Abstract
Interstrand cross-links (ICLs) prevent DNA strand separation and, therefore, transcription and replication, making them extremely cytotoxic. The precise mechanism by which ICLs are removed from mammalian genomes largely remains elusive. Genetic evidence implicates ATR, the Fanconi anemia proteins, proteins required for homologous recombination, translesion synthesis, and at least two endonucleases, MUS81-EME1 and XPF-ERCC1. ICLs cause replication-dependent DNA double-strand breaks (DSBs), and MUS81-EME1 facilitates DSB formation. The subsequent repair of these DSBs occurs via homologous recombination after the ICL is unhooked by XPF-ERCC1. Here, we examined the effect of the loss of either nuclease on FANCD2 monoubiquitination to determine if the nucleolytic processing of ICLs is required for the activation of the Fanconi anemia pathway. FANCD2 was monoubiquitinated in Mus81−/−, Ercc1−/−, and XPF-deficient human, mouse, and hamster cells exposed to cross-linking agents. However, the monoubiquitinated form of FANCD2 persisted longer in XPF-ERCC1-deficient cells than in wild-type cells. Moreover, the levels of chromatin-bound FANCD2 were dramatically reduced and the number of ICL-induced FANCD2 foci significantly lower in XPF-ERCC1-deficient cells. These data demonstrate that the unhooking of an ICL by XPF-ERCC1 is necessary for the stable localization of FANCD2 to the chromatin and subsequent homologous recombination-mediated DSB repair.
The XPF-ERCC1 heterodimer is a structure-specific endonuclease that incises double-strand DNA immediately adjacent to a 3′-single-stranded region, removing 3′ overhangs or opening bubbles (12, 69). ERCC1 is required for DNA binding (74), and XPF harbors the catalytic domain (17). XPF-ERCC1 makes the incision 5′ to the lesion during nucleotide excision repair (NER), the pathway responsible for removing helix-distorting DNA lesions (69). Defects in NER cause xeroderma pigmentosum (XP), a syndrome characterized by photosensitivity and a dramatically increased risk of skin cancers due to failure to repair UV photolesions. Cells from all XP complementation groups (XP-A to XP-G) and the recently reported ERCC1-deficient patient (33) are hypersensitive to UV irradiation. However, cells deficient in XPF-ERCC1 differ from other XP cells in that they also are exquisitely sensitive to chemicals that induce DNA interstrand cross-links (ICLs) (13, 28, 54). ICLs are extremely cytotoxic lesions formed when bifunctional agents covalently link both strands of DNA, preventing strand separation, which is necessary for replication or transcription (46). Cross-linking agents such as nitrogen mustards (HN2) (37) and mitomycin C (MMC) (31) produce a mixture of monoadducts and ICLs. However, cytotoxicity correlates with the number of ICLs formed rather than monoadducts (60, 62).
ICLs present a unique challenge to cells, in that they affect both strands of DNA and therefore cannot be repaired by a simple excision and resynthesis mechanism. The mechanism of ICL repair in Escherichia coli is well characterized. It involves the NER complex UvrABC, the recombination repair machinery, or DNA polymerase II-dependent translesion synthesis (6, 7, 9, 75). In Saccharomyces cerevisiae, ICL repair requires both the NER and the homologous recombination (HR) machinery (32). There also is evidence for the involvement of PSO2/SNM1 (25, 26, 66), as well as base excision repair (45), mismatch repair (16), and translesion polymerases (67). ICL repair in mammalian cells, however, remains poorly understood. Genetic evidence implicates XPF-ERCC1 (10), HR proteins such as XRCC2 and XRCC3 (13, 42), MutSβ (82), RPA, PCNA (40, 81), the PSO4 complex (PSO4/PRP19, CDC5L, PLRG1, SPF27) (80), WRN (80), BRCA2 (79), MUS81-EME1 (1, 47), SNM1a, SNM1b (5, 15, 24, 30), and the Fanconi anemia (FA) proteins (4, 18, 68, 73). Of these, the members of the FA family of proteins are unique, in that they appear to be conserved only in higher eukaryotes (11, 34).
FA is a rare and clinically heterogeneous disease characterized by congenital skeletal abnormalities, growth retardation, bone marrow failure, aplastic anemia, genomic instability, and susceptibility to cancer (11, 34). The patients have been grouped into 13 distinct complementation groups (FANCA, FANCB, FANCC, FANCD1, FANCD2, FANCE, FANCF, FANCG, FANCI, FANCJ, FANCL, FANCM, and FANCN), each representing the deficiency of one protein in the FA pathway. Of these, FANCA, FANCB, FANCC, FANCE, FANCF, FANCG, FANCL, FANCM, and FAAP100 and FAAP24, newly discovered components without complementation groups, form the FA core complex (8, 19, 41, 48-51). FANCL is an E3 ubiquitin ligase that monoubiquitinates FANCD2 in response to genotoxic stress (49) during the S phase of the cell cycle (72). FANCI, a newly discovered paralog of FANCD2, which also is monoubiquitinated, forms a chromatin-bound complex with FANCD2. The monoubiquitination of both proteins is required for complex stability and ICL repair (70). FANCD2 monoubiquitination commonly is used as a readout to define which FA proteins act upstream and therefore participate in damage signaling, or are downstream and therefore are more likely to directly contribute to ICL repair. Monoubiquitinated FANCD2 colocalizes with proteins involved in HR, including RAD51 and BRCA1 (14, 20, 72). FANCD1 is identical to BRCA2 (27) and interacts with XRCC3 along with FANCG (29) and RAD51 (43). These data indicate a direct physical interaction between the FA proteins and the HR machinery (78).
The emerging model for the mechanism of ICL repair in mammals suggests that the FA pathway is activated in response to ICL damage in an ATR-dependent manner (3, 64). ATR is activated by blocked replication forks (71) and also phosphorylates numerous downstream effectors, including FA proteins (3, 51, 64) and H2AX (77). Before a replication fork that is blocked by an ICL can be restarted, the ICL must be completely unhooked from one strand of DNA, requiring two incisions 5′ and 3′ of the lesion and potentially producing a DNA double-strand break (DSB) at the fork. MUS81-EME1 is thought to be responsible for the first incision, creating a DSB (22, 23). The second incision, or the unhooking of the cross-link, has been proposed to be a key function of XPF-ERCC1 in ICL repair (56). Once the ICL is completely unhooked from one strand, translesion synthesis can fill the gap, albeit in an error-prone fashion. The repaired strand now can act as a template for reestablishing the replication fork via HR (55).
To further elucidate the mechanism of ICL repair in mammals, it is necessary to define the relationship between the nucleolytic cleavage of cross-linked DNA and the FA pathway. Here, we provide evidence that XPF-ERCC1 nuclease participates in the same ICL repair mechanism as the FA proteins, and that the nucleolytic processing of damaged DNA is not required for FA pathway activation but is required for the stable localization of monoubiquitinated FANCD2 on chromatin.
MATERIALS AND METHODS
Cell culture and drug treatment.
Human fibroblast lines immortalized by the stable expression of human telomerase, C5RO (normal), XP51RO (XFE), XP42RO (XP-F), and XP25RO (XP-A) (54) were cultured in Ham's F-10 with 10% fetal bovine serum, antibiotics, and nonessential amino acids. Cells were treated for 12 h with 0.3 μM MMC or for 1 h with 3 μM MMC diluted in medium. At the end of the treatment, cells were washed twice with phosphate-buffered saline (PBS), the medium was replaced, and the cells were incubated at 5% CO2 at 37°C under humidity. Time points were calculated with this cell status as 0 h. Wild-type and Ercc1−/− mouse embryonic fibroblasts (MEFs), transformed with simian virus 40 large T antigen, were cultured in a 1:1 mixture of Ham's F-10 and Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum, antibiotics, and nonessential amino acids. Wild-type, Ercc1−/− (53), and Mus81−/− (23) mouse embryonic stem (ES) cells were cultured on gelatin-coated plates in a 1:1 mixture of DMEM and buffalo rat liver (BRL) cell-conditioned medium with 10% FBS, antibiotics, nonessential amino acids, 0.1 mM 2-mercaptoethanol, and leukemic inhibitory factor (1,000 U/ml; Gibco). Wild-type (AA8), UV47 (Xpf mutant), UV96 (Ercc1 mutant), and UV135 (Xpg mutant) CHO cells were grown as a monolayer in Ham's F-12 HEPES medium (Sigma) supplemented with 5 mM glutamine and 10% fetal calf serum (FCS). Cells were grown at 37°C in a 5% CO2 humidified incubator. Cells were treated with 1 μM MMC for 12 h or for 1 h with the stated doses of nitrogen mustard (mechlorethamine hydrochloride; Aldrich) dissolved in culture medium (without FCS) immediately prior to use. Following nitrogen mustard or MMC treatment, media were removed, and cells were washed twice with PBS and returned to the incubator in drug-free complete medium for the stated times.
Immunoblotting.
Cells were trypsinized, washed with PBS, and lysed with 1 ml NETT buffer (100 mM NaCl, 50 mM Tris base, pH 7.5, 5 mM EDTA, pH 8.0, 0.5% Triton X-100) containing Complete mini-protease inhibitor cocktail and phosphatase inhibitor cocktail (Roche Molecular Biochemicals). From each sample, 50 μg protein was resolved on sodium dodecyl sulfate-6% polyacrylamide gel electrophoresis gels. FANCD2 was detected with rabbit anti-FANCD2 (for human cells; 1:10,000; Novus Biologicals), rabbit anti-murine FANCD2 (for mouse cells; 1:1,000), or rabbit anti-FANCD2 (for CHO cells; 1:1,000; Abcam). The loading control used for whole-cell extracts (WCEs) was tubulin (rabbit antitubulin; 1:1,000 or 1:15,000; Abcam and Sigma, respectively). The loading control for cytoplasmic protein from HeLa cells was GRB2 (anti-GRB2; 1:5,000; BD Biosciences). The loading control for chromatin and nuclear fractions of ES cells was TATA binding protein (mouse anti-TBP; 1:500; Abcam) and for hamster cells was ORC2 (mouse anti-ORC2; 1:800; BD-Pharmingen). The loading controls for human fibroblasts and MEFs were nucleophosmin (mouse anti-nucleophosmin; 1:5,000; Millipore) for the soluble nuclear fraction and histone H3 (rabbit anti-H3; 1:20,000; Abcam) for the chromatin fraction. Secondary antibodies used were mouse AP (1:7,500; Promega), rabbit AP (1:1,000; Promega), rat horseradish peroxidase (HRP; 1:5,000; Abcam), rabbit HRP (1:1200; Dako), and mouse HRP (1:1,500 or 1:2,500; Promega).
Depletion of ERCC1 by siRNA.
HeLa cells were obtained from Cancer Research UK Clare Hall Cell Services and maintained in RPMI medium supplemented with 10% FCS and glutamine without antibiotics. Cells were transfected with short interfering RNA (siRNA) duplexes using Hiperfect transfection reagent (Qiagen). Cells were seeded at 50% confluence immediately before the transfection complex (containing 5 nM siRNA oligonucleotides) was added. This transfection step was repeated 24 h later. siRNA-transfected cells were harvested for subsequent drug treatment 72 h after the first transfection. The siRNA duplexes were purchased from Qiagen, and the sequence used to deplete Ercc1 was 5′-GCCCUUAUUCCGAUCUACATT-3′, which has been characterized previously in detail (39). For negative controls, HeLa cells were transfected with Hiperfect with Qiagen AllStars RNA interference negative control duplex.
Cell cycle analysis.
Cells were plated at 50% confluence and then 16 h later were exposed to 0.3 μM MMC for 12 h or were left untreated. At 12, 24, 48, and 72 h after exposure, the cells were fixed with ice-cold 70% ethanol and stored at 4°C overnight. CHO cells were treated with 5 μM HN2 as described above, and samples were removed at the stated times for fixing. The fixed cells were washed with PBS, treated with RNase A (Roche) at a final concentration of 100 U/ml for 30 min at room temperature, and then stained with 1 μg/ml propidium iodide in PBS. Cell cycle analysis was performed on a Dako MoFlo flow cytometer or a Becton Dickinson FACSCalibur.
Modified comet assay.
The modified version of the comet assay for the detection of ICLs was performed exactly as previously described (13).
Cell fractionation.
Cells were fractionated into cytosolic, nuclear, and chromatin fractions with a modification of a published protocol (35). In brief, cells were trypsinized, pelleted, and washed twice with PBS. The pellet was vortexed at full speed for 15 s with 200 μl of CERI reagent from the Pierce NE-PER fractionation kit (Pierce Biotech) with Complete mini-protease inhibitor cocktail and then was incubated on ice for 10 min. This was followed by the addition of 11 μl of CERII, vortexing for 5 s, and incubation on ice for 1 min. The mixture was spun down at 13,000 rpm for 5 min, and the supernatant was collected as the cytosolic fraction. The nuclei were suspended in extraction buffer (15 mM Tris-HCl, pH 7.3, 1 mM EDTA, 0.4 M NaCl, 1 mM MgCl2, 10% glycerol, 10 mM β-mercaptoethanol, and Complete mini-protease inhibitor cocktail), mixed for 1 h at 4°C, and spun down at 13,000 rpm for 10 min. The supernatant was collected as the soluble nuclear fraction, and the pellet was suspended in micrococcal nuclease buffer (20 mM Tris·HCl, pH 7.5, 100 mM KCl, 2 mM MgCl2, 1 mM CaCl2, 0.3 M sucrose, 0.1% Triton X-100, and Complete mini-protease inhibitor cocktail) and digested with 400 U/ml S7 micrococcal nuclease (Roche) at 37°C for 30 min. The reaction was spun down at 13,000 rpm for 10 min, and supernatant was collected as the chromatin fraction. For CHO and HeLa cells, fractionation was performed exactly as described previously (65).
Immunodetection of FANCD2 foci.
C5RO and XP51RO human fibroblasts (54) were plated on glass coverslips at 50% confluence and then 16 h later were exposed to 3 μM MMC for 1 h or left untreated. At 6, 12, 24, and 48 h following exposure, cells were fixed as described previously (76) with some modifications. Briefly, cells were washed with PBS, permeabilized with ice-cold 0.5% Triton X-100 in PBS, and then fixed with 2% paraformaldehyde and blocked with 5% bovine serum albumin at room temperature. FANCD2 was detected by incubation with rabbit polyclonal anti-FANCD2 antibody (1:1,000; Novus Biologicals) for 90 min at room temperature and then with goat anti-rabbit antibody-Alexa-488 (1:1,000; Invitrogen). Images were taken of 20 fields of cells for each genotype. The total number of nuclei and the number of nuclei with foci were counted in each field. The sum from all fields was used to calculate the percent of nuclei with foci. The experiment was done in triplicate. Student's t test was used to probe significant differences between cell lines.
RESULTS
FANCD2 is monoubiquitinated in XPF-ERCC1- and MUS81-EME1-deficient cells.
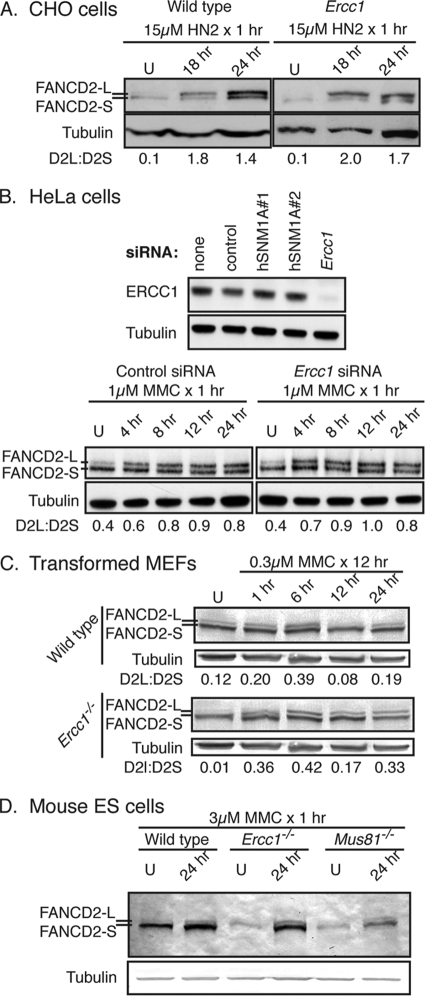
To determine if XPF-ERCC1 deficiency affects the activation of the FA pathway, FANCD2-L (monoubiquitinated form) and FANCD2-S (nonubiquitinated form) were measured in mutant cells exposed to cross-linking agents. Wild-type and Ercc1 mutant CHO cells were exposed to the cross-linking agent nitrogen mustard (HN2) for 1 h, and then WCEs were collected 18 or 24 h later. The levels of FANCD2-L were increased similarly in both cell types after exposure to HN2 relative to the levels for untreated samples (Fig. 1A). This was confirmed in an isogenic pair of human cell lines by knocking down ERCC1 expression in HeLa cells using siRNA and exposing them to a different cross-linking agent, MMC (Fig. 1B and data not shown). Comparable results were obtained with Ercc1−/− MEFs exposed to MMC (Fig. 1C). Since XPF-ERCC1 nuclease appeared not to be required for FA pathway activation, we also screened cells deficient in MUS81, a second nuclease required for ICL repair (23). Mouse ES cells genetically deleted for either Ercc1 or Mus81 were exposed to MMC for 1 h, and then WCEs were collected 24 h later. The immunodetection of FANCD2 revealed increased levels of FANCD2-L in all cells exposed to the cross-linking agents relative to those of the untreated cells (Fig. 1D). These data demonstrate that FANCD2 is monoubiquitinated in response to cross-link damage in the absence of XPF-ERCC1 or MUS81-EME1. This indicates that the nucleolytic processing of cross-links by MUS81-EME1 or XPF-ERCC1 is not necessary for FA pathway activation, as defined by FANCD2 monoubiquitination.
FIG. 1.
FANCD2 is monoubiquitinated in XPF-ERCC1- and MUS81-EME1-deficient cells after cross-link damage. (A) Wild-type and Ercc1 mutant CHO cells were exposed to 15 μM nitrogen mustard (HN2) for 1 h, and then WCEs were collected 18 or 24 h later for the immunodetection of FANCD2. FANCD2-L is the monoubiquitinated protein, and FANCD2-S is the nonubiquitinated form. U indicates cells that were untreated. (B) Wild-type HeLa cells were transfected with control siRNA, with one of two siRNAs against hSNM1A as further controls for specificity, or with siRNA against Ercc1. After confirming the specific knockdown of ERCC1 by immunoblotting, cells were treated with 1 μM MMC for 1 h, and WCEs were collected at multiple time points after exposure for the immunodetection of FANCD2. (C) Wild-type and Ercc1−/− MEFs were exposed to 0.3 μM MMC for 12 h, and then WCEs were collected at multiple time points after exposure for the immunodetection of FANCD2. (D) Wild-type, Mus81−/−, and Ercc1−/− mouse ES cells were exposed to 3 μM MMC for 1 h, and then WCEs were collected 24 h later for the immunodetection of FANCD2.
FA pathway activation persists in XPF-ERCC1-deficient cells.
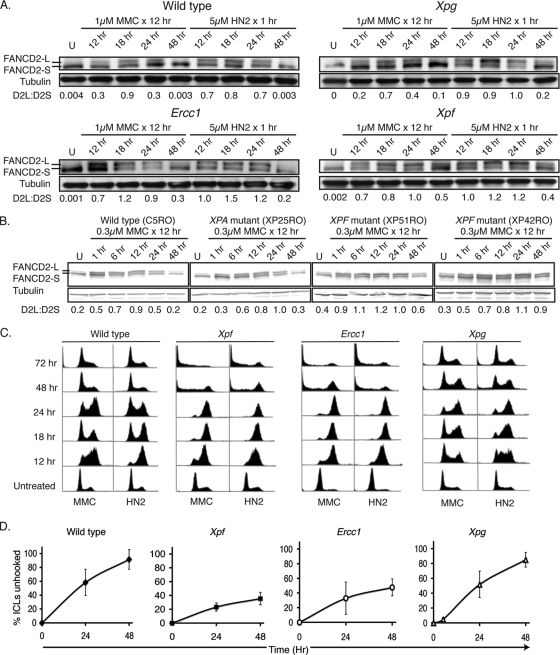
Although FANCD2 is monoubiquitinated in XPF-ERCC1-deficient cells, initial data from the MEFs suggested that FANCD2-L levels remained elevated longer in Ercc1−/− cells exposed to MMC than in wild-type cells (Fig. 1C). To investigate this further, wild-type and mutant CHO cells and human fibroblasts were exposed to MMC or HN2, and FANCD2 was visualized by immunoblotting at multiple time points following DNA damage. Both MMC and HN2 induced an increase in the ratio of FANCD2-L to FANCD2-S in wild-type and NER-deficient Xpg mutant CHO cells (Fig. 2A). By 48 h after HN2 or MMC exposure, FANCD2-L levels returned to near normal in these cell lines. In contrast, the ratio of FANCD2-L to FANCD2-S still was elevated in both Xpf and Ercc1 mutant CHO cells at 48 h after exposure to HN2 or MMC (Fig. 2A). The results were recapitulated in cells derived from human XP patients (Fig. 2B). In wild-type and NER-deficient XP-A human fibroblasts, MMC induced a rapid increase in the ratio of FANCD2-L to FANCD2-S that persisted for 24 h. The levels of monoubiquitinated FANCD2 returned to those of untreated cells by 48 h. In two different XP-F cell lines, the ratio of FANCD-L to FANCD2-S was increased by MMC, as was the case in wild-type cells. However, in the XPF-ERCC1-deficient cells, the ratio of FANCD2-L to FANCD-S remained elevated at 48 h after exposure to MMC (Fig. 2B). This suggests that in the absence of XPF-ERCC1, the FA pathway remains activated in response to cross-link damage.
FIG. 2.
Monoubiquitination of FANCD2 persists in XPF-ERCC1-deficient cells. (A) AA8 (wild type), UV135 (Xpg mutant), UV96 (Ercc1 mutant), and UV47 (Xpf mutant) CHO cells were exposed to 1 μM MMC for 12 h or 5 μM nitrogen mustard (HN2) for 1 h, and then WCEs were collected at multiple time points following exposure for the immunodetection of FANCD2. U indicates cells that were untreated. (B) Wild-type, XP-A (XP25RO), and XP-F (XP51RO and XP42RO) immortalized human fibroblasts were exposed to 0.3 μM MMC for 12 h, and then WCEs were collected at multiple time points after exposure for the immunodetection of FANCD2. (C) CHO cells were treated as described for panel A, fixed at the same time points, and stained with propidium iodide for cell cycle analysis by flow cytometry. (D) The efficiency of the unhooking of interstrand cross-links, as determined by modified comet assay following 1 h of treatment with 5 μM HN2 in Xpg, Xpf, and Ercc1 mutant and wild-type parent cell lines.
Because FANCD2 monoubiquitination is cell cycle dependent (36), the cell cycle profile of the CHO cell lines was measured at multiple time points after exposure to cross-linking agents (Fig. 2C). HN2 and MMC caused an accumulation of cells in S and G2/M phases at 12 and 18 h in all cells. By 48 h following MMC treatment, some wild-type and Xpg mutant cells had escaped G2/M arrest, but XPF-ERCC1-defective cells remained arrested in late S and G2/M. Therefore, persistent FANCD2-L correlates with G2/M accumulation. The Xpg mutant cells were somewhat slower to escape the G2/M arrest than wild-type cells, and this might be attributable to the role of NER in repairing the monoadducts produced by cross-linking agents. The persistence of FANCD2-L also correlates with the impaired unhooking of cross-link damage (Fig. 2D). The modified comet assay (13) was used to measure the nucleolytic processing of cross-link damage in CHO cells following HN2 exposure. In wild-type and Xpg mutant cells, cross-link unhooking was greater than 85% at 48 h. In contrast, in the Ercc1 and Xpf mutant cells in which FANCD2 monoubiquitination persisted, cross-link unhooking was only approximately 40% at 48 h. These data indicate that the nucleolytic processing of ICLs coincides with the deactivation of the FA pathway, which was marked by a decrease in the FANCD2-L/FANCD2-S ratio and is associated with escape from G2/M.
FANCD2 foci are reduced in XPF-ERCC1-deficient human fibroblasts.
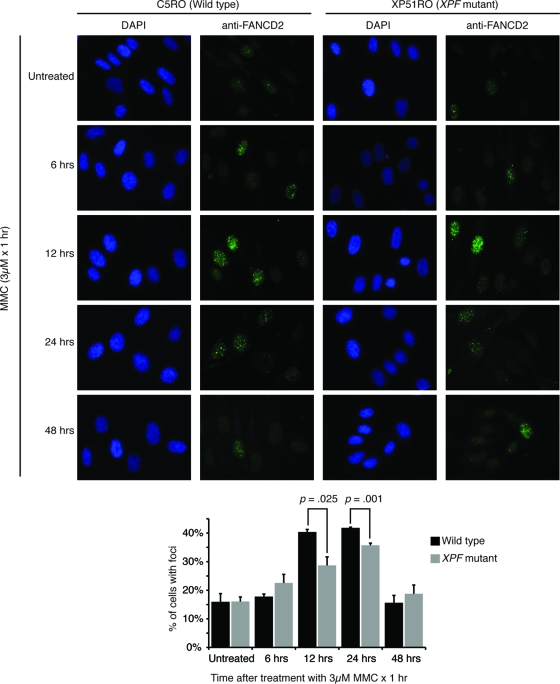
Another way to quantify FA pathway activation is to measure chromatin-bound FANCD2 (61). Surprisingly, in human fibroblasts the number of cells with FANCD2 foci was significantly reduced in the XPF-ERCC1-deficient cell line (Fig. 3). We measured FANCD2 foci in human fibroblasts exposed to MMC after the detergent extraction of soluble proteins. FANCD2 foci were detected in approximately 15% of untreated wild-type and XPF mutant cells. Twelve hours after exposure to MMC, 40% of wild-type cells had FANCD2 foci, whereas 29% of XPF mutant cells had foci (P = 0.025), and 24 h after the exposure 42% of wild-type cells and 36% of XPF mutant cells had FANCD2 foci (P = 0.001). By 48 h after cross-link damage, the fraction of cells with FANCD2 foci had returned to normal in both cell types. These data suggest that although FANCD2 is efficiently ubiquitinated in XPF-ERCC1-deficient cells in response to cross-link damage, the localization of FANCD2 to the chromatin is impaired, a critical step for the subsequent HR-mediated repair of replication-induced DSBs.
FIG. 3.
FANCD2 focus formation is impaired in XPF-ERCC1-deficient cells. Wild-type (C5RO) and XP-F (XP51RO) human fibroblasts were seeded on glass coverslips. Sixteen hours later, the cells were exposed to 3 μM MMC for 1 h and then fixed at multiple time points. FANCD2 was detected by immunofluorescence. Cells with FANCD2 foci were counted. Representative images are shown, and the averages from three experiments are plotted. A paired Student's t test was used to calculate significance. DAPI, 4′,6′-diamidino-2-phenylindole.
Chromatin-bound FANCD2 is reduced in XPF-ERCC1-deficient cells.
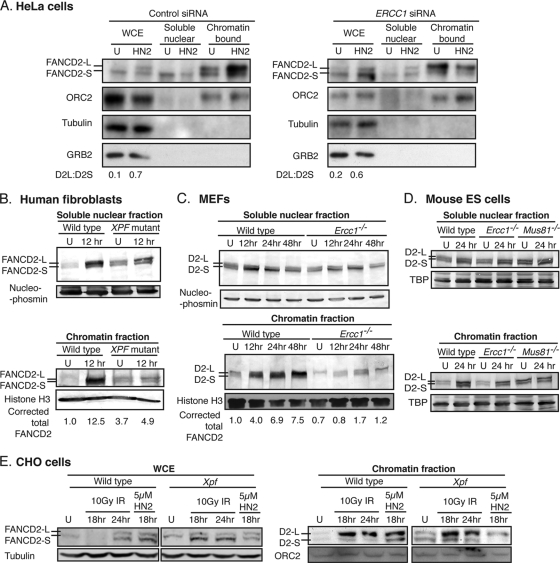
FANCD2 levels were normal in XPF-ERCC1-deficient cells exposed to cross-linking agents, yet chromatin-bound FANCD2 focus formation was diminished, suggesting the aberrant subcellular localization of FANCD2 in the absence of this nuclease. To further explore this, ERCC1 was depleted in HeLa cells by siRNA; the cells then were exposed to HN2 and fractionated. The immunodetection of FANCD2 in WCEs revealed that, as previously observed (Fig. 1B), FANCD2 was efficiently monoubiquitinated in both control and ERCC1-depleted cells (Fig. 4A). In control cells, the level of FANCD2-L in the chromatin fraction increased dramatically following the exposure of the cells to HN2 (Fig. 4A). In contrast, in ERCC1-depleted cells, the amount of FANCD2-L in the chromatin fraction did not increase following the exposure of cells to HN2 but instead was elevated in the soluble nuclear fraction (Fig. 4A).
FIG. 4.
Chromatin localization of FANCD2 is impaired in XPF-ERCC1-deficient cells exposed to cross-linking agents but not ionizing radiation. (A) HeLa cells depleted for ERCC1 or mock depleted with a control siRNA duplex were treated with 1 μM HN2 for 1 h. At 18 h after exposure, WCEs were prepared or cells were fractionated to isolate the soluble nuclear and chromatin-bound protein fractions. Samples were immunoblotted for FANCD2. The immunodetection of ORC2 (origin recognition complex 2) was used as a loading control for chromatin-bound protein and GRB2 for cytoplasmic protein. U indicates cells that were untreated. (B) Wild-type (C5RO) and XP-F (XP51RO) human fibroblasts were exposed to 3 μM MMC for 1 h and then fractionated 12 h later. The fractions were immunoblotted for FANCD2. Nucleophosmin and histone H3 were used as loading controls for the soluble nuclear and chromatin fractions, respectively. (C) Wild-type and Ercc1−/− MEFs were exposed to 3 μM MMC for 1 h and fractionated at multiple time points after exposure. Fractions were immunoblotted for FANCD2 and the loading controls. (D) Wild-type (IB10) and Ercc1−/− (clone 49) mouse ES cells were exposed to 3 μM MMC for 1 h and fractionated at 24 h after exposure. Fractions were immunoblotted for FANCD2 with TATA binding protein (TBP) as the loading control. (E) Wild-type and Xpf mutant CHO cells were exposed to 5 μM HN2 for 1 h or 10 Gy of ionizing radiation (IR). Cells were processed as described for panel A at 18 and 24 h after radiation, and samples were immunoblotted for FANCD2. Tubulin was used as a loading control for WCE, and ORC2 was used as a loading control for the chromatin fraction.
In human fibroblasts, the level of FANCD2-L was comparable for XP-F and wild-type cells before and after exposure to MMC (Fig. 4B). However, the amount of FANCD2 detected in the chromatin fraction of XP-F fibroblasts treated with MMC was substantially lower than that detected in normal human fibroblasts (Fig. 4B). This is consistent with the observation that XP-F cells had significantly fewer FANCD2 foci than wild-type cells after MMC treatment (Fig. 3). Similar results were obtained using congenic Ercc1−/− MEFs exposed to MMC (Fig. 4C), where again there was less chromatin-bound FANCD2 in the knockout cells than in the wild type in response to cross-link damage. The subcellular localization of FANCD2 was less dramatically affected in ES cells (Ercc1−/− and Mus81−/−) than fibroblasts (Fig. 4D). One possible explanation is that ES cells differ dramatically from fibroblasts in their cell cycle profile and checkpoint responses to cross-link damage (data not shown). Finally, an equivalent level of FANCD2-L was detected in wild-type and Xpf mutant CHO cells exposed to HN2 (Fig. 4E). However, substantially less FANCD2-L was detected in the chromatin fraction of the mutant cells. In contrast, the chromatin localization of FANCD2-L was not impaired in Xpf mutant cells exposed to ionizing radiation (Fig. 4E). This indicates that the aberrant subcellular localization of FANCD2-L seen in XPF-ERCC1-deficient cells was specific for ICLs and not other forms of DNA damage, such as direct DSBs, where XPF-ERCC1 is not required prior to HR-mediated repair (2).
Taken together, these data, obtained from three mammalian cell systems exposed to two different cross-linking agents, support the conclusion that FANCD2 is not stably localized to the chromatin in the absence of XPF-ERCC1. This suggests that the nucleolytic processing of cross-link damage is required for the stable accumulation of FANCD2 at or near the sites of damage. The data also imply that FANCD2 monoubiquitination does not always correlate with chromatin localization and should be considered distinct steps in the response to cross-link damage.
DISCUSSION
Two sets of proteins that are critical for the response to and repair of ICLs are the FA family of proteins and the XPF-ERCC1 complex (46, 55). Whether or not these proteins function in the same DNA damage detection and repair mechanism has not been established. A significant body of biochemical and cellular data indicates an essential role for XPF-ERCC1 endonuclease in the unhooking of ICLs (13, 38). However, it is not known if the unhooking of ICLs must occur prior to the activation of the FA pathway. Our studies revealed that XPF-ERCC1 nuclease is not required for the activation of the FA pathway, as defined by the monoubiquitination of FANCD2. In the same experiments, we confirmed that both XPF and ERCC1 are required for efficient ICL unhooking by using the modified comet assay. Thus, the nucleolytic processing of ICLs by XPF-ERCC1 is not a prerequisite for FA pathway activation. This is consistent with the results of numerous other laboratories, indicating that replisome stalling at DNA lesions and secondary structures, as occurs with ICLs, is sufficient to generate the signal that ultimately leads to FANCD2 ubiquitination (11, 21, 52). Given that ATR activation also is required for the timely ubiquitination of FANCD2 in response to ICLs (3), it is tempting to speculate that tracts of single-stranded DNA that occur at stalled replisomes are what trigger ATR-dependent FANCD2 monoubiquitination. In the case of ICLs, single-stranded DNA likely results from the nucleolytic processing of stalled forks rather than the uncoupling of leading and lagging strand synthesis (63), which is what activates ATR when replication is blocked by lesions that affect only one strand of DNA.
Although the XPF-ERCC1-dependent nucleolytic processing of ICLs is not required for FA pathway activation, our data did reveal a difference in the kinetics of FANCD2 modification in XPF-ERCC1-deficient cells. These nuclease-deficient cells show a persistence of monoubiquitinated FANCD2 in WCEs following ICL induction. In contrast to our results, McCabe et al. reported the decreased monoubiquitination of FANCD2 in cells depleted of ERCC1 by siRNA (44). However, like us, these authors did observe that MMC causes an induction of FANCD2 monoubiquitination in ERCC1-depleted cells, and that the relative increase in the ratio of FANCD2-L to FANCD2-S was similar in ERCC1-depleted and control cells upon cross-link damage. We observed persistent FANCD2 monoubiquitination in XPF-ERCC1-deficient cells from three species in response to two different cross-linking agents. Prolonged FANCD2 monoubiquitination has not been reported in other cross-link sensitive mutants previously, suggesting that the incision of cross-linked DNA by XPF-ERCC1 is essential for the repair of ICLs and the termination of the DNA damage response.
In addition, the chromatin localization of FANCD2 in response to cross-link damage is impaired in XPF-ERCC1-deficient cells, as demonstrated by cellular fractionation and FANCD2 foci formation. This suggests that the stable association of monoubiquitinated FANCD2 with chromatin requires (at a minimum) an unhooked ICL. Indeed, the persistent FANCD2-L observed in Xpf and Ercc1 mutant WCEs might reflect continued attempts to target FANCD2 to sites of damage in chromatin, which are nonproductive when cross-linked DNA is not incised.
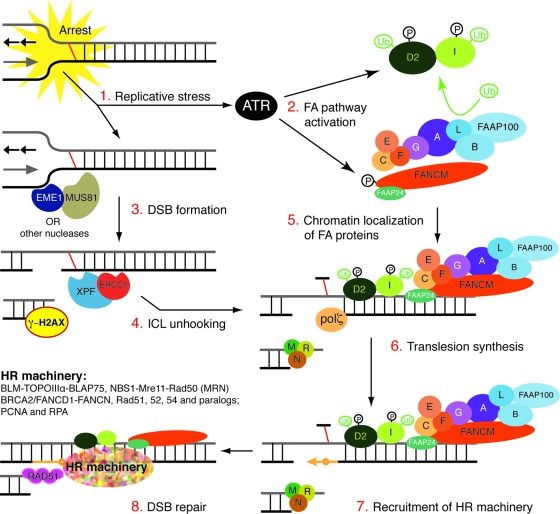
The XPF-ERCC1-related nuclease MUS81-EME1 also is implicated in ICL repair by virtue of the hypersensitivity of Mus81−/− and Eme1−/− mouse cells to cross-linking agents (23). Specifically, MUS81 is required for the cleavage of replication forks stalled by ICLs, producing DSBs with one accessible end (22, 23). However, the ICL-induced monoubiquitination of FANCD2 is normal in Mus81−/− cells (Fig. 1), and the genetic disruption of FANCB and MUS81 does not render cells hypersensitive to cross-linking agents relative to the deletion of FANCB alone (59). These data support the conclusion that the generation of DSBs at ICLs during S phase by MUS81-EME1 is not a prerequisite for the activation of FANCD2. Taken together, these data suggest a model for ICL repair (Fig. 5; also see a recent review by Thompson and Hinz [73]) in which the stalling of a replication fork at an ICL leads to the formation of a DSB created by MUS81-EME1 nuclease. Fork stalling, but not the generation of a DSB, is sufficient for the activation of ATR and the monoubiquitination of FANCD2. However, in the absence of the unhooking of the ICL by XPF-ERCC1, FANCD2 and, therefore, presumably the HR machinery do not stably associate at the sites of damage. This results in failure to repair ICL-dependent DSBs in XPF-ERCC1-deficient cells (56).
FIG. 5.
Model of how XPF-ERCC1 and MUS81-EME1 nucleases function in the same S-phase-specific DNA interstrand cross-link repair mechanism as the FA proteins. See the text for more details.
The data presented herein demonstrate that there is not an absolute correlation between the level of monoubiquitinated FANCD2 and its chromatin association. In XPF-ERCC1-deficient cells in which ICL-dependent DSBs are not repaired (56), the levels of monoubiquitinated FANCD2 persist longer than in wild-type cells after cross-link damage, but the chromatin localization of FANCD2 is reduced. This suggests that in the absence of ICL repair, damage signaling persists. In accordance, it was reported recently that FANCD2 deubiquitination by USP1 (58) is required for efficient ICL repair (61). Thus, when the chromatin association of FANCD2 is compromised, for instance in the absence of ICL unhooking by XPF-ERCC1, the deubiquitination of FANCD2 by USP1 does not occur. This is supported by the observation that FANCD2 deubiquitination is concurrent with the release of FANCD2 from chromatin (61). The eviction of FANCD2 from chromatin likely coincides with the recruitment of factors that are undertaken after unhooking steps of ICL repair, including HR-mediated DSB repair and translesion synthesis to fill the gap created by ICL unhooking (57, 76).
Acknowledgments
L.J.N. and N.R.B. were supported by The Ellison Medical Foundation (AG-NS-0303-05) and NCI (CA111525-03). A.O. was supported by ECMC, and A.T.W. was supported by a New Zealand Top Achiever Scholarship. Research in P.M.'s laboratory is funded by Cancer Research UK. Work in the R.K. laboratory was supported by The Netherlands Genomics Initiative/Netherlands Organization for Scientific Research (NWO) and the European Commission (Integrated Project 512113).
Footnotes
Published ahead of print on 5 October 2009.
REFERENCES
- 1.Abraham, J., B. Lemmers, M. P. Hande, M. E. Moynahan, C. Chahwan, A. Ciccia, J. Essers, K. Hanada, R. Chahwan, A. K. Khaw, P. McPherson, A. Shehabeldin, R. Laister, C. Arrowsmith, R. Kanaar, S. C. West, M. Jasin, and R. Hakem. 2003. Eme1 is involved in DNA damage processing and maintenance of genomic stability in mammalian cells. EMBO J. 22:6137-6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmad, A., A. R. Robinson, A. Duensing, E. van Drunen, H. B. Beverloo, D. B. Weisberg, P. Hasty, J. H. Hoeijmakers, and L. J. Niedernhofer. 2008. ERCC1-XPF endonuclease facilitates DNA double-strand break repair. Mol. Cell. Biol. 28:5082-5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andreassen, P. R., A. D. D'Andrea, and T. Taniguchi. 2004. ATR couples FANCD2 monoubiquitination to the DNA-damage response. Genes Dev. 18:1958-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Auerbach, A. D., and S. R. Wolman. 1976. Susceptibility of Fanconi's anaemia fibroblasts to chromosome damage by carcinogens. Nature 261:494-496. [DOI] [PubMed] [Google Scholar]
- 5.Bae, J. B., S. S. Mukhopadhyay, L. Liu, N. Zhang, J. Tan, S. Akhter, X. Liu, X. Shen, L. Li, and R. J. Legerski. 2008. Snm1B/Apollo mediates replication fork collapse and S Phase checkpoint activation in response to DNA interstrand cross-links. Oncogene 27:5045-5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berardini, M., P. L. Foster, and E. L. Loechler. 1999. DNA polymerase II (polB) is involved in a new DNA repair pathway for DNA interstrand cross-links in Escherichia coli. J. Bacteriol. 181:2878-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng, S., A. Sancar, and J. E. Hearst. 1991. RecA-dependent incision of psoralen-crosslinked DNA by (A)BC excinuclease. Nucleic Acids Res. 19:657-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ciccia, A., C. Ling, R. Coulthard, Z. Yan, Y. Xue, A. R. Meetei, H. Laghmani el, H. Joenje, N. McDonald, J. P. de Winter, W. Wang, and S. C. West. 2007. Identification of FAAP24, a Fanconi anemia core complex protein that interacts with FANCM. Mol. Cell 25:331-343. [DOI] [PubMed] [Google Scholar]
- 9.Cole, R. S. 1973. Repair of DNA containing interstrand crosslinks in Escherichia coli: sequential excision and recombination. Proc. Natl. Acad. Sci. USA 70:1064-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins, A. R. 1993. Mutant rodent cell lines sensitive to ultraviolet light, ionizing radiation and cross-linking agents: a comprehensive survey of genetic and biochemical characteristics. Mutat. Res. 293:99-118. [DOI] [PubMed] [Google Scholar]
- 11.D'Andrea, A. D., and M. Grompe. 2003. The Fanconi anaemia/BRCA pathway. Nat. Rev. Cancer 3:23-34. [DOI] [PubMed] [Google Scholar]
- 12.de Laat, W. L., E. Appeldoorn, N. G. Jaspers, and J. H. Hoeijmakers. 1998. DNA structural elements required for ERCC1-XPF endonuclease activity. J. Biol. Chem. 273:7835-7842. [DOI] [PubMed] [Google Scholar]
- 13.De Silva, I. U., P. J. McHugh, P. H. Clingen, and J. A. Hartley. 2000. Defining the roles of nucleotide excision repair and recombination in the repair of DNA interstrand cross-links in mammalian cells. Mol. Cell. Biol. 20:7980-7990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Digweed, M., S. Rothe, I. Demuth, R. Scholz, D. Schindler, M. Stumm, M. Grompe, A. Jordan, and K. Sperling. 2002. Attenuation of the formation of DNA-repair foci containing RAD51 in Fanconi anaemia. Carcinogenesis 23:1121-1126. [DOI] [PubMed] [Google Scholar]
- 15.Dronkert, M. L., J. de Wit, M. Boeve, M. L. Vasconcelos, H. van Steeg, T. L. Tan, J. H. Hoeijmakers, and R. Kanaar. 2000. Disruption of mouse SNM1 causes increased sensitivity to the DNA interstrand cross-linking agent mitomycin C. Mol. Cell. Biol. 20:4553-4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Durant, S. T., M. M. Morris, M. Illand, H. J. McKay, C. McCormick, G. L. Hirst, R. H. Borts, and R. Brown. 1999. Dependence on RAD52 and RAD1 for anticancer drug resistance mediated by inactivation of mismatch repair genes. Curr. Biol. 9:51-54. [DOI] [PubMed] [Google Scholar]
- 17.Enzlin, J. H., and O. D. Scharer. 2002. The active site of the DNA repair endonuclease XPF-ERCC1 forms a highly conserved nuclease motif. EMBO J. 21:2045-2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujiwara, Y., and M. Tatsumi. 1977. Cross-link repair in human cells and its possible defect in Fanconi's anemia cells. J. Mol. Biol. 113:635-649. [DOI] [PubMed] [Google Scholar]
- 19.Garcia-Higuera, I., Y. Kuang, D. Naf, J. Wasik, and A. D. D'Andrea. 1999. Fanconi anemia proteins FANCA, FANCC, and FANCG/XRCC9 interact in a functional nuclear complex. Mol. Cell. Biol. 19:4866-4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia-Higuera, I., T. Taniguchi, S. Ganesan, M. S. Meyn, C. Timmers, J. Hejna, M. Grompe, and A. D. D'Andrea. 2001. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol. Cell 7:249-262. [DOI] [PubMed] [Google Scholar]
- 21.Gregory, R. C., T. Taniguchi, and A. D. D'Andrea. 2003. Regulation of the Fanconi anemia pathway by monoubiquitination. Semin. Cancer Biol. 13:77-82. [DOI] [PubMed] [Google Scholar]
- 22.Hanada, K., M. Budzowska, S. L. Davies, E. van Drunen, H. Onizawa, H. B. Beverloo, A. Maas, J. Essers, I. D. Hickson, and R. Kanaar. 2007. The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nat. Struct. Mol. Biol. 14:1096-1104. [DOI] [PubMed] [Google Scholar]
- 23.Hanada, K., M. Budzowska, M. Modesti, A. Maas, C. Wyman, J. Essers, and R. Kanaar. 2006. The structure-specific endonuclease Mus81-Eme1 promotes conversion of interstrand DNA crosslinks into double-strands breaks. EMBO J. 25:4921-4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hazrati, A., M. Ramis-Castelltort, S. Sarkar, L. J. Barber, C. J. Schofield, J. A. Hartley, and P. J. McHugh. 2008. Human SNM1A suppresses the DNA repair defects of yeast pso2 mutants. DNA Repair 7:230-238. [DOI] [PubMed] [Google Scholar]
- 25.Henriques, J. A., and E. Moustacchi. 1980. Isolation and characterization of pso mutants sensitive to photo-addition of psoralen derivatives in Saccharomyces cerevisiae. Genetics 95:273-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henriques, J. A., E. J. Vicente, K. V. Leandro da Silva, and A. C. Schenberg. 1989. PSO4: a novel gene involved in error-prone repair in Saccharomyces cerevisiae. Mutat. Res. 218:111-124. [DOI] [PubMed] [Google Scholar]
- 27.Howlett, N. G., T. Taniguchi, S. Olson, B. Cox, Q. Waisfisz, C. De Die-Smulders, N. Persky, M. Grompe, H. Joenje, G. Pals, H. Ikeda, E. A. Fox, and A. D. D'Andrea. 2002. Biallelic inactivation of BRCA2 in Fanconi anemia. Science 297:606-609. [DOI] [PubMed] [Google Scholar]
- 28.Hoy, C. A., L. H. Thompson, C. L. Mooney, and E. P. Salazar. 1985. Defective DNA cross-link removal in Chinese hamster cell mutants hypersensitive to bifunctional alkylating agents. Cancer Res. 45:1737-1743. [PubMed] [Google Scholar]
- 29.Hussain, S., J. B. Wilson, E. Blom, L. H. Thompson, P. Sung, S. M. Gordon, G. M. Kupfer, H. Joenje, C. G. Mathew, and N. J. Jones. 2006. Tetratricopeptide-motif-mediated interaction of FANCG with recombination proteins XRCC3 and BRCA2. DNA Repair 5:629-640. [DOI] [PubMed] [Google Scholar]
- 30.Ishiai, M., M. Kimura, K. Namikoshi, M. Yamazoe, K. Yamamoto, H. Arakawa, K. Agematsu, N. Matsushita, S. Takeda, J. M. Buerstedde, and M. Takata. 2004. DNA cross-link repair protein SNM1A interacts with PIAS1 in nuclear focus formation. Mol. Cell. Biol. 24:10733-10741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iyer, V. N., and W. Szybalski. 1963. A molecular mechanism of mitomycin action: linking of complementary DNA strands. Proc. Natl. Acad. Sci. USA 50:355-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jachymczyk, W. J., R. C. von Borstel, M. R. Mowat, and P. J. Hastings. 1981. Repair of interstrand cross-links in DNA of Saccharomyces cerevisiae requires two systems for DNA repair: the RAD3 system and the RAD51 system. Mol. Gen. Genet. 182:196-205. [DOI] [PubMed] [Google Scholar]
- 33.Jaspers, N. G., A. Raams, M. C. Silengo, N. Wijgers, L. J. Niedernhofer, A. R. Robinson, G. Giglia-Mari, D. Hoogstraten, W. J. Kleijer, J. H. Hoeijmakers, and W. Vermeulen. 2007. First reported patient with human ERCC1 deficiency has cerebro-oculo-facio-skeletal syndrome with a mild defect in nucleotide excision repair and severe developmental failure. Am. J. Hum. Genet. 80:457-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Joenje, H., and K. J. Patel. 2001. The emerging genetic and molecular basis of Fanconi anaemia. Nat. Rev. Genet. 2:446-457. [DOI] [PubMed] [Google Scholar]
- 35.Kapetanaki, M. G., J. Guerrero-Santoro, D. C. Bisi, C. L. Hsieh, V. Rapic-Otrin, and A. S. Levine. 2006. The DDB1-CUL4ADDB2 ubiquitin ligase is deficient in xeroderma pigmentosum group E and targets histone H2A at UV-damaged DNA sites. Proc. Natl. Acad. Sci. USA 103:2588-2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim, J. M., Y. Kee, A. Gurtan, and A. D. D'Andrea. 2008. Cell cycle dependent chromatin loading of the Fanconi anemia core complex by FANCM/FAAP24. Blood 111:5215-5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kohn, K. W., C. L. Spears, and P. Doty. 1966. Inter-strand crosslinking of DNA by nitrogen mustard. J. Mol. Biol. 19:266-288. [DOI] [PubMed] [Google Scholar]
- 38.Kuraoka, I., W. R. Kobertz, R. R. Ariza, M. Biggerstaff, J. M. Essigmann, and R. D. Wood. 2000. Repair of an interstrand DNA cross-link initiated by ERCC1-XPF repair/recombination nuclease. J. Biol. Chem. 275:26632-26636. [DOI] [PubMed] [Google Scholar]
- 39.Lan, L., T. Hayashi, R. M. Rabeya, S. Nakajima, S. Kanno, M. Takao, T. Matsunaga, M. Yoshino, M. Ichikawa, H. Riele, S. Tsuchiya, K. Tanaka, and A. Yasui. 2004. Functional and physical interactions between ERCC1 and MSH2 complexes for resistance to cis-diamminedichloroplatinum(II) in mammalian cells. DNA Repair 3:135-143. [DOI] [PubMed] [Google Scholar]
- 40.Li, L., C. A. Peterson, X. Zhang, and R. J. Legerski. 2000. Requirement for PCNA and RPA in interstrand crosslink-induced DNA synthesis. Nucleic Acids Res. 28:1424-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ling, C., M. Ishiai, A. M. Ali, A. L. Medhurst, K. Neveling, R. Kalb, Z. Yan, Y. Xue, A. B. Oostra, A. D. Auerbach, M. E. Hoatlin, D. Schindler, H. Joenje, J. P. de Winter, M. Takata, A. R. Meetei, and W. Wang. 2007. FAAP100 is essential for activation of the Fanconi anemia-associated DNA damage response pathway. EMBO J. 26:2104-2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu, N., J. E. Lamerdin, R. S. Tebbs, D. Schild, J. D. Tucker, M. R. Shen, K. W. Brookman, M. J. Siciliano, C. A. Walter, W. Fan, L. S. Narayana, Z. Q. Zhou, A. W. Adamson, K. J. Sorensen, D. J. Chen, N. J. Jones, and L. H. Thompson. 1998. XRCC2 and XRCC3, new human Rad51-family members, promote chromosome stability and protect against DNA cross-links and other damages. Mol. Cell 1:783-793. [DOI] [PubMed] [Google Scholar]
- 43.Marmorstein, L. Y., T. Ouchi, and S. A. Aaronson. 1998. The BRCA2 gene product functionally interacts with p53 and RAD51. Proc. Natl. Acad. Sci. USA 95:13869-13874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCabe, K. M., A. Hemphill, Y. Akkari, P. M. Jakobs, D. Pauw, S. B. Olson, R. E. Moses, and M. Grompe. 2008. ERCC1 is required for FANCD2 focus formation. Mol. Genet. Metab. 95:66-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McHugh, P. J., R. D. Gill, R. Waters, and J. A. Hartley. 1999. Excision repair of nitrogen mustard-DNA adducts in Saccharomyces cerevisiae. Nucleic Acids Res. 27:3259-3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McHugh, P. J., V. J. Spanswick, and J. A. Hartley. 2001. Repair of DNA interstrand crosslinks: molecular mechanisms and clinical relevance. Lancet Oncol. 2:483-490. [DOI] [PubMed] [Google Scholar]
- 47.McPherson, J. P., B. Lemmers, R. Chahwan, A. Pamidi, E. Migon, E. Matysiak-Zablocki, M. E. Moynahan, J. Essers, K. Hanada, A. Poonepalli, O. Sanchez-Sweatman, R. Khokha, R. Kanaar, M. Jasin, M. P. Hande, and R. Hakem. 2004. Involvement of mammalian Mus81 in genome integrity and tumor suppression. Science 304:1822-1826. [DOI] [PubMed] [Google Scholar]
- 48.Medhurst, A. L., P. A. Huber, Q. Waisfisz, J. P. de Winter, and C. G. Mathew. 2001. Direct interactions of the five known Fanconi anaemia proteins suggest a common functional pathway. Hum. Mol. Genet. 10:423-429. [DOI] [PubMed] [Google Scholar]
- 49.Meetei, A. R., J. P. de Winter, A. L. Medhurst, M. Wallisch, Q. Waisfisz, H. J. van de Vrugt, A. B. Oostra, Z. Yan, C. Ling, C. E. Bishop, M. E. Hoatlin, H. Joenje, and W. Wang. 2003. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat. Genet. 35:165-170. [DOI] [PubMed] [Google Scholar]
- 50.Meetei, A. R., M. Levitus, Y. Xue, A. L. Medhurst, M. Zwaan, C. Ling, M. A. Rooimans, P. Bier, M. Hoatlin, G. Pals, J. P. de Winter, W. Wang, and H. Joenje. 2004. X-linked inheritance of Fanconi anemia complementation group B. Nat. Genet. 36:1219-1224. [DOI] [PubMed] [Google Scholar]
- 51.Meetei, A. R., A. L. Medhurst, C. Ling, Y. Xue, T. R. Singh, P. Bier, J. Steltenpool, S. Stone, I. Dokal, C. G. Mathew, M. Hoatlin, H. Joenje, J. P. de Winter, and W. Wang. 2005. A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat. Genet. 37:958-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Montes de Oca, R., P. R. Andreassen, S. P. Margossian, R. C. Gregory, T. Taniguchi, X. Wang, S. Houghtaling, M. Grompe, and A. D. D'Andrea. 2005. Regulated interaction of the Fanconi anemia protein, FANCD2, with chromatin. Blood 105:1003-1009. [DOI] [PubMed] [Google Scholar]
- 53.Niedernhofer, L. J., J. Essers, G. Weeda, B. Beverloo, J. de Wit, M. Muijtjens, H. Odijk, J. H. Hoeijmakers, and R. Kanaar. 2001. The structure-specific endonuclease Ercc1-Xpf is required for targeted gene replacement in embryonic stem cells. EMBO J. 20:6540-6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Niedernhofer, L. J., G. A. Garinis, A. Raams, A. S. Lalai, A. R. Robinson, E. Appeldoorn, H. Odijk, R. Oostendorp, A. Ahmad, W. van Leeuwen, A. F. Theil, W. Vermeulen, G. T. van der Horst, P. Meinecke, W. J. Kleijer, J. Vijg, N. G. Jaspers, and J. H. Hoeijmakers. 2006. A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature 444:1038-1043. [DOI] [PubMed] [Google Scholar]
- 55.Niedernhofer, L. J., A. S. Lalai, and J. H. Hoeijmakers. 2005. Fanconi anemia (cross)linked to DNA repair. Cell 123:1191-1198. [DOI] [PubMed] [Google Scholar]
- 56.Niedernhofer, L. J., H. Odijk, M. Budzowska, E. van Drunen, A. Maas, A. F. Theil, J. de Wit, N. G. Jaspers, H. B. Beverloo, J. H. Hoeijmakers, and R. Kanaar. 2004. The structure-specific endonuclease Ercc1-Xpf is required to resolve DNA interstrand cross-link-induced double-strand breaks. Mol. Cell. Biol. 24:5776-5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Niedzwiedz, W., G. Mosedale, M. Johnson, C. Y. Ong, P. Pace, and K. J. Patel. 2004. The Fanconi anaemia gene FANCC promotes homologous recombination and error-prone DNA repair. Mol. Cell 15:607-620. [DOI] [PubMed] [Google Scholar]
- 58.Nijman, S. M., T. T. Huang, A. M. Dirac, T. R. Brummelkamp, R. M. Kerkhoven, A. D. D'Andrea, and R. Bernards. 2005. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol. Cell 17:331-339. [DOI] [PubMed] [Google Scholar]
- 59.Nomura, Y., N. Adachi, and H. Koyama. 2007. Human Mus81 and FANCB independently contribute to repair of DNA damage during replication. Genes Cells 12:1111-1122. [DOI] [PubMed] [Google Scholar]
- 60.O'Connor, P. M., and K. W. Kohn. 1990. Comparative pharmacokinetics of DNA lesion formation and removal following treatment of L1210 cells with nitrogen mustards. Cancer Commun. 2:387-394. [DOI] [PubMed] [Google Scholar]
- 61.Oestergaard, V. H., F. Langevin, H. J. Kuiken, P. Pace, W. Niedzwiedz, L. J. Simpson, M. Ohzeki, M. Takata, J. E. Sale, and K. J. Patel. 2007. Deubiquitination of FANCD2 is required for DNA crosslink repair. Mol. Cell 28:798-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Palom, Y., G. Suresh Kumar, L. Q. Tang, M. M. Paz, S. M. Musser, S. Rockwell, and M. Tomasz. 2002. Relative toxicities of DNA cross-links and monoadducts: new insights from studies of decarbamoyl mitomycin C and mitomycin C. Chem. Res. Toxicol. 15:1398-1406. [DOI] [PubMed] [Google Scholar]
- 63.Paulsen, R. D., and K. A. Cimprich. 2007. The ATR pathway: fine-tuning the fork. DNA Repair 6:953-966. [DOI] [PubMed] [Google Scholar]
- 64.Pichierri, P., and F. Rosselli. 2004. The DNA crosslink-induced S-phase checkpoint depends on ATR-CHK1 and ATR-NBS1-FANCD2 pathways. EMBO J. 23:1178-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Qiao, F., A. Moss, and G. M. Kupfer. 2001. Fanconi anemia proteins localize to chromatin and the nuclear matrix in a DNA damage- and cell cycle-regulated manner. J. Biol. Chem. 276:23391-23396. [DOI] [PubMed] [Google Scholar]
- 66.Ruhland, A., M. Kircher, F. Wilborn, and M. Brendel. 1981. A yeast mutant specifically sensitive to bifunctional alkylation. Mutat. Res. 91:457-462. [DOI] [PubMed] [Google Scholar]
- 67.Sarkar, S., A. A. Davies, H. D. Ulrich, and P. J. McHugh. 2006. DNA interstrand crosslink repair during G1 involves nucleotide excision repair and DNA polymerase zeta. EMBO J. 25:1285-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sasaki, M. S., and A. Tonomura. 1973. A high susceptibility of Fanconi's anemia to chromosome breakage by DNA cross-linking agents. Cancer Res. 33:1829-1836. [PubMed] [Google Scholar]
- 69.Sijbers, A. M., W. L. de Laat, R. R. Ariza, M. Biggerstaff, Y. F. Wei, J. G. Moggs, K. C. Carter, B. K. Shell, E. Evans, M. C. de Jong, S. Rademakers, J. de Rooij, N. G. Jaspers, J. H. Hoeijmakers, and R. D. Wood. 1996. Xeroderma pigmentosum group F caused by a defect in a structure-specific DNA repair endonuclease. Cell 86:811-822. [DOI] [PubMed] [Google Scholar]
- 70.Smogorzewska, A., S. Matsuoka, P. Vinciguerra, E. R. McDonald III, K. E. Hurov, J. Luo, B. A. Ballif, S. P. Gygi, K. Hofmann, A. D. D'Andrea, and S. J. Elledge. 2007. Identification of the FANCI Protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell 129:289-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stiff, T., S. A. Walker, K. Cerosaletti, A. A. Goodarzi, E. Petermann, P. Concannon, M. O'Driscoll, and P. A. Jeggo. 2006. ATR-dependent phosphorylation and activation of ATM in response to UV treatment or replication fork stalling. EMBO J. 25:5775-5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Taniguchi, T., I. Garcia-Higuera, P. R. Andreassen, R. C. Gregory, M. Grompe, and A. D. D'Andrea. 2002. S-phase-specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood 100:2414-2420. [DOI] [PubMed] [Google Scholar]
- 73.Thompson, L. H., and J. M. Hinz. 2009. Cellular and molecular consequences of defective Fanconi anemia proteins in replication-coupled DNA repair: mechanistic insights. Mutat. Res. 668:54-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tsodikov, O. V., J. H. Enzlin, O. D. Scharer, and T. Ellenberger. 2005. Crystal structure and DNA binding functions of ERCC1, a subunit of the DNA structure-specific endonuclease XPF-ERCC1. Proc. Natl. Acad. Sci. USA 102:11236-11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Van Houten, B., H. Gamper, S. R. Holbrook, J. E. Hearst, and A. Sancar. 1986. Action mechanism of ABC excision nuclease on a DNA substrate containing a psoralen crosslink at a defined position. Proc. Natl. Acad. Sci. USA 83:8077-8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang, X., P. R. Andreassen, and A. D. D'Andrea. 2004. Functional interaction of monoubiquitinated FANCD2 and BRCA2/FANCD1 in chromatin. Mol. Cell. Biol. 24:5850-5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ward, I. M., and J. Chen. 2001. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J. Biol. Chem. 276:47759-47762. [DOI] [PubMed] [Google Scholar]
- 78.Wilson, J. B., K. Yamamoto, A. S. Marriott, S. Hussain, P. Sung, M. E. Hoatlin, C. G. Mathew, M. Takata, L. H. Thompson, G. M. Kupfer, and N. J. Jones. 2008. FANCG promotes formation of a newly identified protein complex containing BRCA2, FANCD2 and XRCC3. Oncogene 27:3641-3652. [DOI] [PubMed] [Google Scholar]
- 79.Yu, V. P., M. Koehler, C. Steinlein, M. Schmid, L. A. Hanakahi, A. J. van Gool, S. C. West, and A. R. Venkitaraman. 2000. Gross chromosomal rearrangements and genetic exchange between nonhomologous chromosomes following BRCA2 inactivation. Genes Dev. 14:1400-1406. [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang, N., R. Kaur, X. Lu, X. Shen, L. Li, and R. J. Legerski. 2005. The Pso4 mRNA splicing and DNA repair complex interacts with WRN for processing of DNA interstrand cross-links. J. Biol. Chem. 280:40559-40567. [DOI] [PubMed] [Google Scholar]
- 81.Zhang, N., X. Lu, and R. J. Legerski. 2003. Partial reconstitution of human interstrand cross-link repair in vitro: characterization of the roles of RPA and PCNA. Biochem. Biophys. Res. Commun. 309:71-78. [DOI] [PubMed] [Google Scholar]
- 82.Zhang, N., X. Lu, X. Zhang, C. A. Peterson, and R. J. Legerski. 2002. hMutSβ is required for the recognition and uncoupling of psoralen interstrand cross-links in vitro. Mol. Cell. Biol. 22:2388-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]