, M. Wilm, P. Bork, and E. Izaurralde. 2000. REF, an evolutionarily conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA 6:638-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams, B. J. L., J. R. Boyne, D. J. Goodwin, L. Roaden, G. M. Hautbergue, S. A. Wilson, and A. Whitehouse. 2005. The prototype γ-2 herpesvirus nucleocytoplasmic shuttling protein, ORF57, transports viral RNA through the cellular mRNA export pathway. Biochem. J. 387:295-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang, J., H. Bogerd, P. Wang, D. Page, and B. Cullen. 2001. Two closely related human nuclear export factors utilize entirely distinct export pathways. Mol. Cell 8:397-406. [DOI] [PubMed] [Google Scholar]