Abstract
Interleukin-2 tyrosine kinase (Itk) is a Tec family tyrosine kinase that mediates signaling processes after T cell receptor engagement. Activation of Itk requires recruitment to the membrane via its pleckstrin homology domain, phosphorylation of Itk by the Src kinase, Lck, and binding of Itk to the SLP-76/LAT adapter complex. After activation, Itk phosphorylates and activates phospholipase C-γ1 (PLC-γ1), leading to production of two second messengers, DAG and IP3. We have previously shown that phosphorylation of PLC-γ1 by Itk requires a direct, phosphotyrosine-independent interaction between the Src homology 2 (SH2) domain of PLC-γ1 and the kinase domain of Itk. We now define this docking interface using a combination of mutagenesis and NMR spectroscopy and show that disruption of the Itk/PLCγ1 docking interaction attenuates T cell signaling. The binding surface on PLCγ1 that mediates recognition by Itk highlights a nonclassical binding activity of the well-studied SH2 domain providing further evidence that SH2 domains participate in important signaling interactions beyond recognition of phosphotyrosine.
Keywords: substrate recognition, T cell signaling, Tec kinases
Interleukin-2 tyrosine kinase (Itk) is a nonreceptor protein tyrosine kinase that is expressed in T cells, mast cells and NK cells (1–5). Previous studies have shown that activation of Itk after T cell receptor engagement requires Itk recruitment to PIP3 in the membrane via its PH domain, binding of Itk to the SLP-76/LAT adapter complex, and phosphorylation of Itk by Lck at the activation loop tyrosine in its kinase domain (6). Activated Itk then phosphorylates its substrate, phospholipase C-γ1 (PLC-γ1), resulting in activation of PLCγ1 lipase activity and subsequent hydrolysis of PIP2 to IP3 and DAG. IP3 and DAG stimulate the release of calcium ions from the endoplasmic reticulum and activate Protein Kinase C, respectively (7–11). The overall signaling pathway has been clearly delineated but many of the precise molecular details of the protein–protein interactions and enzyme/substrate interactions that control signal transduction after TCR engagement remain to be determined.
Itk belongs to the Tec family of nonreceptor tyrosine kinases that also includes Btk, Tec, Rlk, and Bmx (12). The Tec kinases, like the larger protein kinase superfamily, control numerous cellular signaling networks by phosphorylating target amino acid side chains in a stringently specific manner. Based on results of combinatorial peptide library screens and structures of kinase/peptide substrate complexes, the view has emerged that the active sites of most kinases tolerate different sequences and are therefore not necessarily stringently specific for short peptide sequences (13, 14). Stringent specificity is, however, a strict requirement of cellular signaling cascades and so these enzymes must have mechanisms to control substrate fidelity (14–22).
The physiological substrate of Itk, PLCγ1, is a phospholipase that contains an amino-terminal PH domain, EF hand motif, a split catalytic domain, a split PH domain, two tandem Src homology 2 (SH2) domains (SH2N for amino-terminal and SH2C for carboxy-terminal, respectively), a 33-amino acid linker, an SH3 domain, and a C2 domain. The tyrosine at position 783, located within the 33-amino acid linker region between the SH2C and SH3 domains, is the site of PLCγ1 phosphorylation by Itk (23–25). How does Itk achieve selectivity for this particular tyrosine when multiple potential sites of phosphohrylation are present in the substrate molecule?
We have previously shown that the PLCγ1 SH2C domain (spanning residues 659–756 within full-length PLCγ1) binds directly to the Itk kinase domain and is required for efficient phosphorylation of Y783 by Itk (26). Fragments of PLCγ1 that contain Y783 but not the SH2C domain are not efficiently phosphorylated by Itk. Moreover, phosphorylation of PLCγ1 substrate fragments that contain both SH2C and Y783 (spanning 659–789, hereafter referred to as PLCγ1 SH2C-linker) can be inhibited by titration with isolated PLCγ1 SH2C domain (26). The excess, free SH2C domain binds to the Itk kinase domain preventing association between Itk kinase domain and the PLCγ1 SH2C-linker substrate. Competition by free SH2C domain is specific since the SH2 domains from Grb2 and PI3K have no effect on Itk mediated phosphorylation of PLCγ1 (26). Thus, the PLCγ1 SH2C domain contains a recognition motif or docking site for Itk that mediates specific phosphorylation of the target tyrosine that is located outside of the SH2C domain at position 783.
Despite their fame as phosphotyrosine recognition modules, we find that mutation of the conserved phosphotyrosine binding pocket in PLCγ1 SH2C (R694A/R696A) has no effect on the PLCγ1/Itk docking interaction. Equal levels of Y783 phosphorylation by Itk are measured for both wildtype and the R694A/R696A mutant PLCγ1 SH2C-linker substrates suggesting that the specific SH2C residues involved in docking onto the Itk kinase are located outside of the classical SH2 ligand-binding surface (26).
We now define the Itk-docking element within the PLCγ1 SH2C domain and confirm that the site of interaction is distinct from the classical phosphotyrosine ligand binding surface common to all SH2 domains. Mutation of this nonclassical binding surface in the SH2C domain within full-length PLCγ1 leads to loss of Y783 phosphorylation by Itk and attenuation of calcium flux upon TCR stimulation in T cells. A comparison of these data to other SH2 mediated protein kinase docking interactions characterized to date suggests a wide versatility among SH2 binding interactions.
Results
Mapping the Docking Site on PLCγ1.
To identify the PLCγ1 SH2C residues involved in substrate recognition by the Itk kinase domain, we compared the primary sequences of the PLCγ1 SH2C domain derived from several species with the primary sequences of two SH2 domains that do not compete with Itk mediated substrate phosphorylation (PI3K and Grb2). Surface exposed residues that are conserved across the PLCγ1 SH2 domains from different species but differ in either PI3K or Grb2 (or both) were targeted for mutation. A total of 23 PLCγ1 SH2C candidate residues (see Fig. S1) were identified as potential substrate docking site residues. We expect that mutation of the docking motif that mediates the direct Itk/PLCγ1 substrate interaction will result in loss or reduction of substrate phosphorylation by Itk.
In the context of the PLCγ1 SH2C-linker substrate, the 23 conserved SH2C residues were mutated to alanine either separately or in groups. Wild-type SH2C-linker substrate and the resulting panel of mutant substrates were screened in an in vitro phosphorylation assay using full-length Itk enzyme (Fig. 1A). Phosphorylation levels for each substrate were assessed by Western blot using the anti-pY783 antibody. As shown in Fig. 1A, mutation of E709, K711, R748, K749, K751, or R753 to alanine (lanes 19–27) either completely abolishes or significantly diminishes Itk mediated phosphorylation of Y783 in the PLCγ1 SH2C-linker substrate. These data suggest that these SH2 domain residues are required for the interaction with Itk and subsequent phosphorylation of the remote Y783 site. Mutation of the other residues in the SH2C domain had little or no effect on the phosphorylation level of the PLCγ1 SH2C-linker substrate (Fig. 1A; lanes 4–18 and 28–39). Substrate phosphorylation levels that are equal to wild-type substrate in this functional assay indicate that the mutated residues do not contribute to the docking interaction between PLCγ1 SH2C domain and Itk kinase domain.
Fig. 1.
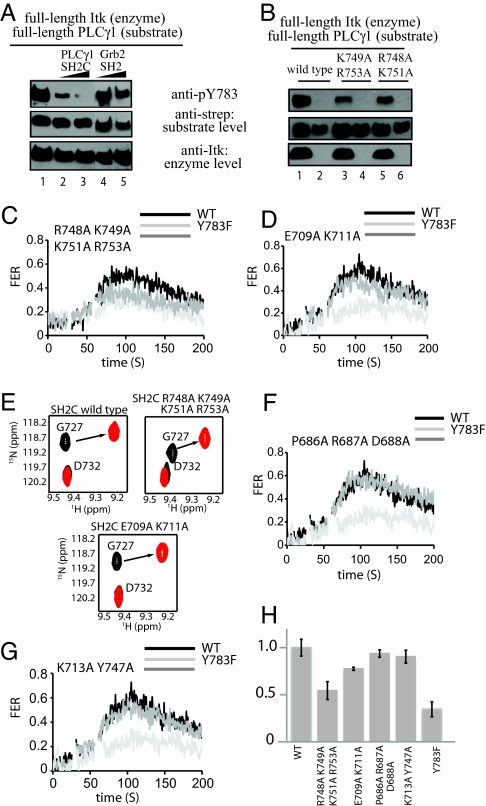
PLCγ1 SH2C domain mediates phosphorylation of remote tyrosine. (A) One and five micromolar PLCγ1 SH2C-linker substrate (wild-type: lanes 1–3) or indicated mutants (lanes 4–39) were subjected to in vitro phosphorylation by 475 nM full-length FLAG-tagged Itk enzyme. Lane 3, 6, 9, 12, 15, 18, 21, 24, 27, 30, 33, 36, and 39 are no Itk enzyme controls for the wild-type substrate or indicated mutants. Anti-pY783 blot indicates extent of PLCγ1 substrate phosphorylation by Itk; Coomassie stain shows substrate levels and the anti-FLAG blot shows the Itk enzyme levels. (B) Full-length Itk enzyme (158 nM) was incubated with 5 μM PLCγ1 SH2C-linker wild-type (lane 2), SH2C-linker K749A/R753A (lane 3), SH2C-linker R748A/K751A (lane 4), SH2C-linker E709A/K711A (lane 5) in an in vitro kinase assay and initial velocity is measured as described in refs. 26 and 43. Lane 1 is enzyme alone that has been subjected to the same assay conditions. (C) CD spectra of purified substrates (wild-type PLCγ1 SH2C domain and mutants shown in A).
To quantify the effect of the PLCγ1 SH2C domain mutations on Y783 phosphorylation, the substrate mutants that have the most pronounced effect in the assay shown in Fig. 1A were subjected to an in vitro kinase assay using purified full length Itk. The initial velocity for Y783 phosphorylation was determined for wild-type and PLCγ1 SH2C-linker mutants (Fig. 1B). Consistent with the Western blot data, Y783 phosphorylation is decreased to background levels in PLCγ1 SH2C-linker fragments in which E709, K711, R748, K749, K751, or R753 are mutated to alanine. To ensure that the mutations in the SH2C domain do not cause large structural changes to the SH2C-linker substrate that could account for the loss of phosphorylation on Y783, we compared the circular dichroism (CD) spectra for each mutant with wild-type PLCγ1 SH2C-linker (Fig. 1C). The CD spectra acquired for wild-type and each of the purified mutant substrates indicate that none of the mutations caused changes in secondary structure. Thus, the results of the mutational analyses indicate a set of surface residues on the PLCγ1 SH2C domain that mediate formation of an enzyme/substrate complex between Itk and PLCγ1 that is required for Y783 phosphorylation in vitro.
Mutation of SH2C Surface Residues in Full-Length PLCγ1 Diminishes Itk-Mediated Phosphorylation of Y783.
We next examined the extent to which the Itk/PLCγ1 docking interaction mediates phosphorylation of Y783 in the full-length PLCγ1 substrate instead of the short SH2C-linker substrate fragment used thus far. Given the previous observation that free PLCγ1 SH2C domain can compete with Itk mediated phosphorylation of Y783 in the PLCγ1 SH2C-linker substrate (26), we carried out the same experiment using full-length PLCγ1 as substrate. First, we find that full-length Itk enzyme readily phosphorylates Y783 in full-length PLCγ1 (Fig. 2A; lane 1). Consistent with the earlier experiments using the PLCγ1 substrate fragment (26), we find that addition of isolated PLCγ1 SH2C domain to the kinase assay competes effectively with Itk mediated phosphorylation of Y783 within full-length PLCγ1 (Fig. 2A; lanes 2 and 3). We also find that the Grb2 SH2 domain is not effective in competing with docking of full-length PLCγ1 onto Itk (Fig. 2A; lanes 4 and 5). These data provide further evidence that the SH2C domain within full length PLCγ1 is mediating interactions with Itk that are required for productive phosphorylation on Y783.
Fig. 2.
SH2 docking in full length PLCγ1 (A) Full-length PLCγ1 was subjected to an in vitro kinase assay using Itk full-length enzyme alone (lane 1), or with PLCγ1 SH2C domain (10 and 20 μM; lanes 2 and 3, respectively), or the Grb2 SH2 domain (10 and 20 μM; lanes 4 and 5). Anti-pY783 antibody is used to detect levels of Itk induced phosphorylation of Y783, anti-strep tag antibody indicates the level of the full-length PLCγ1 substrate and anti-Itk antibody is used to confirm Itk enzyme levels. (B) In lane 1: Strep-tagged, wild-type full-length PLCγ1 is incubated with Itk enzyme. Lanes 2, 4, and 6 are no Itk enzyme controls. Lanes 3 and 5 are the same experiment as lane 1 using either the K749A/R753A mutant of full-length PLCγ1 (lane 3) or the R748A/K751A mutant of full-length PLCγ1 (lane 5) as substrate. Blots are probed as described in A. (C, D, F, and G) Jurkat Jγ1 cells were transfected with full-length PLCγ1 wild-type or different mutants; histograms of GFP fluorescence after sorting for all cell lines are provided as (Fig. S2). Jurkat Jγ1 cells transfected with the indicated PLCγ1 constructs [wild-type PLCγ1, PLCγ1 mutants (C: R748A/K749A/K751A/R753A; D: E709A/K711A; F: P686A/R687A/D688A; G: K713A/Y747A and Y783F control)]. The two variants that target the basic C-terminal region of SH2C (see data in B) were combined in subsequent experiments (see C). After stimulation [Ca2+]i change is measured as fluorescence emission ratio (FER) at 398 nm (Ca2+ bound Indo-1) and 490 nm (Ca2+ unbound Indo-1) over a 200-s period. In C, D, F, and G [Ca2+]i change for Jurkat Jγ1 cells transfected with PLCγ1 wild-type after TCR stimulation is shown in black, [Ca2+]i change for Jurkat Jγ1 cells transfected with PLCγ1 Y783F after TCR stimulation is shown in light gray, and [Ca2+]i change for Jurkat Jγ1 cells transfected with the indicated mutation is shown in medium gray. The [Ca2+]i change for Jurkat Jγ1 cells transfected with PLCγ1 wild-type after mouse IgG isotype stimulation is provided as (Fig. S2). (E) Superposition of NMR data for wild-type or specified mutant PLCγ1 SH2C domain before (spectra of free SH2C shown in black in A–H) and after addition of phospholigand (red in each). The 15N-labeled wild-type SH2C, R784A/K749A/K751A/R753A SH2C mutant and E709A/K711A SH2C mutant were all concentrated to 0.3 mM (black spectrum) and an equimolar amount of phosphopeptide (PGF(pY)VEAN) was added before acquisition of the second spectrum in each case (red). Arrow indicates chemical shift change induced by phospholigand binding to each SH2C domain. NMR data here and in Fig. 3 were acquired on a Bruker AV700 spectrometer. (H) Histogram summarizing the data in C, D, F. and G, which are representative of at least three independent experiments. Data are normalized as described in SI Text. The Y783F data represents the extent of calcium flux with no phosphorylation at position 783 in PLCγ1.
To directly examine the Itk/PLCγ1 docking interaction in full-length PLCγ1, we first used an in vitro assay and targeted the highly basic cluster on SH2C. R748, K749, K751, and R753 were mutated to alanine in the context of full-length PLCγ1 and these full-length mutants were subjected to phosphorylation by Itk and compared to wild-type substrate. Eliminating the positively charged residues in the SH2C domain of full-length PLCγ1 results in diminished Itk mediated Y783 phosphorylation when compared to pY783 levels in the wild-type PLCγ1 substrate (Fig. 2B). These preliminary in vitro data prompted us to next examine the substrate-docking role of the PLCγ1 SH2C domain in vivo.
Mutation of the SH2C Domain in Full-Length PLCγ1 Attenuates Calcium Mobilization After TCR Stimulation.
It has been established previously that phosphorylation of PLCγ1 Y783 by Itk is important for T cell receptor (TCR)-stimulation dependent calcium mobilization (8, 27–30). Following this earlier work, we tested the extent to which mutations in the SH2C domain of PLCγ1 that diminish phosphorylation of Y783 affect calcium mobilization in T cells. The PLCγ1-deficient Jurkat T cell line, Jγ1, was transiently transfected with wild-type or mutant PLCγ1 constructs that had been cloned into a plasmid (pMSCV2.2-IRES-GFP) that encodes GFP on the same transcript to ensure that wild-type and mutant PLCγ1 are expressed at comparable levels (see Fig. S2 a and b). Sorted GFP positive cells were then stimulated with anti-CD3 and calcium signaling was measured as the fluorescence emission ratio (FER) of the Ca2+-bound and -unbound forms of Indo-1 (Fig. 2 C, D, F, and G). In addition to testing the role of specific PLCγ1 SH2C domain side chains in mediating T cell signaling, we included the Y783F mutant of PLCγ1 as a control for complete loss of phosphorylation at this position.
We first compared calcium flux after stimulation of Jγ1 cells transfected with wild-type PLCγ1 and the Y783F mutant, as well as isotype stimulation of nontransfected cells. Consistent with previous work (29), mutation of Y783 to phenylalanine in PLCγ1 results in a decrease in receptor-stimulation induced calcium mobilization compared to wild-type PLCγ1 (Fig. 2 C, D, F, and G) and the isotype stimulation control results in no measurable calcium flux (see Fig. S2c). The magnitude of the difference in calcium concentration between wild-type PLCγ1 and the Y783F mutant after TCR stimulation provides sufficient dynamic range to measure the effect of mutations in the SH2C domain of PLCγ1 on T cell signaling. Thus, calcium mobilization data for the mutants, PLCγ1 R748A/K749A/K751A/R753A and E709A/K711A, were acquired and compared to wild-type PLCγ1 and the Y783F mutant (Fig. 2 C and D). Consistent with the in vitro data presented in Figs. 1 B and C and 2 A and B, the SH2C domain mutations that disrupt the substrate docking interaction with the Itk kinase domain significantly attenuate calcium mobilization (Fig. 2H).
Because changes in the canonical phospholigand binding of the PLCγ1 SH2C domain could also affect calcium signaling via a distinct mechanism, we were careful to compare the CD spectra and phospholigand binding behavior of the wild-type PLCγ1 SH2C domain and the R748A/K749A/K751A/R753A and E709A/K711A SH2C mutants. The mutant and wild-type PLCγ1 SH2C domains exhibit similar CD spectra (Fig. 1C and Fig. S2d) and identical phospholigand binding in an NMR binding assay (Fig. 2E) providing further support that the reduced signaling observed for the PLCγ1 mutants in T cells is due to reduced interaction between PLCγ1 and Itk and not due to altered phospholigand binding or an altered structure of the mutated PLCγ1 SH2C domain.
PLCγ1 SH2C Structure, the Substrate Docking Site.
The six SH2C residues in PLCγ1 that affect Y783 phosphorylation and calcium mobilization (R748, K749, K751, R753, E709, and K711) map to a contiguous surface on the structure of the PLCγ1 SH2C domain that is located outside of the canonical phosphotyrosine binding site (Fig. 3A). K711 and E709 reside on the CD loop of the SH2C domain and R748, K749, K751, and R753 are located at the C terminus of the domain. Residues for which mutation to alanine has no detectable effect on Y783 phosphorylation in the Itk phosphorylation assay (Fig. 1B) are outside of this binding site and are depicted in orange in Fig. 3A.
Fig. 3.
Structural features of the SH2C docking surface. (A) Results of the mutational analyses (Fig. 1A) are mapped onto the structure of PLCγ1 SH2C domain (45) (PDB ID code 2PLD). Blue corresponds to amino acids for which mutation to alanine disrupts docking onto the Itk kinase domain and subsequent phosphorylation of Y783 and calcium mobilization in T cell signaling. Docking site residues are labeled and are located on the CD loop and the C terminus of the SH2C domain. Orange indicates residues that, when mutated, have no effect on Y783 phosphorylation and/or calcium mobilization in T cell signaling. (B) Residues peripheral to the binding site are shown in orange and labeled. Structure figures are created using PyMol (46). (C) Overlay of HSQC spectra acquired for a 50 μM sample of PLCγ1 SH2N-SH2C alone (red) and in the presence of 30 μM unlabeled Itk kinase domain (black). Extensive line broadening is evident for a subset of the PLCγ1 SH2C residues after addition of the Itk kinase domain. Three broadened resonances are shown (K751, E708, and M750) and one representative peak that does not change upon addition of kinase is also shown (R684) for comparison. (D) PLCγ1 SH2C residues for which NMR resonances broaden on addition of Itk kinase domain are shown in red and labeled on the SH2C structure. The docking site region (as determined by mutagenesis) is circled as is the phosphotyrosine binding pocket (dashed circle).
In the structural view that emerges in Fig. 3A, there are several additional residues (K713, Y747, P686, R687, and D688) that surround the substrate-docking surface (Fig. 3B) but were not tested in our initial mutant screen (Fig. 1A). We therefore tested how mutation of these side chains to alanine affects calcium mobilization in Jγ1 cells to further delineate the substrate-docking site on SH2C. For these mutants we find a minimal effect on calcium mobilization upon TCR stimulation when compared with wild-type PLCγ1 (Fig. 2 F and G). These data therefore suggest that the primary docking surface on the PLCγ1 SH2C domain is confined to the largely basic patch of amino acids clustered in the C terminus and the CD loop.
To further probe the interaction between the PLCγ1 SH2C domain and Itk, we expressed and purified a fragment of PLCγ1 containing SH2C (SH2N-SH2C) and acquired NMR data both with and without addition of purified Itk kinase domain (Fig. 3C). We find that the NMR resonances corresponding to R675, G689, L692, V693, R696, I704, E709, K713, R748, M750, K751, and R753 showed significant line broadening after addition of Itk kinase while all other SH2C resonances in the spectrum remain unchanged (Fig. 3C). Half of the broadened resonances correspond to the substrate-docking site on SH2C that has been mapped by mutagenesis (G689, E709, K713, R748, M750, K751, and R753) and the remainder cluster within the interior beta strands of the SH2C domain and the phosphotyrosine binding pocket (Fig. 3D). The fact that spectral changes in the phosphotyrosine (pY) binding pocket are observed is interesting but our previously published mutational data in this site unequivocally show that substrate docking is not directly mediated by this region of SH2C (26). Thus, the results of this NMR binding experiment provide further support for the direct interaction between the PLCγ1 SH2C domain and the Itk kinase domain that facilitates Y783 phosphorylation. Whether allosteric perturbations of the phosphotyrosine binding pocket (via core SH2C residues) also plays a role in controlling PLCγ1 activation by Itk remains to be determined.
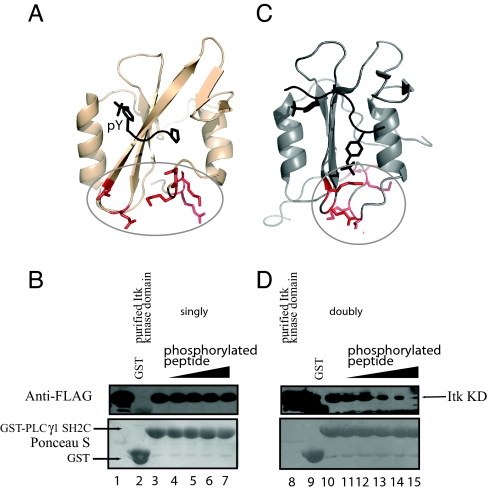
Visualizing the docking site on the structure of the PLCγ1 SH2C domain bound to a phosphopeptide ligand (Fig. 4A) illustrates why mutations in the pY binding pocket that disrupt classical phosphotyrosine binding of the SH2C domain have no measurable effect on the substrate docking interaction between PLCγ1 and the Itk kinase domain (26). The NMR spectral changes in the pY pocket of SH2C described above prompted us to test whether the interaction between PLCγ1 SH2C and its classical phospholigand might alter binding of PLCγ1 SH2C to the Itk kinase domain. Using the binding assay developed previously to monitor the PLCγ1 SH2C/Itk kinase interaction (26), we find that increasing concentrations of the tyrosine phosphorylated peptide, PGF(pY)VEAN, has no effect on the SH2C/kinase interaction (Fig. 4B). This result suggests that the substrate docking interaction between PLCγ1 and the Itk kinase domain can occur while the PLCγ1 SH2C domain is bound to its canonical ligand target.
Fig. 4.
Doubly phosphorylated peptide alters docking surface. (A) Structure of the PLCγ1 SH2C domain bound to a classical phosphotyrosine containing ligand. Ligand is depicted in black and the docking site residues that bind the Itk kinase domain are red and circled. (B) Binding assay showing the interaction between purified Itk kinase domain (0.22 μM) and the GST-PLCγ1 SH2C fusion protein (3.8 μM added to beads). Anti-flag is used to detect Itk kinase domain that binds to immobilized SH2C (Top). Lane 1 is purified Itk kinase domain, lane 2 is the GST control, lane 3 shows the SH2C/kinase interaction with no added peptide, lanes 4–7 are the same experiment with increasing concentration of phosphopeptide, PGF(pY)VEAN (peptide concentrations are: 0.19, 0.38, 3.84, and 19.22 μM in lanes 4–7, respectively). The Bottom shows GST and GST-SH2C levels. (C) Structure of the PLCγ1 SH2C domain bound to a doubly phosphorylated peptide derived from Syk showing the structural rearrangement within SH2C (31). Peptide ligand is shown in black and the SH2C residues required for binding to Itk kinase domain are in red and circled. (D) Same binding experiment as in (B). Increasing concentration of the doubly phosphorylated peptide ligand, DTEV(pY)ESP(pY)ADPE (peptide concentrations are: 0.38, 0.96, 1.92, 3.84, and 19.22 μM in lanes 11–15, respectively).
The PLCγ1 SH2C domain has been structurally well characterized and an unusual structure of this domain involves binding of a doubly tyrosine phosphorylated peptide, DTEV(pY)ESP(pY)ADPE, derived from Syk (31). Binding of the doubly phosphorylated peptide to the PLCγ1 SH2C domain induces a significant conformational change in the SH2C domain structure (Fig. 4C). The largest structural changes are localized to the CD loop, β strand D (βD), and the C terminus (Fig. 4C), coinciding precisely with the region of the SH2C domain that has been mapped in this study as the binding site for the Itk kinase domain. We therefore reasoned that, unlike the classical phospholigand (Fig. 4 A and B), the doubly phosphorylated ligand derived from Syk might alter the SH2C/kinase docking interaction by altering the structure of the SH2C domain in the region required for binding to Itk. Using the binding assay described in Fig. 4B, we find that binding of the Itk kinase domain to immobilized PLCγ1 SH2C domain is inhibited by addition of increasing concentration of the doubly phosphorylated peptide ligand (Fig. 4D). This result further confirms the importance of the C terminus and CD loop residues on the PLCγ1 SH2C domain for a productive interaction with the Itk kinase domain leading to phosphorylation on Y783 and a normal T cell response.
Discussion
SH2 domains are well known as binding modules for phosphotyrosine (pY) containing sequences (32). The classical SH2/pY interaction plays an essential role in a large number of cell signaling pathways including the T cell signaling pathway involving Itk and PLCγ1 (9, 27, 33–36). The substrate recognition/docking role of the PLCγ1 SH2C domain that we describe here involves an interaction surface on the PLCγ1 SH2C domain that is distinct from, and does not overlap with, the classical phosphotyrosine ligand-binding pocket. Disrupting the PLCγ1 SH2C domain/Itk kinase domain interaction interface by mutagenesis causes decreased Y783 phosphorylation in vitro and attenuated calcium mobilization in T cells.
In addition to the Y783 target in PLCγ1, Itk also phosphorylates a tyrosine (Y180) within its own SH3 domain and like PLCγ1, this autophosphorylation is mediated by a direct docking interaction between the Itk kinase domain and the Itk SH2 domain that is adjacent to the target tyrosine (26, 37). Thus, both the PLCγ1 and Itk SH2 domains bind directly to the Itk kinase domain to facilitate specific tyrosine phosphorylation elsewhere in the substrate molecule. Like PLCγ1 SH2C, the Itk SH2 docking site contains a cluster of basic amino acids that when mutated disrupt phsophorylation of the neighboring Y180 (38). The chemical similarities between the docking sites on the PLCγ1 and Itk SH2 domains, and the fact that both domains are the target of the same kinase, suggest the possibility that both substrates interact with the same or closely related surfaces on the Itk kinase domain. However, a comparison of the Itk and PLCγ1 SH2 structures (Fig. 5 A and B) shows that the two docking sites are located on distinct regions of the SH2 domain structure, likely reflecting different geometrical requirements of the two substrates. The target tyrosine within PLCγ1, Y783, is located in a linker region that is C-terminal to the SH2C domain while the autophosphorylation site within Itk, Y180, is located in the domain that is N-terminal to the Itk SH2 domain.
Fig. 5.
Structural comparisons of the PLCγ1 SH2C docking site. (A) Itk binding site on PLCγ1 SH2C is highlighted in red. (B) Itk binding site on Itk SH2 (Pdb: 1LUN) in cyan (38). In (A) and (B) the orientation of the PLCγ1 and Itk SH2 domains is the same permitting comparison of the respective kinase docking sites. (C) Structure of the SAP SH2/Fyn SH3/SLAM complex (PDB ID code 1M27). The site of Fyn SH3 binding is circled (red dashes) on the SAP SH2 domain (DE loop and B helix). (D) Structure of the PLCγ1 SH2N domain (blue) bound to the FGFR1 kinase domain (black) (PDB ID code 3GQI). The classical phosphotyrosine (pY) binding of SH2N to the tail of FGFR1 is shown. The secondary site of SH2N/kinase interaction is circled (red dashes) and involves the BC and DE loops. In (C) and (D) the SH2 domains are shown in the same orientation as the PLCγ1 SH2C domain in Fig. 4A to facilitate comparison.
Two additional SH2 domains that exploit binding regions outside of the classical pY binding pocket to colocalize a kinase and substrate are shown in Fig. 5 C and D. The SAP SH2 domain (Fig. 5C) binds simultaneously to the SH3 domain of the Fyn kinase and the cytoplasmic tyrosine-based motifs of SLAM family receptors resulting in colocalization of Fyn with its SLAM substrate (39, 40). Structural data show how the single SH2 domain of SAP binds to both a canonical ligand (SLAM) via its phosphotyrosine binding pocket and the Fyn SH3 domain in a phosphotyrosine-independent manner (41). The Fyn binding site on SAP SH2 is outside of the phosphotyrosine binding region and involves the DE loop and part of the B helix, structural elements that do not mediate the PLCγ1/Itk docking interaction (Figs. 4A and 5C). Another example of SH2 mediated kinase recognition comes from the interaction between the fibroblast growth factor receptor kinase, FGFR1, and the amino-terminal SH2 domain (SH2N) of PLCγ1 (42). In this case, the SH2 domain binds to the pY containing tail of the kinase domain and makes additional contacts with the kinase domain via a secondary binding site on the SH2 domain consisting of the BC and DE loops (Fig. 5D). Thus, in these examples it is evident that SH2 domains harbor a diverse set of interaction surfaces outside of the well-defined phospholigand binding pockets (Figs. 4A and 5 C and D). Notably, in each of these examples the docking interactions can occur simultaneously with phospholigand binding to the SH2 domain underscoring the diverse and multivalent nature of the SH2 domain.
The preponderance of positively charged residues on the mapped docking surface of the PLCγ1 SH2C domain suggests that the docking site on the Itk kinase domain likely consists of a complementary cluster of acidic residues. The Itk kinase domain contains several acidic clusters that are located outside of the kinase active site but confirmation of the precise binding residues on the kinase domain must await experimental verification. It is interesting to note that for other kinases [reviewed in Remenyi et al. (15)], distinctly different docking locations have been described on the catalytic domains (all outside of the catalytic cleft) suggesting that different kinases solve the problem of substrate fidelity using distinct docking platforms. It will also be important to evaluate the interplay between substrate docking onto the Itk kinase domain and allosteric control of Itk catalytic activity (43). As details of substrate recognition and activation mechanisms are further characterized for the Itk/PLCγ1 and other kinase/substrate pairs, opportunities should arise to use small molecules to modulate phosphorylation events, and in turn particular signaling cascades, in a highly specific manner that would not require the small molecule/drug to discriminate between similar kinase active sites.
Methods
Reagent Production.
Description of DNA and baculoviral constructs is provided as SI Text. Protein production and purification for full-length Itk and PLCγ1 fragments have been described in detail (see refs. 26 and 43). For purification of full-length PLCγ1, insect cells were infected with PLCγ1 baculovirus and harvested 72 h postinfection, rinsed with phosphate-buffer saline (PBS), and stored at −80 °C. Before purification, cell pellets were resuspended in lysis buffer [20 mM Hepes (pH 7.2), 150 mM NaCl, 2 mM EDTA, 0.1% Triton X-100, mixture protease inhibitor] and lysed by sonication. The total lysate was cleared by spinning at 14 K at 4 °C for 20 min. The supernatant was incubated with Strep Tactin resin (Novagen) for a minimum of 2 h at 4 °C. The protein-bound resin was then washed four times with PBS and two times with Hepes buffer [50 mM Hepes (pH 7.0), 10 mM MgCl2]. Purified, phosphorylated peptides were purchased from Genscript.
Assays.
Kinase assays, initial velocity measurements and Western blot procedures have been described in refs. 26, 38, and 43. Circular dichroism (CD) measurements of the PLCγ1 SH2C-linker variants were carried out as described in ref. 38 and NMR data acquisition followed procedures described in ref. 44.
For calcium flux measurements Jurkat Jγ1 (PLCγ1 deficient Jurkat) were maintained in RPMI medium 1640 supplemented with 10% FBS (HyClone), 10 mM Hepes (Invitrogen), 1 mM sodium pyruvate (Invitrogen), penicillin (100 units/mL)/streptomycin (100 μg/mL, Invitrogen), and 2 mM l-glutamine (Invitrogen). Jurkat Jγ1 cells were harvested and resuspended at 2 × 107 cells/mL in RPMI MEDIUM 1640 without supplements. Cells (0.4 mL per sample) were incubated with 20 μg desired DNA for 20 min, then cells were transfected by electroporation on a BTX 600 at 260 V, 1050 mF and 720 ohms with a disposable cuvette P/N 640. GFP positive cells were enriched by sorting on a FacsAria (Becton Dickinson). Sorting was done with a 70-μm nozzle under high pressure (70 psi) with a single cell mask purity setting. Live cells were gated using a Forward scatter × side (orthogonal) scatter plot and GFP was filtered using a 530/15 filter. Sorted GFP positive cells were resuspended in HBSS supplemented with 10 mM Hepes (pH7.0) at 1 × 106 cells/mL. After loading with 2 μM Indo-1-AM and 4 mM probenecid, the cell suspension was incubated at 37 °C for 30 min. The Indo-1-loaded cells were washed with HBSS supplemented with 10 mM Hepes (pH 7.0) and 2 mg/mL BSA. After washing, cells were resuspended in HBSS supplemented with 10 mM Hepes (pH 7.0) and rested at room temperature for 20 min in the dark. Before conducting the assay, cells were prewarmed at 37 °C for 10 min. Calcium flux was measured on a Cary Eclipse fluorescence spectrophotometer (Varian, Inc.) at an excitation wavelength of 352 nm (5 nm slit) while being imaged simultaneously at emission wavelengths of 398 and 490 nm (10 nm slit). Cells were stimulated with 5 μg/mL mouse anti-human anti-CD3 at 25 s and then cross-linked with 10 μg/mL goat-anti-mouse IgG at 55 s. [Ca2+]i concentration change induced by TCR stimulation and T cell activation are presented as the fluorescence emission ratio (FER) of the Ca2+-bound and -unbound forms of Indo-1.
Supplementary Material
Acknowledgments.
We thank Dr. Bruce Pesch, of the National Center for Animal Health in Ames, Iowa for assistance using the flow cytometer, Leslie Berg, Bob Geahlen, and Carol Post for discussions during preparation of the manuscript. This work was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0911309106/DCSupplemental.
References
- 1.Gibson S, et al. Identification, cloning, and characterization of a novel human T-cell-specific tyrosine kinase located at the hematopoietin complex on chromosome 5q. Blood. 1993;82:1561–1572. [PubMed] [Google Scholar]
- 2.Heyeck SD, Berg LJ. Developmental regulation of a murine T-cell-specific tyrosine kinase gene, Tsk. Proc Natl Acad Sci USA. 1993;90:669–673. doi: 10.1073/pnas.90.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siliciano JD, Morrow TA, Desiderio SV. itk, a T-cell-specific tyrosine kinase gene inducible by interleukin 2. Proc Natl Acad Sci USA. 1992;89:11194–11198. doi: 10.1073/pnas.89.23.11194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanaka N, Asao H, Ohtani K, Nakamura M, Sugamura K. A novel human tyrosine kinase gene inducible in T cells by interleukin 2. FEBS Lett. 1993;324:1–5. doi: 10.1016/0014-5793(93)81520-a. [DOI] [PubMed] [Google Scholar]
- 5.Yamada N, et al. Structure and expression of novel protein-tyrosine kinases, Emb and Emt, in hematopoietic cells. Biochem Biophys Res Commun. 1993;192:231–240. doi: 10.1006/bbrc.1993.1404. [DOI] [PubMed] [Google Scholar]
- 6.Berg LJ, Finkelstein LD, Lucas JA, Schwartzberg PL. Tec family kinases in T lymphocyte development and function. Annu Rev Immunol. 2005;23:549–600. doi: 10.1146/annurev.immunol.22.012703.104743. [DOI] [PubMed] [Google Scholar]
- 7.Lee SB, Rhee SG. Significance of PIP2 hydrolysis and regulation of phospholipase C isozymes. Curr Opin Cell Biol. 1995;7:183–189. doi: 10.1016/0955-0674(95)80026-3. [DOI] [PubMed] [Google Scholar]
- 8.Liu KQ, Bunnell SC, Gurniak CB, Berg LJ. T cell receptor-initiated calcium release is uncoupled from capacitative calcium entry in Itk-deficient T cells. J Exp Med. 1998;187:1721–1727. doi: 10.1084/jem.187.10.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perez-Villar JJ, Kanner SB. Regulated association between the tyrosine kinase Emt/Itk/Tsk and phospholipase-C gamma 1 in human T lymphocytes. J Immunol. 1999;163:6435–6441. [PubMed] [Google Scholar]
- 10.Schaeffer EM, et al. Requirement for Tec kinases Rlk and Itk in T cell receptor signaling and immunity. Science. 1999;284:638–641. doi: 10.1126/science.284.5414.638. [DOI] [PubMed] [Google Scholar]
- 11.Wilcox HM, Berg LJ. Itk phosphorylation sites are required for functional activity in primary T cells. J Biol Chem. 2003;278:37112–37121. doi: 10.1074/jbc.M304811200. [DOI] [PubMed] [Google Scholar]
- 12.Schwartzberg PL, Finkelstein LD, Readinger JA. TEC-family kinases: Regulators of T-helper-cell differentiation. Nat Rev Immunol. 2005;5:284–295. doi: 10.1038/nri1591. [DOI] [PubMed] [Google Scholar]
- 13.Bose R, Holbert MA, Pickin KA, Cole PA. Protein tyrosine kinase-substrate interactions. Curr Opin Struct Biol. 2006;16:668–675. doi: 10.1016/j.sbi.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 14.Miller WT. Determinants of substrate recognition in nonreceptor tyrosine kinases. Acc Chem Res. 2003;36:393–400. doi: 10.1021/ar020116v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Remenyi A, Good MC, Lim WA. Docking interactions in protein kinase and phosphatase networks. Curr Opin Struct Biol. 2006;16:676–685. doi: 10.1016/j.sbi.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 16.Biondi RM, Nebreda AR. Signalling specificity of Ser/Thr protein kinases through docking-site-mediated interactions. Biochem J. 2003;372:1–13. doi: 10.1042/BJ20021641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mayer BJ, Hirai H, Sakai R. Evidence that SH2 domains promote processive phosphorylation by protein-tyrosine kinases. Curr Biol. 1995;5:296–305. doi: 10.1016/s0960-9822(95)00060-1. [DOI] [PubMed] [Google Scholar]
- 18.Lewis LA, et al. The Lck SH2 phosphotyrosine binding site is critical for efficient TCR-induced processive tyrosine phosphorylation of the zeta-chain and IL-2 production. J Immunol. 1997;159:2292–2300. [PubMed] [Google Scholar]
- 19.Pellicena P, Stowell KR, Miller WT. Enhanced phosphorylation of Src family kinase substrates containing SH2 domain binding sites. J Biol Chem. 1998;273:15325–15328. doi: 10.1074/jbc.273.25.15325. [DOI] [PubMed] [Google Scholar]
- 20.Brunati AM, et al. Molecular features underlying the sequential phosphorylation of HS1 protein and its association with c-Fgr protein-tyrosine kinase. J Biol Chem. 1999;274:7557–7564. doi: 10.1074/jbc.274.11.7557. [DOI] [PubMed] [Google Scholar]
- 21.Lee S, Ayrapetov MK, Kemble DJ, Parang K, Sun G. Docking-based substrate recognition by the catalytic domain of a protein tyrosine kinase, C-terminal Src kinase (Csk) J Biol Chem. 2006;281:8183–8189. doi: 10.1074/jbc.M508120200. [DOI] [PubMed] [Google Scholar]
- 22.Lee S, Lin X, Nam NH, Parang K, Sun G. Determination of the substrate-docking site of protein tyrosine kinase C-terminal Src kinase. Proc Natl Acad Sci USA. 2003;100:14707–14712. doi: 10.1073/pnas.2534493100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Houtman JC, Houghtling RA, Barda-Saad M, Toda Y, Samelson LE. Early phosphorylation kinetics of proteins involved in proximal TCR-mediated signaling pathways. J Immunol. 2005;175:2449–2458. doi: 10.4049/jimmunol.175.4.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller AT, Wilcox HM, Lai Z, Berg LJ. Signaling through Itk promotes T helper 2 differentiation via negative regulation of T-bet. Immunity. 2004;21:67–80. doi: 10.1016/j.immuni.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 25.Lucas JA, Miller AT, Atherly LO, Berg LJ. The role of Tec family kinases in T cell development and function. Immunol Rev. 2003;191:119–138. doi: 10.1034/j.1600-065x.2003.00029.x. [DOI] [PubMed] [Google Scholar]
- 26.Joseph RE, Min L, Xu R, Musselman ED, Andreotti AH. A remote substrate docking mechanism for the tec family tyrosine kinases. Biochemistry. 2007;46:5595–5603. doi: 10.1021/bi700127c. [DOI] [PubMed] [Google Scholar]
- 27.Bonvini E, et al. On the mechanism coupling phospholipase Cgamma1 to the B- and T-cell antigen receptors. Adv Enzyme Regul. 2003;43:245–269. doi: 10.1016/s0065-2571(02)00033-x. [DOI] [PubMed] [Google Scholar]
- 28.Park D, Rho HW, Rhee SG. CD3 stimulation causes phosphorylation of phospholipase C-gamma 1 on serine and tyrosine residues in a human T-cell line. Proc Natl Acad Sci USA. 1991;88:5453–5456. doi: 10.1073/pnas.88.12.5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Serrano CJ, et al. A new tyrosine phosphorylation site in PLC gamma 1: The role of tyrosine 775 in immune receptor signaling. J Immunol. 2005;174:6233–6237. doi: 10.4049/jimmunol.174.10.6233. [DOI] [PubMed] [Google Scholar]
- 30.Humphries LA, et al. Tec kinases mediate sustained calcium influx via site-specific tyrosine phosphorylation of the phospholipase Cgamma Src homology 2-Src homology 3 linker. J Biol Chem. 2004;279:37651–37661. doi: 10.1074/jbc.M311985200. [DOI] [PubMed] [Google Scholar]
- 31.Groesch TD, Zhou F, Mattila S, Geahlen RL, Post CB. Structural basis for the requirement of two phosphotyrosine residues in signaling mediated by Syk tyrosine kinase. J Mol Biol. 2006;356:1222–1236. doi: 10.1016/j.jmb.2005.11.095. [DOI] [PubMed] [Google Scholar]
- 32.Pawson T, Gish GD, Nash P. SH2 domains, interaction modules, and cellular wiring. Trends Cell Biol. 2001;11:504–511. doi: 10.1016/s0962-8924(01)02154-7. [DOI] [PubMed] [Google Scholar]
- 33.Smith-Garvin JE, Koretzky GA, Jordan MS. T Cell Activation. Annu Rev Immunol. 2009;27:591–619. doi: 10.1146/annurev.immunol.021908.132706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Irvin BJ, Williams BL, Nilson AE, Maynor HO, Abraham RT. Pleiotropic contributions of phospholipase C-gamma1 (PLC-gamma1) to T-cell antigen receptor-mediated signaling: Reconstitution studies of a PLC-gamma1-deficient Jurkat T-cell line. Mol Cell Biol. 2000;20:9149–9161. doi: 10.1128/mcb.20.24.9149-9161.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stoica B, et al. The amino-terminal Src homology 2 domain of phospholipase C gamma 1 is essential for TCR-induced tyrosine phosphorylation of phospholipase C gamma 1. J Immunol. 1998;160:1059–1066. [PubMed] [Google Scholar]
- 36.Braiman A, Barda-Saad M, Sommers CL, Samelson LE. Recruitment and activation of PLCgamma1 in T cells: A new insight into old domains. EMBO J. 2006;25:774–784. doi: 10.1038/sj.emboj.7600978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joseph RE, Fulton DB, Andreotti AH. Mechanism and functional significance of itk autophosphorylation. J Mol Biol. 2007;373:1281–1292. doi: 10.1016/j.jmb.2007.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Joseph RE, Severin A, Min L, Fulton DB, Andreotti AH. SH2-Dependent Autophosphorylation within the Tec Family Kinase Itk. J Mol Biology. 2009;391:164–177. doi: 10.1016/j.jmb.2009.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Latour S, et al. BiBinding of SAP SH2 domain to FynT SH3 domain reveals a novel mechanism of receptor signaling in immune regulation. Nat Cell Biol. 2003;5:149–154. doi: 10.1038/ncb919. [DOI] [PubMed] [Google Scholar]
- 40.Chen R, Latour S, Shi X, Veillette A. Association between SAP and FynT: Inducible SH3 domain-mediated interaction controlled by engagement of the SLAM receptor. Mol Cell Biol. 2006;26:5559–5568. doi: 10.1128/MCB.00357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chan B, et al. SAP couples Fyn to SLAM immune receptors. Nat Cell Biol. 2003;5:155–160. doi: 10.1038/ncb920. [DOI] [PubMed] [Google Scholar]
- 42.Bae JH, Lew ED, Yuzawa S, Tomé F, Lax I, Schlessinger J. The Selectivity of Receptor Tyrosine Kinase Signaling Is Controlled by a Secondary SH2 Domain Binding Site. Cell. 2009;138:514–524. doi: 10.1016/j.cell.2009.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Joseph RE, Min L, Andreotti AH. The Linker between SH2 and kinase domains positively regulates catalysis of the Tec family kinases. Biochemistry. 2007;46:5455–5462. doi: 10.1021/bi602512e. [DOI] [PubMed] [Google Scholar]
- 44.Brazin KN, Fulton DB, Andreotti AH. A specific intermolecular association between the regulatory domains of a Tec family kinase. J Mol Biol. 2000;302:607–623. doi: 10.1006/jmbi.2000.4091. [DOI] [PubMed] [Google Scholar]
- 45.Pascal SM, et al. Nuclear magnetic resonance structure of an SH2 domain of phospholipase C-gamma 1 complexed with a high affinity binding peptide. Cell. 1994;77:461–472. doi: 10.1016/0092-8674(94)90160-0. [DOI] [PubMed] [Google Scholar]
- 46.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific, Palo Alto, CA. 2002. [Accessed September 28, 2008]. http://www.pymol.org.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.