Abstract
It has been well established that amino acid availability can control gene expression. Previous studies have shown that amino acid depletion induces transcription of the ATF3 (activation transcription factor 3) gene through an amino acid response element (AARE) located in its promoter. This event requires phosphorylation of activating transcription factor 2 (ATF2), a constitutive AARE-bound factor. To identify the signaling cascade leading to phosphorylation of ATF2 in response to amino acid starvation, we used an individual gene knockdown approach by small interfering RNA transfection. We identified the mitogen-activated protein kinase (MAPK) module MEKK1/MKK7/JNK2 as the pathway responsible for ATF2 phosphorylation on the threonine 69 (Thr69) and Thr71 residues. Then, we progressed backwards up the signal transduction pathway and showed that the GTPase Rac1/Cdc42 and the protein Gα12 control the MAPK module, ATF2 phosphorylation, and AARE-dependent transcription. Taken together, our data reveal a new signaling pathway activated by amino acid starvation leading to ATF2 phosphorylation and subsequently positively affecting the transcription of amino acid-regulated genes.
In mammals, amino acids exhibit two important characteristics: first, nine amino acids are essential in healthy adult humans, and second, there is no proper storage of amino acids, which means that essential amino acids must be obtained from the diet. Consequently, amino acid homeostasis may be altered in response to malnutrition (4, 40, 59) and also by various forms of pathology leading to a negative nitrogen balance (chronic pathology, AIDS, and cancer, etc.) (41, 86, 89). Very often, in one of these situations, the availability of one or several essential amino acids is dramatically affected. Consequently, individuals have to adjust several physiological functions involved in the defense/adaptation response to amino acid limitation. In such a situation, it has been shown that amino acids by themselves are involved in a variety of regulatory processes (26, 47, 67). For all these reasons, the role of amino acids as signaling molecules that regulate gene expression and physiological functions has received considerable attention in recent years. However, the molecular mechanisms involved in this process are not completely understood for mammals at present (44, 47, 52).
Up to now, two ubiquitous amino acid-sensing processes have been described to occur in mammals. They involve protein kinases mTORC1, activated by amino acid supplementation, and GCN2, activated by amino acid starvation. These two kinases play a major role in the control of protein synthesis (71), transcription, and mRNA turnover of specific genes (24, 46, 69). Although the mechanisms involved in the regulation of gene expression by the mTORC1 pathway are not yet identified, the role of the GCN2 pathway has been widely studied using the experimental model of limitation with one essential amino acid. This model has been used to characterize the cellular transcriptional response to nutritional stress. At a molecular level, most of the results have been obtained by studying the transcriptional regulation of the activating transcription factor 3 (ATF3), C/EBP homologous protein (CHOP), and asparagine synthetase (ASNS) genes. An amino acid response element (AARE) was identified in the promoters of these genes and can confer amino acid responsiveness to a heterologous promoter (5, 13, 68). Subsequently, functional sequences that share a high level of similarity with the described AARE were identified in other amino acid-regulated genes, such as the system A amino acid transporter (SNAT2) (66) and arginine/lysine transporter cat-1 (cat-1) genes (55). The sequences of these AAREs are related to C/EBP and ATF/cis-acting replication element binding sites. Most of these AAREs were described as binding a combination of several transcription factors and regulatory proteins to precisely modulate the rate of transcription (ATF4, ATF2, C/EBPβ, ATF3, TRB3, PCAF, and JDP2) (for a review, see reference 47). Of these regulatory proteins, two transcription factors, ATF4 and ATF2, have been shown to play an essential role in the amino acid regulation of a large number of genes (2, 12).
Changes in ATF4 protein level are induced in response to amino acid starvation by a pathway that involves activation of the GCN2 kinase by the accumulation of free tRNA, which in turn catalyzes phosphorylation of the alpha subunit of eukaryotic initiation factor 2 (eIF-2α). An immediate consequence of eIF-2α phosphorylation is a general decrease in protein synthesis and enhanced translation of ATF4, due to the presence of upstream open reading frames in the 5′ untranslated region of its mRNA (56, 81). Subsequently, ATF4 induces the expression of a large number of specific target genes (2, 31, 75).
ATF2 is the other transcription factor essential for amino acid control of gene transcription. ATF2 functions either as a homodimer or as a heterodimer with other members of the ATF family, as well as other bZIP proteins, to bind to specific DNA sequences and activate gene expression (8, 82). One major role of ATF2 is to regulate the response of cells to stress signals (30, 34). The transactivation capacity of the N-terminal domain of this transcription factor can be enhanced through phosphorylation of two N-terminal threonine residues, Thr69 and Thr71 (human ATF2). Phosphorylation at these residues has been shown to be induced in response to several stress signals in a mitogen-activated protein kinase (MAPK)-dependent pathway (3, 53, 60, 65).
In the context of gene regulation by amino acid starvation, we previously studied the role of ATF2 by using leucine-regulated transcription of the CHOP and ATF3 genes as a working model (2, 12). It was shown that (i) in cells devoid of ATF2, the induction of CHOP or ATF3 transcription upon amino acid starvation is lost; (ii) ATF2 binds in vivo to the AARE under starved and unstarved conditions; (iii) ATF2 is phosphorylated on Thr71 in response to amino acid starvation; and then, (iv) ATF2 promotes modification of the chromatin structure to enhance transcription (12). Whereas the molecular events leading to ATF4 regulation have been well identified, the signaling pathway responsible for ATF2 phosphorylation in response to amino acid starvation is not known.
This study was designed to identify the signaling pathway leading to phosphorylation of ATF2 in response to amino acid starvation. Using individual gene knockdown experiments, we demonstrated that c-Jun NH2-terminal kinase 2 (JNK2) is essential for regulation of ATF2 phosphorylation. Then, we progressed backwards up the signal transduction pathway to identify the different steps required. Taken together, our data reveal a new signaling pathway activated by amino acid starvation and leading to ATF2 phosphorylation.
MATERIALS AND METHODS
Cell culture and treatment conditions.
HeLa cells, HEK293 cells, and mouse embryonic fibroblasts (MEF) were cultured at 37°C in Dulbecco's modified Eagle's medium (DMEM)-Ham's F-12 medium (Sigma) containing 10% fetal bovine serum. When indicated, DMEM-Ham's F-12 medium lacking leucine was used. In all experiments involving amino acid starvation, 10% dialyzed fetal calf serum (A15-107) (PAA laboratories) was used.
Antibodies.
The following antibodies were purchased from Santa Cruz Biotechnology: those against ATF2 (sc-187), ATF4 (sc-200), JNK1 (sc-571), MEKK1 (sc-252), Gα12 (sc-409), and Gα13 (sc-410). Antibodies against ATF2 phosphorylated on Thr71 (P-Thr71 ATF2) (1268-1), P-Tyr185 JNK (2155-1), JNK2 (2037-1), MKK4 (1650-1), MKK7 (1949-1), MEKK2 (1662-1), and MEKK3 (1672-1) were obtained from Epitomics. The anti-P-Thr69+Thr71 ATF2 (9225), anti-P-Thr389 S6K (9205), and anti-P-Ser408 MEF2A (9737) antibodies were from Cell Signaling Technology. The Rac1 (610650), P-Thr202+Tyr204 extracellular signal-regulated kinase (ERK) (612358), and Cdc42 (610928) antibodies were provided by BD Biosciences.
Immunoblot analysis.
Cells were lysed in a buffer containing 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% Triton X-100, 0.1% sodium dodecyl sulfate, 50 mM NaF, 2 mM Na3VO4, 100 nM okadaic acid, 25 mM β-glycerophosphate, 1 mM phenylmethylsulfonyl fluoride, and a protease inhibitor cocktail from Sigma. Proteins (40 μg) were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto a Hybond-P polyvinylidene difluoride membrane (Amersham Biosciences). Membranes were incubated in blocking solution (5% nonfat milk powder in Tris-buffered saline, 0.1% Tween 20) for 1 h at room temperature. The blots were then incubated with primary antibody in blocking solution overnight at 4°C. Antibodies were diluted in accordance with the manufacturer's instructions. The blots were washed three times in Tris-buffered saline, 0.1% Tween 20 and incubated with horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (1:5,000) (Santa Cruz, CA) in blocking buffer for 1 h at room temperature. After three washes, the blots were developed using an enhanced chemiluminescence detection system (Amersham Biosciences).
Oligonucleotides.
Oligonucleotides were from Sigma-Aldrich. When double-stranded oligonucleotides were required, equal numbers of moles of complementary strands were heated to 90°C for 1 min and annealed by slow cooling to room temperature.
Plasmid construction and reagents.
TATA-Tk-Luc, containing the minimum herpes simplex virus promoter for thymidine kinase (positions −40 to +50), was generated as previously described (13). The 2xATF3AARE-Tk-Luc plasmid was constructed by inserting SacI-XhoI double-stranded oligonucleotides containing two copies of the ATF3 AARE sequence into the Tk-Luc plasmid. The 2xCHOPAARE-Tk-Luc plasmid was constructed as described in reference 13. The MAPK inhibitors SB20350 (tlrl-sb20) and U0126 (tlrl-u0126) were obtained from InvivoGen. The JNK inhibitor 8: JI8 (420135) and rapamycin (553211) were from Calbiochem. The JNK inhibitor SP600125 (S5567-10MG) was from Sigma-Aldrich.
siRNA transfection.
Small interfering RNA (siRNA) corresponding to MKK4 (SI02655079), MKK7 (SI02660588), MEKK1 (SI02659965), MEKK2 (SI02224166), MEKK3 (SI00605619), Rac1 (SI02655051), Cdc42 (SI02757328), Gα12 (SI00096565) and Gα13 (SI00089761) mRNA and to a control (1027280) were from Qiagen. siRNA against JNK1 (12936-42) and JNK2 (12936-44) were purchased from Invitrogen. The JNK1/2 siRNA primers (5′-AAAGAAUGUCCUACCUUCUdTdT-3′ and 5′-AGAAGGUAGGACAUUCUUUdTdT-3′) target a common sequence in both JNK1 and JNK2 mRNA and was synthesized by Qiagen. One day before transfection with siRNA, HeLa cells were plated in six-well plates at 25% confluence. Then, 60 pmol of siRNA was transfected into the cells by using the calcium phosphate precipitation method as previously described (45). Seventy-two hours after transfection, the amino acid starvation experiment was performed and the expression level of target mRNA was analyzed by Western blotting.
Transient transfection and Luc assay.
Cells were plated in 12-well dishes and transfected by the calcium phosphate coprecipitation method. Twenty-four hours after siRNA transfection (30 pmol), 1 microgram of luciferase (Luc) plasmid (2xATF3AARE-Tk-Luc or 2xCHOPAARE-Tk-Luc) was transfected into the cells, along with 0.1 μg of pCMV-ßGal as an internal control. This plasmid carries the bacterial ß-galactosidase gene fused to the human cytomegalovirus immediate-early enhancer/promoter region. Relative Luc activity was given as the ratio of relative Luc units/relative ß-galactosidase units. All values are the means calculated from the results of at least three independent experiments performed in triplicate.
Analysis of gene expression by use of real-time reverse transcription-PCR (RT-PCR).
Total RNA was prepared using an RNeasy minikit (Qiagen) and treated with DNase I, amplification grade (Invitrogen), prior to cDNA synthesis. RNA integrity was electrophoretically verified by ethidium bromide staining. RNA (0.5 mg) was reverse transcribed with 100 U of Superscript II plus RNase H-reverse transcriptase (Invitrogen), using 100 μM random hexamer primers (Amersham Biosciences), in accordance with the manufacturer's instructions. We used the following primers: those targeting mouse ATF3 (forward, 5′-CGCCATCCAGAATAAACACC-3′; reverse, 5′-GCAGGCACTCTGTCTTCTCC-3′), human ATF3 (forward, 5′-GCCATTGGAGAGCTGTCTTC-3′; reverse, 5′-GGGCCATCTGGAACATAAGA-3′), human ASNS (forward, 5′-ATCACTGTCGGGATGTACCC-3′; reverse, 5′-CTTCAACAGAGTGGCAGCAA-3′), human β-actin (forward, 5′-TCCCTGGAGAAGAGCTACGA-3′; reverse, 5′-AGCACTGTGTTGGCGTACAG-3′), and mouse β-actin (forward, 5′-TACAGCTTCACCACCACAGC-3′; reverse, 5′-AAGGAAGGCTGGAAAAGAGC-3′). Real-time quantitative PCR was carried out using a LightCycler system (Roche Applied Science) as described previously (2). Relative results were displayed as relative levels of ATF3 per ß-actin. Each experiment was repeated at least four times.
ChIP analysis.
Chromatin immunoprecipitation (ChIP) analysis was performed as previously described (12). The following antibodies were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA): those against ATF2 (sc-187) and ATF4 (sc-200). Anti P-thr71 ATF2 (9221) was obtained from Cell Signaling Technology (Beverly, MA). The primers used to amplify the mouse ATF3 AARE sequence were 5′-GGTCTCCACCCACCTTTTG-3′ and 5′-CTCGCTGAGTGAGACTGTGG-3′. The results are expressed as the percentage of antibody binding versus the amount of PCR product obtained using a standardized aliquot of input chromatin. Sample results are the means from at least three independent immunoprecipitations.
RESULTS
ATF2 is phosphorylated in response to amino acid starvation.
We first determined the role of ATF2 and ATF4 in the amino acid regulation of ATF3. This gene was chosen as a working model since previous studies have shown that (i) ATF3 is dramatically induced upon amino acid starvation and that (ii) it contains a genomic cis-acting AARE (ATF3 AARE) that has been shown to be involved in the binding of several transcription factors (12, 68). To determine whether ATF2 or ATF4 is required to mediate ATF3 mRNA induction during a short period of leucine starvation, we used MEF deficient in ATF2 or ATF4. Figure 1A shows that ATF3 expression is dramatically enhanced in response to a 4-hour period of leucine starvation. In contrast, lack of either ATF4 or ATF2 resulted in a loss of ATF3 mRNA inducibility.
FIG. 1.
Role of ATF2 in the transcriptional regulation of ATF3 in response to amino acid starvation. (A) ATF2−/−, ATF4−/−, and wild-type (WT) MEF were incubated for 4 h in either control or leucine-free (−Leu) medium and then harvested. Total RNA was extracted and analyzed by quantitative RT-PCR for ATF3 mRNA content as described in Materials and Methods. Each point represents the mean value of results from three independent experiments. We obtained the same results using the wild-type counterparts of the ATF2−/− and the ATF4−/− MEF (not shown). (B) ChIP analysis was performed using antibodies specific for ATF4, ATF2, and phosphorylated ATF2 (Thr71). Quantitative RT-PCR was performed with immunoprecipitated DNA and a dilution of input DNA samples, using primers to amplify the AARE region of the ATF3 promoter. Data were plotted as percentages of PCR product from immunoprecipitated DNA versus input DNA. Each point represents the mean value of results from three independent experiments, and the error bars represent the standard errors of the means (* indicates statistical significance at P values of <0.05). IgG, immunoglobulin G. (C) Cells were incubated in either control or leucine-free medium and harvested after the indicated incubation times. Western blot analysis of phospho-ATF2 (Thr71 and Thr69+Thr71) and total ATF2 was performed with nuclear extract as described in Materials and Methods. (D) Cells were incubated for 4 h either in control medium (Ctrl) or in a medium devoid of one individual amino acid (leucine, lysine, methionine, or glutamine), and then analysis of phospho-ATF2 (Thr69+Thr71), ATF2, and ATF4 was performed.
To investigate the binding of these factors on the ATF3 AARE, cells were incubated in control or leucine-free medium for 4 h and ChIP assays were performed with primer sets covering the ATF3 AARE (Fig. 1B). For wild-type cells, ChIP experiments showed an increase in ATF4 binding to the AARE following leucine deprivation whereas binding of ATF2 remained constitutive. ChIP analysis using an antibody that specifically recognizes ATF2 phosphorylated on threonine 71 revealed that binding of the phosphorylated form of ATF2 was significantly increased after removal of leucine. In ATF2-deficient cells, neither ATF2 nor phosphorylated ATF2 bound to the ATF3 AARE was detected. However, the increase in ATF4 binding remained. In cells lacking ATF4, the levels of bound ATF2 and the increase of bound phospho-ATF2 were similar to those in the wild-type cells. Taken together, these results demonstrate that ATF4 binding and ATF2 phosphorylation are two independent events that are associated with ATF3 induction upon amino acid starvation.
In the next experiment, we investigated the kinetics of ATF2 phosphorylation on Thr71 and on a combination of Thr69 and Thr71 (Thr69+Thr71) in response to leucine starvation. The phosphorylated form of ATF2 was estimated using two specific antibodies recognizing either phosphothreonine 71 or double phosphorylation on Thr69 and Thr71. Figure 1C shows that leucine deprivation resulted in a rapid and sustained phosphorylation of ATF2 on both Thr69 and Thr71 that appeared at least 30 min after the beginning of leucine starvation and persisted for up to 4 h. Furthermore, we checked the effects on ATF2 phosphorylation of starvation for other individual amino acids. Figure 1D shows that starvation for essential amino acids such as lysine and methionine enhanced ATF2 phosphorylation but that glutamine starvation (glutamine being a nonessential amino acid) had no significant effects. This experiment also showed that ATF4 expression is induced by lysine or methionine starvation, confirming that the GCN2/ATF4 pathway is activated following essential-amino-acid starvation.
GCN2 and mTORC1 pathways are not required for ATF2 phosphorylation in response to amino acid starvation.
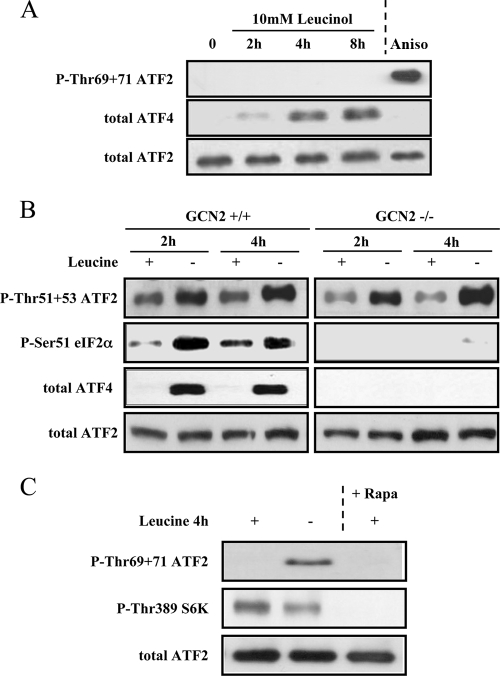
The simplest hypothesis for explaining phosphorylation of ATF2 by amino acid starvation would be the involvement of one of the two ubiquitous pathways previously described. The first one involves the protein kinase GCN2, activated by the accumulation of free tRNA resulting from the lack of amino acid. We checked whether uncharged tRNAleu accumulation resulting from leucine starvation is involved in phosphorylation of ATF2 on Thr69+Thr71. We treated the cells for 2, 4, and 8 h with 10 mM of leucinol, an alcohol derivative of leucine. Leucinol inhibits leucyl-tRNA-synthetase and then increases the uncharged tRNAleu content of the cells. Figure 2A shows that leucinol treatment did not affect the phosphorylation of ATF2 but induced ATF4 expression. This result demonstrates that accumulation of uncharged tRNAleu alone is not able to induce ATF2 phosphorylation. We also measured the phosphorylation of ATF2 in response to leucine starvation in GCN2−/− and GCN2+/+ MEF cells (Fig. 2B). Measurement of eIF-2α phosphorylation and ATF4 expression indicates that the GCN2 pathway is not activated in−/− cells. Our results clearly show that ATF2 is phosphorylated upon leucine starvation in both GCN2+/+ and GCN2−/− cells, demonstrating that the GCN2 kinase is not involved in this process.
FIG. 2.
ATF2 phosphorylation upon leucine starvation does not involve the GCN2 and mTORC1 pathways. (A) Cells were incubated with 10 mM leucinol for the indicated times, and then ATF2 phosphorylation and ATF4 expression were analyzed as previously described. A 30-minute incubation with 20 μg/ml anisomycin (Aniso) was used as a positive control for the measurement of ATF2 phosphorylation. (B) GCN2+/+ or GCN2−/− MEF were incubated in either control or leucine-free medium for 2 and 4 h. Cells were then harvested for analysis of ATF4 expression and phosphorylation of ATF2 and eIF-2α. (C) Cells were leucine starved for 4 h or treated with 50 nM rapamycin (Rapa) for 4 h, and then the phosphorylations of ATF2 and S6K were measured.
We next addressed the contribution of mTORC1 inhibition in phosphorylation of ATF2. We treated the cells with rapamycin, a pharmacological inhibitor of mTORC1. Both amino acid starvation and rapamycin treatment are known to inhibit mTORC1 activity. Figure 2C shows that rapamycin treatment did not affect the phosphorylation level of ATF2. The efficiency of rapamycin treatment was checked by measuring the dephosphorylation of S6K1, a direct target of mTORC1. However, it has been shown that rapamycin does not inhibit all the function of mTORC1 (83); therefore, we cannot totally exclude the role of mTORC1 in this process. Taken together, our data show that ATF2 phosphorylation in response to amino acid starvation does not involve the GCN2 pathway and the rapamycin-sensitive component of mTORC1 activity.
JNK2 inhibition abolishes amino acid starvation-induced ATF2 phosphorylation.
Numerous studies have shown that, in response to various stress signals, ATF2 is phosphorylated on Thr69 and Thr71 by the JNK and p38 pathways (11, 30, 80). In response to mitogens, such as insulin, ATF2 is phosphorylated via a two-step mechanism that involves the cooperation of two different kinases (ERK1/2 and p38 or JNK), which phosphorylate ATF2-Thr71 and then ATF2-Thr69, sequentially (3, 65). These data prompted us to analyze the role of the MAPK pathways in amino acid regulation of ATF2 phosphorylation. For that purpose, we used pharmacological inhibitors of these kinases. Figure 3 shows that inhibition of p38 by use of SB203580 (19) and prevention of ERK1/2 phosphorylation by MEK1/2 (MAPK/ERK kinase) inhibitor U0126 (19) had no effect on the induction of ATF2 phosphorylation on Thr69 and Thr71 by 4 h of leucine starvation. The efficiency of these inhibitors was checked by measuring ERK (78) and MEF2A (64, 88) phosphorylation (see Fig. S1 in the supplemental material). In contrast, inhibition of JNK by either JI8 (63, 77) or SP600125 (6) completely cancelled induction of ATF2 phosphorylation in response to leucine starvation. These results suggest that JNK rather than p38 or ERK is responsible for amino acid regulation of ATF2 phosphorylation. It is notable that ATF2 phosphorylations on Thr71 and on Thr69+Thr71 are similarly regulated by leucine starvation in the presence and absence of inhibitor, suggesting that these two residues are phosphorylated by a single protein kinase. Similar conclusions had previously been drawn for ATF2 phosphorylation in response to other stress inducers, such as UV, methyl methane sulfonate, or tumor necrosis factor alpha (65). For these reasons, only the phosphorylation on ATF2-Thr71 will be shown in subsequent studies.
FIG. 3.
Pharmacological inhibition of JNK prevents ATF2 phosphorylation by leucine starvation. Cells were incubated in either control or leucine-free medium for 4 hours in the presence of various MAPK inhibitors (20 μM SB203580 inhibits p38, 50 μM U0126 inhibits MEK1/2 and thus ERK, and 20 μM JI8 and 50 μM SP600125 inhibit JNK). The phosphorylation of ATF2 was then measured. For this experiment, cells were first preincubated for 1 h with MAPK inhibitors or vehicle (dimethyl sulfoxide [DMSO]).
It has been well demonstrated that JNK proteins are regulated by dual phosphorylation of threonine and tyrosine in a TXY motif (21). To determine whether amino acid starvation induces JNK phosphorylation, we measured the phosphorylated form of JNK on Tyr185 proteins upon amino acid starvation by using a specific antibody. Figure 4A shows that phosphorylation of JNK proteins was enhanced by 1 to 4 h of leucine starvation, demonstrating that JNK phosphorylation matches the time course of ATF2 phosphorylation.
FIG. 4.
Silencing of JNK2 by siRNA prevents ATF2 phosphorylation by leucine starvation. (A) Cells were incubated in either control or leucine-free medium and harvested after the indicated incubation times. The phosphorylated forms of JNK (Tyr185) and ATF2 were measured. Each blot was obtained from a representative experiment among three independent experiments. (B) HeLa cells were transfected with control siRNA, JNK1 siRNA, JNK2 siRNA, or siRNA targeting both JNK1 and JNK2, as described in Materials and Methods. At 72 h posttransfection, cells were incubated in either DMEM or leucine-free DMEM for 4 h and then harvested for analysis of ATF2, JNK1, JNK2, ATF4, and the phosphorylated form of ATF2. Each blot was obtained from a representative experiment among three independent experiments.
JNK proteins are encoded by three genes. The jnk1 and jnk2 genes are expressed ubiquitously, whereas the jnk3 gene has a limited pattern of expression, mainly restricted to the brain, heart, and testis (20). To examine which JNK isoform is involved in amino acid regulation of ATF2 phosphorylation, HeLa cells were transfected with siRNA to silence JNK1, JNK2, or both JNK1 and JNK2. We monitored the effectiveness of the siRNA action by immunoblot analysis, using antibodies that recognize JNK1 or JNK2 specifically. Figure 4B shows that siRNA treatment was effective in dramatically reducing the corresponding target expression relative to the level for the control siRNA. These results reveal that knockdown of JNK2 but not JNK1 abolished ATF2 phosphorylation on Thr71 in response to leucine starvation but that ATF4 induction was not affected. We obtained similar results when measuring ATF2 phosphorylation on ATF2 Thr69+Thr71 (data not shown).
A cascade of protein kinases including MKK7 and MEKK1 mediates leucine starvation-induced ATF2 phosphorylation.
We next analyzed which known JNK2 activator(s) (MKK4, MKK7, or both) was involved in response to leucine starvation. Cells were transfected with siRNA targeting MKK4, MKK7, or both, and the experiment was performed as described for Fig. 4B. Figure 5A shows that siRNA treatment (i) was effective in reducing the expression of the corresponding target and (ii) did not prevent ATF4 induction upon leucine starvation. Moreover, it was clear that MKK7 knockdown completely abolished ATF2 phosphorylation in response to leucine starvation. Conversely, in MKK4-silenced cells, leucine-dependent phosphorylation of ATF2 was maintained.
FIG. 5.
Knockdown of MKK7 or MEKK1 inhibits leucine starvation-induced ATF2 phosphorylation. (A) HeLa cells were transfected with either control siRNA, MKK4 siRNA, MKK7 siRNA, or both MKK4 siRNA and MKK7 siRNA. At 72 h posttransfection, cells were incubated in either DMEM or leucine-free DMEM for 4 h. Cells were then harvested for analysis of MKK4, MKK7, ATF4, and the phosphorylated form of ATF2 as previously described. (B) MEKK1, MEKK2, MEKK3, or all three proteins were silenced as previously described. Then, the expression levels of MEKK1, MEKK2, MEKK3, ATF4, and the phosphorylated form of ATF2 were analyzed. (C) Protein extracts from cells transfected with either control, MKK7, or MEKK1 siRNA were analyzed for JNK phosphorylation. Each blot was obtained from a representative experiment among three independent experiments.
MKK7 activity was increased following phosphorylation at Ser and Thr residues within a SKAKT motif in its activation loop by multiple kinases, including MEKK1 to -4, the protein kinase of the mixed-lineage family (MLK1 to -3), apoptosis signal-regulated kinase 1 (ASK1), dual-leucine-zipper-bearing kinase, and transforming growth factor β-activated kinase 1 (TAK1). According to the literature, the properties of certain kinases meant we did not consider them to be potential candidates for activation of MKK7 by amino acid starvation. First, PAK1, dual-leucine-zipper-bearing kinase, MLK1, and MLK2 expressions are restricted to a limited number of tissues and are either not expressed or poorly expressed in HeLa cells (9, 36). Second, several studies showed that MLK2, PAK1, ASK1, TAK1, or MEKK4 activity predominantly phosphorylates MKK4 rather than MKK7 (1, 15, 18, 38, 48, 51). Therefore, we first investigated whether MEKK1, MEKK2, and MEKK3 were involved in the signaling pathway linking leucine deprivation to ATF2 phosphorylation. Cells were transfected with specific siRNA to silence MEKK1, MEKK2, MEKK3, or all three proteins. Figure 5B shows that MEKK1 knockdown abolishes ATF2 phosphorylation resulting from leucine starvation while the GCN2/ATF4 pathway is not affected. Conversely, MEKK2 or MEKK3 silencing had no effect. Finally, we next confirm the role of JNK in this amino acid-regulated pathway. Figure 5C shows that JNK phosphorylation in response to leucine starvation is prevented by MKK7 and MEKK1 silencing.
Taken together, our results demonstrate that amino acid starvation triggered a cascade of protein kinases that includes MKK7 and MEKK1 and mediated JNK2 activation and subsequent ATF2 phosphorylation.
Leucine starvation-induced ATF2 phosphorylation requires both Cdc42 and Rac1.
Several lines of evidence indicate that the Rho family of GTPases mediates activation of the JNK pathway in response to various stimuli (10, 20). Particularly, it has been shown that Rac1 and Cdc42 play a critical role in controlling the JNK signaling pathway (17). In addition, these small GTPases appear to transmit signals to the JNK module via MEKK1 (for a review, see reference 29). Taken together, these data prompted us to investigate the role of Cdc42 and Rac1 in our model. Cells were transfected with specific siRNA targeting either Cdc42 or Rac1 or both Cdc42 and Rac1. We first checked that each siRNA had silenced its target gene (Fig. 6). Our results show that silencing of Rac1 or Cdc42 dramatically decreased but did not abolish ATF2 phosphorylation upon leucine starvation. However, knockdown of both Cdc42 and Rac1 completely abolished the response to leucine deprivation with respect to ATF2 phosphorylation while ATF4 induction was not affected. From these experiments, we can conclude that (i) these two GTPases are required to trigger full phosphorylation of ATF2 upon leucine starvation and that (ii) there is partial functional redundancy between Cdc42 and Rac1.
FIG. 6.
Silencing of Cdc42 and Rac1 prevents ATF2 phosphorylation in response to leucine starvation. Rac1, Cdc42, or both were silenced as previously described. The phosphorylated form of ATF2 and the expression of Rac1, Cdc42, and ATF4 were measured. Each blot was obtained from a representative experiment among four independent experiments.
Gα12 knockdown prevents leucine starvation-induced ATF2 phosphorylation.
In mammalian cells, members of the Rho GTPase family are positioned at the center of a complex signaling network. Diverse upstream signals can regulate Rho GTPase through activation of various receptors, such as G protein-coupled receptor (GPCR) or receptor tyrosine kinase (73, 74). Since amino acids have already been shown to activate GPCR (16), we favor the hypothesis that a GPCR could be involved in the sensing of leucine deficiency. So we focused our investigation on the role of G proteins. Among the G protein alpha or beta-gamma subunits, Gα12 and Gα13 have been shown to be able to activate the JNK pathway (29, 76). For Fig. 7A, we investigated the role of Gα12 and Gα13 in the regulation of ATF2 phosphorylation by leucine starvation. We silenced Gα12, Gα13, or both Gα12 and Gα13 by transfecting appropriate siRNA. First, we observed that Gα12 protein silencing decreased the basal level of ATF4 protein expression. Further work beyond the scope of this study will be necessary to identify the role of Gα12 in the regulation of ATF4 expression. More importantly, Fig. 7A shows that knockdown of Gα12 prevents ATF2 phosphorylation but that knockdown of Gα13 has no effect. Furthermore, data from literature showed that the expression of an activated mutant of Gα12 (Gα12 QL) leads to phosphorylation and activation of JNKs specifically through the stimulation of the JNK-specific upstream kinase MKK7 (23, 70). In Fig. 7B, we show that transfection of Gα12 QL in HEK293 cells activates the phosphorylation of ATF2 and JNK. Taken together, these results show that the Gα12 protein is involved in this pathway, suggesting that a GPCR could be at the origin of the sensing of leucine starvation by cells.
FIG. 7.
Involvement of the Gα12 protein in leucine starvation-induced ATF2 phosphorylation. HeLa cells were transfected with either control siRNA, Gα12 siRNA, Gα13 siRNA, or both Gα12 siRNA and Gα13 siRNA. At 72 h posttransfection, cells were incubated in either DMEM or leucine-free DMEM for 4 h. Cells were then harvested for analysis of Gα12, Gα13, ATF4, and the phosphorylated form of ATF2 as previously described. Each blot was obtained from a representative experiment among three independent experiments. (B) HEK293 cells were transfected with an activated mutant of Gα12 (Gα12 QL) or empty vector (pcDNA3). At 48 h posttransfection, cells were lysed, then protein was harvested for analysis of Gα12, and the phosphorylated forms of ATF2 and JNK were measured as previously described.
Inhibition of the pathway leading to ATF2 phosphorylation affects the regulation of ATF3 expression in response to amino acid starvation.
To assess the role of the JNK pathway in amino acid regulation of gene expression, we first measured the effect of leucine starvation on ATF3 mRNA content in the context of JNK2 or JNK1 silencing. The experiment was performed as described in the legend to Fig. 4. Our results (Fig. 8A) show that JNK2-siRNA transfection decreased ATF3 mRNA induction significantly by leucine starvation but that JNK1-siRNA and control siRNA had no effect. It was previously shown that ASNS is transcriptionally regulated by leucine starvation through an AARE that does not require ATF2 binding (this point will be evoked in Discussion) (12). As negative control, we show that the amino acid regulation of ASNS is not affected by JNK2-siRNA treatment. Second, we focused our investigation on the impact of JNK1 or JNK2 silencing on AARE-dependent transcription. Cells were transiently transfected with one siRNA together with a vector encoding Luc driven by two copies of the ATF3 AARE. Figure 8B shows that the increase of the ATF3 AARE-dependent transcription upon leucine starvation was affected by the JNK2-siRNA but not by the JNK1 or control siRNA. Our previous article (12) demonstrates that, like ATF3, the amino acid-regulated transcription of the CHOP gene requires the expression of ATF2. Figure 8B shows that the CHOP AARE-dependent transcription is also affected by transfection of JNK2-siRNA, demonstrating that the necessity of ATF2 phosphorylation is not restricted to the regulation of ATF3 expression.
FIG. 8.
Role of JNK2 and Rac1/Cdc42 in the transcriptional regulation of ATF3 in response to amino acid starvation. (A) HeLa cells were transfected with control siRNA, JNK1 siRNA, or JNK2 siRNA and then incubated in a control medium or leucine-free medium (−Leu) for 4 h as described for Fig. 4A. Total RNA was then extracted and analyzed by real-time RT-PCR for ATF3 or ASNS mRNA content. (B) Cells were transfected with the appropriate siRNA, together with a reporter plasmid containing two copies of the ATF3 or CHOP AARE inserted 5′ to the thymidine kinase promoter driving the Luc gene (2xAARE-Tk-Luc). A plasmid encoding β-galactosidase, driven by a cytomegalovirus promoter, was cotransfected to normalize transfection (see Materials and Methods). At 48 h posttransfection, cells were incubated in either DMEM or leucine-free DMEM for 16 h and then harvested to measure Luc activity. (C) HeLa cells were transfected with control siRNA or with Rac1 siRNA and Cdc42 siRNA. Cells were then treated and analyzed as described for panel B. All values are means calculated from the results for at least three independent experiments performed in triplicate. Data are expressed as mean ± standard error of the mean. Statistical analyses were performed using a Student test. Asterisks indicate that −Leu/siRNA-treated cells had statistical significance at P values of <0.05 for comparison with the −Leu/control siRNA-treated cells.
Data from Fig. 4B and 5A show that silencing JNK1 or MKK4 leads to a slight decrease in induction of ATF2 phosphorylation upon leucine starvation; however, it does not affect AARE-dependent transcription and ATF3 expression (Fig. 8) (data not shown for MKK4 silencing). The latter observation shows that the signaling pathway connecting amino acid starvation to phosphorylation of ATF2 protein bound on the AARE involves MKK7 rather than MKK4 and JNK2 rather than JNK1.
Finally, we investigated whether inhibition of one of the upstream steps of the signaling pathway could affect the response to leucine starvation. Since Gα12 protein silencing decreases the basal level of ATF4 protein, it could influence AARE-dependent transcription (Fig. 7). In consequence, we chose to measure the impact of Rac1-Cdc42 knockdown on regulation of ATF3 expression upon leucine starvation. The experiment was performed as described for Fig. 8A and B. Our results (Fig. 8C) show that silencing of Rac1 together with Cdc42 affects induction of ATF3 mRNA in response to leucine starvation.
These data clearly show that inhibition of ATF2 phosphorylation by siRNA targeting of upstream components of the signaling pathway impairs induction of ATF3 by amino acid starvation. However, Fig. 1 and previous results (12) show that ATF2 knockout almost abolishes the regulation of ATF3 upon amino acid deprivation. The most plausible hypothesis by which to explain these data could be that inhibition of the Gα12/JNK pathway by siRNA does not totally inhibit phosphorylation of the ATF2 molecules bound on the AARE.
Taken together, these results suggest that a signaling pathway involving the Gα12 protein, Rac1/Cdc42, and the JNK module is necessary to fully activate the AARE-dependent transcription in response to amino acid starvation.
DISCUSSION
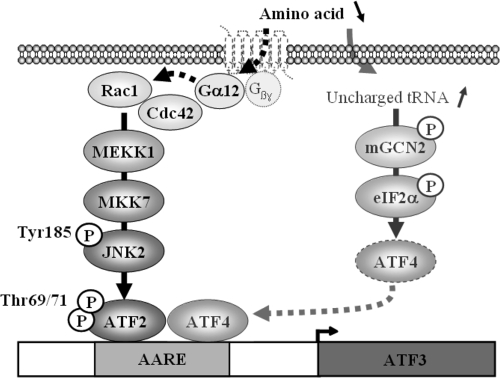
Cells respond to the stress of amino acid starvation by activating a gene expression program that either protects them from stress or leads to apoptosis (42, 57, 58). The regulation of ATF3 gene expression represents a mechanistic model for investigating how changes in amino acid availability regulate transcription. ATF3 is a member of the ATF/CREB transcription factor family, which also includes members of the C/EBP family and Jun/Fos. Normally expressed at low levels in cells, ATF3 expression is rapidly induced in response to various stress signals and is likely to be involved in the control of a number of stress-related responses, including nutrient stress (33, 43). For example, ATF3 is expressed in the islets of mice that have developed diabetes and in human patients with type 1 or type 2 diabetes. It was also shown that ATF3 is a regulator of stress-induced pancreatic β-cell apoptosis (32). The literature clearly shows that two events are required for ATF3 induction in response to amino acid starvation: ATF4 induction and ATF2 phosphorylation. The data described in our study include several novel observations concerning the signaling pathway by which ATF2 is phosphorylated upon amino acid starvation and then activates gene transcription. We demonstrated that a Gα12 protein-, Rac1/Cdc42-, MEKK1-, MKK7-, and JNK2-dependent pathway is essential for regulation of ATF2 phosphorylation and then ATF3 induction. The results of this study, together with earlier data, suggest a model where two independent pathways are involved for the activation of AARE-dependent transcription by amino acid availability (Fig. 9). One pathway leads to the induction of ATF4, and the other one leads to the phosphorylation of ATF2.
FIG. 9.
Model of amino acid starvation-induced AARE-dependent transcription in human cells. In response to amino acid starvation, two pathways are activated. The first one is activated by uncharged tRNA and leads to ATF4 induction. The second one involves the Gα12 protein and leads to ATF2 phosphorylation.
It was previously shown that the AARE-regulated genes (dependent on ATF4 expression) can be divided into two classes, according to the degree of their ATF2 dependence in eukaryotic cells (2, 68). The first class is composed of genes that are totally ATF2 dependent, such as ATF3, CHOP, SARS, or 4EBP1. In response to amino acid starvation, phosphorylation of ATF2, subsequent stimulation of the associated histone acetyltransferase activity, and modification of the chromatin structure are a general mechanism involved in transcriptional regulation of the ATF2-dependent genes (12). The second class, consisting of genes such as ASNS and YARS, is partially ATF2 dependent since the lack of ATF2 decreases but does not abolish amino acid regulation. For these genes, it appears that the increase in histone acetylation resulting from amino acid starvation involves an ATF2-independent histone acetyltransferase mechanism (14). Such differences in the mechanism of histone acetylation would permit flexibility between amino acid-regulated genes with regard to the rapidity and magnitude of the transcriptional response despite the same initial signal.
ATF2 phosphorylation results from the activation of a specific signaling cascade by amino acid starvation.
Among the upper components of the signaling pathway, we identified the Gα12 protein and two membrane-associated GTPases, Cdc42 and Rac1. It is notable that knockdown of both Cdc42 and Rac1 is necessary to completely abolish the response to leucine deprivation. Similar data have been reported for other models (17, 62). For example, epidermal growth factor activates both Rac1 and Cdc42 in a synergistic manner to regulate the downstream pathway (50). The observation that multiple GTPases contribute to activation of the JNK module either indicates a simple redundancy or suggests a specific role for each GTPase, depending on the different cellular contexts.
Once activated, Rac1 and Cdc42 can interact with numerous downstream effectors to regulate a diverse set of cellular functions. Our data show that, in response to amino acid starvation, MEKK1 is the downstream effector of Rac1 and Cdc42 in the pathway leading to ATF2 phosphorylation. According to the literature, the mechanisms by which the interactions between Rac1/Cdc42 and their effectors activate the downstream signal transduction pathway are still unclear. For example, multiple Cdc42 downstream effectors (including MEKK1) (27) have the same interaction motif, with similar affinities for binding to active Cdc42. It has been hypothesized that guanine nucleotide exchange factors are a component of the Rac1- or Cdc42-signaling complex and could direct Rac1/Cdc42 to select the specific downstream effector (35, 39, 74).
Downstream from MEKK1, the signaling between amino acid starvation and ATF2 phosphorylation requires MKK7 and JNK2 kinases. Previous data already mentioned the specificity of a given kinase for a given downstream effector leading to the specificity of a signaling pathway (22). For example, MKK7 activates only JNK, whereas MKK4 activates both JNK and p38 (79, 84). Similarly, the substrate specificities of JNK1 and JNK2 were different. For example, c-Jun was preferentially phosphorylated by JNK1, while ATF2 was preferentially phosphorylated by JNK2 (20). The specificity of the signaling cascade involves the affinity of the kinase for its substrate as well as tissue-specific factors and scaffold proteins (84). For example, it has been shown that JNK-interacting protein 1 (JIP-1) forms a complex with JNK, MKK7, and certain MAPK kinase kinases (25). So the phosphorylation cascade occurs within this protein complex. The factor(s) responsible for the specificity of the pathway at each step has not yet been identified. In particular, the activity of various guanine nucleotide exchange factors such as guanine exchange factor (GEF), GTPase activating protein (GAP), or guanine dissociation inhibitor (GDI) or the role of scaffold proteins (JIP and POSH proteins) in the model of the JNK pathway activation in response to amino acid starvation could be investigated (35, 85, 87).
Recently, it has been shown that, in addition to the JNK pathway, amino acid limitation also activates the kinases MEK and ERK (78). The authors described a complex cross talk between the MEK-ERK signaling pathway and the GCN2/ATF4 pathway. Indeed, activation of the ERK pathway requires eIF-2α phosphorylation and ATF4 induction, which, in turn, is regulated by ERK activity. Taken together, these data demonstrate that, in addition to the well-described amino acid-regulated kinases (GCN2 and mTORC1), several components of the MAPK pathways are involved in the control of particular cellular functions following amino acid deprivation. The regulatory role of MAPK could depend on the tissue or cell type (78).
Amino acid starvation-dependent ATF2 phosphorylation involves an unidentified sensor initiated at the cell membrane.
Our results suggest that the amino acid starvation sensor could involve an unidentified GPCR. GPCRs constitute one of the largest and most diverse protein families in mammalian genomes (49). In a recent analysis, more than 800 GPCRs were listed in the human genome (72). Some members of GPCR class 3 are described as broad-spectrum amino acid sensors that couple changes in extracellular-amino-acid levels to the activation of intracellular signaling pathways (16). They include the heterodimeric taste receptor (T1R1 and T1R3), GPCR6A, and the extracellular calcium sensing receptor (CaR). In yeast, a signaling pathway from the plasma membrane to the nucleus is involved in sensing amino acid availability in the environment. Extracellular amino acids are recognized by a receptor at the plasma membrane, the integral membrane protein Ssy1 (7, 28). In association with two other peripheral membrane proteins, Ptr3 and Ssy5, it activates transcription factors Stp1 and Stp2, which regulate the expression of target genes (54).
In mammals, the nature of the upstream amino acid sensor that could activate the JNK pathway in the context of low amino acid concentrations remains to be identified. Several articles report that binding of particular amino acids to their respective transporter proteins could also serve as an effective means of sensing amino acid availability at the cell surface (37). For example, it was reported that leucine and glutamine transporter activity was necessary for the regulation of mTORC1 by these amino acids (61). However, in this article the authors did not determine the origin of the signal: either the transporter behaves as a receptor (and turns on the pathway) or it allows a high intracellular concentration of amino acid to be maintained, which is sensed by the cell. It is notable that, in addition to leucine starvation, ATF2 phosphorylation is enhanced by starvation for lysine or methionine (Fig. 1D). These amino acids are not transported by a common transporter, suggesting that either (i) the amino acid sensor is not an amino acid carrier or (ii) several amino acid transporters could be connected to a common protein complex that could turn on the downstream signaling pathway. Further work will be necessary to identify the amino acid sensor.
In conclusion, we have identified a new signal transduction pathway activated by amino acid starvation and leading to ATF2 phosphorylation. Our results demonstrate that, in mammalian cells, at least two signaling pathways are activated by amino acid deprivation. (i) The first one senses the intracellular lack of amino acid through the activation of GCN2 and leads to the induction of ATF4. (ii) The other sensor is located at the plasma membrane and leads to the phosphorylation of ATF2 through the activation of the JNK cascade. These two pathways regulate the AARE-dependent transcription and control a set of genes involved in adaptation to amino acid limitation. However, we cannot exclude that these signaling pathways could regulate biological functions other than AARE-dependent transcription. In particular, the role of the JNK module in the adaptation to an amino acid limitation remains to be investigated. Defining the precise cascade of molecular events by which the cellular concentration of an individual amino acid regulates gene expression will be an important contribution to our understanding of metabolite control in mammalian cells. These studies will provide insight into the role of amino acids in the regulation of cellular functions like cell division, protein synthesis, and proteolysis.
Supplementary Material
Acknowledgments
We thank A. Conigrave (University of Sydney, NSW, Australia) for comments and advice. We thank D. Ron (Skirball Institute of Biomolecular Medicine, New York, NY) for giving us the MEF deficient in GCN2 and ATF4, N. Jones (Paterson Institute for Cancer, Manchester, United Kingdom) for the MEF deficient in ATF2, and D. N. Dhanasekaran (Fels Institute for Cancer Research and Molecular Biology, Philadelphia, PA) for the Gα12 QL plasmid.
This work was supported by INRA, CNRS, and Association pour la Recherche sur le Cancer.
Footnotes
Published ahead of print on 12 October 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Almeida, E. A., D. Ilic, Q. Han, C. R. Hauck, F. Jin, H. Kawakatsu, D. D. Schlaepfer, and C. H. Damsky. 2000. Matrix survival signaling: from fibronectin via focal adhesion kinase to c-Jun NH(2)-terminal kinase. J. Cell Biol. 149:741-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Averous, J., A. Bruhat, C. Jousse, V. Carraro, G. Thiel, and P. Fafournoux. 2004. Induction of CHOP expression by amino acid limitation requires both ATF4 expression and ATF2 phosphorylation. J. Biol. Chem. 279:5288-5297. [DOI] [PubMed] [Google Scholar]
- 3.Baan, B., H. van Dam, G. C. van der Zon, J. A. Maassen, and D. M. Ouwens. 2006. The role of c-Jun N-terminal kinase, p38, and extracellular signal-regulated kinase in insulin-induced Thr69 and Thr71 phosphorylation of activating transcription factor 2. Mol. Endocrinol. 20:1786-1795. [DOI] [PubMed] [Google Scholar]
- 4.Baertl, J. M., R. P. Placko, and G. G. Graham. 1974. Serum proteins and plasma free amino acids in severe malnutrition. Am. J. Clin. Nutr. 27:733-742. [DOI] [PubMed] [Google Scholar]
- 5.Barbosa-Tessmann, I. P., C. Chen, C. Zhong, F. Siu, S. M. Schuster, H. S. Nick, and M. S. Kilberg. 2000. Activation of the human asparagine synthetase gene by the amino acid response and the endoplasmic reticulum stress response pathways occurs by common genomic elements. J. Biol. Chem. 275:26976-26985. [DOI] [PubMed] [Google Scholar]
- 6.Bennett, B. L., D. T. Sasaki, B. W. Murray, E. C. O'Leary, S. T. Sakata, W. Xu, J. C. Leisten, A. Motiwala, S. Pierce, Y. Satoh, S. S. Bhagwat, A. M. Manning, and D. W. Anderson. 2001. SP600125, an anthrapyrazolone inhibitor of Jun N-terminal kinase. Proc. Natl. Acad. Sci. USA 98:13681-13686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernard, F., and B. Andre. 2001. Genetic analysis of the signalling pathway activated by external amino acids in Saccharomyces cerevisiae. Mol. Microbiol. 41:489-502. [DOI] [PubMed] [Google Scholar]
- 8.Bhoumik, A., and Z. Ronai. 2008. ATF2: a transcription factor that elicits oncogenic or tumor suppressor activities. Cell Cycle 7:2341-2345. [DOI] [PubMed] [Google Scholar]
- 9.Bisson, N., M. Tremblay, F. Robinson, D. R. Kaplan, S. P. Trusko, and T. Moss. 2008. Mice lacking both mixed-lineage kinase genes Mlk1 and Mlk2 retain a wild type phenotype. Cell Cycle 7:909-916. [DOI] [PubMed] [Google Scholar]
- 10.Boutros, T., E. Chevet, and P. Metrakos. 2008. Mitogen-activated protein (MAP) kinase/MAP kinase phosphatase regulation: roles in cell growth, death, and cancer. Pharmacol. Rev. 60:261-310. [DOI] [PubMed] [Google Scholar]
- 11.Breitwieser, W., S. Lyons, A. M. Flenniken, G. Ashton, G. Bruder, M. Willington, G. Lacaud, V. Kouskoff, and N. Jones. 2007. Feedback regulation of p38 activity via ATF2 is essential for survival of embryonic liver cells. Genes Dev. 21:2069-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bruhat, A., Y. Cherasse, A. C. Maurin, W. Breitwieser, L. Parry, C. Deval, N. Jones, C. Jousse, and P. Fafournoux. 2007. ATF2 is required for amino acid-regulated transcription by orchestrating specific histone acetylation. Nucleic Acids Res. 35:1312-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bruhat, A., C. Jousse, V. Carraro, A. M. Reimold, M. Ferrara, and P. Fafournoux. 2000. Amino acids control mammalian gene transcription: activating transcription factor 2 is essential for the amino acid responsiveness of the CHOP promoter. Mol. Cell. Biol. 20:7192-7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen, H., Y. X. Pan, E. E. Dudenhausen, and M. S. Kilberg. 2004. Amino acid deprivation induces the transcription rate of the human asparagine synthetase gene through a timed program of expression and promoter binding of nutrient-responsive bZIP transcription factors as well as localized histone acetylation. J. Biol. Chem. 279:50829-50839. [DOI] [PubMed] [Google Scholar]
- 15.Chi, H., M. R. Sarkisian, P. Rakic, and R. A. Flavell. 2005. Loss of mitogen-activated protein kinase kinase kinase 4 (MEKK4) results in enhanced apoptosis and defective neural tube development. Proc. Natl. Acad. Sci. USA 102:3846-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conigrave, A. D., and D. R. Hampson. 2006. Broad-spectrum L-amino acid sensing by class 3 G-protein-coupled receptors. Trends Endocrinol. Metab. 17:398-407. [DOI] [PubMed] [Google Scholar]
- 17.Coso, O. A., M. Chiariello, J. C. Yu, H. Teramoto, P. Crespo, N. Xu, T. Miki, and J. S. Gutkind. 1995. The small GTP-binding proteins Rac1 and Cdc42 regulate the activity of the JNK/SAPK signaling pathway. Cell 81:1137-1146. [DOI] [PubMed] [Google Scholar]
- 18.Cuenda, A., and D. S. Dorow. 1998. Differential activation of stress-activated protein kinase kinases SKK4/MKK7 and SKK1/MKK4 by the mixed-lineage kinase-2 and mitogen-activated protein kinase kinase (MKK) kinase-1. Biochem. J. 333:11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davies, S. P., H. Reddy, M. Caivano, and P. Cohen. 2000. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem. J. 351:95-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davis, R. J. 2000. Signal transduction by the JNK group of MAP kinases. Cell 103:239-252. [DOI] [PubMed] [Google Scholar]
- 21.Derijard, B., M. Hibi, I. H. Wu, T. Barrett, B. Su, T. Deng, M. Karin, and R. J. Davis. 1994. JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell 76:1025-1037. [DOI] [PubMed] [Google Scholar]
- 22.Derijard, B., J. Raingeaud, T. Barrett, I. H. Wu, J. Han, R. J. Ulevitch, and R. J. Davis. 1995. Independent human MAP-kinase signal transduction pathways defined by MEK and MKK isoforms. Science 267:682-685. [DOI] [PubMed] [Google Scholar]
- 23.Dermott, J. M., J. H. Ha, C. H. Lee, and N. Dhanasekaran. 2004. Differential regulation of Jun N-terminal kinase and p38MAP kinase by Galpha12. Oncogene 23:226-232. [DOI] [PubMed] [Google Scholar]
- 24.Deval, C., C. Chaveroux, A. C. Maurin, Y. Cherasse, L. Parry, V. Carraro, D. Milenkovic, M. Ferrara, A. Bruhat, C. Jousse, and P. Fafournoux. 2009. Amino acid limitation regulates the expression of genes involved in several specific biological processes through GCN2-dependent and GCN2-independent pathways. FEBS J. 276:707-718. [DOI] [PubMed] [Google Scholar]
- 25.Dickens, M., J. S. Rogers, J. Cavanagh, A. Raitano, Z. Xia, J. R. Halpern, M. E. Greenberg, C. L. Sawyers, and R. J. Davis. 1997. A cytoplasmic inhibitor of the JNK signal transduction pathway. Science 277:693-696. [DOI] [PubMed] [Google Scholar]
- 26.Fafournoux, P., A. Bruhat, and C. Jousse. 2000. Amino acid regulation of gene expression. Biochem. J. 351:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fanger, G. R., P. Gerwins, C. Widmann, M. B. Jarpe, and G. L. Johnson. 1997. MEKKs, GCKs, MLKs, PAKs, TAKs, and tpls: upstream regulators of the c-Jun amino-terminal kinases? Curr. Opin. Genet. Dev. 7:67-74. [DOI] [PubMed] [Google Scholar]
- 28.Forsberg, H., C. F. Gilstring, A. Zargari, P. Martinez, and P. O. Ljungdahl. 2001. The role of the yeast plasma membrane SPS nutrient sensor in the metabolic response to extracellular amino acids. Mol. Microbiol. 42:215-228. [DOI] [PubMed] [Google Scholar]
- 29.Goldsmith, Z. G., and D. N. Dhanasekaran. 2007. G protein regulation of MAPK networks. Oncogene 26:3122-3142. [DOI] [PubMed] [Google Scholar]
- 30.Gupta, S., D. Campbell, B. Derijard, and R. J. Davis. 1995. Transcription factor ATF2 regulation by the JNK signal transduction pathway. Science 267:389-393. [DOI] [PubMed] [Google Scholar]
- 31.Harding, H. P., Y. Zhang, H. Zeng, I. Novoa, P. D. Lu, M. Calfon, N. Sadri, C. Yun, B. Popko, R. Paules, D. F. Stojdl, J. C. Bell, T. Hettmann, J. M. Leiden, and D. Ron. 2003. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 11:619-633. [DOI] [PubMed] [Google Scholar]
- 32.Hartman, M. G., D. Lu, M. L. Kim, G. J. Kociba, T. Shukri, J. Buteau, X. Wang, W. L. Frankel, D. Guttridge, M. Prentki, S. T. Grey, D. Ron, and T. Hai. 2004. Role for activating transcription factor 3 in stress-induced β-cell apoptosis. Mol. Cell. Biol. 24:5721-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hashimoto, Y., C. Zhang, J. Kawauchi, I. Imoto, M. T. Adachi, J. Inazawa, T. Amagasa, T. Hai, and S. Kitajima. 2002. An alternatively spliced isoform of transcriptional repressor ATF3 and its induction by stress stimuli. Nucleic Acids Res. 30:2398-2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hayakawa, J., S. Mittal, Y. Wang, K. S. Korkmaz, E. Adamson, C. English, M. Ohmichi, M. McClelland, and D. Mercola. 2004. Identification of promoters bound by c-Jun/ATF2 during rapid large-scale gene activation following genotoxic stress. Mol. Cell 16:521-535. [DOI] [PubMed] [Google Scholar]
- 35.Heasman, S. J., and A. J. Ridley. 2008. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat. Rev. Mol. Cell Biol. 9:690-701. [DOI] [PubMed] [Google Scholar]
- 36.Hirai, S., A. Kawaguchi, J. Suenaga, M. Ono, D. F. Cui, and S. Ohno. 2005. Expression of MUK/DLK/ZPK, an activator of the JNK pathway, in the nervous systems of the developing mouse embryo. Gene Expr. Patterns 5:517-523. [DOI] [PubMed] [Google Scholar]
- 37.Hundal, H. S., and P. M. Taylor. 2009. Amino acid transceptors: gate keepers of nutrient exchange and regulators of nutrient signaling. Am. J. Physiol. Endocrinol Metab. 296:E603-E613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ichijo, H., E. Nishida, K. Irie, P. ten Dijke, M. Saitoh, T. Moriguchi, M. Takagi, K. Matsumoto, K. Miyazono, and Y. Gotoh. 1997. Induction of apoptosis by ASK1, a mammalian MAPKKK that activates SAPK/JNK and p38 signaling pathways. Science 275:90-94. [DOI] [PubMed] [Google Scholar]
- 39.Itoh, R. E., E. Kiyokawa, K. Aoki, T. Nishioka, T. Akiyama, and M. Matsuda. 2008. Phosphorylation and activation of the Rac1 and Cdc42 GEF Asef in A431 cells stimulated by EGF. J. Cell Sci. 121:2635-2642. [DOI] [PubMed] [Google Scholar]
- 40.Jackson, A. A., and M. S. Grimble. 1990. The malnourished child, vol. 19. Raven Press, Vevey, Switzerland.
- 41.Jeevanandam, M., G. D. Horowitz, S. F. Lowry, and M. F. Brennan. 1984. Cancer cachexia and protein metabolism. Lancet i(8392):1423-1426. [DOI] [PubMed] [Google Scholar]
- 42.Jiang, H. Y., and R. C. Wek. 2005. Phosphorylation of the alpha-subunit of the eukaryotic initiation factor-2 (eIF2alpha) reduces protein synthesis and enhances apoptosis in response to proteasome inhibition. J. Biol. Chem. 280:14189-14202. [DOI] [PubMed] [Google Scholar]
- 43.Jiang, H. Y., S. A. Wek, B. C. McGrath, D. Lu, T. Hai, H. P. Harding, X. Wang, D. Ron, D. R. Cavener, and R. C. Wek. 2004. Activating transcription factor 3 is integral to the eukaryotic initiation factor 2 kinase stress response. Mol. Cell. Biol. 24:1365-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jousse, C., J. Averous, A. Bruhat, V. Carraro, S. Mordier, and P. Fafournoux. 2004. Amino acids as regulators of gene expression: molecular mechanisms. Biochem. Biophys. Res. Commun. 313:447-452. [DOI] [PubMed] [Google Scholar]
- 45.Jousse, C., C. Deval, A. C. Maurin, L. Parry, Y. Cherasse, C. Chaveroux, R. Lefloch, P. Lenormand, A. Bruhat, and P. Fafournoux. 2007. TRB3 inhibits the transcriptional activation of stress-regulated genes by a negative feedback on the ATF4 pathway. J. Biol. Chem. 282:15851-15861. [DOI] [PubMed] [Google Scholar]
- 46.Kawai, T., J. Fan, K. Mazan-Mamczarz, and M. Gorospe. 2004. Global mRNA stabilization preferentially linked to translational repression during the endoplasmic reticulum stress response. Mol. Cell. Biol. 24:6773-6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kilberg, M. S., Y. X. Pan, H. Chen, and V. Leung-Pineda. 2005. Nutritional control of gene expression: how mammalian cells respond to amino acid limitation. Annu. Rev. Nutr. 25:59-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim, E. K., K. T. Noh, J. H. Yoon, J. H. Cho, K. W. Yoon, G. Dreyfuss, and E. J. Choi. 2007. Positive regulation of ASK1-mediated c-Jun NH(2)-terminal kinase signaling pathway by the WD-repeat protein Gemin5. Cell Death Differ. 14:1518-1528. [DOI] [PubMed] [Google Scholar]
- 49.Kroeze, W. K., D. J. Sheffler, and B. L. Roth. 2003. G-protein-coupled receptors at a glance. J. Cell Sci. 116:4867-4869. [DOI] [PubMed] [Google Scholar]
- 50.Kurokawa, K., R. E. Itoh, H. Yoshizaki, Y. O. Nakamura, and M. Matsuda. 2004. Coactivation of Rac1 and Cdc42 at lamellipodia and membrane ruffles induced by epidermal growth factor. Mol. Biol. Cell 15:1003-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li, M. G., K. Katsura, H. Nomiyama, K. Komaki, J. Ninomiya-Tsuji, K. Matsumoto, T. Kobayashi, and S. Tamura. 2003. Regulation of the interleukin-1-induced signaling pathways by a novel member of the protein phosphatase 2C family (PP2Cepsilon). J. Biol. Chem. 278:12013-12021. [DOI] [PubMed] [Google Scholar]
- 52.Liao, X. H., A. Majithia, X. Huang, and A. R. Kimmel. 2008. Growth control via TOR kinase signaling, an intracellular sensor of amino acid and energy availability, with crosstalk potential to proline metabolism. Amino Acids 35:761-770. [DOI] [PubMed] [Google Scholar]
- 53.Livingstone, C., G. Patel, and N. Jones. 1995. ATF-2 contains a phosphorylation-dependent transcriptional activation domain. EMBO J. 14:1785-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ljungdahl, P. O. 2009. Amino-acid-induced signalling via the SPS-sensing pathway in yeast. Biochem. Soc. Trans. 37:242-247. [DOI] [PubMed] [Google Scholar]
- 55.Lopez, A. B., C. Wang, C. C. Huang, I. Yaman, Y. Li, K. Chakravarty, P. F. Johnson, C. M. Chiang, M. D. Snider, R. C. Wek, and M. Hatzoglou. 2007. A feedback transcriptional mechanism controls the level of the arginine/lysine transporter cat-1 during amino acid starvation. Biochem. J. 402:163-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu, P. D., H. P. Harding, and D. Ron. 2004. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J. Cell Biol. 167:27-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lu, P. D., C. Jousse, S. J. Marciniak, Y. Zhang, I. Novoa, D. Scheuner, R. J. Kaufman, D. Ron, and H. P. Harding. 2004. Cytoprotection by pre-emptive conditional phosphorylation of translation initiation factor 2. EMBO J. 23:169-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marciniak, S. J., C. Y. Yun, S. Oyadomari, I. Novoa, Y. Zhang, R. Jungreis, K. Nagata, H. P. Harding, and D. Ron. 2004. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 18:3066-3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maurin, A. C., C. Jousse, J. Averous, L. Parry, A. Bruhat, Y. Cherasse, H. Zeng, Y. Zhang, H. P. Harding, D. Ron, and P. Fafournoux. 2005. The GCN2 kinase biases feeding behavior to maintain amino acid homeostasis in omnivores. Cell Metab. 1:273-277. [DOI] [PubMed] [Google Scholar]
- 60.Morton, S., R. J. Davis, and P. Cohen. 2004. Signalling pathways involved in multisite phosphorylation of the transcription factor ATF-2. FEBS Lett. 572:177-183. [DOI] [PubMed] [Google Scholar]
- 61.Nicklin, P., P. Bergman, B. Zhang, E. Triantafellow, H. Wang, B. Nyfeler, H. Yang, M. Hild, C. Kung, C. Wilson, V. E. Myer, J. P. MacKeigan, J. A. Porter, Y. K. Wang, L. C. Cantley, P. M. Finan, and L. O. Murphy. 2009. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell 136:521-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nicola, C., P. K. Lala, and C. Chakraborty. 2008. Prostaglandin E2-mediated migration of human trophoblast requires RAC1 and CDC42. Biol. Reprod. 78:976-982. [DOI] [PubMed] [Google Scholar]
- 63.Ogino, T., M. Ozaki, M. Hosako, M. Omori, S. Okada, and A. Matsukawa. 2009. Activation of c-Jun N-terminal kinase is essential for oxidative stress-induced Jurkat cell apoptosis by monochloramine. Leuk. Res. 33:151-158. [DOI] [PubMed] [Google Scholar]
- 64.Ornatsky, O. I., D. M. Cox, P. Tangirala, J. J. Andreucci, Z. A. Quinn, J. L. Wrana, R. Prywes, Y. T. Yu, and J. C. McDermott. 1999. Post-translational control of the MEF2A transcriptional regulatory protein. Nucleic Acids Res. 27:2646-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ouwens, D. M., N. D. de Ruiter, G. C. van der Zon, A. P. Carter, J. Schouten, C. van der Burgt, K. Kooistra, J. L. Bos, J. A. Maassen, and H. van Dam. 2002. Growth factors can activate ATF2 via a two-step mechanism: phosphorylation of Thr71 through the Ras-MEK-ERK pathway and of Thr69 through RalGDS-Src-p38. EMBO J. 21:3782-3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Palii, S. S., H. Chen, and M. S. Kilberg. 2004. Transcriptional control of the human sodium-coupled neutral amino acid transporter system A gene by amino acid availability is mediated by an intronic element. J. Biol. Chem. 279:3463-3471. [DOI] [PubMed] [Google Scholar]
- 67.Palii, S. S., C. E. Kays, C. Deval, A. Bruhat, P. Fafournoux, and M. S. Kilberg. 2009. Specificity of amino acid regulated gene expression: analysis of genes subjected to either complete or single amino acid deprivation. Amino Acids 37:79-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pan, Y. X., H. Chen, M. M. Thiaville, and M. S. Kilberg. 2007. Activation of the ATF3 gene through a co-ordinated amino acid-sensing response programme that controls transcriptional regulation of responsive genes following amino acid limitation. Biochem. J. 401:299-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Peng, T., T. R. Golub, and D. M. Sabatini. 2002. The immunosuppressant rapamycin mimics a starvation-like signal distinct from amino acid and glucose deprivation. Mol. Cell. Biol. 22:5575-5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Prasad, M. V., J. M. Dermott, L. E. Heasley, G. L. Johnson, and N. Dhanasekaran. 1995. Activation of Jun kinase/stress-activated protein kinase by GTPase-deficient mutants of G alpha 12 and G alpha 13. J. Biol. Chem. 270:18655-18659. [DOI] [PubMed] [Google Scholar]
- 71.Proud, C. G. 2007. Signalling to translation: how signal transduction pathways control the protein synthetic machinery. Biochem. J. 403:217-234. [DOI] [PubMed] [Google Scholar]
- 72.Regard, J. B., I. T. Sato, and S. R. Coughlin. 2008. Anatomical profiling of G protein-coupled receptor expression. Cell 135:561-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schiller, M. R. 2006. Coupling receptor tyrosine kinases to Rho GTPases—GEFs what's the link. Cell. Signal. 18:1834-1843. [DOI] [PubMed] [Google Scholar]
- 74.Sinha, S., and W. Yang. 2008. Cellular signaling for activation of Rho GTPase Cdc42. Cell. Signal. 20:1927-1934. [DOI] [PubMed] [Google Scholar]
- 75.Siu, F., P. J. Bain, R. LeBlanc-Chaffin, H. Chen, and M. S. Kilberg. 2002. ATF4 is a mediator of the nutrient-sensing response pathway that activates the human asparagine synthetase gene. J. Biol. Chem. 277:24120-24127. [DOI] [PubMed] [Google Scholar]
- 76.Suzuki, N., N. Hajicek, and T. Kozasa. 2009. Regulation and physiological functions of G12/13-mediated signaling pathways. Neurosignals 17:55-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Szczepankiewicz, B. G., C. Kosogof, L. T. Nelson, G. Liu, B. Liu, H. Zhao, M. D. Serby, Z. Xin, M. Liu, R. J. Gum, D. L. Haasch, S. Wang, J. E. Clampit, E. F. Johnson, T. H. Lubben, M. A. Stashko, E. T. Olejniczak, C. Sun, S. A. Dorwin, K. Haskins, C. Abad-Zapatero, E. H. Fry, C. W. Hutchins, H. L. Sham, C. M. Rondinone, and J. M. Trevillyan. 2006. Aminopyridine-based c-Jun N-terminal kinase inhibitors with cellular activity and minimal cross-kinase activity. J. Med. Chem. 49:3563-3580. [DOI] [PubMed] [Google Scholar]
- 78.Thiaville, M. M., Y. X. Pan, A. Gjymishka, C. Zhong, R. J. Kaufman, and M. S. Kilberg. 2008. MEK signaling is required for phosphorylation of eIF2alpha following amino acid limitation of HepG2 human hepatoma cells. J. Biol. Chem. 283:10848-10857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tournier, C., A. J. Whitmarsh, J. Cavanagh, T. Barrett, and R. J. Davis. 1997. Mitogen-activated protein kinase kinase 7 is an activator of the c-Jun NH2-terminal kinase. Proc. Natl. Acad. Sci. USA 94:7337-7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.van Dam, H., D. Wilhelm, I. Herr, A. Steffen, P. Herrlich, and P. Angel. 1995. ATF-2 is preferentially activated by stress-activated protein kinases to mediate c-jun induction in response to genotoxic agents. EMBO J. 14:1798-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vattem, K. M., and R. C. Wek. 2004. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. USA 101:11269-11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vlahopoulos, S. A., S. Logotheti, D. Mikas, A. Giarika, V. Gorgoulis, and V. Zoumpourlis. 2008. The role of ATF-2 in oncogenesis. Bioessays 30:314-327. [DOI] [PubMed] [Google Scholar]
- 83.Wang, X., A. Beugnet, M. Murakami, S. Yamanaka, and C. G. Proud. 2005. Distinct signaling events downstream of mTOR cooperate to mediate the effects of amino acids and insulin on initiation factor 4E-binding proteins. Mol. Cell. Biol. 25:2558-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang, X., A. Destrument, and C. Tournier. 2007. Physiological roles of MKK4 and MKK7: insights from animal models. Biochim. Biophys. Acta 1773:1349-1357. [DOI] [PubMed] [Google Scholar]
- 85.Whitmarsh, A. J. 2006. The JIP family of MAPK scaffold proteins. Biochem. Soc. Trans. 34:828-832. [DOI] [PubMed] [Google Scholar]
- 86.Wolfe, R. R., F. Jahoor, and W. H. Hartl. 1989. Protein and amino acid metabolism after injury. Diabetes Metab. Rev. 5:149-164. [DOI] [PubMed] [Google Scholar]
- 87.Xu, Z., N. V. Kukekov, and L. A. Greene. 2003. POSH acts as a scaffold for a multiprotein complex that mediates JNK activation in apoptosis. EMBO J. 22:252-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhao, M., L. New, V. V. Kravchenko, Y. Kato, H. Gram, F. di Padova, E. N. Olson, R. J. Ulevitch, and J. Han. 1999. Regulation of the MEF2 family of transcription factors by p38. Mol. Cell. Biol. 19:21-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ziegler, T. R., C. Gatzen, and D. W. Wilmore. 1994. Strategies for attenuating protein-catabolic responses in the critically ill. Annu. Rev. Med. 45:459-480. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.