Abstract
The Mpk1 mitogen-activated protein kinase (MAPK) of the cell wall integrity signaling pathway uses a noncatalytic mechanism to activate the SBF (Swi4/Swi6) transcription factor. Active Mpk1 forms a complex with Swi4, the DNA-binding subunit of SBF, conferring the ability to bind DNA. Because SBF activation is independent of Mpk1 catalytic activity but requires Mpk1 to be in an active conformation, we sought to understand how Mpk1 interacts with Swi4. Mutational analysis revealed that binding and activation of Swi4 by Mpk1 requires an intact D-motif-binding site, a docking surface common to MAPKs that resides distal to the phosphorylation loop but does not require the substrate-binding site, revealing a novel mechanism for MAPK target regulation. Additionally, we found that Mpk1 binds near the autoinhibitory C terminus of Swi4, suggesting an activation mechanism in which Mpk1 substitutes for Swi6 in promoting Swi4 DNA binding. Finally, we show that caffeine is an atypical activator of cell wall integrity signaling, because it induces phosphorylation of the Mpk1 C-terminal extension at Ser423 and Ser428. These phosphorylations were dependent on the DNA damage checkpoint kinases, Mec1/Tel1 and Rad53. Phosphorylation of Ser423 specifically blocked SBF activation by preventing Mpk1 association with Swi4, revealing a novel mechanism for regulating MAPK target specificity.
The cell wall of the budding yeast Saccharomyces cerevisiae is required to maintain cell shape and integrity (12, 28). The cell must remodel this rigid structure during vegetative growth and during pheromone-induced morphogenesis. Wall remodeling is monitored and regulated by the cell wall integrity (CWI) signaling pathway controlled by the Rho1p GTPase (reviewed in reference 31). Two essential functions have been identified for Rho1. First, it serves as an integral regulatory subunit of the 1,3-β-glucan synthase complex (GS), stimulating GS activity in a GTP-dependent manner. A second essential function of Rho1 is to bind and activate protein kinase C, which is encoded by PKC1. Loss of Pkc1 function, or of any of the components of the mitogen-activated protein kinase (MAPK) cascade under its control, results in a cell lysis defect that is attributable to a deficiency in cell wall construction. The MAPK cascade is a linear pathway comprised of a MEK kinase (Bck1), a pair of redundant MEKs (Mkk1/2), and a MAPK (Mpk1/Slt2). Mpk1 is a functional homolog of human extracellular signal-regulated kinase 5 (ERK5) (48), a MAPK that is activated in response to growth factors as well as physical and chemical stresses (1, 57).
CWI signaling is induced in response to a variety of cell wall stressors. First, signaling is activated persistently in response to growth at elevated temperatures (e.g., 37 to 39°C; 23), consistent with the finding that strains with null mutations in many of the pathway components display cell lysis defects only when cultivated at high temperatures. Second, hypoosmotic shock induces a rapid but transient activation of signaling (13, 23). Third, treatment with mating pheromone stimulates signaling at a time that is coincident with the onset of morphogenesis (9). Finally, CWI signaling is also stimulated by agents that interfere with cell wall biogenesis, such as the chitin antagonist calcofluor white (25), Congo red, caffeine, or zymolyase (14, 35).
One consequence of CWI signaling is activation of the Rlm1 transcription factor (15, 55) through phosphorylation by Mpk1 (21). Rlm1 regulates the transcription of a wide array of cell wall metabolism genes (17, 22, 40). However, the FKS2 gene, which encodes one of two alternative catalytic subunits of the GS, is an unusual transcriptional target of CWI signaling, because it is induced independently of Rlm1 (22, 60).
A second transcription factor that plays a role in CWI signaling is SBF (6, 33). SBF is a dimeric transcriptional regulator, comprised of Swi4 and Swi6, which is essential to normal regulation of G1-specific transcription (reviewed in reference 8). Swi4 is the sequence-specific DNA-binding subunit (47), but Swi6 is required for binding to cell cycle-regulated promoters (4, 5, 43). Swi6 allows Swi4 to bind DNA by relieving an intramolecular association of the Swi4 C terminus with its DNA-binding domain. The fact that SBF has a second function related to CWI signaling was established with the finding that Mpk1 associates with SBF in vivo (33) and with Swi4 (but not Swi6) in vitro (6). Additionally, Swi6 is phosphorylated in vivo and in vitro by Mpk1 in response to cell wall stress (33).
We reported previously that CWI signaling drives expression of the FKS2 gene through SBF (27). Mpk1 and its pseudokinase paralog, Mlp1, use a noncatalytic mechanism to activate transcription of FKS2 that is dependent on SBF and on an activating signal to Mpk1. Activated (phosphorylated) Mpk1, or Mlp1, form a complex with Swi4 that associates with an SBF-binding site in the FKS2 promoter independently of Swi6. However, Swi6 is recruited to this complex to activate transcription. Because transcriptional activation of FKS2 through SBF is independent of Mpk1 catalytic activity but nevertheless requires Mpk1 to be in an active (phosphorylated) conformation, we were interested in how Mpk1 interacts with Swi4. MAPKs bind to their substrates and regulators through two distinct protein-protein interaction sites. A docking sequence (D motif) on the substrate or regulator engages in a high-affinity interaction with the D-motif-binding (DB) site of the MAPK, which is remote from the active site and is comprised of an acidic patch called the common docking site and a hydrophobic docking groove (2, 59). The bound protein wraps around the MAPK to make additional contacts with a second surface called the substrate-binding (SB) site, which is positioned near the catalytic center (59).
Here, we demonstrate that binding and activation of Swi4 by phosphorylated Mpk1 requires an intact DB site, but not the SB site, revealing a novel mechanism by which a MAPK activates a target. This finding also indicates that DB site interactions are regulated by the phosphorylation state of the MAPK. Additionally, Mpk1 binds near the C terminus of Swi4, adjacent to the Swi6-binding site, suggesting a Swi4 activation mechanism in which Mpk1 substitutes for Swi6 in promoting Swi4 DNA binding. Finally, we show that caffeine is an atypical activator of CWI signaling, because it induces phosphorylation of Mpk1 on two serine residues in its C-terminal extension. These modifications are dependent on the DNA damage checkpoint kinases Mec1/Tel1 and Rad53, and phosphorylation specifically at Ser423 blocks Mpk1 activation of SBF by preventing its association with Swi4.
MATERIALS AND METHODS
Strains, growth conditions, and transformations.
The Saccharomyces cerevisiae strains used in this study are listed in Table 1. Yeast cultures were grown in YPD (1% Bacto yeast extract, 2% Bacto peptone, 2% glucose) with or without 10% sorbitol or in SD (0.67% yeast nitrogen base, 2% glucose) supplemented with the appropriate nutrients to select for plasmids and gene replacements. Escherichia coli DH5α was used to propagate all plasmids. E. coli cells were cultured in Luria broth medium (1% Bacto tryptone, 0.5% Bacto yeast extract, 1% NaCl) and transformed to carbenicillin resistance by standard methods. Two-hybrid tests were conducted using yeast strains PJ694a and PJ694α (gift of Stanley Fields) transformed with Gal4 activation domain (AD) fusion plasmids of Swi4 or a Gal4 DNA-binding domain (DBD) fusion plasmid of Mpk1 (26), respectively. Transformed strains were crossed under selective conditions to generate diploids expressing the desired pair of fusions. Diploid strains were grown to mid-log phase for extract preparation. Promoter-lacZ expression experiments were carried out as described previously (27). β-Galactosidase assays for promoter-lacZ expression experiments and for two-hybrid analyses were conducted as described previously (60). Because MPK1 mutants display an osmotically remedial cell lysis defect under conditions of cell wall stress (31), it was necessary to conduct gene expression experiments with these mutants in the presence of sorbitol for osmotic support, which diminished the stress responses somewhat.
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Relevant genotypea | Source or reference |
|---|---|---|
| 1788 | MATa/MATα EG123 leu2-3,112 trp1-1 ura3-52 his4 can1r | I. Herskowitz |
| DL100 | MATa EG123 leu2-3,112 trp1-1 ura3-52 his4 can1r | I. Herskowitz |
| DL456 | MATa/MATα EG123 mpk1Δ::TRP1/mpk1Δ::TRP1 | 23 |
| DL2649 | MATa EG123 mlp1Δ::LEU2 | This study |
| DL3145 | MATa/MATα EG123 swi4Δ::TRP1/swi4Δ::TRP1 | 27 |
| DL3195 | MATa/MATα S288c mpk1Δ::KanMX4/mpk1Δ::KanMX4 | 27 |
| DL3196 | MATa/MATα S288c mpk1Δ::KanMX4/mpk1Δ::KanMX4 mlp1Δ::KanMX4/mlp1Δ::KanMX4 | 27 |
| DL3929 | MATa EG123 mlp1Δ::LEU2 mpk1::MPK1-FLAG-HphMX4 | This study |
| DL3930 | MATa EG123 mlp1Δ::LEU2 mpk1::mpk1-S423A-FLAG-HphMX4 | This study |
| DL3931 | MATa EG123 mlp1Δ::LEU2 mpk1::mpk1-S428A-FLAG-HphMX4 | This study |
| DL3932 | MATa EG123 mlp1Δ::LEU2 mpk1::mpk1-S423A, S428A-FLAG-HphMX4 | This study |
| DL3950 | MATα RDK2669 sml1Δ::TRP1 | M. Smolka |
| DL3953 | RDK2669 sml1Δ::TRP1 rad53Δ::HIS3 | M. Smolka |
| DL3954 | MATa/MATα RDK2669 sml1Δ::TRP1/SML1 mec1Δ::HIS3/MEC1 tel1Δ::URA3/TEL1 | M. Smolka |
| DL3955 | RDK2669 sml1Δ::TRP1 mec1Δ::HIS3 tel1Δ::URA3 (p2704) | This study |
| DL3956 | RDK2669 sml1Δ::TRP1 mec1Δ::HIS3 tel1Δ::URA3 (p2824) | This study |
| DL3957 | RDK2669 sml1Δ::TRP1 mec1Δ::HIS3 tel1Δ::URA3 (p2825) | This study |
| PJ694a | MATatrp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ LYS2::GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ | S. Fields |
| PJ694α | MATα trp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ LYS2::GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ | S. Fields |
Strain background EG123 is described by Siliciano and Tatchell (45).
Plasmid and strain construction.
Plasmids used in this study are listed in Table 2. Plasmid p2704 contains the MPK1 coding sequence and its own promoter (residues −1 to −453) cloned with a C-terminal FLAG epitope by SalI/NotI into centromeric plasmid pRS315 (44). This plasmid was used as the basis for construction of the various mutant alleles of MPK1 by QuikChange site-directed mutagenesis (53). Plasmid p2713 contains the SWI4 coding sequence and its promoter (residues −1 to −829), amplified by PCR from genomic yeast DNA (S288c), and cloned by SalI/NotI into centromeric plasmid pRS315. This plasmid was used for construction of mutant SWI4 alleles (swi4-I913A, I915A, and swi4-L966A, L968A; plasmids p2714 and p2715, respectively) by QuikChange mutagenesis. The swi4-I913A, I915A allele was amplified by PCR from p2714 with a C-terminal six-histidine epitope as an SpeI/XhoI fragment and cloned into pUT36 (p2415) (27) for expression of the mutant form of Swi4-6HIS under the control of the MET25 promoter (p2716).
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Reference or source |
|---|---|---|
| pRS315 | LEU2-based centromeric plasmid | 44 |
| p1172 | pGBT9; two-hybrid vector with Gal4-DBD | Clontech |
| p1173 | pGAD424; two-hybrid vector with Gal4-AD | Clontech |
| p1366 | PRM5 (YIL117c)-lacZ with URA3 marker | 21 |
| p2052 | FKS2 (−540 to −375)-CYC1-lacZ with URA3 marker | 27 |
| p2066 | CLN2 (−600 to −400)-CYC1-lacZ with URA3 marker | 27 |
| p2266 | pGBT9 MPK1(1-328) | 26 |
| p2313 | YEp351 MPK1-FLAG | 26 |
| p2316 | YEp351 mpk1-T190A, Y192F-FLAG | 27 |
| p2317 | YEp351 mpk1-K54R-FLAG | 27 |
| p2352 | pGAD424 SWI4(1-1093) | 27 |
| p2415 | pUT36; URA3-based MET25 promoter | 27 |
| p2418 | pUT36 SWI4-6HIS | 27 |
| p2704 | pRS315 MPK1-FLAG | This study |
| p2705 | pRS315 mpk1-H116A-FLAG | This study |
| p2706 | pRS315 mpk1-R196A-FLAG | This study |
| p2707 | pRS315 mpk1-K234A-FLAG | This study |
| p2708 | pRS315 mpk1-D237A-FLAG | This study |
| p2709 | pRS315 mpk1-K83A-FLAG | This study |
| p2710 | pRS315 mpk1-Q166A-FLAG | This study |
| p2711 | pRS315 mpk1-W321A-FLAG | This study |
| p2712 | pRS315 mpk1-K54R-FLAG | This study |
| p2713 | pRS315 SWI4 | This study |
| p2714 | pRS315 swi4-(I913A, I915A) | This study |
| p2715 | pRS315 swi4-(L966A, L968A) | This study |
| p2716 | pUT36 swi4-(I913A, I915A)-6HIS | This study |
| p2717 | pGAD424 SWI4(1-671) | This study |
| p2718 | pGAD424 SWI4(672-1093) | This study |
| p2824 | YEp351 mpk1-S423A-FLAG | This study |
| p2825 | YEp351 mpk1-S428A-FLAG | This study |
| p2826 | YEp351 mpk1-S423A, S428A-FLAG | This study |
Gal4 AD fusions to Swi4 for two-hybrid analysis were constructed in pGAD424 (Clontech Technologies; p1173) by PCR amplification of the coding region using primers that include a SalI site in the upstream primer and a BglII site in the downstream primer. The Gal4 DBD fusion to the catalytic domain of Mpk1 [pGBT9-Mpk1(1-328) (pGBT9-Mpk1 with amino acid residues 1 to 328); p2266] was a gift of M. Molina.
Phospho-site mutants of Mpk1 were constructed by QuikChange mutagenesis of MPK1 in plasmid YEp351 (MPK1-FLAG) (p2313) to create mpk1-S423A-FLAG (p2824), mpk1-S428A-FLAG (p2825), and mpk1-S423A, S428A-FLAG (p2826). For construction of strains expressing integrated phospho-site mutants of Mpk1, MPK1-FLAG was amplified by PCR with BamHI/AscI overhangs and inserted into BamHI/AscI-cut vector pAG32 (contains the Hph gene, which confers resistance to hygromycin B) (18) (p2823), creating pAG32-MPK1-FLAG (p2819). MPK1 was then mutated at either Ser423 or Ser428 to Ala by QuikChange mutagenesis, resulting in pAG32-mpk1-S423-FLAG (p2820), mpk1-S428A-FLAG (p2821), and mpk1-S423A, S428A-FLAG (p2822). The MPK1-FLAG-Hph cassettes from these alleles were then PCR amplified using primers for the 5′ end of MPK1 and the 3′ end of the Hph cassette. The 5′ primers contained overhangs to the promoter of MPK1, whereas the Hph cassette primer contained overhangs to sequence directly downstream of the MPK1 gene. The resulting cassettes were purified and transformed into yeast strain DL2649 (mlp1Δ) on YPD containing 300 μg/ml hygromycin B. Colonies were checked by colony PCR for proper integration of the cassette. The resulting strains contained the MPK1-FLAG alleles replacing the native MPK1 gene. All PCR-amplified sequences and site-directed mutations were confirmed by DNA sequence analysis across the entire amplified region or the complete gene, respectively. Primer sequences are available upon request.
The mec1Δ tel1Δ sml1Δ strains (DL3955, DL3956, and DL3957) were isolated as segregants from the triply heterozygous diploid strain, DL3954 (MBS115; gift of M. Smolka), which had been transformed with plasmids that express mutant forms of Mpk1-FLAG. The strain construction was done in this way because mec1Δ tel1Δ sml1Δ strains are genetically unstable and refractory to transformation.
Coimmunoprecipitation.
Protein extraction and coimmunoprecipitation of Mpk1-FLAG from yeast strain DL456 (mpk1Δ) or DL3145 (swi4Δ) expressing Mpk1-FLAG with or without Swi4-6HIS (from 300 μg protein) were conducted as described previously (23), using mouse monoclonal anti-FLAG (M2) affinity beads (Sigma) or protein A affinity beads (no-antibody controls; Sigma). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (on 7.5% gels) of samples representing 100 μg of initial protein was followed by immunoblot detection of either Mpk1-FLAG (M2 mouse monoclonal anti-FLAG antibody; Sigma) at a dilution of 1:10,000 or Swi4-6HIS (mouse monoclonal anti-tetra-His antibody; Qiagen) at a dilution of 1:3,000. Secondary antibodies (goat anti-mouse antibodies) were used at a dilution of 1:5,000. Input (lysate) controls represented 50 μg of initial protein.
ChIP.
Chromatin immunoprecipitation (ChIP) experiments to detect association of Swi4-6HIS with the CLN2 promoter or the FKS2 promoter were carried out as described previously (27) with the following modifications. Swi4-6HIS was precipitated with Ni-nitrilotriacetic acid Superflow beads (30 μl; Qiagen). Primer sequences for amplifying the FKS2 promoter were GGCTAAAGAAATTTGGGCGG (upstream) and CGTGCTGGGTAGACTTTTAG (downstream), amplifying a 346-bp region. Primers for the CLN2 promoter were TATCCTCCGCACTTTTACCC (upstream) and CTACGCCAAATGTGCTCTTC (downstream), amplifying a 273-bp region.
Immunoblot detection of Swi6 and phosphorylated Mpk1.
Activated Mpk1 was detected with rabbit polyclonal anti-phospho-p44/p42 MAPK (Thr202/Tyr204) antibodies (New England Biolabs) as described previously (50), with the following modifications. Mpk1-FLAG was immunoprecipitated from extracts (100 μg protein extract) prior to SDS-PAGE and immunoblot analysis using 25% of the sample. The anti-phospho-p44/p42 antibody was used at a dilution of 1:3,000, and the secondary antibody (donkey anti-rabbit antibody; GE Healthcare) was used at a dilution of 1:4,000. Serine-phosphorylated Mpk1 was also detected from immunoprecipitated Mpk1-FLAG (as described above) using mouse monoclonal antiphosphoserine antibodies (Sigma) at a dilution of 1:2,000 and the secondary antibody (goat anti-mouse antibody; Jackson Immunoresearch) at a dilution of 1:5,000. The antiphosphoserine antibody is reported by the manufacturer to detect specifically phosphoserine.
RESULTS
Mutations in the SB site and DB site of Mpk1 define separation-of-function alleles.
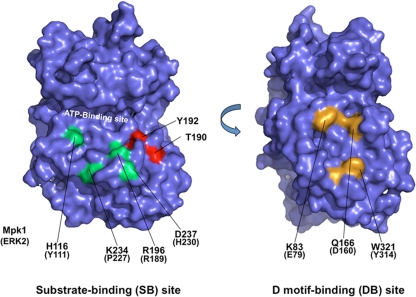
Rlm1 is an Mpk1 target whose activity depends on phosphorylation by Mpk1 (21, 55). In contrast, Swi4 is an Mpk1 target whose activity in response to Mpk1 activation is independent of phosphorylation by Mpk1 (27). To understand how Mpk1 interacts with and activates Swi4, we created a set of mutations in Mpk1 that were predicted to ablate protein binding at either the DB site or the SB site. The mutations created were based on the residues important for the mammalian MAPK ERK2 association with its substrates and regulators (Elk1 and MKP; 59). Mutations in the Mpk1 DB site (K83A, Q166A, and W321A; Fig. 1), located on the backside of the protein relative to the catalytic pocket, were predicted to interfere with high-affinity docking interactions but to still allow activation of phosphorylation targets through interaction with the SB site. In contrast, mutations in the Mpk1 SB site (H116A, R196A, K234A, and D237A; Fig. 1) were predicted to block activation of targets that must interact with the catalytic pocket, while still allowing docking interactions at the DB site.
FIG. 1.
Positions of substrate-binding site mutations and D-motif-binding site mutations within the predicted structure of Mpk1. The structure was modeled against known MAPK structures using the SWISS-MODEL program (http://swissmodel.expasy.org/SWISS-MODEL.html). The two images are rotated 180° with respect to each other. Mutated substrate-binding site residues are indicated in green, and mutated D-motif-binding site residues are indicated in yellow. Residues phosphorylated in the activation loop (T190 and Y192) are indicated in red.
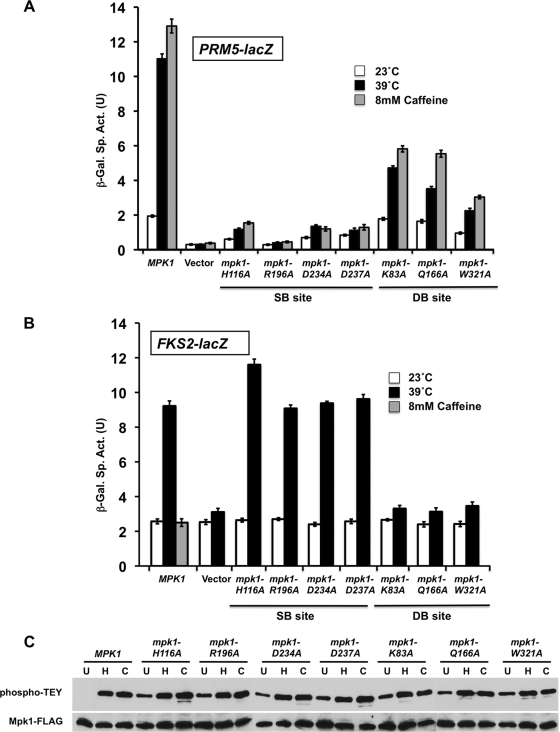
We used two transcriptional readouts that distinguish between Mpk1 activation of Rlm1 by phosphorylation (PRM5-lacZ; 21) and Mpk1 activation of Swi4 by its noncatalytic mechanism (FKS2-lacZ; 27). All of the mutations within the Mpk1 SB site completely blocked Rlm1-driven transcription in response to CWI signaling induced by either thermal stress (39°C) or caffeine treatment (Fig. 2A). In contrast, these mutant forms were able to activate Swi4-dependent transcription normally in response to thermal stress (Fig. 2B). On the other hand, all of the Mpk1 DB site mutants were completely defective for activation of Swi4-dependent transcription, but two of these mutants (K83A and Q166A mutants) retained some Rlm1-dependent transcription. The mpk1-W321A mutant appears to be defective for both functions. None of the mutations impaired phosphorylation of Mpk1 by its activating kinases (Mkk1/2) in response to cell wall stress (Fig. 2C), indicating that the observed deficits in Mpk1 function do not result from an inability to interact with its activators. In fact, all of these mutants displayed an increased basal level of Mpk1 phosphorylation, likely reflecting cell wall stress associated with their functional deficits. Interestingly, caffeine failed to activate FKS2-lacZ transcription through wild-type Mpk1 (Fig. 2B) even though it stimulates phosphorylation of Mpk1 (Fig. 2C) (35, 49), its protein kinase activity in vitro against an artificial substrate (21), and Rlm1-driven transcription (Fig. 2A) (21). This failure to activate Swi4-dependent transcription is unusual among cell wall stressors. We shall return to this point.
FIG. 2.
Mutations in the substrate-binding site and the D-motif-binding site of Mpk1 cause differential effects on cell wall stress induction of FKS2 and PRM5 expression. (A) Cell wall stress-induced PRM5 transcription. A PRM5-lacZ reporter plasmid (p1366) was cotransformed with centromeric plasmids expressing MPK1-FLAG (p2704), plasmids with the indicated point mutants in MPK1, or empty vector (pRS315) into an mpk1Δ mutant (yeast strain DL3195). Transformants were grown to saturation at 23°C in SD medium with 10% sorbitol lacking Ura and Leu. Cultures were diluted into 100 ml of medium so that subsequent incubation at 23°C, 39°C, or 23°C with 8 mM caffeine for 2 h resulted in mid-log-phase cultures (A600 of 1.0). β-Galactosidase activity was measured in crude extracts. The specific activity of β-galactosidase (in units) [β-Gal Sp. Act. (U)] is shown on the y axis. Each value represents the mean ± standard deviation (error bar) from three independent transformants. (B) Cell wall stress-induced FKS2 transcription. An FKS2-lacZ reporter plasmid (p2052) was cotransformed with centromeric plasmids expressing MPK1 (p2704), plasmids with the indicated point mutations in MPK1, or empty vector (pRS315) into an mpk1Δ mlp1Δ mutant (DL3196). The mpk1Δ mlp1Δ mutant was used for this experiment because the Mlp1 pseudokinase contributes to FKS2 expression. Transformants were grown as described above and diluted into 100 ml of medium so that subsequent incubation at 23°C, 39°C, or 23°C with 8 mM caffeine (MPK1 only) for 15 h resulted in mid-log-phase cultures (A600 of 1.0 to 1.5). (C) Point mutations in Mpk1 are not compromised for phosphorylation by their activating protein kinases (Mkk1/2). Protein extracts from cells treated as in panel A (left untreated at 23°C [lanes U], heated to 39°C [lanes H], or treated with 8 mM caffeine [lanes C]) were subjected to immunoprecipitation of Mpk1-FLAG with anti-FLAG antibodies followed by SDS-PAGE. Dual phosphorylation of Mpk1 was detected with anti-phospho-p44/p42 MAPK antibodies; total Mpk1 was detected with anti-FLAG antibodies.
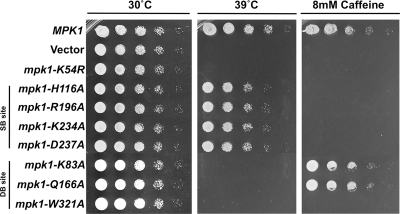
MPK1 null mutants are sensitive to cell wall stress (31). For example, they lyse at elevated growth temperatures (37 to 39°C; 31) and in response to caffeine treatment (34). In contrast, an rlm1Δ mutant is caffeine sensitive, but not temperature sensitive (54), a finding that suggests separation of Mpk1 functions may be discernible through differential growth phenotypes. To determine whether the strains with separation-of-function mutations in MPK1 described above display different growth phenotypes, we tested their sensitivity to heat stress and caffeine treatment. Figure 3A shows that all of the SB site mutants are caffeine sensitive, but not temperature sensitive for growth. This parallels the inability of these mutants to activate Rlm1-driven transcription and is consistent with rlm1Δ mutant phenotypes (54). However, an ATP-binding site mutant of Mpk1 (mpk1-K54R), which is devoid of detectable catalytic activity (33, 58), is both caffeine sensitive and temperature sensitive (Fig. 3). This suggests that although the Mpk1 SB site mutants cannot activate Rlm1, they retain the ability to phosphorylate one or more additional targets. Conversely, all of the DB site mutants, which cannot activate Swi4, are temperature sensitive, but the two that retain some ability to activate Rlm1-driven transcription (with K83A and Q166A mutations) are not caffeine sensitive (Fig. 3). This lends further support to the notion that caffeine sensitivity is a consequence of diminished Rlm1 activity.
FIG. 3.
Mutations in the substrate-binding site and the D-motif-binding site of Mpk1 cause differential phenotypes. An mpk1Δ strain (yeast strain DL456) was transformed with centromeric plasmids expressing the indicated alleles of MPK1 or empty vector (pRS315). Transformants were grown overnight to saturation in selective medium (SD lacking Leu), and serial 10-fold dilutions were plated by pin plating from 96-well plates onto YPD alone or YPD plus 8 mM caffeine and incubated at the indicated temperature for 2 days.
Mpk1 binds to Swi4 through its DB site.
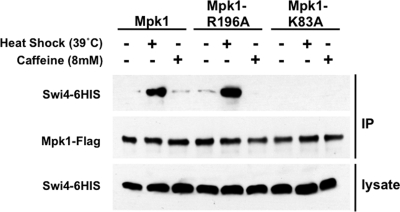
We next tested by coimmunoprecipitation the effects of mutations in the DB site and SB site on cell wall stress-activated Mpk1 binding to Swi4 (27). For these experiments, we chose Mpk1-R196A as a representative of the SB site mutants, and Mpk1-K83A as a representative of the DB site mutants. Epitope-tagged forms of Mpk1 (Mpk1-FLAG) were immunoprecipitated from extracts of cells that had been exposed to cell wall stress (Fig. 4). When mild heat shock was used to activate signaling to Mpk1, epitope-tagged Swi4 (Swi4-6HIS) coprecipitated with both wild-type Mpk1-FLAG and the SB site mutant (Mpk1-R196A-FLAG), but not the DB site mutant (Mpk1-K83A-FLAG), supporting the conclusion that Mpk1 engages Swi4 through its DB site. This is an interesting finding, because it suggests that the phosphorylation state of Mpk1 alters the conformation of the DB site, which resides on the opposite face of the protein from the phosphorylation loop, in such a way that it enables binding to Swi4.
FIG. 4.
Coimmunoprecipitation of Swi4 with mutant forms of Mpk1 in response to heat shock and caffeine treatment. Yeast strain DL456 (mpk1Δ) was cotransformed with plasmids bearing the indicated FLAG-tagged MPK1 allele (wild-type MPK1 [p2704], mpk1-R196A [p2706], and mpk1-K83A [p2709]) and HIS-tagged SWI4 (p2418). Transformants were cultivated to mid-log phase in selective medium lacking methionine (to induce expression of Swi4-6HIS) and either maintained at 25°C or subjected to cell wall stress by changing the temperature to 39°C (+) or by adding 8 mM caffeine for 2 h (+). Cell extracts (lysate) and immunoprecipitates (IP) with anti-FLAG M2 affinity gel were subjected to SDS-PAGE and analyzed by immunoblotting with anti-FLAG antibodies to detect Mpk1 or with anti-HIS antibodies to detect Swi4.
Mpk1 binds to a D motif in the C-terminal region of Swi4.
The results presented above reveal that Mpk1 binding to and activation of Swi4 require an intact DB site but are completely independent of the SB site. Therefore, we used this information to identify the Mpk1 interaction site on Swi4. The canonical D motif found in MAPK substrates and regulators possesses both basic and hydrophobic residues in the arrangement (Arg/Lys)1-2-X2-6-ΦA-x-ΦB (where Φ indicates hydrophobic residues Leu, Ile, and Val) (2, 39, 42, 59). The basic residues interact with an acidic patch within the MAPK DB site called the common docking site (59), and the hydrophobic residues bind to a hydrophobic groove that is adjacent to the common docking site (11, 19).
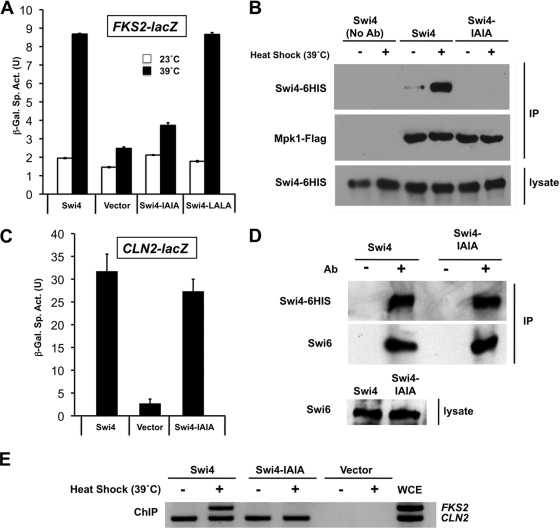
We first narrowed the region of interest in Swi4 using a two-hybrid assay. We reported previously that Mpk1 displays a two-hybrid interaction with Swi4 under activating conditions (27). An N-terminal AD-Swi4 fusion [Gal4AD-Swi4(1-671)] failed to interact with a DBD-Mpk1 fusion [Gal4DBD-Mpk1(1-328)] (Fig. 5). However, a C-terminal Swi4 fusion [Gal4AD-Swi4(672-1093)] displayed a signal-dependent interaction with Mpk1, thereby limiting our search to the C-terminal half of Swi4. Within that region of Swi4 are two sequences that resemble the canonical D motif, 962-KSQALKL-1068 and 909-KKRLITI-915. We mutated the two hydrophobic residues (underlined) in each of these potential Swi4 D motifs to Ala (Swi4-LALA or Swi4 IAIA, respectively). The swi4-LALA mutant was able to activate FKS2-lacZ transcription normally in response to heat stress (Fig. 6A). However, the swi4-IAIA mutant was severely compromised for FKS2-lacZ transcription, suggesting that its ability to dock with Mpk1 was impaired. Additionally, an epitope-tagged form of the Swi4-IAIA mutant protein (Swi4-IAIA-6HIS) failed to coimmunoprecipitate with heat shock-activated Mpk1-FLAG (Fig. 6B), supporting this conclusion.
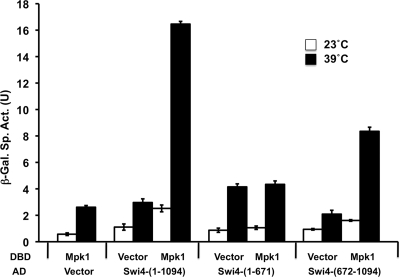
FIG. 5.
Two-hybrid association of Mpk1 with the C-terminal region of Swi4. A Gal4 DNA-binding domain (DBD) fusion to the catalytic domain of Mpk1 (residues 1 to 328; p2266) was tested for interaction with Gal4 activation domain (AD) fusions to full-length Swi4 (residues 1 to 1093; p2352), the N-terminal region (residues 1 to 671; p2717), and the C-terminal region (residues 672 to 1093; p2718) in yeast two-hybrid strain SFY526. Transformants were cultivated in selective medium for 15 h at either 23°C or 39°C. Vector controls (Gal4BDB vector [p1172] and Gal4AD vector [p1173]) are included for each fusion. β-Galactosidase (β-Gal) activity was measured in crude extracts. The specific activity of β-galactosidase (in units) [β-Gal Sp. Act. (U)] is shown on the y axis. Each value represents the mean and standard deviation (error bar) from three independent transformants.
FIG. 6.
Behavior of a strain with a mutation in the Mpk1-binding site of Swi4. (A) Cell wall stress-induced FKS2 transcription is impaired in the swi4-IAIA mutant. An FKS2-lacZ reporter plasmid (p2052) was cotransformed with centromeric plasmids expressing SWI4 (p2713), plasmids with the indicated point mutations in SWI4 (I913A/I915A [Swi4-IAIA] [p2714] or L966A/L968A [Swi4-LALA] [p2715]), or empty vector (pRS315) into a swi4Δ mutant (yeast strain DL3145). Transformants were grown as described in the legend to Fig. 2A and subjected to cell wall stress at 39°C or maintained at 23°C. β-Galactosidase activity was measured in crude extracts. The specific activity of β-galactosidase (in units) [β-Gal Sp. Act. (U)] is shown on the y axis. Each value represents the mean and standard deviation (error bar) from three independent transformants. (B) The Swi4-IAIA mutant protein fails to coimmunoprecipitate with activated Mpk1. A swi4Δ mutant (DL3145) was cotransformed with plasmids bearing FLAG-tagged MPK1 (p2704) and a HIS-tagged SWI4 allele (wild-type SWI4 [p2418] or swi4-IAIA [p2716]). Transformants were cultivated to mid-log phase in selective medium lacking methionine (to induce expression of Swi4-6HIS) and either maintained at 25°C or subjected to cell wall stress by heat shock (changing the temperature to 39°C) for 2 h (+). Cell extracts (lysate) and immunoprecipitates (IP) with anti-FLAG M2 affinity gel were subjected to SDS-PAGE and analyzed by immunoblotting with anti-FLAG antibodies to detect Mpk1 or anti-HIS antibodies to detect Swi4. No Ab, no antibody. (C) CLN2 transcription is not impaired in the swi4-IAIA mutant. A CLN2-lacZ reporter plasmid (p2066) was cotransformed with centromeric plasmids expressing SWI4 (p2713), the swi4-IAIA mutant (p2714), or empty vector (pRS315) into a swi4Δ mutant (DL3145). Transformants were grown as described above but were maintained at 23°C. (D) The Swi4-IAIA mutant is not impaired for Swi4 binding. A swi4Δ mutant (DL3145) was transformed with a plasmid bearing a HIS-tagged SWI4 allele (wild-type SWI4 [p2418] or swi4-IAIA [p2716]). Transformants were cultivated to mid-log phase at 30°C in selective medium lacking methionine (to induce expression of Swi4-6HIS). Cell extracts (lysate) and immunoprecipitates (IP) with Ni-nitrilotriacetic acid beads were subjected to SDS-PAGE and analyzed by immunoblotting with anti-HIS antibodies to detect Swi4 or anti-Swi6 antibodies to detect coprecipitated endogenous Swi6. Ab, antibody. (E) The Swi4-IAIA mutant is defective for FKS2, but not CLN2, promoter occupancy. Transformants from panel D and a vector control (p2415) were cultivated in YPD medium or subjected to heat shock at 39°C for 1 h prior to ChIP analysis using primers designed to detect the endogenous FKS2 or CLN2 promoter. PCRs of whole-cell extract (WCE) from the unstressed wild-type Swi4 sample is also shown.
In contrast, the Swi4-IAIA mutant was not compromised for its interaction with Swi6, which is required for the cell cycle functions of SBF, because it displayed nearly normal transcription of the cell cycle-regulated CLN2-lacZ reporter (Fig. 6C) and it bound in vivo to Swi6 (Fig. 6D). We have shown previously by ChIP that wild-type Swi4 binds to the FKS2 promoter in an Mpk1-dependent but Swi6-independent manner in response to cell wall stress (27). On the other hand, Swi4 binds to cell cycle promoters only when in complex with Swi6, which relieves autoinhibition of DNA binding caused by the Swi4 C terminus (4, 5, 43). Therefore, as a final demonstration that the Swi4-IAIA mutant is specifically defective for its Mpk1-associated function, we tested the ability of this mutant to occupy the CLN2 and FKS2 promoters. Figure 6E shows that the Swi4-IAIA mutant is defective for FKS2 promoter binding but is competent to bind the CLN2 promoter. These findings support a model in which Mpk1 binds to the D motif near the C terminus of Swi4, relieving autoinhibition of DNA binding in a manner similar to that of Swi6, which binds to a site in Swi4 immediately C terminal to the D motif.
Mpk1 activated by caffeine treatment fails to bind Swi4 and undergoes additional phosphorylation on serine.
Noting that activation of Mpk1 by caffeine treatment failed to induce FKS2 transcription, we asked whether this treatment impaired the ability of Mpk1 to bind Swi4. Intriguingly, when caffeine was used to activate Mpk1, this stress failed to induce Mpk1 binding to Swi4 (Fig. 4). This likely explains the failure of caffeine to induce Swi4-dependent transcription and indicates that Mpk1 can be directed selectively to one target over another depending on the activating signal. Nevertheless, caffeine-activated Mpk1 is capable of phosphorylating Swi6 in vivo (K.-Y. Kim, unpublished results).
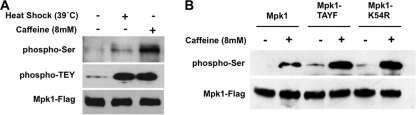
To begin to explore the difference between Mpk1 activated by caffeine or by heat stress, we sought to detect additional phosphorylation events on Mpk1 using a phosphoserine-specific antibody. The rationale for using this antibody was that the activating dual phosphorylations of Mpk1 are on threonyl and tyrosyl residues within the activation loop (TEY), which should not interfere with detection of phosphoserine. Immunoprecipitated Mpk1-FLAG activated either by heat shock or caffeine was subjected to immunodetection with phospho-p42/p44 antibody to measure Mpk1 activation or with a phosphoserine-specific antibody to detect additional modifications (Fig. 7A). Although both the caffeine and heat shock treatments resulted in similar levels of activated Mpk1, caffeine-activated Mpk1 displayed strong serine phosphorylation, which was only weakly detected in Mpk1 from unstressed cells and heat-shocked cells. This new modification is unlikely to be caused by autophosphorylation of Mpk1, because there are no Ser-Pro MAPK target sites within Mpk1. This conclusion was confirmed by the finding that caffeine treatment induced serine phosphorylation on mutant forms of Mpk1 (Fig. 7B) that are either blocked for catalysis (mpk1-K54R) or for phosphorylation by the upstream MEKs (mpk1-TAYF). Interestingly, serine phosphorylation appeared to be slightly enhanced in the inactive forms of Mpk1, suggesting the possibility of negative-feedback regulation of this phosphorylation by Mpk1 activity. Along similar lines, Mpk1 is known to phosphorylate its other regulators, Msg5 and Mkk2 (16, 20).
FIG. 7.
Mpk1 undergoes serine phosphorylation in response to caffeine treatment. (A) Caffeine induces serine phosphorylation of Mpk1. Yeast strain DL456 (mpk1Δ), transformed with a plasmid expressing FLAG-tagged Mpk1 (p2313), was either maintained at 25°C or subjected to cell wall stress (heat shock) by changing the temperature to 39°C (+) or by adding 8 mM caffeine (+) for 2 h. Cell extracts were subjected to immunoprecipitation of Mpk1-FLAG, followed by immunodetection of total Mpk1 with anti-FLAG antibodies, activated Mpk1 with phospho-p42/p44 antibodies (phospho-TEY), and serine phosphorylation of Mpk1 with antiphosphoserine antibodies. (B) Caffeine-induced serine phosphorylation of Mpk1 is independent of Mpk1 activation state or catalytic activity. Yeast strain DL456 (mpk1Δ), transformed with a plasmid expressing FLAG-tagged Mpk1 (p2313), nonactivatable Mpk1-TAYF (p2316), or catalytically inactive Mpk1-K54R (p2317), were subjected to caffeine stress (+) and processed as described above for detection of Mpk1 phosphorylated on serine and total Mpk1.
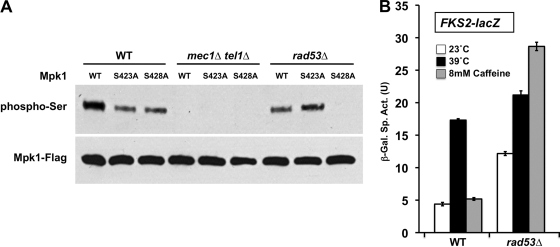
Caffeine induces phosphorylation of Mpk1 on Ser423 and Ser428.
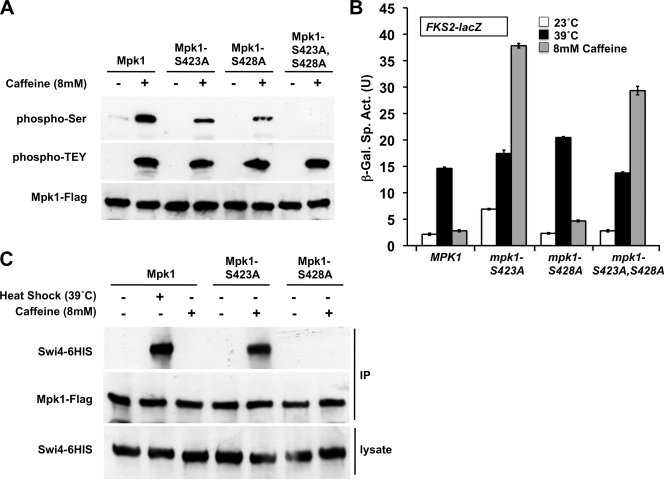
A previous report of phospho-proteomic sites in yeast (3) revealed that Mpk1 is phosphorylated on Ser423 and Ser428 in response to treatment with the mutagen, methyl methanesulfonate. Because these are the only known phospho-sites on Mpk1 other than those in the activation loop and because caffeine treatment has been reported to induce frameshift mutations in E. coli (38), we mutated these residues individually and in combination to determine whether they are the sites phosphorylated in response to caffeine treatment. Figure 8A shows that caffeine induced serine phosphorylation of Mpk1 at both residues Ser423 and Ser428. The phosphoserine-specific antibody did not detect the Mpk1-S423A, S428A double mutant form even upon overexposure of the film (not shown), suggesting that these are the only serine phosphorylations that occur in Mpk1. A reduced level of serine phosphorylation was detected in both the Mpk1-S423A and Mpk1-S428A mutant forms. Because both single mutants were detected by the phosphoserine antibody, phosphorylation at Ser423 and Ser428 can occur independently of each other. It should also be noted that neither one of the single mutants nor the double mutant affected the ability of Mpk1 to acquire its traditional activating phosphorylations at Thr190 or Tyr192. This is important because the Mpk1-Swi4 interaction requires these phosphorylations.
FIG. 8.
Behavior of Mpk1 mutants that block serine phosphorylation induced by caffeine. (A) Ser423 and Ser428 of Mpk1 are phosphorylated in response to caffeine. Yeast strain DL456 (mpk1Δ), transformed with plasmids expressing FLAG-tagged Mpk1 (p2313) or Ser-to-Ala mutant forms (S423A [p2824], S428A [p2825], or S423A S428A [p2826]) were subjected to caffeine stress (+), and extracts were processed for detection of serine-phosphorylated Mpk1 (phospho-Ser), activated Mpk1 (phospho-TEY), and total Mpk1 (Mpk1-Flag) as described in the legend to Fig. 7. (B) Blocking phosphorylation of Mpk1 at Ser423 allows caffeine-induced cell wall stress to drive FKS2 transcription. An FKS2-lacZ reporter plasmid (p2052) was transformed into yeast strains with the indicated MPK1 alleles integrated into the genome (MPK1 [DL3929], mpk1-S423A [DL3930], mpk1-S428A [DL3931], and mpk1-S423A S428A [DL3932]). Transformants were grown as described in the legend to Fig. 2A and subjected to thermal or caffeine stress or maintained at 23°C. β-Galactosidase activity was measured in crude extracts. β-Galactosidase specific activity (in units) [β-Gal Sp. Act. (U)] is shown on the y axis. Each value represents the mean and standard deviation (error bar) from three independent transformants. (C) Blocking phosphorylation of Mpk1 on Ser423 allows caffeine-activated Mpk1 to bind Swi4. Yeast strain DL456 (mpk1Δ), cotransformed with plasmids expressing FLAG-tagged Mpk1 (p2313) or Ser-to-Ala mutant forms (Mpk1-S423A [p2824] or Mpk1-S428A [p2825]) and Swi4-6HIS (p2418) were subjected to heat shock or caffeine stress (+), and extracts were processed for coimmunoprecipitation of Swi6 with Mpk1 as described in the legend to Fig. 6B.
To assess the impact of caffeine-induced serine phosphorylation of Mpk1 on the behavior of this MAPK, we tested the mpk1-S423A and mpk1-S428A mutants for their ability to induce FKS2-lacZ transcription in response to heat stress and caffeine treatment. In contrast to the behavior of wild-type Mpk1, caffeine strongly induced FKS2-lacZ transcription in the mpk1-S423A mutant (Fig. 8B), suggesting that phosphorylation of Ser423 normally inhibits a function of Mpk1 associated with FKS2-lacZ transcription. The mpk1-S428A mutant was not appreciably affected, leaving unanswered the importance of phosphorylation of this residue. Neither mutation had an appreciable affect on the ability of heat stress to induce FKS2-lacZ transcription.
As noted above, although caffeine treatment induces activation of Mpk1, the MAPK activated in this manner does not bind to Swi4. Because the S423A mutation conferred upon Mpk1 the capacity to drive FKS2-lacZ transcription in response to caffeine treatment, we examined the ability of the Mpk1-S423A mutant form to bind Swi4 under this condition. Figure 8C shows that the Mpk1-S423A mutant acquired the ability to associate with Swi4 in response to caffeine. Thus, caffeine treatment induces a novel Ser phosphorylation on Mpk1, which imposes a shift in the target specificity of this MAPK.
Caffeine-induced phosphorylation of Mpk1 on Ser423 is dependent on Rad53.
Phosphorylation of Mpk1 on Ser423 and Ser428 is provoked by DNA damage (3). In this regard, it is interesting to note the context of these phosphorylation sites. Specifically, Ser428 resides at a Ser Gln site, a target motif for the partially redundant Mec1/Tel1 DNA damage checkpoint kinases (46). Additionally, Ser423 resides at a Ser Phe site, which fits the S/T-ψ consensus motif (where ψ is a hydrophobic residue, such as F, I, V, or L) for the Rad53 DNA damage checkpoint kinase, whose activity is under the control of Mec1/Tel1 (46). Therefore, we asked whether the novel caffeine-induced Mpk1 phosphorylations were dependent on Mec1/Tel1 or Rad53. Because these genes are essential, the mec1Δ/tel1Δ and rad53Δ mutants were tested in a background that suppresses their lethality (sml1Δ) (46, 61). Serine phosphorylation of Mpk1-S428A (but not wild-type Mpk1) was abolished in the rad53Δ mutant (Fig. 9A), strongly suggesting that Ser423 is a direct target of Rad53. Phosphorylation of both Ser423 and Ser428 was abolished in the mec1Δ/tel1Δ mutant. This, together with the rad53Δ result, strongly suggests that Ser428 is a direct target of Mec1/Tel1 and Ser423 is an indirect target of these kinases through Rad53. We reasoned that a rad53Δ mutant should confer upon Mpk1 the capacity to drive FKS2-lacZ transcription in response to caffeine treatment in the same manner as the Mpk1-S423A mutant. Figure 9B shows that this prediction was borne out, revealing that caffeine-induced modification of Mpk1 target specificity is regulated by the Rad53 kinase.
FIG. 9.
Dependence of Mpk1 serine phosphorylation on Mec1/Tel1 and Rad53 and its role in FKS2 transcription. (A) Caffeine-induced phosphorylation of Mpk1 Ser423 is blocked in a rad53Δ mutant, and phosphorylation of both Ser423 and Ser428 are blocked in a Mec1/Tel1 mutant. Yeast strains with the sml1Δ gene (wild type [WT] [strain DL3950]), mec1Δ tel1Δ sml1Δ gene (strains DL3955 to DL3957), and rad53Δ sml1Δ gene (strain DL3953) that had been transformed with plasmid expressing Mpk1-FLAG (p2704), Mpk1-S423A-FLAG (p2824), or Mpk1-S428A-FLAG (p2825) were subjected to caffeine stress. The cell extracts were processed for detection of serine-phosphorylated Mpk1 (phospho-Ser) and total Mpk1 (Mpk1-Flag) as described in the legend to Fig. 7. (B) Blocking Ser423 phosphorylation in a rad53Δ mutant allows caffeine-induced cell wall stress to drive FKS2 transcription. An FKS2-lacZ reporter plasmid (p2052) was transformed into yeast strains with the sml1Δ gene (WT; DL3950) and rad53Δ sml1Δ gene (DL3953). Transformants were grown as described in the legend to Fig. 2A and subjected to thermal or caffeine stress or maintained at 23°C. β-Galactosidase activity was measured in crude extracts. β-Galactosidase specific activity (in units) [β-Gal Sp. Act. (U)] is shown on the y axis. Each value represents the mean and standard deviation (error bar) from three independent transformants.
DISCUSSION
MAPK substrates and regulators bind to the kinase through a bipartite recognition mechanism (11, 59). A kinase interaction site (D motif), which is located on the backside of the protein kinase relative to the catalytic pocket (Fig. 1), engages in high-affinity associations with the DB site. The substrate-binding region, located near the kinase active site, binds to other structural elements in the interacting protein that are C terminal to the D motif and is required for phosphorylation of substrates (59). In this study, we sought to understand how the CWI pathway MAPK, Mpk1, interacts with the Swi4 transcription factor. Dually phosphorylated Mpk1 activates Swi4-driven transcription through a noncatalytic mechanism that involves the formation of an Mpk1-Swi4 complex on the promoter of the FKS2 gene (27). The ability of Mpk1 to activate Swi4 without phosphorylating it raised an interesting mechanistic question. Because formation of the Swi4-Mpk1 complex is strictly dependent upon activation of Mpk1 by its cognate MEKs (Mkk1/2), we wondered whether Mpk1 interacts with Swi4 as it would a phosphorylation substrate. To address this question, we constructed a set of mutant forms of Mpk1 that were predicted to ablate binding of proteins at either the DB site or the SB site.
Interaction of Swi4 with the DB site of Mpk1.
Our study revealed that Mpk1 binds to and activates Swi4 in a manner that requires an intact DB site but is independent of the SB site. This result was somewhat surprising, because of the dependence of this interaction on phosphorylation of Mpk1 at its activation loop, suggesting that the affinity of Swi4 for the Mpk1 DB site is altered by the phosphorylation state of Mpk1. Comparison of the structures of phosphorylated and unphosphorylated ERK2 (10), a mammalian MAPK to which Mpk1 is closely related, reveals that the greatest conformational changes are in the phosphorylation loop and little change is evident within the DB site or the regions that surround it. On the other hand, binding of D-motif peptides derived from ERK2 regulators to the DB site of unphosphorylated ERK2 induces large conformational changes in the activation loop and elsewhere (62), revealing that there is communication between the two sites. Specifically, binding of peptides from the HePTP tyrosine phosphatase or the MEK2 kinase induce the dual phosphorylation sites of ERK2, which are buried in the unphosphorylated form, to become solvent exposed as in active ERK2. In fact, the peptide-induced conformational changes result in an ERK2 structure that is overall more similar to the phosphorylated form than to the unphosphorylated form (62). This observation suggests that regulators and substrates may bind preferentially to the DB site of activated MAPKs, although this has not been explored directly in ERK2 or other mammalian MAPKs. On the other hand, the unphosphorylated yeast MAPK Kss1 binds to the transcriptional repressors Dig1 and Dig2 through its DB site, blocking transcriptional activation of filamentous growth genes (30).
We are aware of only one prior precedent for a protein binding specifically or preferentially to the DB site of an active form of a MAPK. The Ptp2 protein phosphatase, which inactivates Hog1, binds only to the phosphorylated form of this MAPK (56). This interaction was shown recently to be predominantly through the Hog1 DB site (37).
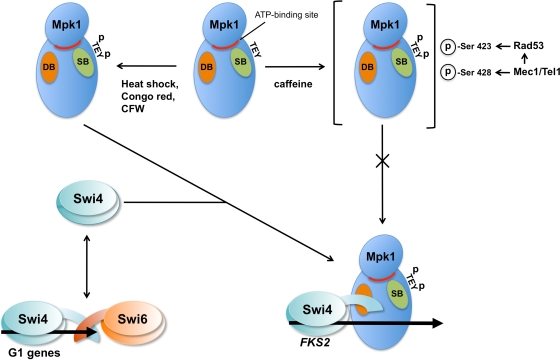
We identified the D motif (kinase interaction site) in Swi4, which resides near its C terminus but N terminal to its Swi6 interaction site. The C terminus of Swi4 is an autoinhibitory domain that prevents its binding to DNA, but association of Swi6 to the Swi4 C terminus relieves autoinhibition and allows the heterodimer to bind SCB sites in cell cycle-regulated genes (4, 5, 43). In contrast, under conditions of cell wall stress, Mpk1 binds to Swi4 and allows this complex to bind an SCB in the FKS2 promoter (27). Our finding that Mpk1 interacts with Swi4 at a D motif that neighbors its Swi6-binding site suggests that Mpk1 serves to relieve autoinhibition of Swi4 DNA binding in a manner that is similar to that of Swi6 (Fig. 10).
FIG. 10.
Model for interaction of Mpk1 with Swi4. Mpk1 activated by cell wall stressors other than caffeine is capable of activating Swi4-dependent transcription. Swi4 binds Mpk1 through the D motif near the C terminus of Swi4, which allows Swi4 to bind the FKS2 promoter. This interaction is proposed to relieve the autoinhibitory interaction of the Swi4 C terminus that prevents DNA binding. In this regard, Mpk1 serves the same function as Swi6 in allowing Swi4 to bind DNA. Swi6 is recruited to the Mpk1-Swi4 complex and binds to Swi4 through their respective C termini (not shown), in a manner similar to their interaction on G1-induced cell cycle genes (e.g., CLN2). Mpk1 activated by caffeine is not capable of Swi4-driven transcription. Under these conditions, activated Mpk1 is incapable of interacting with Swi4. Additional phosphorylations on Mpk1 Ser423 and Ser428 are provoked by caffeine treatment through the DNA damage checkpoint kinases Mec1/Tel1 and Rad53. Phosphorylation of Ser423, which is dependent on Rad53, specifically prevents Mpk1 binding to Swi4. DB, D-motif-binding site; SB, substrate-binding site; p, phosphate group; CFW, calcofluor white.
In contrast to the requirements for Swi4 activation, the Rlm1 transcription factor, which is activated through phosphorylation by Mpk1 (21, 55), was inactive in SB site mutants of MPK1. Although Rlm1 also binds to Mpk1 through a D motif (21), we found that ablation of the Mpk1 DB site did not completely block Rlm1 activation. Consequently, the DB site mutants and SB site mutants of Mpk1 displayed different phenotypic defects that reflected differential activation of Rlm1 and Swi4.
Caffeine-activated Mpk1 does not bind Swi4 because it undergoes a Rad53-dependent phosphorylation at Ser423.
Caffeine has been used for many years to induce cell wall stress (21, 34, 35), yet the mechanism by which it acts is not understood (29). We found that caffeine is an unusual cell wall stress agent, because although it induces activation of Mpk1 and consequent transcriptional activation through Rlm1 (21), it failed to induce Swi4-driven transcription. This failure was accompanied by a failure of caffeine-activated Mpk1 to form a complex with Swi4.
We found that caffeine treatment provokes novel phosphorylations on Ser423 and Ser428 of Mpk1 independently of the standard dual phosphorylation associated with MAPK activation. We found additionally that phosphorylation at Ser423 was responsible for blocking association of active Mpk1 with Swi4, specifically preventing Mpk1 from driving transcription of the FKS2 gene. This phosphorylation was dependent on the DNA damage checkpoint kinase, Rad53, and its activating kinases, Tel1 and Mec1. In contrast, phosphorylation of Ser428 was dependent only on Tel1/Mec1. Taken together with the sequence motifs at these phosphorylation sites, our findings suggest that Ser423 is a direct target of Rad53 and Ser428 is a direct target of Mec1/Tel1 (Fig. 10). These results reveal a novel mechanism by which signal-specific targeting of a MAPK can be achieved. Ser423 resides outside of the catalytic domain in the long, C-terminal extension of Mpk1, which is shared only with its mammalian ortholog, ERK5. We do not yet know how this modification interferes with the binding of Mpk1 to Swi4. Although both Ser423 and Ser428 become phosphorylated under caffeine stress, Ser428 has no apparent impact on the Mpk1-Swi4 interaction or on consequent induction of FKS2. Perhaps phosphorylation of Ser428 by Mec1/Tel1 is important for regulating the interaction of Mpk1 with other targets.
The primary target of caffeine in Saccharomyces cerevisiae appears to be the TORC1 complex (29, 32). This conclusion derives both from the identification of TORC1 components (Tor1, Tco89, and Kog1) in a global screen of heterozygous deletion mutants for enhanced caffeine sensitivity (32) and from a global gene expression analysis in which the transcriptomic response to caffeine was found to overlap 25% with that of rapamycin, an inhibitor of TORC1 (29). However, several observations reveal the existence of additional caffeine targets. First, caffeine induces a nonuniform growth arrest (29), whereas rapamycin induces arrest specifically at G1 phase as a result of inhibition of protein synthesis initiation (7). Second, the sensitivity of strains with mutations in the CWI pathway to caffeine is the consequence of cell lysis, as judged by microscopic observation of nonrefractile ghosts (D. E. Levin, unpublished results) and suppression of the growth defect by inclusion of sorbitol in the growth medium for osmotic support (29; Levin, unpublished). In contrast, caffeine sensitivity of a tor1Δ mutant is not suppressed by sorbitol (29). Finally, the ATM and ATR DNA damage checkpoint kinases are inhibited by caffeine in Saccharomyces cerevisiae, fission yeast, and mammalian cells (24, 36, 41, 51, 52). In this regard, it is difficult to explain why caffeine induces Tel1/Mec1-dependent phosphorylations on Mpk1, considering that Mec1 and Tel1 are the yeast homologs of ATR and ATM, respectively (46). Clearly, additional connections between the action of caffeine on these kinases and CWI signaling await discovery.
Acknowledgments
We thank Melanie Cobb, Betsy Goldsmith, and Haruo Saito for valuable discussions, S. Fields for the two-hybrid strains, and M. Smolka for DNA damage checkpoint mutants.
This work was supported by a grant from the NIH (GM48533) to D.E.L. A.W.T. was supported by a Dimitri V. d'Arbeloff postdoctoral fellowship from the Millipore Corporation.
Footnotes
Published ahead of print on 5 October 2009.
REFERENCES
- 1.Abe, J., M. Kusuhara, R. J. Ulevitch, B. C. Berk, and J. D. Lee. 1996. Big mitogen-activated protein kinase 1 (BMK1) is a redox-sensitive kinase. J. Biol. Chem. 271:16586-16590. [DOI] [PubMed] [Google Scholar]
- 2.Akella, R., T. M. Moon, and E. J. Goldsmith. 2008. Unique MAP kinase binding sites. Biochim. Biophys. Acta 1784:48-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albuquerque, C. P., M. B. Smolka, S. H. Payne, V. Bafna, J. Eng, and H. Zhou. 2008. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol. Cell. Proteomics 7:1389-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrews, B. J., and L. A. Moore. 1992. Interaction of the yeast Swi4 and Swi6 cell cycle regulatory proteins in vitro. Proc. Natl. Acad. Sci. USA 89:11852-11856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baetz, K., and B. Andrews. 1999. Regulation of the cell cycle transcription factor Swi4 through autoinhibition of DNA binding. Mol. Cell. Biol. 19:6729-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baetz, K., J. Moffat, J. Haynes, M. Chang, and B. Andrews. 2001. Transcriptional coregulation by the cell integrity mitogen-activated protein kinase Slt2 and the cell cycle regulator Swi4. Mol. Cell. Biol. 21:6515-6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barbet, N. C., U. Schneider, S. B. Helliwell, I. Stansfield, M. F. Tuite, and M. N. Hall. 1996. TOR controls translation initiation and early G1 progression in yeast. Mol. Biol. Cell 7:25-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Breeden, L. L. 2003. Periodic transcription: a cycle within a cycle. Curr. Biol. 13:R31-R38. [DOI] [PubMed] [Google Scholar]
- 9.Buehrer, B. M., and B. Errede. 1997. Coordination of the mating and cell integrity mitogen-activated protein kinase pathways in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:6517-6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Canagarajah, B. J., A. Khokhlatchev, M. H. Cobb, and E. J. Goldsmith. 1997. Activation mechanism of the MAP kinase ERK2 by dual phosphorylation. Cell 90:859-869. [DOI] [PubMed] [Google Scholar]
- 11.Chang, C.-I., B. Xu, R. Akella, M. H. Cobb, and E. J. Goldsmith. 2002. Crystal structure of MAP kinase p38 complexed to the docking sites on its nuclear substrate MEF2A and activator MKK3b. Mol. Cell 9:1241-1249. [DOI] [PubMed] [Google Scholar]
- 12.Cid, V. J., A. Duran, F. Rey, M. P. Snyder, C. Nombela, and M. Sanchez. 1995. Molecular basis of cell integrity and morphogenesis in Saccharomyces cerevisiae. Microbiol. Rev. 59:345-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davenport, K. R., M. Sohaskey, Y. Kamada, D. E. Levin, and M. C. Gustin. 1995. A second osmosensing signal transduction pathway in yeast. Hypotonic shock activates the PKC1 protein kinase-regulated cell integrity pathway. J. Biol. Chem. 27:30157-30161. [DOI] [PubMed] [Google Scholar]
- 14.de Nobel, H., C. Ruiz, H. Martin, W. Morris, S. Brul, M. Molina, and F. M. Klis. 2000. Cell wall perturbation in yeast results in dual phosphorylation of the Slt2/Mpk1 MAP kinase and in an Slt2-mediated increase in FKS2-lacZ expression, glucanase resistance and thermotolerance. Microbiology 146:2121-2132. [DOI] [PubMed] [Google Scholar]
- 15.Dodou, E., and R. Treisman. 1997. The Saccharomyces cerevisiae MADS-box transcription factor Rlm1 is a target for the Mpk1 mitogen-activated protein kinase pathway. Mol. Cell. Biol. 17:1848-1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flández, M., I. C. Cosano, C. Nombela, H. Martín, and M. Molina. 2004. Reciprocal regulation between Slt2 MAPK and isoforms of Msg5 dual-specificity protein phosphatase modulates the yeast cell integrity pathway. J. Biol. Chem. 279:11027-11034. [DOI] [PubMed] [Google Scholar]
- 17.Garcia, R., C. Bermejo, C. Grau, R. Perez, J. M. Rodriquez-Pena, J. Francois, C. Nombela, and J. Arroyo. 2004. The global transcriptional response to transient cell wall damage in Saccharomyces cerevisiae and its regulation by the cell integrity signaling pathway. J. Biol. Chem. 279:15183-15195. [DOI] [PubMed] [Google Scholar]
- 18.Goldstein, A. L., and J. H. McCusker. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15:1541-1553. [DOI] [PubMed] [Google Scholar]
- 19.Heo, Y. S., S. K. Kim, C. I. Seo, Y. K. Kim, B. J. Sung, H. S. Lee, J. I. Lee, S. Y. Park, J. H. Kim, K. Y. Hwang, Y. L. Hyun, Y. H. Jeon, S. Ro, J. M. Cho, T. G. Lee, and C. H. Yang. 2004. Structural basis for the selective inhibition of JNK1 by the scaffolding protein JIP1 and SP600125. EMBO J. 23:2185-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiménez-Sánchez, M., V. J. Cid, and M. Molina. 2007. Retrophosphorylation of Mkk1 and Mkk2 MAPKKs by the Slt2 MAPK in the yeast cell integrity pathway. J. Biol. Chem. 282:31174-31185. [DOI] [PubMed] [Google Scholar]
- 21.Jung, U. S., A. K. Sobering, M. J. Romeo, and D. E. Levin. 2002. Regulation of the yeast Rlm1 transcription factor by the Mpk1 cell wall integrity MAP kinase. Mol. Microbiol. 46:781-789. [DOI] [PubMed] [Google Scholar]
- 22.Jung, U. S., and D. E. Levin. 1999. Genome-wide analysis of gene expression regulated by the yeast cell wall integrity signalling pathway. Mol. Microbiol. 34:1049-1057. [DOI] [PubMed] [Google Scholar]
- 23.Kamada, Y., U. S. Jung, J. Piotrowski, and D. E. Levin. 1995. The protein kinase C-activated MAP kinase pathway of Saccharomyces cerevisiae mediates a novel aspect of the heat shock response. Genes Dev. 9:1559-1571. [DOI] [PubMed] [Google Scholar]
- 24.Kaufmann, W. K., T. P. Heffernan, L. M. Beaulieu, S. Doherty, A. R. Frank, Y. Zhou, M. F. Bryant, T. Zhou, D. D. Luche, N. Nikolaishvili-Feinberg, D. A. Simpson, and M. Cordeiro-Stone. 2003. Caffeine and human DNA metabolism: the magic and the mystery. Mutat. Res. 532:85-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ketela, T., R. Green, and H. Bussey. 1999. Saccharomyces cerevisiae Mid2p is a potential cell wall stress sensor and upstream activator of the PKC1-MPK1 cell integrity pathway. J. Bacteriol. 181:3330-3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim, K.-Y., I. C. Cosano, D. E. Levin, M. Molina, and H. Martin. 2007. Dissecting the transcriptional activation function of the cell wall integrity MAP kinase. Yeast 24:335-342. [DOI] [PubMed] [Google Scholar]
- 27.Kim, K.-Y., A. W. Truman, and D. E. Levin. 2008. Yeast Mpk1 mitogen-activated protein kinase activates transcription through Swi4/Swi6 by a noncatalytic mechanism that requires upstream signal. Mol. Cell. Biol. 28:2579-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klis, F. M. 1994. Review: cell wall assembly in yeast. Yeast 10:851-869. [DOI] [PubMed] [Google Scholar]
- 29.Kuranda, K., V. Leberra, S. Sokol, G. Palamarczyk, and J. Francois. 2006. Investigating the caffeine effects in the yeast Saccharomyces cerevisiae brings new insight into the connection between TOR, PKC, and Ras/cAMP signaling pathways. Mol. Microbiol. 61:1147-1166. [DOI] [PubMed] [Google Scholar]
- 30.Kusari, A. B., D. M. Molina, W. Sabbagh, Jr., C. S. Lau, and L. Bardwell. 2004. A conserved protein interaction network involving the yeast MAP kinases Fus3 and Kss1. J. Biol. Chem. 164:267-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levin, D. E. 2005. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69:262-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lum, P. Y., C. D. Armour, S. B. Stepaniants, G. Cavet, M. K. Wolf, J. S. Butler, J. C. Hinshaw, P. Garnier, G. D. Prestwich, A. Leonardson, P. Garrett-Engele, C. M. Rush, M. Bard, G. Schimmack, J. W. Phillips, C. J. Roberts, and D. D. Shoemaker. 2004. Discovering modes of action for therapeutic compounds using a genome-wide screen of yeast heterozygotes. Cell 116:121-137. [DOI] [PubMed] [Google Scholar]
- 33.Madden, K., Y. J. Sheu, K. Baetz, B. Andrews, and M. Snyder. 1997. SBF cell cycle regulator as a target of the yeast PKC-MAP kinase pathway. Science 275:1781-1784. [DOI] [PubMed] [Google Scholar]
- 34.Martin, H., M. C. Castellanos, R. Cenamor, M. Sanchez, M. Molina, and C. Nombela. 1996. Molecular and functional characterization of a mutant allele of the mitogen-activated protein kinase gene SLT2 (MPK1) rescued from yeast autolytic mutants. Curr. Genet. 29:516-522. [DOI] [PubMed] [Google Scholar]
- 35.Martin, H., J. M. Rodriguez-Pachon, C. Ruiz, C. Nombela, and M. Molina. 2000. Regulatory mechanisms for modulation of signaling through the cell integrity Slt2-mediated pathway in Saccharomyces cerevisiae. J. Biol. Chem. 275:1511-1519. [DOI] [PubMed] [Google Scholar]
- 36.Moser, B. A., J.-M. Brondello, B. Baber-Furnari, and P. Russell. 2000. Mechanism of caffeine-induced checkpoint override in fission yeast. Mol. Cell. Biol. 20:4288-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murakami, T., K. Tatebayashi, and H. Saito. 2008. Two adjacent docking sites in the yeast Hog1 mitogen-activated protein (MAP) kinase differentially interact with the Pbs2 MAP kinase kinase and the Ptp2 protein tyrosine phosphatase. Mol. Cell. Biol. 28:2481-2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pons, F. W., and P. Müller. 1990. Induction of frameshift mutations by caffeine in Escherichia coli K12. Mutagenesis 5:173-178. [DOI] [PubMed] [Google Scholar]
- 39.Remenyi, A., M. C. Good, R. P. Bhattacharyya, and W. A. Lim. 2005. The role of docking interactions in mediating signaling input, output, and discrimination in the yeast MAPK network. Mol. Cell 29:951-962. [DOI] [PubMed] [Google Scholar]
- 40.Roberts, C. J., B. Nelson, M. J. Marton, R. Stoughton, M. R. Meyer, H. A. Bennett, Y. D. He, H. Dai, W. L. Walker, T. R. Hughes, M. Tyers, C. Boone, and S. H. Friend. 2000. Signaling and circuitry of multiple MAP kinase pathways revealed by a matrix of global gene expression profiles. Science 287:873-880. [DOI] [PubMed] [Google Scholar]
- 41.Sarkaria, J. N., E. C. Busby, R. S. Tibbetts, P. Roos, Y. Taya, L. M. Karnitz, and R. T. Abraham. 1999. Inhibition of ATM and ATR kinase activities by the radiosensing agent, caffeine. Cancer Res. 59:4375-4382. [PubMed] [Google Scholar]
- 42.Sharrocks, A. D., S.-H. Yang, and A. Galanis. 2000. Docking domains and substrate-specificity determination for MAP kinases. Trends Biochem. Sci. 25:448-453. [DOI] [PubMed] [Google Scholar]
- 43.Sidorova, J. M., and L. L. Breeden. 1993. Analysis of the SWI4/SWI6 protein complex, which directs G1/S-specific transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 13:1069-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siliciano, P. G., and K. Tatchell. 1984. Transcription and regulatory signals at the mating type locus in yeast. Cell 37:969-978. [DOI] [PubMed] [Google Scholar]
- 46.Smolka, M. B., C. P. Albuquerque, S.-H. Chen, and H. Zhou. 2007. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc. Natl. Acad. Sci. USA 104:10364-10369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taylor, I. A., P. B. McIntosh, P. Pala, M. K. Treiber, S. Howell, A. N. Lane, and S. J. Smerdon. 2000. Characterization of the DNA-binding domains from the yeast cell-cycle transcription factors Mbp1 and Swi4. Biochemistry 39:3943-3954. [DOI] [PubMed] [Google Scholar]
- 48.Truman, A. W., S. H. Millson, J. M. Nuttall, V. King, M. Mollapour, C. Prodromou, L. H. Pearl, and P. W. Piper. 2006. Expressed in the yeast Saccharomyces cerevisiae, human ERK5 is a client of the Hsp90 chaperone that complements loss of the Slt2 (Mpk1) cell integrity stress-activated protein kinase. Eukaryot. Cell 5:1914-1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Truman, A. W., S. H. Millson, J. M. Nuttall, M. Mollapour, C. Prodromou, and P. Piper. 2007. In the yeast heat shock response, Hsf1-directed induction of Hsp90 facilitates the activation of the Slt2 (Mpk1) mitogen-activated protein kinase required for cell integrity. Eukaryot. Cell 6:744-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vay, H. A., B. Philip, and D. E. Levin. 2004. Mutational analysis of the cytoplasmic domain of the Wsc1 cell wall stress sensor. Mol. Microbiol. 150:3281-3288. [DOI] [PubMed] [Google Scholar]
- 51.Vaze, M. B., A. Pallicioli, S. E. Lee, G. Ira, G. Liberi, A. Arbel-Eden, M. Foiani, and J. E. Haber. 2002. Recovery from checkpoint-mediated arrest after repair of a double-strand break requires Srs2 helicase. Mol. Cell 10:373-385. [DOI] [PubMed] [Google Scholar]
- 52.Wang, S.-W., C. Norbury, A. L. Harris, and T. Toda. 1999. Caffeine can override the S-M checkpoint in fission yeast. J. Cell Sci. 112:927-937. [DOI] [PubMed] [Google Scholar]
- 53.Wang, W., and B. A. Malcolm. 1999. Two-stage PCR protocol allowing introduction of multiple mutations, deletions and insertions using QuikChange site-directed mutagenesis. BioTechniques 26:680-682. [DOI] [PubMed] [Google Scholar]
- 54.Watanabe, Y., K. Irie, and K. Matsumoto. 1995. Yeast RLM1 encodes a serum response factor-like protein that may function downstream of the Mpk1 (Slt2) mitogen-activated protein kinase pathway. Mol. Cell. Biol. 15:5740-5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watanabe, Y., G. Takaesu, M. Hagiwara, K. Irie, and K. Matsumoto. 1997. Characterization of a serum response factor-like protein in Saccharomyces cerevisiae, Rlm1, which has transcriptional activity regulated by the Mpk1 (Slt2) mitogen-activated protein kinase pathway. Mol. Cell. Biol. 17:2615-2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wurgler-Murphy, S. M., T. Maeda, E. A. Witten, and H. Saito. 1997. Regulation of the Saccharomyces cerevisiae HOG1 mitogen-activated protein kinase by the PTP2 and PTP3 protein tyrosine phosphatases. Mol. Cell. Biol. 17:1289-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yan, C., H. Luo, J. D. Lee, J. Abe, and B. C. Berk. 2001. Molecular cloning of mouse ERK5/BMK1 splice variants and characterization of ERK5 functional domains. J. Biol. Chem. 276:10870-10878. [DOI] [PubMed] [Google Scholar]
- 58.Zarzov, P., C. Mazzoni, and C. Mann. 1996. The SLT2 (MPK1) MAP kinase is activated during periods of polarized cell growth in yeast. EMBO J. 15:83-91. [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang, J., B. Zhou, C.-F. Zheng, and Z.-Y. Zhang. 2003. A bipartite mechanism for ERK2 recognition by its cognate regulators and substrates. J. Biol. Chem. 278:29901-29912. [DOI] [PubMed] [Google Scholar]
- 60.Zhao, C., U. S. Jung, P. Garrett-Engele, T. Roe, M. S. Cyert, and D. E. Levin. 1998. Temperature-induced expression of yeast FKS2 is under the dual control of protein kinase C and calcineurin. Mol. Cell. Biol. 18:1013-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao, X., E. G. Muller, and R. Rothstein. 1998. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell 2:329-340. [DOI] [PubMed] [Google Scholar]
- 62.Zhou, T., L. Sun, J. Humphreys, and E. J. Goldsmith. 2006. Docking interactions induce exposure of activation loop in the MAP kinase ERK2. Structure 14:1011-1019. [DOI] [PubMed] [Google Scholar]