Abstract
Limiting genome replication to once per cell cycle is vital for maintaining genome stability. Inhibition of cyclin-dependent kinase 1 (CDK1) with the specific inhibitor RO3306 is sufficient to trigger multiple rounds of genome reduplication. We demonstrated that although anaphase-promoting complex/cyclosome (APC/C) remained inactive during the initial G2 arrest, it was activated upon prolonged inhibition of CDK1. Using cellular biosensors and live-cell imaging, we provide direct evidence that genome reduplication was associated with oscillation of APC/C activity and nuclear-cytoplasmic shuttling of CDC6 even in the absence of mitosis at the single-cell level. Genome reduplication was abolished by ectopic expression of EMI1 or depletion of CDC20 or CDH1, suggesting the critical role of the EMI1-APC/C axis. In support of this, degradation of EMI1 itself and genome reduplication were delayed after downregulation of PLK1 and β-TrCP1. In the absence of CDK1 activity, activation of APC/C and genome reduplication was dependent on cyclin A2 and CDK2. Genome reduplication was then promoted by a combination of APC/C-dependent destruction of geminin (thus releasing CDT1), accumulation of cyclin E2-CDK2, and CDC6. Collectively, these results underscore the crucial role of cyclin A2-CDK2 in regulating the PLK1-SCFβ-TrCP1-EMI1-APC/C axis and CDC6 to trigger genome reduplication after the activity of CDK1 is suppressed.
Limiting genome replication to once per cell cycle is critical for maintaining genome stability and suppressing tumorigenesis (reviewed in reference 18). DNA replication is a biphasic process consisting of origin licensing and origin firing. During late mitosis to early G1 phase, origins are licensed by orderly loading of prereplicative complex components, including ORC, CDT1, CDC6, and MCM2-7 (reviewed in reference 3). Origin licensing occurs in a biochemical environment characterized by low activity of cyclin-cyclin-dependent kinase (CDK) and high activity of anaphase-promoting complex/cyclosome (APC/C) (reviewed in reference 51). Firing of the origins is coordinated by phosphorylation carried out by cyclin-CDK and DBF4-CDC7 and the binding of other replication factors, including CDC45, MCM10, RPA, and DNA polymerase (reviewed in reference 58).
Critical roles in replication have been attributed to both cyclin A and cyclin E, but the distinct roles performed by the two cyclins remain incompletely understood. Cyclin A is especially interesting among the cyclins because of its association with multiple CDKs (CDK1 and CDK2) and its proposed functions in multiple points of the cell cycle (S phase and mitosis). In S phase, cyclin A is believed to be involved in the loading of CDC45 onto origins. Cyclin A is also involved in blocking the reloading of fired origins (reviewed in reference 43). CDT1 is targeted to degradation after phosphorylation by cyclin-CDK in SCFSKP2- or CUL4-DDB1-mediated mechanisms (15). Geminin accumulates during S phase and inactivates the remaining CDT1 (32). Moreover, CDC6 is phosphorylated by cyclin-CDK and translocated out of the nucleus. Finally, ORC1 (the largest subunit of the ORC) is inactivated either by polyubiquitination by SCFSKP2 and degradation by the proteasome (38) or by monoubiquitination and dissociation from the chromatin (29). Thus, the high cyclin-CDK and low APC/C activities during S phase prevent the formation of the prereplicative complex and reduplication. The system is reset during the next mitosis, when APC/C is activated and degrades cyclins and geminin, allowing the prereplicative complex to form again.
Genome reduplication generates polyploid cells. A growing body of evidence indicates that polyploidization can initiate chromosomal instability and aneuploidy. A seminal study by Fujiwara et al. (16) indicates that tetraploids can be generated by transient blocking of cytokinesis in p53-null mouse mammary epithelial cells. Importantly, tetraploidization promotes aneuploidy and tumorigenesis (16). Another study reported that chromosome nondisjunction (both copies of a chromosome segregate to the same daughter cells) leads to binucleated tetraploids by promoting cleavage furrow regression; the tetraploid cells then become aneuploidy through further divisions (48). This and other studies provide strong evidence of the importance of tetraploidization as an early step in tumorigenesis (reviewed in reference 53).
While rereplication is stringently prevented in the normal cell cycle, multiple rounds of genome reduplication, called endoreduplication, occur in cell types such as megakaryocytes, trophoblast giant cells, numerous plant cells (26), and in the salivary glands of Drosophila melanogaster (49). In yeasts, different levels of a single CDK are believed to allow origin licensing and firing and prevent relicensing and mitosis (reviewed in reference 45). In contrast, the complex interplay between different cyclin-CDK complexes and licensing factors to prevent genome reduplication in higher eukaryotes remains to be fully characterized.
Cyclin E is required for the endoreduplication cycles in Drosophila cells (14, 31, 57), trophoblasts, and megakaryocytes (20, 40). In fact, ectopic expression of cyclin E can promote endoreduplication in megakaryocytes (19). In contrast, although a decrease in cyclin A promotes endoreduplication in plant cells (24, 62) and Drosophila cells (39), it does not appear to be the case for megakaryocytic cell lines (19, 65, 66). A decrease in cyclin B1, but not cyclin A2, has been reported to be required for endoreduplication in megakaryocytes (66). Precisely which cyclin-CDK complexes are involved in safeguarding rereplication remains largely unresolved. Rereplication induced by EMI1 depletion is correlated with a reduction of cyclin A2 and cyclin B1, which can be rescued with nondestructible cyclin A2 (34). However, rereplication in HeLa cells is induced only weakly by cyclin A2 depletion, but it occurs more efficiently after codepletion of geminin (34). Conversely, expression of cyclin A2 (but not cyclin E) potentiates the rereplication induced by CDC6 and CDT1 in mammalian cells (56).
In contrast to the complexity and uncertainty about the different cyclins in DNA reduplication, the central role of inactivation of mitotic CDK1 is generally accepted. This has been observed in a wide range of endoreduplication cycles, including those in maize endosperm (21), Drosophila cells (49), trophoblasts (54), and megakaryocytes (65). Likewise, extensive DNA reduplication can be triggered by disruption of CDK1 in a mammalian cell line (7, 25, 28). The molecular basis of how CDK1 inactivation contributes to genome reduplication remains to be defined.
The prevailing view is that APC/C plays a salient role in preventing rereplication. The orthologs of CDH1 in plant cells (Ccs52A) and in Drosophila cells (fzr) are both essential for endoreduplication (26, 49). Unscheduled activation of APC/C reduces the concentrations of mitotic cyclins and geminin, resulting in rereplication (12, 34). An extra level of regulation provided by the APC/C inhibitor EMI1 has been uncovered recently. EMI1 begins to accumulate at the G1/S transition, thereby inactivating APC/C and allowing the accumulation of cyclins and geminin (46). Accordingly, depletion of EMI1 with RNA interference promotes unscheduled APC/C activation and rereplication (12, 34).
In this study, we utilized a specific CDK1 chemical inhibitor to induce whole-genome reduplication in cancer cells. We found that genome reduplication was associated with spontaneous oscillation of APC/C activity and nuclear-cytoplasmic shuttling of CDC6 even in the absence of mitosis. Moreover, the PLK1-SCFβ-TrCP1-EMI1 axis and cyclin A2-CDK2 were inextricably linked to the APC/C activation and genome reduplication. These data extended our understanding of the role of the different cyclin-CDK complexes in coordinating genome reduplication.
MATERIALS AND METHODS
Materials.
All reagents were obtained from Sigma-Aldrich (St. Louis, MO) unless stated otherwise.
DNA constructs and siRNA.
Plasmids expressing small hairpin RNA (shRNA) against cyclin A2 and cyclin B1 were constructed as described previously (17). A puromycin-resistant cassette was inserted into the HindIII site of mU6pro vector (a gift from David Turner, University of Michigan, MI) (63) to create mU6pro/PUR. The following pairs of oligonucleotides were annealed into BbsI-XbaI-cut mU6pro/PUR: 5′TTTGCCCCTCATCAAGAGCTATCTTCAAGAGAGATAGCTCTTGATGAGGGGTTTTT3′ and 5′CTAGAAAAACCCCTCATCAAGAGCTATCTCTCTTGAAGATAGCTCTTGATGAGGGG3′ (shRNA corresponded to positions 304 to 322 of human CDK2 open reading frame); 5′TTTGTGCGAGCAATTCTTCTGGATTCAAGAGATCCAGAAGAATTGCTCGCATTTTT3′ and 5′CTAGAAAAATGCGAGCAATTCTTCTGGATCTCTTGAATCCAGAAGAATTGCTCGCA3′ (shRNA corresponded to positions 385 to 403 of human cyclin E2 open reading frame). The NcoI-XbaI fragment from pEGFP-N3 (Clontech, Palo Alto, CA) was ligated into pUHD-P3 (previously called pUHD-P1/3C [17]) to obtain FLAG-3C-enhanced green fluorescent protein (EGFP). Glutathione S-transferase-cyclin B1 in pGEX-KG (60) was amplified with PCR analysis using a vector forward primer and 5′GCTTCCATGGTCATAGGCATTTTGG3′; the PCR product was cut with NcoI and ligated into FLAG-3C-EGFP to obtain FLAG-3C-cyclin B1(CΔ62)-EGFP in pUHD-P3. The red fluorescent protein (mRFP1) cDNA was a gift from Roger Tsien (University of California San Diego, CA). The NcoI site in mRFP1 was first disrupted with site-directed mutagenesis, and the cDNA was amplified with PCR analysis using the primers 5′GGCTAGCATGGCCTCCTCCGAGGAC3′ and 5′GACCATGGAGGCGCCGGTGGAGTGGC3′; the PCR products were cut with NheI and NcoI and ligated into pUHD-P3T. A puromycin resistance cassette was inserted into the HindIII site to create mRFP1 in pUHD-P3T (a modified version of pUHD-P3 created by replacing the PvuI-StuI fragment with that of pTRETight [Clontech]). CDC6 cDNA (IMAGE cDNA clone 5457941) was amplified by PCR analysis with the primers 5′TGGATCCATGCCTCAAACCCGATCCC3′ and 5′GGGATCCTTAAGGCAATCCAGTAGC3′; the PCR products were cut with BamHI and ligated into mRFP1 in pUHD-P3T/PUR to create mRFP1-CDC6 in pUHD-P3T/PUR. EMI1 in pcDNA3 was a gift from Michele Pagano (New York University, NY). EMI1 was amplified by PCR analysis with 5′TGCCATGGGCCGGCGCCCCTGCAGCT3′ and 5′TGGATCCTCACAATCTTCGTAAATTCTT3′; the PCR products were cut with NcoI-BamHI and ligated into pUHD-P1 (59) to generate FLAG-EMI1 in pUHD-P1. Site-directed mutagenesis of EMI1 (S145A and S149A) was carried out with the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) with 5′ATGAAGACGCTGGCTATTCCGCATTTTCTC3′ and its antisense. Stealth small interfering RNAs (siRNAs) targeting human CDC20, CDH1, CDK1, cyclin A2, cyclin B1, PLK1, β-TrCP1, and control siRNA were obtained from Invitrogen Life Technologies (Carlsbad, CA).
Cell culture.
The HeLa cell line (human cervical carcinoma) used in this study is a clone that expressed the tetracycline transactivator-tetracycline repressor chimera (61). Stable cell lines expressing histone H2B-GFP (8) and coexpressing cyclin A2 shRNA and cyclin A2 (33) were generated as described previously. To generate stable APC/C reporter cells, HeLa cells were transfected with the cyclin B1(CΔ62)-EGFP-expressing construct and selected with puromycin. Single colonies were isolated after about 2 weeks of selection. Cells were propagated in Dulbecco's modified Eagle's medium supplemented with 10% (vol/vol) calf serum (Invitrogen) in a humidified incubator at 37°C in 5% CO2. Unless stated otherwise, cells were treated with the following reagents at the following final concentrations: blasticidin (3 μg/ml), caspase inhibitor Z-VAD(OMe)-FMK (Alexis, Lausen, Switzerland) (10 μM), cycloheximide (10 μg/ml), doxycycline (2 μg/ml), GW843682X (10 μM), nocodazole (0.33 μM), and RO3306 (Alexis) (10 μM). During long-term treatment, RO3306 was replenished every 24 h. Cells were blocked in S phase with 2 mM of thymidine for 14 h and then release into thymidine-free medium supplemented with 24 μM of deoxycytidine. Double-thymidine synchronization (1) and preparation of cell extracts (42) were performed as described. Cells were transfected with the calcium phosphate precipitation method (2). To enrich the transfected cells, cells were subjected to a transient selection with blasticidin as outlined previously (17). Transfection of siRNA was carried out using Lipofectamine RNAiMAX (Invitrogen).
Live-cell imaging.
Cells were imaged using a TE2000E-PFS inverted fluorescent microscope (Nikon, Tokyo, Japan) equipped with a Spot Boost EMCCD camera (Diagnostic Instrument, Sterling Heights, MI) and an INU-NI-F1 temperature, humidity, and CO2 control system (Tokai Hit, Shizuoka, Japan). Data acquisition and analysis were carried out using the Metamorph 7.5 software (Molecular Devices, Downingtown, PA).
Flow cytometry analysis.
Flow cytometry analysis after propidium iodide staining was performed as previously described (11).
Kinase assays.
Histone H1 kinase assays were performed as previously described (44). Phosphorylation was detected and quantified with a phosphorimager (Pharos FX Plus molecular imager; Bio-Rad, Hercules, CA). Treatment of immunoprecipitates with glutathione S-transferase-CDC25A prior to kinase assay was performed as described previously (11).
Antibodies and immunological methods.
Immunoblotting and immunoprecipitation were performed as previously described (42). Monoclonal antibodies against β-actin (9), CDK1 (50), cyclin A2 (60), and cyclin B1 (8) and polyclonal antibodies against CDK1 (30), phospho-CDK1Tyr15 (17), and CDK2 (50) were obtained from sources as described previously. Monoclonal antibodies against CDC6 (sc-9964) and cyclin E2 (sc-247) and polyclonal antibodies against CDC20 (sc-8358), geminin (sc-13015), phospho-histone H3Ser10 (sc-8656R), and securin (sc-22772) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Monoclonal antibodies against EMI1 (Invitrogen Life Technologies, Carlsbad, CA) and CDH1 (Thermo Fisher Scientific, Waltham, MA) were obtained from the respective manufacturers.
RESULTS
CDK1 inhibition triggers a G2 arrest followed by whole-genome reduplication.
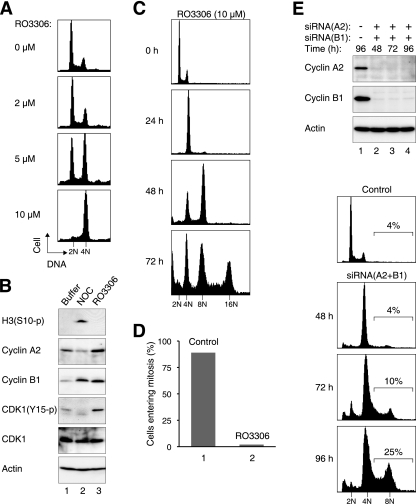
To evaluate the role of cyclin-CDK1 in regulating DNA reduplication in cancer cell lines, we utilized the specific CDK1 inhibitor RO3306, which displays high specificity toward CDK1 (55). RO3306 is 10-fold more potent against cyclin B1-CDK1 than against cyclin E-CDK2. Flow cytometry analysis verified that RO3306 triggered a G2/M arrest in HeLa cells in a dose-dependent manner (Fig. 1A). The cells were arrested in G2 phase instead of mitosis because CDK1 was present in an inactive Tyr15-phosphorylated state (Fig. 1B). In support of this, the mitotic marker histone H3Ser10 phosphorylation was absent in RO3306-treated cells. As a control, a mitotic block induced by nocodazole elicited CDK1Tyr15 dephosphorylation and histone H3Ser10 phosphorylation.
FIG. 1.
Inhibition of CDK1 with RO3306 induces G2 arrest followed by whole-genome reduplication. (A) Dose-dependent cell cycle arrest at G2/M by RO3306. HeLa cells were treated with either dimethyl sulfoxide (DMSO) or the indicated concentration of RO3306 for 18 h. The cells were fixed, stained with propidium iodide, and analyzed with flow cytometry. (B) RO3306 triggers an arrest in G2 but not mitosis. Cells were treated with DMSO, nocodazole (NOC), or RO3306 (10 μM) for 18 h. Cell extracts were prepared, and the expression of the indicated proteins was detected by immunoblotting. Equal loading of lysates was confirmed by immunoblotting for actin. (C) Prolonged RO3306 treatment induces genome reduplication. HeLa cells were treated with RO3306 and harvested for flow cytometry analysis at the indicated time points. Fresh medium and RO3306 were replenished every 24 h. A pan-caspase inhibitor was included to inhibit apoptosis. The positions of DNA contents up to 16N are indicated. (D) RO3306 treatment suppresses mitosis. HeLa cells stably expressing histone H2B-GFP were treated with either DMSO control or RO3306. The cells were then monitored with time-lapse microscopy for 48 h. Mitosis in control cells (n = 58) and RO3306-treated cells (n = 632) was scored by roundup morphology, DNA condensation, or cytokinesis. (E) Depletion of mitotic cyclins induces reduplication inefficiently. HeLa cells were transfected with control, cyclin A2, or cyclin B1 siRNAs. Lysates were prepared at the indicated time points and analyzed by immunoblotting. Actin analysis was included to assess protein loading and transfer. Samples were also taken for flow cytometry analysis. Percentages of cells with >4N DNA contents are indicated.
To examine the effects of a more prolonged CDK1 inhibition, cells were exposed to RO3306 for up to 72 h (Fig. 1C). Cells containing DNA contents up to 8N and 16N could be readily detected at 48 h and 72 h, respectively, indicating that the cells underwent rounds of whole-genome reduplication. In agreement with this, time-lapse imaging of cells expressing histone H2B-GFP indicated that while ∼90% of control cells underwent at least one round of mitosis over the 48-h imaging period, less than 5% of RO3306-treated cells entered mitosis over the same period (Fig. 1D). A pan-caspase inhibitor was included in our assays to inhibit apoptosis; this increased the appearance of cells with higher DNA contents. As anticipated, genome reduplication was progressively inhibited at increasing RO3306 concentrations (see Fig. S1 in the supplemental material). This is likely to be due to the fact that CDK2 was also inhibited at higher concentrations of RO3306 (55). Importantly, RO3306-induced genome reduplication was not restricted only to HeLa cells. Mouse fibroblasts (NIH 3T3) containing 8N DNA contents were detected after RO3306 challenge (see Fig. S2 in the supplemental material). Interestingly, transformation of the NIH 3T3 cells with E1A enhanced genome reduplication, suggesting that pathways that counteract genome reduplication may be compromised after transformation. Collectively, these data indicate that abolition of CDK1 activity with RO3306 triggered a transient G2-phase arrest followed by extensive genome reduplication without signs of mitosis.
Genome reduplication occurs inefficiently in the absence of both mitotic cyclins.
Given that genome reduplication could be triggered by CDK1 inhibition, we next examined if the same effect could be obtained by depletion of the cyclin partners of CDK1. Transfection of shRNA-expressing constructs could effectively deplete cyclin A2 and cyclin B1 (see Fig. S3A in the supplemental material). Depletion of cyclin A2 did not significantly affect the total kinase activity of CDK1. In contrast, depletion of cyclin B1 diminished CDK1 activity by ∼75%, verifying that cyclin B1 is a major partner of CDK1 (see Fig. S3B in the supplemental material). To exclude the contribution of inhibitory phosphorylation, the CDK1 immunoprecipitates were treated with purified CDC25A prior to the kinase assays. Codepletion of cyclin A2 and cyclin B1 reduced the kinase activity of CDK1 by >90%.
In agreement with our previous results (17), depletion of cyclin A2 triggered a G2 arrest (see Fig. S3C in the supplemental material). In comparison, knockdown of cyclin B1 did not affect the cell cycle distribution (4, 10, 52, 64). Figure S3C in the supplemental material shows that knockdown of neither cyclin A2 nor cyclin B1 alone resulted in genome reduplication. Codepletion of the two cyclins also induced only low levels of DNA rereplication. To ensure that the results were not due to nonspecific effects of the shRNAs, siRNAs against independent sequences of the cyclins were employed. Figure 1E shows that although both cyclins were effectively downregulated, DNA reduplication was notably less efficient than that after RO3306 treatment (Fig. 1C). These data suggest that unlike the multiple rounds of genome reduplication that occurred after RO3306 treatment, genome reduplication was inefficient in the absence of cyclin A2 and cyclin B1. As shown below, our data indicate that cyclin A2 is in fact required for genome reduplication.
Interestingly, results from siRNA experiments indicated that the effect of CDK1 downregulation was distinct from CDK1 inhibition by RO3306. After downregulation of CDK1, a lower dose of RO3306 was needed to induce cell cycle arrest and genome reduplication (see Fig. S4 in the supplemental material). A simple explanation of this is that the residual CDK1 was sufficient to drive the cell cycle, and consequently a lower concentration RO3306 was needed to suppress all CDK1. There is also the possibility of complex redistribution of other CDKs, such as CDK2. Genome reduplication was notably less efficient in CDK1-depleted cells than in control cells. One possibility is that the effective RO3306 concentration against CDK2 (which was required for genome reduplication [see below]) became higher after downregulation of CDK1.
Spontaneous oscillation of APC/C activity in the absence of mitosis.
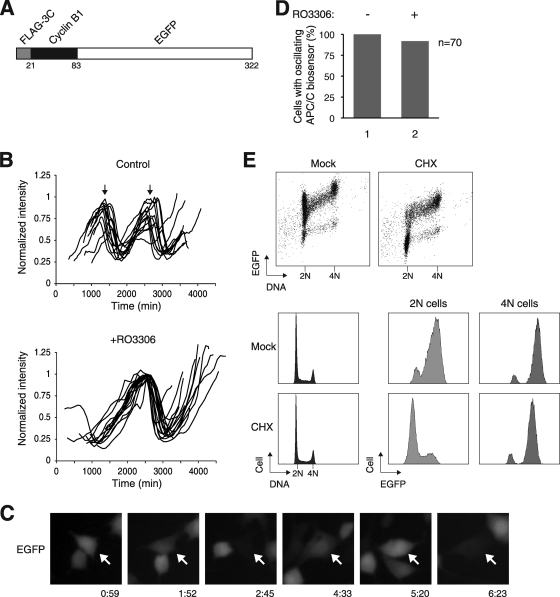
Since the activity of APC/C is integrally linked to CDK1, we next investigated the connection between APC/C and RO3306-induced reduplication. To monitor the activity of APC/C in vivo, we generated cell lines expressing a fluorescence biosensor consisting of the N-terminal fragment of cyclin B1 grafted onto EGFP (Fig. 2A). The destruction box of the cyclin B1 fragment was expected to target the reporter to APC/C-dependent degradation. Stable clones of HeLa cells expressing the biosensor were generated. Furthermore, mRFP1-expressing plasmids were transfected into the cells to serve as an APC/C-insensitive marker. After time-lapse imaging, the EGFP signal of individual cells was quantified and normalized with the mRFP1 signal (Fig. 2B). We found that control cells underwent about two rounds of cell division cycle within the 48-h imaging period. As expected, the APC/C biosensor oscillated in synchrony with the cell cycle.
FIG. 2.
Spontaneous oscillation of APC/C activity after CDK1 inhibition. (A) Schematic diagram of the APC/C biosensor construct. The N-terminal region of cyclin B1 (residues 1 to 62) was fused to EGFP. A FLAG epitope tag and a 3C-protease cleavage site are also placed at the N terminus. (B) Oscillation of APC/C activity after RO3306 treatment. A cell line stably expressing the APC/C biosensor was generated from HeLa cells. The cells were transfected with mRFP before being treated with either dimethyl sulfoxide control or RO3306. The cells were then tracked using time-lapse microscopy for 48 h. The green fluorescence intensity of individual cells was recorded and normalized with the red fluorescence intensity (15 for controls and 17 for RO3306-treated cells). The arrows indicate the time of cell division in the control cells as observed using bright-field microscopy. (C) Example of a cell displaying oscillation of the APC/C reporter. APC/C biosensor-expressing cells were treated with RO3306 and tracked using time-lapse microscopy as described for panel B. The time scale is in hours:minutes. (D) APC/C oscillation occurs in the majority of RO3306-treated cells. The experiment was performed as described for panel B. Cells that displayed oscillation of the APC/C biosensor during the imaging period were scored (n = 70). (E) Activation of APC/C in G1 phase. Cells expressing the APC/C biosensor were either mock treated or treated with cycloheximide (CHS) for 3 h. The cells were then fixed and stained with propidium iodide. Bivariate analysis of DNA contents and EGFP was performed with flow cytometry (top). The cell cycle distribution profiles are shown in the bottom left panels. The G1 and G2/M populations were gated, and the EGFP signals were analyzed (bottom right panels).
We next monitored the APC/C activity after CDK1 inhibition with time-lapse imaging (Fig. 2B). Remarkably, the APC/C biosensor still oscillated in the absence of mitosis (a typical example is shown in Fig. 2C). Examples of control and RO3306-treated cells are shown in Movies S1 and S2 in the supplemental material, respectively. Oscillation of APC/C activity was observed in more than 90% of the RO3306-treated cells over the experimental period (Fig. 2D). Compared to the unperturbed cell cycle, APC/C oscillated at a lower frequency after CDK1 inhibition (Fig. 2B).
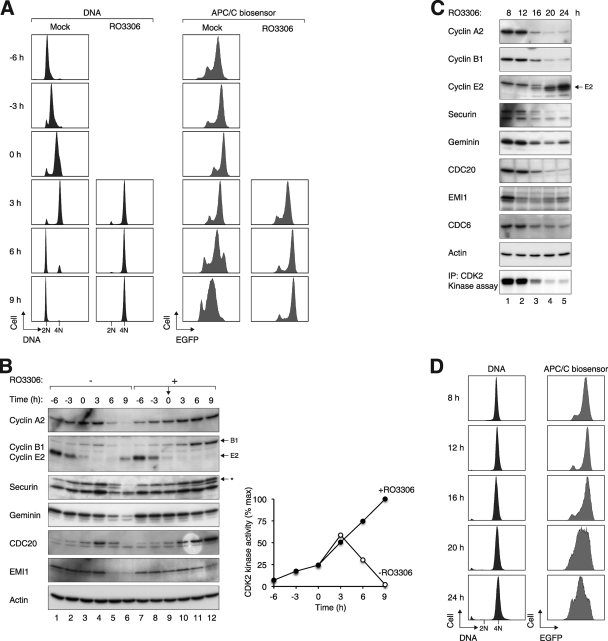
In addition to monitoring the steady states of the APC/C biosensor, APC/C activity was also assessed by the rate of degradation of the biosensor. Accordingly, inhibition of protein synthesis with cycloheximide for 3 h reduced the abundance of the biosensor (Fig. 2E). The brief cycloheximide challenge did not significantly affect cell cycle distribution (Fig. 2E) or survival (data not shown). By gating the fluorescence signals in 2N or 4N DNA cells, we confirmed that the APC/C biosensor was relatively stable in G2/M but was rapidly degraded in G1. To obtain a more detailed picture of the temporal regulation of APC/C after CDK1 inhibition, cells were synchronously released from S phase before being treated with RO3306. Figure 3A shows that while the control cells progressed through the cell cycle after release, RO3306-treated cells were halted with 4N DNA contents. Analysis of the APC/C biosensor indicated that in contrast to its activation as the control cells entered G1, APC/C remained inactive in RO3306-blocked cells. Consistent with these results, several APC/C substrates (including cyclin A2, cyclin B1, securin, and geminin) were stabilized after RO3306 treatment (Fig. 3B). Likewise, CDC20 was degraded when control cells exited mitosis (as CDC20 itself is a proteolytic target of APC/CCDH1) but continued to accumulate after CDK1 inhibition. Finally, while the APC/C inhibitor EMI1 was degraded as the control cells entered mitosis (by SCF-dependent mechanisms), it was stabilized after RO3306 treatment. Collectively, these data indicate that RO3306 triggered an initial G2 delay that was characterized by a suppression of APC/C activity.
FIG. 3.
Activation of APC/C and degradation of endogenous substrates after CDK1 inhibition. (A) APC/C remains inactive during the RO3306-induced G2 block. Cells expressing the APC/C biosensor were synchronized at early S phase with a double-thymidine procedure and released into thymidine-free medium. The cells were either mock treated or treated with RO3306 at 6 h after release (designated as t = 0). At the indicated time points, the cells were fixed, stained with propidium iodide, and analyzed with flow cytometry. (B) APC/C substrates were not destroyed up to 9 h after RO3306 treatment. Cells were synchronized and treated as described for panel A. Cell extracts were prepared at the indicated time points and the abundance of various APC/C substrates was analyzed. Equal loading of lysates was confirmed by immunoblotting for actin. The asterisk indicates the phosphorylated forms of securin. CDK2 was also immunoprecipitated from the lysates, and the kinase activity was measured using histone H1 as a substrate. Phosphorylation was detected and quantified with a phosphorimager (right). (C) APC/C is activated after prolonged RO3306 treatment. Cells expressing the APC/C biosensor were synchronized in S phase with thymidine. The cells were then released into thymidine-free medium for 3 h before being treated with RO3306. At the indicated time points after RO3306 treatment, lysates were prepared and analyzed by immunoblotting. The upper band in the cyclin E2 blot is CDC20 (due to reprobing of the membrane). Actin analysis was included to assess protein loading and transfer. CDK2 was also immunoprecipitated (IP) from the lysates, and the histone H1 kinase activity was assayed (bottom). (D) APC/C is activated at 16 h after inhibition of CDK1. The experiment was performed as described for panel C. At the indicated time points, the cells were analyzed with flow cytometry.
We next examined the behavior of APC/C at later time points when RO3306-treated cells were poised to undergo reduplication. Figure 3C shows that after the initial accumulation, APC/C targets (cyclin A2, cyclin B1, securin, and geminin) were degraded at 16 h after RO3306 treatment. The instability of the APC/C biosensor also confirmed that APC/C was activated (Fig. 3D).
We found that cyclin E2 reaccumulated when the RO3306-treated cells subsequently underwent genome reduplication (Fig. 3C). Depletion of cyclin E2 with shRNA indicated a critical role of cyclin E2 in genome reduplication (see Fig. S5 in the supplemental material). Collectively, these data indicate that after an initial G2 arrest with inactivated APC/C, long-term CDK1 inhibition induced precocious activation of APC/C even in the absence of mitosis, creating a condition with a low abundance of APC/C substrates, such as geminin.
Nuclear-cytoplasmic shuttling of CDC6 in the absence of mitosis.
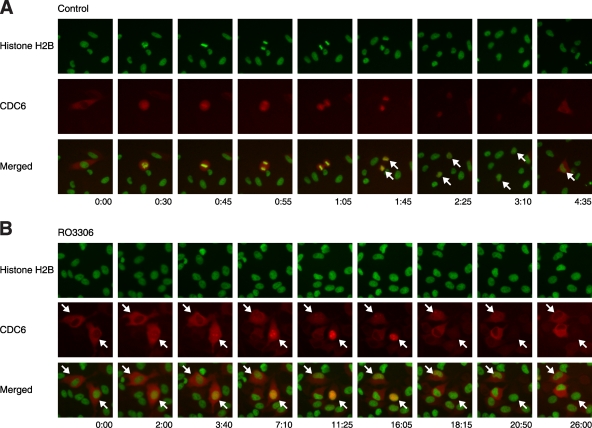
A crucial prerequisite for replication is the accumulation of CDC6 in the nucleus (6). To obtain evidence if CDC6 was regulated during genome reduplication, cells were transfected with mRFP1-tagged CDC6 in conjunction with GFP-tagged histone H2B. The cells were then either mock treated or treated with RO3306 before being analyzed with live-cell imaging. Figure 4A shows the typical behavior of mRFP1-CDC6 during the control cell cycle. CDC6 was present in the cytoplasm during G2 phase and became uniformly distributed after nuclear envelope breakdown (with a slightly stronger affinity for chromatin). After mitosis, CDC6 was initially localized to the nucleus before it was destroyed during late G1 or early S phase. In some cells, a distinct shuttling of CDC6 to the cytoplasm could be detected preceding its proteolysis.
FIG. 4.
Spontaneous nuclear-cytoplasmic translocation of CDC6 after CDK1 inhibition. Plasmids expressing mRFP1-tagged CDC6 (red) were transfected into HeLa cells stably expressing histone H2B-GFP (green). The cells were either mock treated (A) or treated with RO3306 (B) and then subjected to live-cell imaging. Representative cells that undergo a normal mitotic cell cycle (A) and nuclear-cytoplasmic shuttling of CDC6 without cell division (B) are indicated. The time scale is in hours:minutes.
Remarkably, although there was no mitosis after RO3306 treatment, CDC6 still underwent nuclear-cytoplasmic shuttling as well as degradation (Fig. 4B). In some cells, mRFP1-CDC6 shifted from the cytoplasm to the nucleus before it was degraded. In other populations, CDC6 shuttled from the cytoplasm to the nucleus and back to the cytoplasm, without an obvious proteolytic phase. It is noteworthy that similar to the APC/C oscillation, the rate of nuclear-cytoplasmic shuttling and degradation of CDC6 was substantially slower after RO3306 treatment than during the normal cell cycle. Taken together, these results suggest that genome reduplication triggered by CDK1 inhibition was associated with both an oscillation of APC/C and nuclear-cytoplasmic shuttling of CDC6.
Oscillations of APC/CCDC20 and APC/CCDH1 are vital for genome reduplication.
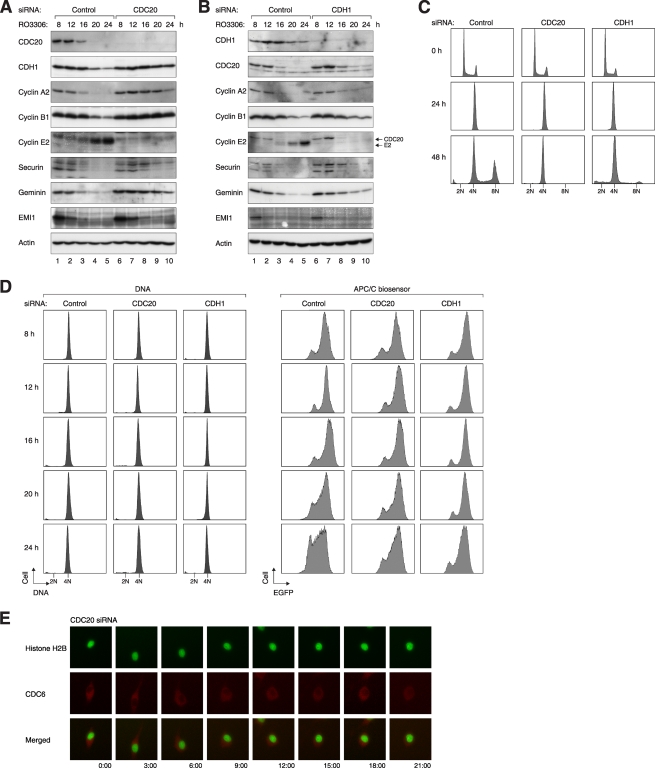
Several lines of evidence from the literature suggest that DNA reduplication may be dependent on APC/C activation (see Introduction). To test this directly for RO3306-induced genome reduplication, APC/C was inactivated by depleting its adaptor proteins CDC20 and CDH1. Effective downregulation of CDC20 and CDH1 by the siRNAs is demonstrated in Fig. 5A and B, respectively. Figure 5C shows that depletion of CDC20 or CDH1 abolished RO3306-mediated reduplication, suggesting a crucial role of APC/C in the process. As expected, APC/C was activated following RO3306 treatment in control cells but was suppressed after CDC20 or CDH1 knockdown (Fig. 5D), verifying the inactivation of APC/C by the siRNAs.
FIG. 5.
APC/CCDC20 and APC/CCDH1 are required for RO3306-mediated genome reduplication. (A) Depletion of CDC20 retards the degradation of APC/C substrates after CDK1 inhibition. Cells were transfected with either control or CDC20 siRNA. At 12 h after transfection, the cells were treated with thymidine for 14 h to synchronize the cells at S phase. After releasing from the block for 3 h, the cells were treated with RO3306 and harvested at the indicated time points. Cell lysates were prepared and analyzed by immunoblotting. Equal loading of lysates was confirmed by immunoblotting for actin. (B) Depletion of CDH1 retards the degradation of APC/C substrates after CDK1 inhibition. Cells were transfected with either control or CDH1 siRNA and treated exactly as described for panel A. (C) CDC20 and CDH1 are required for RO3306-induced reduplication. HeLa cells were transfected with control, CDC20, or CDH1 siRNA. After 24 h, the cells were treated with RO3306. Cells were harvested at the indicated time points and processed for flow cytometry analysis. (D) Activation of APC/C after CDK1 inhibition requires CDC20 and CDH1. APC/C biosensor-expressing cells were transfected with control, CDC20, or CDH1 siRNA and treated as described in panel A. The cells were processed for flow cytometry to analyze the DNA contents and the APC/C reporter. (E) CDC20 is critical for nuclear-cytoplasmic translocation of CDC6 after CDK1 inhibition. Plasmids expressing mRFP1-tagged CDC6 (red) were transfected into HeLa cells stably expressing histone H2B-GFP (green). The cells were then transfected with siRNA against CDC20, treated with RO3306, and subjected to live-cell imaging.
Degradation of most APC/C substrates was delayed in the absence of CDC20 or CDH1 (Fig. 5A and B). In particular, the expression of geminin persisted up to 24 h after the addition of RO3306. Interestingly, cyclin E2 did not accumulate in CDC20- or CDH1-depleted cells. This datum suggests that in addition to the persistence of APC/C targets such as geminin, another possible cause of the attenuation of genome reduplication in CDC20- or CDH1-depleted cells was the lack of cyclin E2.
Finally, the RO3306-mediated nuclear-cytoplasmic shuttling and degradation of CDC6 was disrupted in cells lacking APC/C adaptors. Knockdown of CDC20 resulted in cytoplasmic localization of CDC6 (Fig. 5E). Taken together, these data provide strong evidence that oscillation of APC/CCDC20 and APC/CCDH1 was required for genome reduplication induced by the absence of CDK1 activity.
PLK1-SCFβ-TrCP1-EMI1-APC/C axis is critical for genome reduplication.
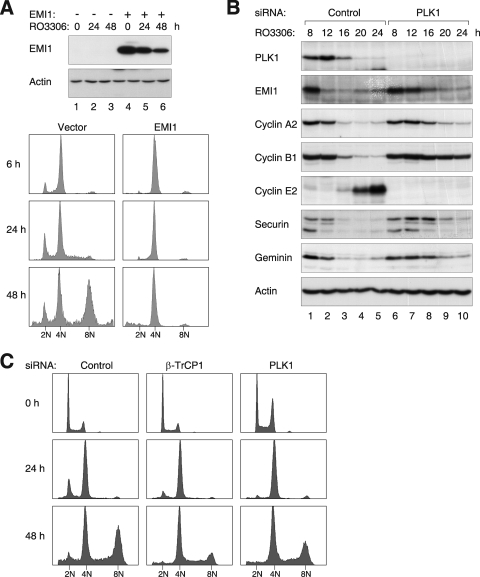
Given that a reduction of EMI1 could be observed after CDK1 inhibition, but not under conditions when genome reduplication was inhibited (e.g., Fig. 3), it is possible that the removal of EMI1 is required for reduplication. Indeed, compelling evidence from Machida and Dutta (34) indicate that EMI1 inhibits rereplication. To test this hypothesis, EMI1 was ectopically expressed before the RO3306 challenge. During normal prophase, EMI1 is targeted to ubiquitin-mediated degradation by SCFβ-TrCP1 (35) via a mechanism involving PLK1-dependent phosphorylation (23). To ensure that the recombinant EMI1 was not destroyed precociously, a stable version (S145A and S149A) that is resistant to SCFβ-TrCP1-mediated degradation (23) was used in this experiment. Figure 6A shows that expression of the recombinant EMI1 abolished RO3306-mediated genome reduplication.
FIG. 6.
RO3306-induced genome reduplication is regulated by the PLK1-SCFβ-TrCP1-EMI1 axis. (A) EMI1 inhibits genome reduplication after RO3306 treatment. HeLa cells were transfected with either control vector or EMI1S145A+S149A-expressing plasmids. At 12 h after transfection, the cells were treated with thymidine for 14 h. After being released from the block for 3 h, the cells were treated with RO3306 and harvested at the indicated time points. Cell lysates were prepared and analyzed by immunoblotting. Samples were also fixed and processed for flow cytometry analysis. (B) Depletion of PLK1 delays the degradation of EMI1 and APC/C substrates. Cells were transfected with either control or PLK1 siRNA. At 12 h after transfection, the cells were treated with thymidine for 14 h to synchronize the cells at S phase. After releasing from the block for 3 h, the cells were treated with RO3306 and harvested at the indicated time points. Cell lysates were prepared and analyzed by immunoblotting. Equal loading of lysates was confirmed by immunoblotting for actin. (C) Downregulation of PLK1 or β-TrCP1 reduces RO3306-induced reduplication. HeLa cells were transfected with control, β-TrCP1, or PLK1 siRNA. After 24 h, the cells were treated with RO3306. Cells were harvested at the indicated time points and analyzed with flow cytometry.
To obtain further evidence of the importance of EMI1 degradation in genome reduplication, β-TrCP1 or PLK1 was downregulated with siRNAs. Depletion of PLK1 (Fig. 6B) or β-TrCP1 (data not shown) stabilized EMI1 after RO3306 treatment. PLK1 depletion also delayed the degradation of all APC/C substrates examined. We found that downregulation of PLK1 or β-TrCP1 reduced genome reduplication (Fig. 6C). It is not clear why PLK1 siRNA was less effective than β-TrCP1 downregulation in impairing genome reduplication. In agreement with this, the PLK1 inhibitor GW843682X (27) was also only partially effective in suppressing genome reduplication (see Fig. S6 in the supplemental material). One possibility is the redundancy within the polo-like kinase family (PLK1 to PLK4). Taken together, these results underscore the critical role of the PLK1-SCFβ-TrCP1-EMI1-APC/C axis in genome reduplication.
Cyclin A2-CDK2 is important for the genome reduplication after CDK1 inhibition.
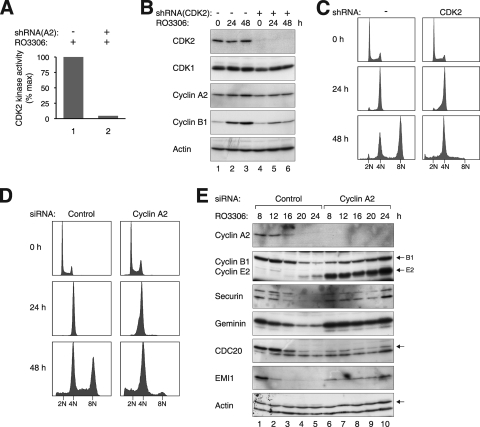
Given the paradoxical results that genome reduplication occurred robustly after RO3306 treatment but was markedly blunted in the absence of both cyclin A2 and cyclin B1, we hypothesized that cyclin A2-CDK2 may contribute to reduplication. As expected, CDK2 kinase activity increased when cells were released from S phase and decreased after cyclin A2 was destroyed (Fig. 3B). In contrast, CDK2 activity remained elevated in cells exposed to RO3306. Depletion of cyclin A2 removed the majority of the CDK2 kinase activity, indicating that the CDK2 activity in RO3306-treated cells was contributed mainly by cyclin A2 complexes (Fig. 7A).
FIG. 7.
Cyclin A2-CDK2 is required for RO3306-induced genome reduplication. (A) The CDK2 activity after RO3306 treatment was abolished by depletion of cyclin A2. HeLa cells were transfected with either control vector or cyclin A2 shRNA-expressing plasmids. Transfected cells were enriched with blasticidin and treated with RO3306 for 16 h before lysates were prepared. CDK2 was immunoprecipitated, and the kinase activity was assayed. (B) CDK2 is required for the genome reduplication after RO3306 treatment. HeLa cells were transfected with either control vector or CDK2 shRNA-expressing plasmids. Transfected cells were enriched with blasticidin and treated with RO3306. At the indicated time points, cell extracts were prepared and analyzed by immunoblotting. (C) Experiment was performed as described for panel B. At the indicated time points, the cells were harvested and analyzed with flow cytometry. (D) Cyclin A2 is required for RO3306-induced reduplication. HeLa cells were transfected with either control siRNA or siRNA against cyclin A2. After 24 h, the cells were treated with RO3306. Cells were harvested at the indicated time points and analyzed with flow cytometry. (E) Cyclin A2 is required for the degradation of APC/C substrates after CDK1 inhibition. HeLa cells were transfected with either control or cyclin A2 siRNA. At 12 h after transfection, the cells were treated with thymidine for 14 h to synchronize at S phase. After releasing from the block for 3 h, the cells were treated with RO3306 and harvested at the indicated time points. Cell lysates were prepared, and the indicated proteins were detected by immunoblotting. Equal loading of lysates was confirmed by immunoblotting for actin.
To address whether CDK2 is important for genome reduplication, cells were transfected with plasmids expressing an shRNA against CDK2. Figure 7B shows that the shRNA downregulated CDK2 without affecting the expression of CDK1. In contrast to control-depleted cells, CDK2-depleted cells did not undergo genome reduplication (Fig. 7C). Analysis of synchronized cells after the knockdown of CDK2 indicated that both endogenous APC/C substrates and the APC/C biosensor were stabilized in the absence of CDK2 (see Fig. S7 in the supplemental material).
We likewise downregulated cyclin A2 to determine if it was essential for genome reduplication. A stable cell line that we have developed previously (33) was used for this purpose. Because both the cyclin A2 shRNA (to deplete the endogenous cyclin A2) and the shRNA-resistant cyclin A2 were expressed in the cell line, knockdown of cyclin A2 could be achieved in an inducible manner by turning off the recombinant cyclin A2 with doxycycline. Figure S8A in the supplemental material verified that cyclin A2 was expressed in the absence but not the presence of doxycycline. Moreover, genome reduplication was induced by RO3306 in the presence but not the absence of cyclin A2 (see Fig. S8B in the supplemental material). The effects of cyclin A2 knockdown were specific because expression of the shRNA-resistant cyclin A2 alone was sufficient in reversing the effects (when the cells were cultured without doxycycline).
To further validate the results described above, cyclin A2 was independently depleted with an siRNA. Figure 7D shows that no genome reduplication was detected upon RO3306 treatment in the absence of cyclin A2. Immunoblot analysis confirmed that cyclin A2 was effectively depleted by the siRNA (Fig. 7E). As with the results obtained from shRNA, while geminin and other APC/C substrates were degraded in control cells, they were not degraded in cells lacking cyclin A2. Interestingly, degradation of cyclin E2 was also abolished in the absence of cyclin A2 (see Fig. S8A and S7E in the supplemental material).
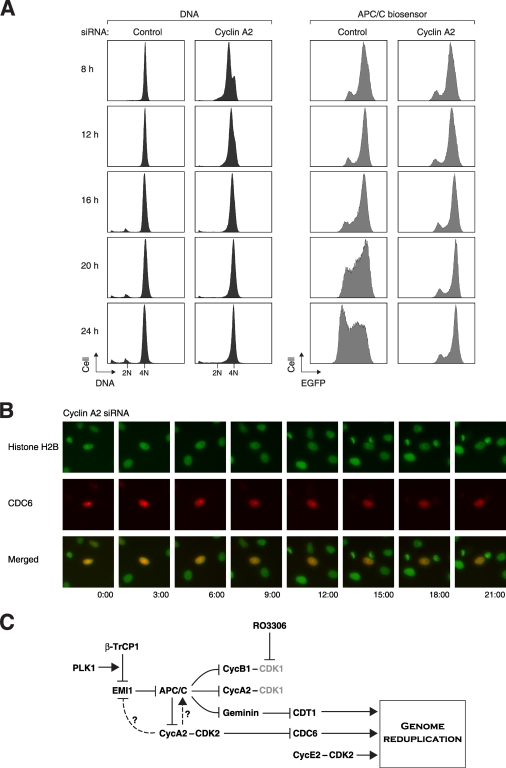
To determine if APC/C activation was compromised in the absence of cyclin A2-CDK2, cells expressing the APC/C biosensor were transfected with either control or cyclin A2 siRNA before they were challenged with RO3306. Figure 8A shows that in contrast to control cells, APC/C remained inactive in cyclin A2-depleted cells after RO3306 exposure (the reporter actually accumulated progressively). Finally, CDC6 remained in the nucleus in the absence of cyclin A2 and did not shuttle during RO3306 block (Fig. 8B). Collectively, these results indicated that the persistence of cyclin A2-CDK2 activity after CDK1 inhibition was required for the subsequent APC/C oscillation and genome reduplication.
FIG. 8.
Cyclin A2 is critical for APC/C oscillation and CDC6 nuclear-cytoplasmic translocation after CDK1 inhibition. (A) Activation of APC/C after CDK1 inhibition requires cyclin A2. APC/C biosensor-expressing cells were transfected with either control or cyclin A2 siRNA. At 12 h after transfection, the cells were treated with thymidine for 14 h to synchronize at S phase. After releasing from the block for 3 h, the cells were treated with RO3306 and harvested at the indicated time points. The DNA contents and APC/C reporter level were analyzed with flow cytometry. (B) Cyclin A2 is critical for nuclear-cytoplasmic translocation of CDC6 after CDK1 inhibition. Plasmids expressing mRFP1-tagged CDC6 (red) were transfected into HeLa cells stably expressing histone H2B-GFP (green). The cells were then transfected with siRNA against cyclin A2, treated with RO3306, and subjected to live-cell imaging. (C) Model of the regulation of genome reduplication after CDK1 inhibition. After treatment with RO3306, cyclin A2/cyclin B1-CDK1 complexes are inactivated and cells are prevented from entering mitosis. APC/C is kept inactivated until PLK1 triggers the SCFβ-TrCP1-dependent degradation of EMI1. In addition to regulating the subcellular localization of CDC6, cyclin A2-CDK2 may also regulate EMI1 degradation and/or APC/C directly in the absence of CDK1. APC/C then destroyed substrates, including cyclins and geminin. Degradation of geminin (which releases CDT1), CDC6, and the accumulation of cyclin E2 trigger genome rereplication.
DISCUSSION
RO3306-treated HeLa cells underwent extensive genome reduplication (Fig. 1C). In this connection, the role of CDK1 in genome reduplication cannot easily be investigated by gene disruption because of early embryonic lethality (47). Moreover, chemically inhibited CDK1 is unlikely to be compensated by redistribution of another CDK, as is believed to be the case for genetic disruption or knockdown approaches. RO3306 also induced genome reduplication in 3T3 mouse fibroblasts (see Fig. S2 in the supplemental material). Interestingly, the same dose of RO3306 did not induce genome reduplication in H1299 and HepG2 cells (our unpublished data). This probably reflects the different ratios of CDK1 versus CDK2 in different cell lines. Careful titration of RO3306 will be needed to test the dose that can induce genome reduplication in different cell lines. Published results also indicated extensive variation of the ability of CDK1 inactivation in inducing genome reduplication in different systems (reviewed in reference 45). Whole-genome reduplication was typically not observed after treatment with CDK inhibitors in mammalian cells (for an example, see reference 55). A possible explanation of the discrepancy is that most CDK inhibitors target CDK1 as well as CDK2. Another point is that genome reduplication was observed only after prolonged treatment of RO3306. Apoptosis may also play a role because suppression of apoptosis increased the portion of cells containing increased DNA contents. It is noteworthy that RO3306 generated cells containing discrete increases in DNA contents, indicating complete rounds of genome reduplication instead of the refiring of some origins. In contrast, owing to the DNA damage generated during unscheduled rereplication (36), the genome is typically not reduplicated in its entirety after the loss of geminin or overexpression of CDT1 or CDC6 (22, 37). Interestingly, the DNA damage-induced apoptotic pathway appears to be turned off during some endoreduplication cycles (36). For this reason, a pan-caspase inhibitor was included in all our genome reduplication assays to suppress apoptosis.
In this study, we have documented the events leading to genome reduplication after the inhibition of CDK1 by RO3306. A model is depicted in Fig. 8C. During the initial G2 cell cycle arrest, APC/C remained inactive (Fig. 3D), and cyclin A2, cyclin B1, securin, and geminin were able to accumulate (Fig. 3B). EMI1 was then degraded at around 12 h after the RO3306 block (Fig. 3B and C), resulting in the activation of APC/C and the degradation of substrates (Fig. 3C and D). Reduplication of the genome was promoted by multiple events, including the release of CDT1 after geminin degradation, translocation of CDC6 into the nucleus (Fig. 4), and the accumulation of cyclin E2 (Fig. 3C).
Both cyclin A2 and cyclin B1 accumulated during the initial arrest induced by RO3306 treatment (Fig. 3B). Although CDK1 associates with both A- and B-type cyclins, depletion of these cyclins induced genome reduplication inefficiently (Fig. 1E and see Fig. S3 in the supplemental material). A minor B-type cyclin, cyclin B2, is present in HeLa cells. Nevertheless, codepletion of cyclin B2 did not generate significantly more polyploids (our unpublished data). An interpretation of these paradoxical data is that while RO3306 inactivated cyclin B1-CDK1 and cyclin A2-CDK1, depletion of the cyclins inactivated the complexes described above as well as cyclin A2-CDK2. Our hypothesis is that cyclin A2-CDK2 is required for the genome reduplication. In agreement with this, we found that CDK2 remained active after RO3306 treatment (Fig. 3B). Furthermore, genome reduplication was abolished after downregulation of cyclin A2 (Fig. 5). Finally, inactivation of CDK2 with shRNA (Fig. 7C) or with a chemical inhibitor (NU6140) (our unpublished data) also inhibited RO3306-induced genome reduplication. It has been shown that endoreduplication in trophoblasts triggered by CDK1-inhibition also requires CDK2 activity (54). Paradoxically, destruction of cyclin A2 is required for induction of rereplication by EMI1 siRNA (34). This may be due to the differential roles of cyclin A2-CDK2 in activation of APC/C and prevention of rereplication.
A mechanistic explanation of the requirement of cyclin A2 for genome reduplication is that oscillation of APC/C was abolished in the absence of cyclin A2. During normal mitosis, APC/CCDC20 is believed to be activated in part by CDK1-dependent phosphorylation (41). In the absence of CDK1 activity, however, we suggest that this function is replaced by cyclin A2-CDK2.
The effects of CDK2 inhibition on genome reduplication was probably not entirely due to cyclin A2-CDK2, because cyclin E2 also accumulated (Fig. 3C). Indeed, genome reduplication was compromised after downregulation of cyclin E2 (see Fig. S5 in the supplemental material). In this connection, cyclin E2 accumulation was dramatically lost in cells lacking CDC20 or CDH1 (Fig. 5) or PLK1 (Fig. 6B), suggesting that cyclin E2 could be a limiting factor for genome reduplication under these circumstances. However, cyclin E2 still accumulated in cells lacking cyclin A2 (Fig. 7E) or CDK2 (see Fig. S7A in the supplemental material), indicating that cyclin E2 is necessary but not sufficient for genome reduplication.
Using cell lines that stably expressed a biosensor of APC/C, we found that the APC/C activity oscillated even in the absence of mitosis (Fig. 2B). Laronne et al. (28) found that the mitotic cyclins can be detected only in some endoreplicating cells, also suggesting that APC/C is periodically activated (28). Several lines of evidence revealed that an intervening mitotic state was not present after RO3306 treatment. Time-lapse imaging of histone H2B-GFP cells indicated that neither chromosome condensation nor cell roundup occurred (Fig. 1D). Phosphorylation of histone H3Ser10 also remained at a background level, and the nuclear envelope was not broken down after RO3306 treatment (data not shown). Using synchronized cells, we found that APC/C was activated at around 16 h after RO3306 treatment (Fig. 3D). Consistent with this, various APC/C substrates, including cyclin A2, cyclin B1, securin, geminin, and CDC20 itself, were degraded at that time (Fig. 3C). As expected, their degradation was inhibited by CDC20 depletion (Fig. 5A). Interestingly, as geminin is mainly a substrate of APC/CCDH1, it is likely that the activation of APC/CCDH1 was also compromised without APC/CCDC20.
APC/C probably regulated multiple components to induce genome reduplication. Several studies have shown that depletion of geminin is important for rereplication (37, 56, 67). Degradation of geminin alone is insufficient to account for genome reduplication, as codepletion of geminin did not rescue the block of genome reduplication by CDC20 siRNA (our unpublished data). CDC6 is likely to be another important component for genome reduplication. Nuclear-cytoplasmic shuttling and proteolysis of CDC6 are regulated by cyclin A2-CDK2-dependent phosphorylation and APC/C, respectively. Indeed, an mRFP1-tagged CDC6 underwent spontaneous nuclear-cytoplasmic shuttling and degradation after CDK1 inhibition (Fig. 4). Degradation of cyclin A2 after RO3306 treatment (Fig. 3C) would inactivate CDK2 and contribute to the nuclear localization of CDC6. As anticipated, downregulation of cyclin A2 or CDC20 abolished the shuttling and degradation of CDC6 (Fig. 5E and 8B). A caveat is that the importance of these mechanisms is contentious even in normal replication, because a fraction of endogenous CDC6 is thought to remain in the nucleus throughout S phase (reviewed in reference 6).
What triggers APC/C activation in the first place? One of the earliest events that destabilized the RO3306-mediated G2 block was the destruction of EMI1 (Fig. 3C). In support of this, removal of cyclin A2 (which also abolished APC/C activation and genome reduplication) inhibited the degradation of EMI1 (Fig. 7E). In contrast, depletion of CDC20 or CDH1 did not affect EMI1 degradation (Fig. 5A and B), raising the probability that EMI1 was an upstream event in genome reduplication. In support of this idea, ectopic expression of a stable EMI1 abolished genome reduplication (Fig. 6A). During normal prophase, the SCFβ-TrCP1-dependent degradation of EMI1 is known to be stimulated by PLK1 (23) and inhibited by PIN1 (5) and EVI5 (13). It is possible that changes in the activities of these regulators during the G2 block set off the degradation of EMI1. Indeed, downregulation of PLK1 or β-TrCP1 delayed EMI1 degradation and genome reduplication (Fig. 6B and C). Since downregulation of cyclin A2 or CDK2 stabilized EMI1 (Fig. 7E and see Fig. S7 in the supplemental material), an intriguing possibility is that EMI1 degradation was regulated by cyclin A2-CDK2.
In conclusion, we have characterized the molecular mechanism of a condition in which CDK1 activity was specifically inhibited by RO3306. We found that after an initial arrest in G2 phase with low APC/C activity, APC/C is kick started by the PLK1-SCFβ-TrCP1-EMI1 pathway. In the absence of CDK1, cyclin A-CDK2 is required for APC/C activation. Degradation of APC/C substrates such as geminin, together with other replication components, including CDC6 and cyclin E2-CDK2, contributes to the whole-genome reduplication. These findings extended our understanding of the molecular mechanisms underlying genome reduplication when CDK1 is inhibited. To what extent this pathway plays in the polyploidization of cancer cells remains to be deciphered. It is conceivable that DNA reduplication can occur in situations where CDK1 activity is inhibited for an extended period of time, such as after DNA damage. According to our model, genome reduplication can occur only if CDK2 remains active, which in fact is believed to be the case in cells with defective p53.
Supplementary Material
Acknowledgments
We thank Michele Pagano, Roger Tsien, and David Turner for generous gifts of reagents. Many thanks are due to Anita Lau, Nelson Lee, and Nicole Ng for technical assistance and members of the Poon laboratory for useful comments.
This work was supported in part by the Research Grants Council grants HKUST6415/05 M, HKUST6439/06 M, and 662007 to R.Y.C.P.
Footnotes
Published ahead of print on 12 October 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Arooz, T., C. H. Yam, W. Y. Siu, A. Lau, K. K. Li, and R. Y. C. Poon. 2000. On the concentrations of cyclins and cyclin-dependent kinases in extracts of cultured human cells. Biochemistry 39:9494-9501. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F., R. Brent, R. Kingston, D. Moore, J. Seidman, J. Smith, and K. Struhl. 1991. Current protocols in molecular biology. Wiley & Sons, Inc., New York, NY.
- 3.Bell, S. P., and A. Dutta. 2002. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71:333-374. [DOI] [PubMed] [Google Scholar]
- 4.Bellanger, S., A. de Gramont, and J. Sobczak-Thepot. 2007. Cyclin B2 suppresses mitotic failure and DNA re-replication in human somatic cells knocked down for both cyclins B1 and B2. Oncogene 26:7175-7184. [DOI] [PubMed] [Google Scholar]
- 5.Bernis, C., S. Vigneron, A. Burgess, J. C. Labbe, D. Fesquet, A. Castro, and T. Lorca. 2007. Pin1 stabilizes Emi1 during G2 phase by preventing its association with SCF(betatrcp). EMBO Rep. 8:91-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borlado, L. R., and J. Mendez. 2008. CDC6: from DNA replication to cell cycle checkpoints and oncogenesis. Carcinogenesis 29:237-243. [DOI] [PubMed] [Google Scholar]
- 7.Cai, D., V. M. J. Latham, X. Zhang, and G. I. Shapiro. 2006. Combined depletion of cell cycle and transcriptional cyclin-dependent kinase activities induces apoptosis in cancer cells. Cancer Res. 66:9270-9280. [DOI] [PubMed] [Google Scholar]
- 8.Chan, Y. W., H. T. Ma, W. Wong, C. C. Ho, K. F. On, and R. Y. C. Poon. 2008. CDK1 inhibitors antagonize the immediate apoptosis triggered by spindle disruption but promote apoptosis following the subsequent rereplication and abnormal mitosis. Cell Cycle 7:1449-1461. [DOI] [PubMed] [Google Scholar]
- 9.Chan, Y. W., K. F. On, W. M. Chan, W. Wong, H. O. Siu, P. M. Hau, and R. Y. C. Poon. 2008. The kinetics of p53 activation versus cyclin E accumulation underlies the relationship between the spindle-assembly checkpoint and the postmitotic checkpoint. J. Biol. Chem. 283:15716-15723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, Q., X. Zhang, Q. Jiang, P. R. Clarke, and C. Zhang. 2008. Cyclin B1 is localized to unattached kinetochores and contributes to efficient microtubule attachment and proper chromosome alignment during mitosis. Cell Res. 18:268-280. [DOI] [PubMed] [Google Scholar]
- 11.Chow, J. P. H., W. Y. Siu, H. T. B. Ho, K. H. T. Ma, C. C. Ho, and R. Y. C. Poon. 2003. Differential contribution of inhibitory phosphorylation of CDC2 and CDK2 for unperturbed cell cycle control and DNA integrity checkpoints. J. Biol. Chem. 278:40815-40828. [DOI] [PubMed] [Google Scholar]
- 12.Di Fiore, B., and J. Pines. 2007. Emi1 is needed to couple DNA replication with mitosis but does not regulate activation of the mitotic APC/C. J. Cell Biol. 177:425-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eldridge, A. G., A. V. Loktev, D. V. Hansen, E. W. Verschuren, J. D. Reimann, and P. K. Jackson. 2006. The evi5 oncogene regulates cyclin accumulation by stabilizing the anaphase-promoting complex inhibitor emi1. Cell 124:367-380. [DOI] [PubMed] [Google Scholar]
- 14.Follette, P. J., R. J. Duronio, and P. H. O'Farrell. 1998. Fluctuations in cyclin E levels are required for multiple rounds of endocycle S phase in Drosophila. Curr. Biol. 8:235-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujita, M. 2006. Cdt1 revisited: complex and tight regulation during the cell cycle and consequences of deregulation in mammalian cells. Cell Div. 1:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujiwara, T., M. Bandi, M. Nitta, E. V. Ivanova, R. T. Bronson, and D. Pellman. 2005. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature 437:1043-1047. [DOI] [PubMed] [Google Scholar]
- 17.Fung, T. K., H. T. Ma, and R. Y. C. Poon. 2007. Specialized roles of the two mitotic cyclins in somatic cells: cyclin A as an activator of M phase-promoting factor. Mol. Biol. Cell 18:1861-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ganem, N. J., Z. Storchova, and D. Pellman. 2007. Tetraploidy, aneuploidy and cancer. Curr. Opin. Genet. Dev. 17:157-162. [DOI] [PubMed] [Google Scholar]
- 19.García, P., J. Frampton, A. Ballester, and C. Cales. 2000. Ectopic expression of cyclin E allows non-endomitotic megakaryoblastic K562 cells to establish re-replication cycles. Oncogene 19:1820-1833. [DOI] [PubMed] [Google Scholar]
- 20.Geng, Y., Q. Yu, E. Sicinska, M. Das, J. E. Schneider, S. Bhattacharya, W. M. Rideout, R. T. Bronson, H. Gardner, and P. Sicinski. 2003. Cyclin E ablation in the mouse. Cell 114:431-443. [DOI] [PubMed] [Google Scholar]
- 21.Grafi, G., and B. A. Larkins. 1995. Endoreduplication in maize endosperm: involvement of M phase-promoting factor inhibition and induction of S phase-related kinases. Science 269:1262-1264. [DOI] [PubMed] [Google Scholar]
- 22.Hall, J. R., H. O. Lee, B. D. Bunker, E. S. Dorn, G. C. Rogers, R. J. Duronio, and J. G. Cook. 2008. Cdt1 and Cdc6 are destabilized by rereplication-induced DNA damage. J. Biol. Chem. 283:25356-25363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hansen, D. V., A. V. Loktev, K. H. Ban, and P. K. Jackson. 2004. Plk1 regulates activation of the anaphase promoting complex by phosphorylating and triggering SCFbetaTrCP-dependent destruction of the APC inhibitor Emi1. Mol. Biol. Cell 15:5623-5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Imai, K. K., Y. Ohashi, T. Tsuge, T. Yoshizumi, M. Matsui, A. Oka, and T. Aoyama. 2006. The A-type cyclin CYCA2;3 is a key regulator of ploidy levels in Arabidopsis endoreduplication. Plant Cell 18:382-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Itzhaki, J. E., C. S. Gilbert, and A. C. Porter. 1997. Construction by gene targeting in human cells of a “conditional” CDC2 mutant that rereplicates its DNA. Nat. Genet. 15:258-265. [DOI] [PubMed] [Google Scholar]
- 26.Kondorosi, E., and A. Kondorosi. 2004. Endoreduplication and activation of the anaphase-promoting complex during symbiotic cell development. FEBS Lett. 567:152-157. [DOI] [PubMed] [Google Scholar]
- 27.Lansing, T. J., R. T. McConnell, D. R. Duckett, G. M. Spehar, V. B. Knick, D. F. Hassler, N. Noro, M. Furuta, K. A. Emmitte, T. M. Gilmer, R. A. J. Mook, and M. Cheung. 2007. In vitro biological activity of a novel small-molecule inhibitor of polo-like kinase 1. Mol. Cancer Ther. 6:450-459. [DOI] [PubMed] [Google Scholar]
- 28.Laronne, A., S. Rotkopf, A. Hellman, Y. Gruenbaum, A. C. Porter, and M. Brandeis. 2003. Synchronization of interphase events depends neither on mitosis nor on cdk1. Mol. Biol. Cell 14:3730-3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, C. J., and M. L. DePamphilis. 2002. Mammalian Orc1 protein is selectively released from chromatin and ubiquitinated during the S-to-M transition in the cell division cycle. Mol. Cell. Biol. 22:105-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, K. K. W., I. O. L. Ng, S. T. Fan, J. H. Albrecht, K. Yamashita, and R. Y. C. Poon. 2002. Activation of cyclin-dependent kinases CDC2 and CDK2 in hepatocellular carcinoma. Liver 22:259-268. [DOI] [PubMed] [Google Scholar]
- 31.Lilly, M. A., and A. C. Spradling. 1996. The Drosophila endocycle is controlled by Cyclin E and lacks a checkpoint ensuring S-phase completion. Genes Dev. 10:2514-2526. [DOI] [PubMed] [Google Scholar]
- 32.Lutzmann, M., D. Maiorano, and M. Mechali. 2006. A Cdt1-geminin complex licenses chromatin for DNA replication and prevents rereplication during S phase in Xenopus. EMBO J. 25:5764-5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma, H. T., K. F. On, Y. H. Tsang, and R. Y. C. Poon. 2007. An inducible system for expression and validation of the specificity of short hairpin RNA in mammalian cells. Nucleic Acids Res. 35:e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Machida, Y. J., and A. Dutta. 2007. The APC/C inhibitor, Emi1, is essential for prevention of rereplication. Genes Dev. 21:184-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Margottin-Goguet, F., J. Y. Hsu, A. Loktev, H. M. Hsieh, J. D. Reimann, and P. K. Jackson. 2003. Prophase destruction of Emi1 by the SCF(betaTrCP/Slimb) ubiquitin ligase activates the anaphase promoting complex to allow progression beyond prometaphase. Dev. Cell 4:813-826. [DOI] [PubMed] [Google Scholar]
- 36.Mehrotra, S., S. B. Maqbool, A. Kolpakas, K. Murnen, and B. R. Calvi. 2008. Endocycling cells do not apoptose in response to DNA rereplication genotoxic stress. Genes Dev. 22:3158-3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Melixetian, M., A. Ballabeni, L. Masiero, P. Gasparini, R. Zamponi, J. Bartek, J. Lukas, and K. Helin. 2004. Loss of geminin induces rereplication in the presence of functional p53. J. Cell Biol. 165:473-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Méndez, J., X. H. Zou-Yang, S. Y. Kim, M. Hidaka, W. P. Tansey, and B. Stillman. 2002. Human origin recognition complex large subunit is degraded by ubiquitin-mediated proteolysis after initiation of DNA replication. Mol. Cell 9:481-491. [DOI] [PubMed] [Google Scholar]
- 39.Mihaylov, I. S., T. Kondo, L. Jones, S. Ryzhikov, J. Tanaka, J. Zheng, L. A. Higa, N. Minamino, L. Cooley, and H. Zhang. 2002. Control of DNA replication and chromosome ploidy by geminin and cyclin A. Mol. Cell. Biol. 22:1868-1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parisi, T., A. R. Beck, N. Rougier, T. McNeil, L. Lucian, Z. Werb, and B. Amati. 2003. Cyclins E1 and E2 are required for endoreplication in placental trophoblast giant cells. EMBO J. 22:4794-4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peters, J. M. 2006. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat. Rev. Mol. Cell Biol. 7:644-656. [DOI] [PubMed] [Google Scholar]
- 42.Poon, R. Y. C., H. Toyoshima, and T. Hunter. 1995. Redistribution of the CDK inhibitor p27 between different cyclin-CDK complexes in the mouse fibroblast cell cycle and in cells arrested with lovastatin or ultraviolet irradiation. Mol. Biol. Cell 6:1197-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poon, R. Y. C., and T. K. Fung. 2 March 2007. Cyclin A2. UCSD—Nature Molecule Pages, Nature Publishing Group, New York, NY. doi: 10.1038/mp.a000717.01. [DOI]
- 44.Poon, R. Y. C., and T. Hunter. 1995. Dephosphorylation of Cdk2 Thr160 by the cyclin-dependent kinase-interacting phosphatase KAP in the absence of cyclin. Science 270:90-93. [DOI] [PubMed] [Google Scholar]
- 45.Porter, A. C. 2008. Preventing DNA over-replication: a Cdk perspective. Cell Div. 3:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reimann, J. D., E. Freed, J. Y. Hsu, E. R. Kramer, J. M. Peters, and P. K. Jackson. 2001. Emi1 is a mitotic regulator that interacts with Cdc20 and inhibits the anaphase promoting complex. Cell 105:645-655. [DOI] [PubMed] [Google Scholar]
- 47.Santamaría, D., C. Barriere, A. Cerqueira, S. Hunt, C. Tardy, K. Newton, J. F. Caceres, P. Dubus, M. Malumbres, and M. Barbacid. 2007. Cdk1 is sufficient to drive the mammalian cell cycle. Nature 448:811-815. [DOI] [PubMed] [Google Scholar]
- 48.Shi, Q., and R. W. King. 2005. Chromosome nondisjunction yields tetraploid rather than aneuploid cells in human cell lines. Nature 437:1038-1042. [DOI] [PubMed] [Google Scholar]
- 49.Sigrist, S. J., and C. F. Lehner. 1997. Drosophila fizzy-related down-regulates mitotic cyclins and is required for cell proliferation arrest and entry into endocycles. Cell 90:671-681. [DOI] [PubMed] [Google Scholar]
- 50.Siu, W. Y., A. Lau, T. Arooz, J. P. Chow, H. T. Ho, and R. Y. C. Poon. 2004. Topoisomerase poisons differentially activate DNA damage checkpoints through ataxia-telangiectasia mutated-dependent and -independent mechanisms. Mol. Cancer Ther. 3:621-632. [PubMed] [Google Scholar]
- 51.Sivaprasad, U., Y. J. Machida, and A. Dutta. 2007. APC/C—the master controller of origin licensing? Cell Div. 2:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soni, D. V., R. M. Sramkoski, M. Lam, T. Stefan, and J. W. Jacobberger. 2008. Cyclin B1 is rate limiting but not essential for mitotic entry and progression in mammalian somatic cells. Cell Cycle 7:1285-1300. [DOI] [PubMed] [Google Scholar]
- 53.Storchova, Z., and C. Kuffer. 2008. The consequences of tetraploidy and aneuploidy. J. Cell Sci. 121:3859-3866. [DOI] [PubMed] [Google Scholar]
- 54.Ullah, Z., M. J. Kohn, R. Yagi, L. T. Vassilev, and M. L. DePamphilis. 2008. Differentiation of trophoblast stem cells into giant cells is triggered by p57/Kip2 inhibition of CDK1 activity. Genes Dev. 22:3024-3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vassilev, L. T., C. Tovar, S. Chen, D. Knezevic, X. Zhao, H. Sun, D. C. Heimbrook, and L. Chen. 2006. Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc. Natl. Acad. Sci. USA 103:10660-10665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vaziri, C., S. Saxena, Y. Jeon, C. Lee, K. Murata, Y. Machida, N. Wagle, D. S. Hwang, and A. Dutta. 2003. A p53-dependent checkpoint pathway prevents rereplication. Mol. Cell 11:997-1008. [DOI] [PubMed] [Google Scholar]
- 57.Weiss, A., A. Herzig, H. Jacobs, and C. F. Lehner. 1998. Continuous cyclin E expression inhibits progression through endoreduplication cycles in Drosophila. Curr. Biol. 8:239-242. [DOI] [PubMed] [Google Scholar]
- 58.Woo, R. A., and R. Y. Poon. 2003. Cyclin-dependent kinases and S phase control in mammalian cells. Cell Cycle 2:316-324. [PubMed] [Google Scholar]
- 59.Yam, C. H., R. W. Ng, W. Y. Siu, A. W. Lau, and R. Y. C. Poon. 1999. Regulation of cyclin A-Cdk2 by SCF component Skp1 and F-box protein Skp2. Mol. Cell. Biol. 19:635-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yam, C. H., W. Y. Siu, D. Kaganovich, J. V. Ruderman, and R. Y. C. Poon. 2001. Cleavage of cyclin A at R70/R71 by the bacterial protease OmpT. Proc. Natl. Acad. Sci. USA 98:497-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yam, C. H., W. Y. Siu, A. Lau, and R. Y. C. Poon. 2000. Degradation of cyclin A does not require its phosphorylation by CDC2 and cyclin-dependent kinase 2. J. Biol. Chem. 275:3158-3167. [DOI] [PubMed] [Google Scholar]
- 62.Yoshizumi, T., Y. Tsumoto, T. Takiguchi, N. Nagata, Y. Y. Yamamoto, M. Kawashima, T. Ichikawa, M. Nakazawa, N. Yamamoto, and M. Matsui. 2006. Increased level of polyploidy1, a conserved repressor of CYCLINA2 transcription, controls endoreduplication in Arabidopsis. Plant Cell 18:2452-2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yu, J. Y., S. L. DeRuiter, and D. L. Turner. 2002. RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc. Natl. Acad. Sci. USA 99:6047-6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yuan, J., A. Kramer, Y. Matthess, R. Yan, B. Spankuch, R. Gatje, R. Knecht, M. Kaufmann, and K. Strebhardt. 2006. Stable gene silencing of cyclin B1 in tumor cells increases susceptibility to taxol and leads to growth arrest in vivo. Oncogene 25:1753-1762. [DOI] [PubMed] [Google Scholar]
- 65.Zhang, Y., Z. Wang, D. X. Liu, M. Pagano, and K. Ravid. 1998. Ubiquitin-dependent degradation of cyclin B is accelerated in polyploid megakaryocytes. J. Biol. Chem. 273:1387-1392. [DOI] [PubMed] [Google Scholar]
- 66.Zhang, Y., Z. Wang, and K. Ravid. 1996. The cell cycle in polyploid megakaryocytes is associated with reduced activity of cyclin B1-dependent cdc2 kinase. J. Biol. Chem. 271:4266-4272. [DOI] [PubMed] [Google Scholar]
- 67.Zhu, W., Y. Chen, and A. Dutta. 2004. Rereplication by depletion of geminin is seen regardless of p53 status and activates a G2/M checkpoint. Mol. Cell. Biol. 24:7140-7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.