Abstract
Background
Hepatocellular carcinoma is a common and aggressive cancer that occurs mainly in men. We examined microRNA expression patterns, survival, and response to interferon alfa in both men and women with the disease.
Methods
We analyzed three independent cohorts that included a total of 455 patients with hepatocellular carcinoma who had undergone radical tumor resection between 1999 and 2003. MicroRNA-expression profiling was performed in a cohort of 241 patients with hepatocellular carcinoma to identify tumor-related microRNAs and determine their association with survival in men and women. In addition, to validate our findings, we used quantitative reverse-transcriptase–polymerase-chain-reaction assays to measure microRNAs and assess their association with survival and response to therapy with interferon alfa in 214 patients from two independent, prospective, randomized, controlled trials of adjuvant interferon therapy.
Results
In patients with hepatocellular carcinoma, the expression of miR-26a and miR-26b in nontumor liver tissue was higher in women than in men. Tumors had reduced levels of miR-26 expression, as compared with paired noncancerous tissues, which indicated that the level of miR-26 expression was also associated with hepatocellular carcinoma. Moreover, tumors with reduced miR-26 expression had a distinct transcriptomic pattern, and analyses of gene networks revealed that activation of signaling pathways between nuclear factor κB and interleukin-6 might play a role in tumor development. Patients whose tumors had low miR-26 expression had shorter overall survival but a better response to interferon therapy than did patients whose tumors had high expression of the microRNA.
Conclusions
The expression patterns of microRNAs in liver tissue differ between men and women with hepatocellular carcinoma. The miR-26 expression status of such patients is associated with survival and response to adjuvant therapy with interferon alfa.
Hepatocellular carcinoma is common globally, with dismal outcomes and an increasing incidence in the United States.1 To date, surgery remains the most effective treatment with curative potential. However, only about 10 to 20% of patients with hepatocellular carcinoma are currently eligible for surgical intervention. In addition, patients who undergo curative resection often have a high rate of relapse. Therapies such as transcatheter arterial chemoembolization (TACE) and interferon alfa may prolong survival in some patients. However, the response is often not satisfactory because of the limited ability to identify patients who are most likely to benefit from such targeted adjuvant therapies.2-5 The recent findings of the Sorafenib Hepatocellular Carcinoma Assessment Randomized Protocol (SHARP) trial (ClinicalTrials.gov number, NCT00105443)6 were encouraging with respect to the use of sorafenib as a therapeutic agent. However, the survival benefit was modest. There is a need for molecular tools that can be used to stratify patients with respect to prognosis and response to therapy.
One of the key features of hepatocellular carcinoma is the higher incidence in men than in women, by a factor of two to six among various ethnic groups.7 Classic in vivo carcinogenesis experiments also reveal a higher susceptibility to hepatocellular carcinoma in male rodents.8-11 Moreover, women with this disease tend to have longer survival than men.12-14 These findings suggest that the biologic features of the tumor and the host microenvironment may differ significantly between the sexes and that sex-related factors may be associated with the prognosis. However, the underlying reasons for the lower incidence of hepatocellular carcinoma in women remain unclear. In a recent study, investigators found that one contributing factor may be that estrogen inhibits the induction of interleukin-6 produced by Kupffer cells.11 The serum level of interleukin-6 is elevated in several cancers, including hepatocellular carcinoma.15,16
MicroRNAs, small noncoding RNAs that regulate the translation of many genes, are excellent biomarkers for cancer diagnosis and prognosis.17-21 Given the therapeutic and prognostic potential of microRNAs as biomarkers in hepatocellular carcinoma, we evaluated microRNA profiles of specimens of hepatocellular carcinomas and paired specimens of noncancerous tissue to search for microRNAs that are associated with sex and with hepatocellular carcinoma.
Methods
Study Oversight
The study was approved by the institutional review board at each study center. All patients from whom tissue samples were obtained provided written informed consent.
Clinical Specimens
We obtained snap-frozen or paraffin-embedded specimens of tumors and surrounding nontumor hepatic tissues from 455 patients with hepatocellular carcinoma who had undergone radical resection between 1999 and 2003 at the Liver Cancer Institute of Fudan University in Shanghai (376 patients) or at the University of Hong Kong Medical Centre in Hong Kong (79 patients).4 Samples of normal liver tissue were obtained from eight disease-free liver donors.22
Our analyses involved three separate cohorts of patients. Cohort 1 consisted of 241 patients with hepatocellular carcinoma (test cohort) for whom microRNA microarray data were available, as described previously.21 Data from these patients were analyzed to search for microRNAs that were associated with sex and survival. In this cohort, data were missing on miR-26 expression for 17 patients and on survival for 9 patients, so we were able to analyze miR-26 expression in 224 patients and survival in 232 patients. Cohorts 2 and 3 consisted of 214 patients from prospective, randomized, controlled trials of adjuvant therapy with interferon alfa in patients with hepatocellular carcinoma.3,4 In cohort 2, data from 135 patients were evaluated in an independent validation analysis to measure microRNA expression with the use of quantitative reverse-transcriptase–polymerase-chain-reaction (qRTPCR) assay. Among these patients, data were missing on miR-26 expression for 6 patients and on survival for 12 patients, leaving 129 patients (60 in the control group and 69 in the group that received interferon alfa) for miR-26-expression analysis and 123 patients for survival analysis. In cohort 3, data from 79 patients (40 in the control group and 39 in the group that received inter-feron alfa) were evaluated to validate the association between miR-26 expression and the response to interferon alfa therapy.
Additional methods (including statistical analysis) are described in detail in the Supplementary Appendix, available with the full text of this article at NEJM.org.
Results
Characteristics of the Patients
Most of the patients in the three cohorts were men (85.1%), were long-term carriers of hepatitis B virus (HBV) (90.5%), and had cirrhosis (88.0%) and an elevated serum level of alpha-fetoprotein (62.2%); 84.4% of the patients had a single tumor nodule at the time of resection (Table 1, and Table 1 in the Supplementary Appendix). Clinical variables were similar between the test and validation cohorts, with the exception of the serum alanine aminotransferase (ALT) level, tumor–node–metastasis (TNM) stage, and status with respect to adjuvant therapy. Liver-inflammation activity in the patients with HBV-related hepato-cellular carcinoma, as indicated by ALT levels, was significantly lower in patients in cohort 2 than in those in either cohort 1 or cohort 3, and more patients in cohort 2 had early-stage tumors. In addition, 39 patients in cohort 1 had received prophylactic adjuvant therapies, but the difference in response did not appear to be significant (P = 0.90 by the log-rank test). In contrast, 53.3% of patients in cohort 2 and 49.4% in cohort 3 had received “intention-to-cure” adjuvant therapy with interferon alfa, which improved overall survival.3,4
Table 1.
Clinical Characteristics of the Patients.*
| Variable | Cohort 1 (N = 241) | Cohort 2 (N = 135) | P Value† |
|---|---|---|---|
| Sex — no. (%) | 0.87 | ||
| Female | 30 (12.4) | 14 (10.4) | |
| Male | 211 (87.6) | 111 (82.2) | |
| Data missing | 0 | 10 (7.4) | |
| Age — yr | 0.29 | ||
| Median | 50 | 50 | |
| Range | 13–83 | 20–77 | |
| Alanine aminotransferase — no. (%) | <0.001 | ||
| Normal, ≤50 U/liter | 145 (60.2) | 107 (79.3) | |
| Elevated, >50 U/liter | 96 (39.8) | 16 (11.9) | |
| Missing data | 0 | 12 (8.9) | |
| Hepatitis B virus — no. (%) | 0.64 | ||
| Negative | 16 (6.6) | 6 (4.4) | |
| Positive | 224 (92.9) | 118 (87.4) | |
| Missing data | 1 (0.4) | 11 (8.1) | |
| Tumor size — no. (%) | 0.91 | ||
| ≤3 cm | 88 (36.5) | 46 (34.1) | |
| >3 cm | 153 (63.5) | 78 (57.8) | |
| Missing data | 0 | 11 (8.1) | |
| Multinodular tumor — no. (%) | 0.50 | ||
| No | 214 (88.8) | 107 (79.3) | |
| Yes | 27 (11.2) | 17 (12.6) | |
| Missing data | 0 | 11 (8.1) | |
| Cirrhosis — no. (%) | 0.08 | ||
| No | 17 (7.1) | 16 (11.9) | |
| Yes | 223 (92.5) | 108 (80.0) | |
| Missing data | 1 (0.4) | 11 (8.1) | |
| TNM stage — no. (%) | <0.001 | ||
| I | 97 (40.2) | 81 (60.0) | |
| II | 90 (37.3) | 29 (21.5) | |
| III or IV | 54 (22.4) | 14 (10.4) | |
| Missing data | 0 | 11 (8.1) | |
| Alpha-fetoprotein — no. (%) | 0.20 | ||
| Negative, ≤20 ng/ml | 77 (32.0) | 49 (36.3) | |
| Positive, >20 ng/ml | 162 (67.2) | 75 (55.6) | |
| Missing data | 2 (0.8) | 11 (8.1) | |
| Adjuvant therapy — no. (%)‡ | <0.001 | ||
| Yes | 39 (16.2) | 72 (53.3) | |
| No | 202 (83.8) | 63 (46.7) | |
| Survival (mo) | 0.89 | ||
| Median | >60 | 67 | |
| Range | 2–67 | 2–82 |
The clinical characteristics of the 79 patients in the validation cohort for interferon alfa therapy (cohort 3) are listed in Table 1 in the Supplementary Appendix. TheTNM denotes tumor–node–metastasis.
A P value of less than 0.05 was considered to indicate statistical significance. P values were calculated with the use of Fisher's exact test, except for age, which was calculated with the unpaired t-test; TNM, which was calculated with the chi-square test; and survival, which was calculated with the log-rank test.
In cohort 1, 34 patients underwent transcatheter arterial chemoembolization, 3 received chemotherapy, 1 received interferon alfa therapy, and 1 received lymphokine-activated killer-cell therapy; in cohort 2, 72 patients underwent interferon alfa therapy.
Association of MicroRNAs with Sex and Clinical Outcome
To search for differences in the expression of microRNAs between liver samples from men and those from women, we globally analyzed the microRNA expression profiles of 241 patients in cohort 1, in which both tumor and nontumor microRNA microarray data were available (Gene Expression Omnibus [GEO] accession number, GSE6857).21 To avoid potential confounding factors, an age-matched and balanced case set was used to identify microRNAs with different expression levels in men and women, consisting of all 30 women and two age-matched control groups of men, each consisting of 31 patients (groups G1 and G2). The clinical characteristics of the women and the men in the two control groups were similar (Table 2 in the Supplementary Appendix).
Class-comparison analysis of nontumor liver tissues revealed that 15 microRNAs were differentially expressed in the comparison between the 30 women and the first control group of men (G1), whereas 45 microRNAs differed in the comparison between the women and the second control group of men (G2). Seven microRNAs showed differential expression between women and men in both comparisons. In contrast, we identified only one microRNA, miR-129-2, that had differential expression in tumor tissue from women and from men in both comparisons (Table 3 in the Supplementary Appendix). Thus, there were more consistent differences in microRNA expression in nontumor hepatic tissue than in tumors.
In humans, there are three miR-26 members: miR-26a-1, miR-26a-2, and miR-26b; miR-26a-1 was chosen for further analysis, since the difference in its level between sexes was most significant, and it was most abundant. Analyses of data from 224 patients in cohort 1 showed that miR-26a-1 expression in normal hepatic tissue was significantly higher in women than in men (Fig. 1A). This finding was validated by the determination of mature miR-26 expression in 26 women and in 56 age-matched men with the use of qRT-PCR (Fig. 1B).
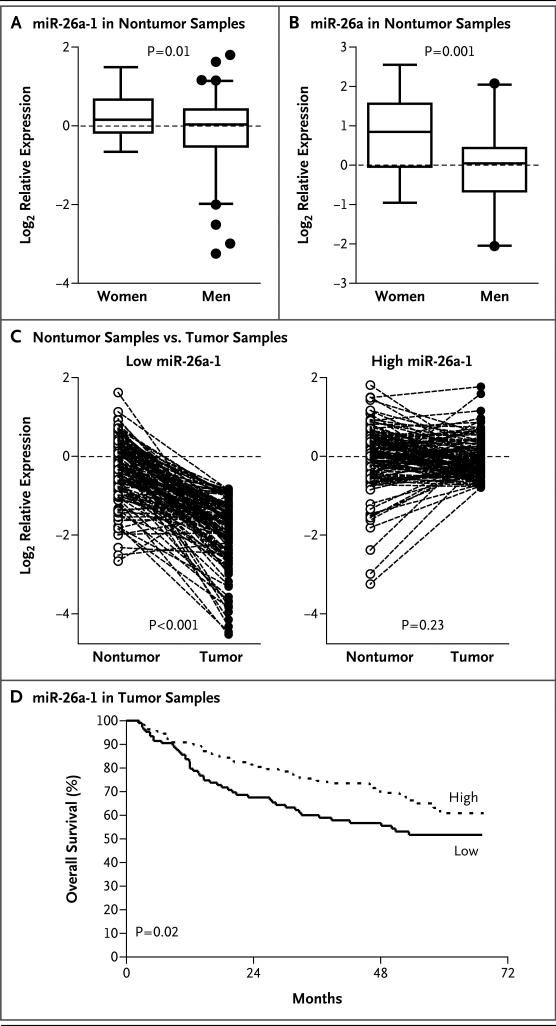
Figure 1. Expression of miR-26 in Hepatic Tumors and Noncancerous Tissue in Men and Women.
Panel A shows miR-26a-1 expression in noncancerous hepatic tissue from 30 women and 194 men, as determined by microarray analysis with the use of an unpaired t-test. Panel B shows miR-26a expression in noncancerous hepatic tissue obtained from 26 women and 56 age-matched men, as determined by quantitative reverse-transcriptase–polymerase-chain-reaction assay with the use of an unpaired t-test. In Panels A and B, the horizontal lines in the box plots represent the median, the boxes represent the interquartile range, and the whiskers represent the 2.5th and 97.5th percentiles. Panel C shows comparisons of relative levels of miR-26a-1 expression in paired tumor and nontumor samples from 224 patients, according to miR-26 status, with the use of paired t-tests. The data in Panels A through C are log2 relative expression levels, normalized to the values in disease-free samples from eight control subjects. Panel D shows survival according to the level of miR-26a-1 expression in tumor samples, as determined by microarray analysis, with the use of the log-rank test. The median expression level was used as the cutoff. Reduced miR-26 expression in 106 patients was classified as the lower 50th percentile (with a mean reduction in tumor tissue, as compared with nontumor tissue, by a factor of 2.69). High miR-26 expression in 111 patients was classified as the upper 50th percentile (with a mean reduction in tumor tissue, as compared with nontumor tissue, by a factor of 0.98).
We then reasoned that miR-26 may act as a tumor-suppressor gene, and if so, the silencing of miR-26 would be a frequent event in tumors. Analyses showed that a significant reduction in miR-26a-1 expression in tumors, as compared with that in nontumor samples, was observed in patients with low levels of miR-26 expression (P<0.001) but not in those with high levels of miR-26 expression (P = 0.23) (Fig. 1C). The median factor change in the expression in tumors, as compared with nontumors, was 0.37 in patients with low miR-26a-1 expression and 0.98 in patients with high miR-26a-1 expression, which suggested that the silencing of miR-26 expression was associated only with low miR-26 expression. Moreover, low miR-26 expression, as compared with high expression, was associated with reduced survival (Fig. 1D). The expression patterns of all three miR-26 members and their associations with survival were similar (Fig. 1 in the Supplementary Appendix).
Tumors with Low miR-26 Expression
To test the hypothesis that tumors with low miR-26 expression may be biologically distinct, we analyzed the 224 matched patients in cohort 1 with available microRNA and messenger RNA (mRNA) microarray data. The mRNA microarray data were based on the expression of approximately 21,000 mRNA genes (GEO accession number, GSE5975).23 Multidimensional scaling analysis on the basis of the first three principal components of all genes revealed that a majority of patients with low miR-26 expression clustered separately from those with high miR-26 expression (Fig. 2A), according to the dichotomized expression status of the three miR-26 genes (Fig. 2A in the Supplementary Appendix). Class-comparison analysis showed that the expression of a significant number of genes differed between tumors with low miR-26 expression and those with high miR-26 expression; the two groups of tumors had 915 significant genes in common (Fig. 2B in the Supplementary Appendix). SLC2A6 and S100P were selected among the differentially expressed genes for validation by means of qRT-PCR (Fig. 3 in the Supplementary Appendix). Furthermore, multivariate analysis showed that mRNA signatures could significantly predict the stratification of patients according to miR-26 expression, with 80.3% overall accuracy. Thus, patients whose tumors had low miR-26 expression had genetic-expression patterns that were distinct from those of tumors in patients with high miR-26 expression.
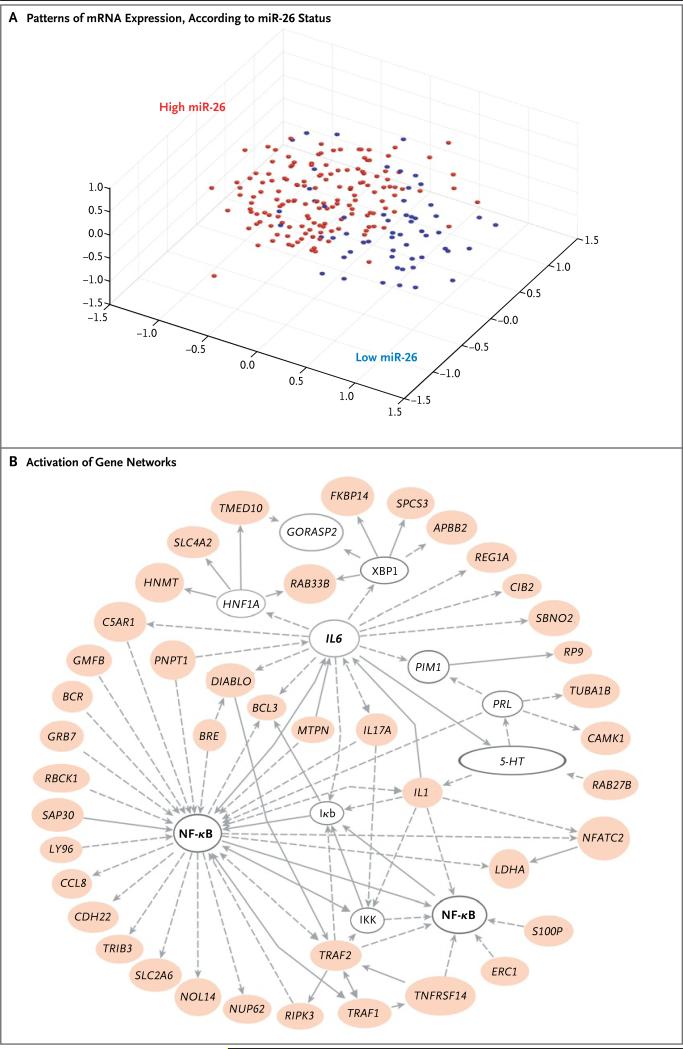
Figure 2. Distinct Transcriptional Activities in Hepatocellular Carcinomas with Low miR-26 Expression.
Panel A shows a multidimensional scaling plot, based on the expression of 11,580 genes, for 224 patients with hepatocellular carcinoma. The level of miR-26 expression was dichotomized on the basis of the median value as low (blue) or high (red). Panel B shows the activation of gene networks of signaling between nuclear factor κB (NF-κB) and interleukin-6 (IL6) in tumors with low miR-26 expression. Colored ovals represent up-regulated genes in tumors with low miR-26 expression, and open ovals represent genes that are not on the list of significant genes but are reported to be associated with the network. The open ovals that are labeled as NF-κB, Iκb, and IKK represent molecular nodes related to protein complexes of NF-κB, inhibitor of NF-κB, and Iκb kinase, respectively. The open oval labeled as 5 = HT (5-hydroxytryptamine) represents a chemical node related to serotonin receptor signaling, as categorized by pathway analysis software (Ingenuity). Arrows represent positive regulation of gene expression, with solid arrows indicating direct regulation, and broken arrows indirect regulation.
Among the 915 overlapping genes, 770 were overexpressed in patients with low miR-26 expression. Gene-network analyses with the use of these 770 genes revealed a series of putative tumorigenesis networks that had a high score (>10) (for details, see Table 4 in the Supplementary Appendix). Examination of the enriched genes in various categories revealed several important signaling networks, the most striking of which showed predominant activation of the signaling pathway between nuclear factor κB (NF-κB) and interleukin-6 in patients with low miR-26 expression (Fig. 2B). To validate which genes were associated with this signaling pathway, we measured the level of the NF-κB target gene, IL6 by qRT-PCR, since it was also related to hepatocellular carcinoma and the disparity in miR-26 expression levels between the sexes. Most of the patients with hepatocellular carcinoma who had reduced miR-26 expression had a concomitant elevation in interleukin-6 expression (Fig. 4 in the Supplementary Appendix). With these findings taken together, we concluded that tumors with low miR-26 expression had a distinct genetic profile.
Validation with Independent Cohorts
To validate the association between the sex-dependent expression of miR-26 and survival, we detected mature miR-26 expression by qRT-PCR in tumor and nontumor tissues obtained from patients in cohort 2. Since adjuvant therapy with interferon alfa altered the survival outcome, we analyzed the control group. Consistent with cohort 1, miR-26 expression was more abundant in nontumor tissues obtained from women, but a significant reduction was observed in tumors, regardless of sex (Fig. 5 in the Supplementary Appendix). Moreover, reduced miR-26 expression in tumors was significantly associated with shorter survival (Fig. 3A, and Fig. 6A in the Supplementary Appendix). The results were similar in another independent cohort (cohort 3) (Fig. 3B, and Fig. 6B in the Supplementary Appendix).
We used Cox proportional-hazards regression to further evaluate the association between miR-26 expression and prognosis in control subjects in cohort 2 (Table 5 in the Supplementary Appendix). In univariate analysis, the level of miR-26a expression in tumors and TNM stage were significantly associated with the prognosis. The final multivariate model revealed that reduced miR-26a expression in tumors was an independent predictor of shorter survival. A similar trend was found for miR-26b. Thus, the dichotomized values for miR-26 expression were independent predictors of prognosis.
Expression of miR-26 and Response to Interferon Alfa
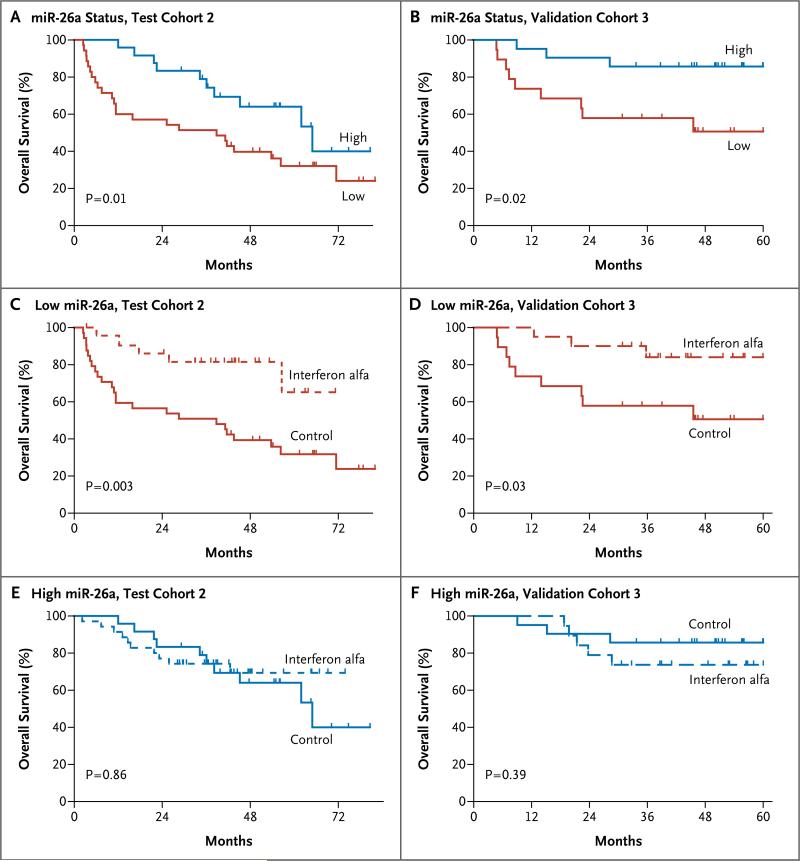
In cohort 2, we analyzed associations between miR-26 expression and the response to interferon alfa. Patients with reduced miR-26a expression in tumors had a significant improvement in overall survival after receiving adjuvant therapy with interferon alfa, as compared with those in the control group (P = 0.003) (Fig. 3C). This finding was validated in cohort 3 (Fig. 3D). In contrast, patients with high miR-26a expression in both cohorts did not have a response to interferon alfa (Fig. 3E and 3F). Similar results were obtained with respect to miR-26b expression (Fig. 6C through 6F in the Supplementary Appendix).
Figure 3. Association between miR-26a Expression in Tumors and Overall Survival in Two Trials of Interferon Alfa.
Panels A and B show the association between miR-26a expression and overall survival in patients with hepatocellular carcinoma from two control groups (cohort 2 in Panel A and cohort 3 in Panel B). In cohort 2, 24 patients had high miR-26a expression and 35 patients had low miR-26a expression. In cohort 3, 21 patients had high miR-26a expression and 19 patients had low miR-26a expression. Panels C and D show the association between adjuvant therapy with interferon alfa and overall survival in patients with tumors with low miR-26a expression. In cohort 2, there were 24 patients in the group that received interferon alfa and 35 patients in the control group. In cohort 3, there were 20 patients in the group that received interferon alfa and 19 patients in the control group. Panels E and F show the association between adjuvant therapy with interferon alfa and overall survival in patients with tumors that had high miR-26a expression. In cohort 2, there were 35 patients in the group that received interferon alfa and 24 patients in the control group. In cohort 3, there were 19 patients in the group that received interferon alfa and 21 patients in the control group.
We used Cox proportional-hazards regression to evaluate the effect of treatment on survival in patients in cohort 2 who had low miR-26 expression (Table 2). In both univariate and multivariate analyses, interferon alfa was associated with significant improvement in survival in patients with reduced miR-26 expression. A significant interaction was observed between miR-26 expression and interferon alfa therapy with respect to the effect on survival (miR-26a, P = 0.004 for interaction; miR-26b, P = 0.02 for interaction). Thus, miR-26 expression emerged as an independent predictor of the response to interferon alfa.
Table 2.
Hazard Ratios for Death among Patients with Low Expression of miR-26a and Patients with Low Expression of miR-26b, According to Univariate and Multivariate Analyses.*
| Variable | Reduced miR-26a Expression (N = 59) | Reduced miR-26b Expression (N = 58) | ||
|---|---|---|---|---|
| Hazard Ratio (95% CI) | P Value† | Hazard Ratio (95% CI) | P Value† | |
| Univariate analysis | ||||
| Interferon alfa vs. control | 0.2 (0.1–0.6) | 0.003 | 0.4 (0.2–0.9) | 0.04 |
| Age (≤50 yr vs. >50 yr) | 1.1 (0.6–2.2) | 0.77 | 0.9 (0.5–1.8) | 0.81 |
| Sex (male vs. female) | 1.3 (0.4–3.7) | 0.65 | 1.4 (0.4–4.7) | 0.57 |
| Alpha-fetoprotein (>20 ng/ml vs. ≤20ng/ml) | 1.8 (0.8–4.1) | 0.15 | 1.9 (0.9–4.0) | 0.10 |
| Cirrhosis (yes vs. no) | 0.7 (0.3–2.1) | 0.57 | 0.7 (0.3–1.8) | 0.43 |
| Alanine aminotransferase (>50 U/liter vs. ≤50 U/liter) | 1.1 (0.4–3.0) | 0.79 | 1.9 (0.8–4.6) | 0.17 |
| Tumor size (>3 cm vs. ≤3cm) | 1.7 (0.8–3.6) | 0.15 | 2.0 (1.0–4.3) | 0.06 |
| Tumor encapsulation (no vs. yes) | 1.8 (0.9–3.6) | 0.12 | 1.6 (0.8–3.1) | 0.20 |
| Multinodular tumor (yes vs. no) | 0.9 (0.3–2.3) | 0.82 | 1.1 (0.4–2.6) | 0.87 |
| TNM staging (II or III vs. I) | 2.7 (1.3–5.5) | 0.005 | 2.7 (1.4–5.3) | 0.004 |
| Multivariate analysis | ||||
| Interferon alfa vs. control | 0.3 (0.1–0.7) | 0.005 | 0.4 (0.2–0.9) | 0.04 |
| TNM staging (II or III vs. I) | 2.4 (1.2–4.9) | 0.02 | 2.6 (1.3–5.1) | 0.007 |
| Sex (male vs. female) | 1.4 (0.5–4.0) | 0.55 | 1.5 (0.5–5.1) | 0.48 |
The analyses were performed with the use of Cox proportional-hazards regression on data from patients with a low level of miR-26 expression in cohort 2.
A P value of less than 0.05 was considered to indicate statistical significance.
Discussion
We analyzed microRNA profiles in predominantly HBV-related hepatocellular carcinomas in men and women and the association of microRNA expression levels with survival and response to therapy with interferon alfa in three independent cohorts. We found that the expression of miR-26a and miR-26b in nontumor hepatic tissues was higher in women than in men, but the expression was significantly down-regulated in tumor samples, as compared with paired samples of noncancerous tissues, regardless of sex. Tumors with reduced miR-26 expression had a distinct gene-expression profile, and patients whose tumors had low miR-26 expression had shorter survival but were more likely to have a response to interferon alfa, as compared with patients whose tumors had high miR-26 expression.
Our results suggest that miR-26 may be a tumor suppressor and that miR-26 silencing in hepatocytes may contribute to the development of a more aggressive form of hepatocellular carcinoma in men. These hypotheses are supported by the following findings: miR-26 was expressed at higher levels in women than in men in the liver, where presumably more anticarcinogenic activities exist in women; miR-26 expression is silenced in a subgroup of patients who have reduced survival; and genes that are activated in tumors with reduced miR-26 expression are selectively enriched in signaling pathways between NF-κB and interleukin-6. It was recently reported that MyD8-dependent interleukin-6 induction by NF-κB differed between male and female mice.11 Intriguingly, estrogens inhibit interleukin-6 promoter activity, which may contribute to a decreased susceptibility to hepatocellular carcinoma in women. Whether such a link is functionally relevant to liver cancer in humans remains to be determined.24 Our results are consistent with these findings, since interleukin-6 expression was inversely correlated with miR-26 expression.
Our analyses revealed that miR-26 expression was an independent predictor of survival. However, when outcomes of therapy with interferon alfa were stratified, only patients whose tumors had reduced miR-26 expression had a favorable response to interferon alfa in two independent, prospective, randomized, controlled trials. These results indicate that miR-26 status in tumors may be a useful tool in estimating prognosis in patients with hepatocellular carcinoma and in assisting in the selection of patients who are likely to benefit from adjuvant therapy with interferon alfa to prevent relapse. In an evaluation of the effects of adjuvant interferon alfa therapy after liver resection or tumor ablation in seven randomized, controlled trials, Clavien5 concluded that all the trials showed modest beneficial effects of the drug. In addition, among multiple experimental agents, only a modest survival benefit has been observed with sorafenib.6 The poor efficacy of current systemic agents may stem from an inability to identify a subpopulation of patients who may have a favorable response to a particular therapy. Prospective studies will be necessary to determine whether interferon alfa might be used as a first-line therapy for patients with hepatocellular carcinoma who have undergone resection and who have tumors with reduced miR-26 expression.
It should be noted that our results involved Chinese patients with a high rate of HBV positivity (approximately 90%). Our findings therefore need to be confirmed by studies of tumors from non-Asian patients and tumors arising from other underlying liver diseases, such as hepatitis C, or associated with alcohol abuse.
The mechanisms behind the sensitivity of tumors with reduced miR-26 expression to therapy with interferon alfa are unclear. One plausible model is that such tumors have specific activation of the signaling pathway that is responsive to interferon alfa. Consistently, tumors with low miR-26 expression were distinct from those with high miR-26 expression in transcriptomic activities and were associated with poor survival. Many of the genes that are overexpressed in tumors with reduced miR-26 expression are related to cell immunity, such as those encoding proinflammatory and antiinflammatory cytokines (i.e., inter-leukin-1, interleukin-2, interleukin-10, and inter-leukin-17). Moreover, many signaling networks that are activated in tumors with low miR-26 expression are immune-associated, such as NF-κB–interleukin-6, interleukin-10, STAT3, and inter-feron alfa inducible factor signaling networks (Table 4 in the Supplementary Appendix). Tumors with low miR-26 expression may be characterized by a unique activation of interferon alfa signaling, possibly through the NF-κB–inter leukin-6 signaling pathway, and thus may be sensitive to growth inhibition mediated by interferon alfa through interleukin-6–STAT3 signaling.25 These hypotheses require evaluation.
In conclusion, we identified systematic differences in patterns of microRNA expression between liver tissues obtained from men and women with hepatocellular carcinoma. Tumors with low miR-26 expression were biologically distinct from those with high expression and were associated with reduced survival but had a favorable response to adjuvant therapy with inter-feron alfa.
Supplementary Material
Acknowledgments
Supported in part by grants (Z01-BC 010313 and Z01-BC 010876) from the Intramural Research Program of the Center for Cancer Research of the National Cancer Institute.
Dr. Sun reports receiving lecture fees from Bayer. No other potential conflict of interest relevant to this article was reported.
We thank Curtis Harris, Elise Kohn, and Aaron Schetter for their critical reading of the manuscript; Suet Yi Leung for advice; Karen MacPherson for bibliographic assistance; Krista Zanetti for statistical assistance; and the patients and clinicians at the Liver Cancer Institute and Zhongshan Hospital, Fudan University, and at the University of Hong Kong for their contributions to this study.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Llovet JM, Bruix J. Systematic review of randomized trials for unresectable hepatocellular carcinoma: chemoembolization improves survival. Hepatology. 2003;37:429–42. doi: 10.1053/jhep.2003.50047. [DOI] [PubMed] [Google Scholar]
- 3.Sun HC, Tang ZY, Wang L, et al. Postoperative interferon alpha treatment postponed recurrence and improved overall survival in patients after curative resection of HBV-related hepatocellular carcinoma: a randomized clinical trial. J Cancer Res Clin Oncol. 2006;132:458–65. doi: 10.1007/s00432-006-0091-y. [DOI] [PubMed] [Google Scholar]
- 4.Lo CM, Liu CL, Chan SC, et al. A randomized, controlled trial of postoperative adjuvant interferon therapy after resection of hepatocellular carcinoma. Ann Surg. 2007;245:831–42. doi: 10.1097/01.sla.0000245829.00977.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clavien PA. Interferon: the magic bullet to prevent hepatocellular carcinoma recurrence after resection? Ann Surg. 2007;245:843–5. doi: 10.1097/SLA.0b013e31805d0788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Llovet JM, Ricci S, Mazzaferro V, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378–90. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 7.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–76. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 8.Ghebranious N, Sell S. Hepatitis B injury, male gender, aflatoxin, and p53 expression each contribute to hepatocarcinogenesis in transgenic mice. Hepatology. 1998;27:383–91. doi: 10.1002/hep.510270211. [DOI] [PubMed] [Google Scholar]
- 9.Nakatani T, Roy G, Fujimoto N, Asahara T, Ito A. Sex hormone dependency of diethylnitrosamine-induced liver tumors in mice and chemoprevention by leuprorelin. Jpn J Cancer Res. 2001;92:249–56. doi: 10.1111/j.1349-7006.2001.tb01089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rogers AB, Theve EJ, Feng Y, et al. Hepatocellular carcinoma associated with liver-gender disruption in male mice. Cancer Res. 2007;67:11536–46. doi: 10.1158/0008-5472.CAN-07-1479. [DOI] [PubMed] [Google Scholar]
- 11.Naugler WE, Sakurai T, Kim S, et al. Gender disparity in liver cancer due to sex differences in MyD88-dependent IL-6 production. Science. 2007;317:121–4. doi: 10.1126/science.1140485. [DOI] [PubMed] [Google Scholar]
- 12.Ng IO, Ng MM, Lai EC, Fan ST. Better survival in female patients with hepatocellular carcinoma: possible causes from a pathologic approach. Cancer. 1995;75:18–22. doi: 10.1002/1097-0142(19950101)75:1<18::aid-cncr2820750105>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 13.Dohmen K, Shigematsu H, Irie K, Ishibashi H. Longer survival in female than male with hepatocellular carcinoma. J Gastroenterol Hepatol. 2003;18:267–72. doi: 10.1046/j.1440-1746.2003.02936.x. [DOI] [PubMed] [Google Scholar]
- 14.Tangkijvanich P, Mahachai V, Suwangool P, Poovorawan Y. Gender difference in clinicopathologic features and survival of patients with hepatocellular carcinoma. World J Gastroenterol. 2004;10:1547–50. doi: 10.3748/wjg.v10.i11.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ashizawa T, Okada R, Suzuki Y, et al. Clinical significance of interleukin-6 (IL-6) in the spread of gastric cancer: role of IL-6 as a prognostic factor. Gastric Cancer. 2005;8:124–31. doi: 10.1007/s10120-005-0315-x. [DOI] [PubMed] [Google Scholar]
- 16.Porta C, De Amici M, Quaglini S, et al. Circulating interleukin-6 as a tumor marker for hepatocellular carcinoma. Ann Oncol. 2008;19:353–8. doi: 10.1093/annonc/mdm448. [DOI] [PubMed] [Google Scholar]
- 17.Calin GA, Ferracin M, Cimmino A, et al. A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 18.Lu J, Getz G, Miska EA, et al. Micro-RNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 19.Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–98. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 20.Schetter AJ, Leung SY, Sohn JJ, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–36. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Budhu A, Jia HL, Forgues M, et al. Identification of metastasis-related micro-RNAs in hepatocellular carcinoma. Hepatology. 2008;47:897–907. doi: 10.1002/hep.22160. [DOI] [PubMed] [Google Scholar]
- 22.Kim JW, Ye Q, Forgues M, et al. Cancer-associated molecular signature in the tissue samples of patients with cirrhosis. Hepatology. 2004;39:518–27. doi: 10.1002/hep.20053. [DOI] [PubMed] [Google Scholar]
- 23.Yamashita T, Forgues M, Wang W, et al. EpCAM and alpha-fetoprotein expression defines novel prognostic subtypes of hepatocellular carcinoma. Cancer Res. 2008;68:1451–61. doi: 10.1158/0008-5472.CAN-07-6013. [DOI] [PubMed] [Google Scholar]
- 24.Wands J. Hepatocellular carcinoma and sex. N Engl J Med. 2007;357:1974–6. doi: 10.1056/NEJMcibr075652. [DOI] [PubMed] [Google Scholar]
- 25.Thyrell L, Arulampalam V, Hjortsberg L, Farnebo M, Grandér D, Pokrovskaja Tamm K. Interferon alpha induces cell death through interference with interleukin 6 signaling and inhibition of STAT3 activity. Exp Cell Res. 2007;313:4015–24. doi: 10.1016/j.yexcr.2007.08.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.