Abstract
On the basis of the results of activity studies, previous reports have suggested that vitamin D binding protein (DBP) is significantly or even completely deglycosylated in cancer patients, eliminating the molecular precursor of the immunologically important Gc macrophage activating factor (GcMAF), a glycosidase-derived product of DBP. The purpose of this investigation was to directly determine the relative degree of O-linked trisaccharide glycosylation of serum-derived DBP in human breast, colorectal, pancreatic, and prostate cancer patients. Results obtained by electrospray ionization-based mass spectrometric immunoassay showed that there was no significant depletion of DBP trisaccharide glycosylation in the 56 cancer patients examined relative to healthy controls. These results suggest that alternative hypotheses regarding the molecular and/or structural origins of GcMAF must be considered to explain the relative inability of cancer patient serum to activate macrophages.
Keywords: vitamin D binding protein, GcMAF, cancer, glycosylation
Introduction
Gc macrophage activating factor (GcMAF) is a naturally derived form of human vitamin D binding protein that has recently shown notable clinical value as a potent immunotherapy against human breast, prostate, and colorectal cancer.1–3 Molecularly, GcMAF is thought to be DBP with a single terminal O-linked GalNAc sugar residue.4–7 In its native molecular state, DBP is a mixture of posttranslationally unmodified (except for disulfide bonds) and O-glycosylated species, the glycosidic degree and nature of which depends on DBP genotype.8–13 For humans, there are three major allelic variants designated Gc1F, Gc1S, and Gc2. Relative to the Gc1F protein, a D416E mutation defines the Gc1S protein, and a T420K mutation defines the Gc2 protein. The Gc2 mutation removes the major site for O-linked trisaccharide glycosylation,10,14 a fact which has recently been verified by mass spectrometric analysis of the intact protein.8,9 All major allelic forms of DBP can be converted to GcMAF (by the action of β-galactosidase of inflammation-activated B cells and, for Gc1 proteins, inflammation-activated T-cell sialidase4–7) despite the fact that Gc2 lacks O-linked trisaccharidic glycosylation,4,5 suggesting the presence of a low occupation, alternative glycosylation site on Gc2 proteins (which must be genetically conserved in Gc1F and Gc1S proteins).4,5,8
Studies in cancer patients have shown that high-serum levels of α-N-acetylgalactosaminidase (nagalase) activity correspond in linear fashion to both tumor burden15,16 (in mice and humans) and, inversely, GcMAF “precursor activity” in humans.2,3 This “precursor activity” has been assigned (without direct structural evidence) as the quantity of O-linked trisaccharide glycosylated DBP available in patient plasma, and is reported to be significantly depleted or even eliminated in cancer patients based on activity studies.1–3,15,17 Since direct structural evidence for this assignment has not previously been obtained, the purpose of this investigation was to assess the relative degree of O-linked trisaccharide glycosylation of DBP in breast, colorectal, pancreatic, and prostate cancer patients compared with healthy individuals.
Results
Charge state deconvoluted ESI mass spectra of immunoaffinity-isolated, intact DBP from the serum or plasma of 56 cancer patients (nine breast cancer, 11 colorectal cancer, nine prostate cancer, and 27 pancreatic cancer patients) and 29 healthy patients (exemplified in Fig. 1) demonstrate no significant differences with regard to the relative degree of O-linked trisaccharide glycosylation compared with healthy individuals (see Fig. 2). The data from the healthy individuals are in agreement with data published recently on the relative degree of DBP trisaccharide glycosylation assessed in the plasma of over 100 noncancerous individuals.8
Figure 1.
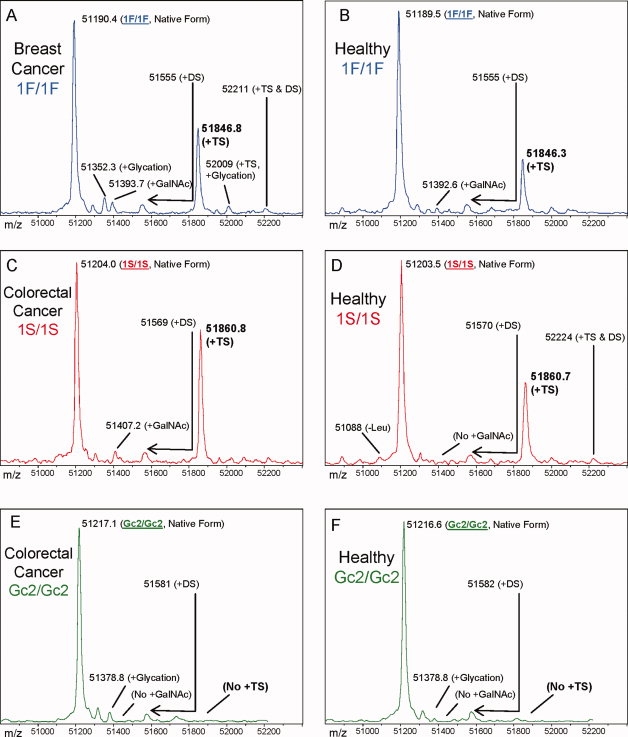
Charge state deconvoluted ESI mass spectra of intact DBP isolated directly from the blood plasma of 3 cancer patients and 3 healthy individuals with matching DBP diploid genotype. The base peaks at approximately 51,200 Da represent unmodified DBP (which varies in exact mass according to DBP genotype), a peak at Δm + 162 Da, if present, represents nonenzymatically glycated DBP, a peak at Δm + 203.2, if present, represents DBP modified by a single GalNAc residue, a broadened peak at approximately Δm + 365.4 likely represents an O-linked disaccharide (DS) glycoform (Gal1GalNAc1),8 and a peak at Δm + 656.6 represents the O-linked trisaccharide glycoform (TS, NeuNAc1Gal1GalNAc1).8,13 Displayed masses correspond to the observed m/z value of an “MH+” species. (A) Homozygous 1F/1F DBP from a breast cancer patient (calculated unmodified 1F DBP m/z = 51,189.2) (B) Homozygous 1F/1F DBP from a healthy individual (C) Homozygous 1S/1S DBP from a colorectal cancer patient (calculated unmodified 1S DBP m/z = 51203.2) (D) Homozygous 1S/1S DBP from a healthy individual (E) Homozygous Gc2/Gc2 DBP from a colorectal cancer patient (calculated unmodified Gc2 DBP m/z = 51216.3) (F) Homozygous Gc2/Gc2 DBP from a healthy individual. Though not shown directly, the mass resolution is sufficient to readily classify the diploid genotype of all heterozygous combinations of DBP allele products.
Figure 2.
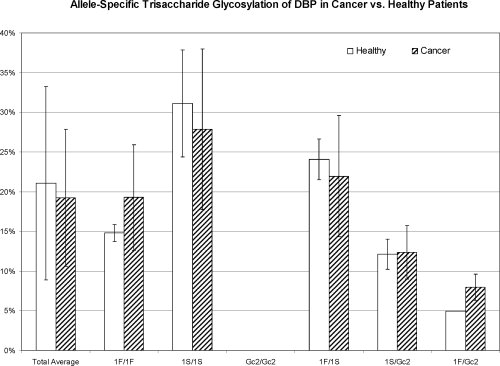
Allele-specific relative abundance of DBP O-linked trisaccharide glycosylation in cancer patients compared to healthy individuals. n-values for each sub-group are as follows: Total Healthy Individuals = 29, Total Cancer Patients = 56; 1F/1F Healthy = 3, 1F/1F Cancer = 4; 1S/1S Healthy = 10, 1S/1S Cancer = 15, Gc2/Gc2 Healthy = 4, Gc2/Gc2 Cancer = 4; 1F/1S Healthy = 3, 1F/1S Cancer = 10, 1S/Gc2 Healthy = 5, 1S/Gc2 Cancer = 18; 1F/Gc2 Healthy = 3, Cancer = 3. A few rare, unclassified DBP genotypic variants were also observed. Error bars indicate sample standard deviation. There are no statistically significant differences between cancer patients and healthy controls for any diploid genotype. As depicted in the plot and shown in raw format in Figure 1, Gc2 allele products completely lack trisaccharide glycosylation; the presence of trisaccharide glycosylated-DBP in heterozygous individuals is unambiguously due to modification of 1F or 1S DBP.8,9
In 1983, Viau et al.13 characterized the O-glycan trisaccharide of DBP as NeuNAc α-(2→3) Gal β-(1→3) GalNAc α-(1→0). Thus, mass shifts of +162 and +203 Da described later in reference to enzymatic glycosylation are assumed to represent Gal and GalNAc, respectively; even though mass shifts alone cannot ab initio distinguish between galactose and mannose or N-acetylgalactosamine and N-acetylglucosamine.
Mass spectra shown in Figure 1 also provide initial evidence, by virtue of a precise +203 Da mass shift (corresponding to the presence of a terminal GalNAc residue8,13), that significant quantities of what might nominally be considered GcMAF exist in the serum of 15 of 29 cancer patients. On the basis of the relative peak areas and the known concentration range of DBP in plasma, the approximate concentration of this molecular species in cancer patients is ∼5–10 mg/L (∼100–200 nM). As compared in Figure 3, the spectra from 21 of 29 age and gender matched healthy patients also exhibited peaks indicating the presence of a single residual GalNAc residue attached to Gc1 DBP. When observed, the relative quantity of Gc1 DBP carrying a single GalNAc residue was found to be greater in cancer patients than healthy individuals (P < 0.02; Fig. 3). Out of 32 observations of Gc2 allele products, no peaks were observed that could possibly represent Gc2 protein molecules carrying this single residual GalNAc residue, even when found paired in heterozygous fashion with Gc1 protein carrying the residual GalNAc residue.
Figure 3.
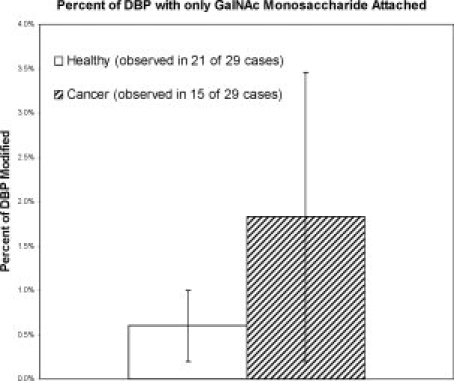
Relative percent of DBP modified with a GalNAc monosaccharide at T420 in cancer patients compared to age and gender matched healthy individuals. Data were tabulated only when a mass spectral peak representing the target species was observed (i.e., samples with a relative percentage of zero are not included). This molecular species was not observed on any of the 32 observed Gc2 allele products-even when found paired in heterozygous fashion with Gc1 protein carrying the residual GalNAc residue-hence the T420 positional assignment. Error bars represent standard deviation. *P < 0.02, Student's t-test.
Discussion
Trisaccharide glycosylated-DBP is not eliminated in cancer patients
The results presented here are in apparent conflict with the proposed mechanism for the inability of cancer patient serum to produce GcMAF on exposure to β-galactosidase and sialidase1–3,15,17: According to the theory, trisaccharide glycosylated-DBP in cancer patients is depleted to the point where it should be undetectable by the method employed in these studies.1–3,15,17 The depletion effect should be dramatic because a small mass spectral peak representing, for example, a 1% relative abundance of trisaccharide glycosylated-DBP would still correspond to relatively large quantities of GcMAF precursor (trisaccharide glycosylated-DBP) in the patient serum, on the order of ∼4 mg/L (based on a total serum concentration of ∼400 mg/L14), a quantity much greater than the ∼100 ng per person quantities of GcMAF employed as an effective cancer therapeutic.1–3 Thus, the observation that there is no depletion of trisaccharide glycosylated-DBP in cancer patients means that trisaccharide glycosylated-DBP cannot be the direct precursor of GcMAF that is eliminated in cancer patients.
Redefining the molecular definition and origin of GcMAF
The presence of relatively abundant quantities (∼5–10 mg/L or ∼100–200 nM) of what might nominally be considered GcMAF (i.e., DBP modified by a single terminal GalNAc residue) in cancer patient serum at first seems to both contradict macrophage activation studies using cancer patient serum1–3,15,17 and present a paradox in which cancer patients with relatively high circulating concentrations of GcMAF can effectively be treated with trace quantities of GcMAF.1–3
These apparent contradictions may be avoided by proposing that the most likely explanation for the apparent observation of GcMAF in the serum of Gc1 allele-carrying cancer patients is that the observed DBP molecules with a single terminal GalNAc at T420 are not GcMAF, that is, are not the same molecular species responsible for the macrophage activation observed by Yamamoto et al.1–3,15,17 This could be the case if, for example, genuine GcMAF carries a T418-linked GalNAc monosaccharide (derived from Gal-GalNAc-T418-modified DBP, as it most likely is for Gc2 allele products4,5), and the GalNAc-modified DBP species directly observed in these studies were derived from T420-linked trisaccharide glycosylated-DBP. The T420 linkage site is suspected for the directly observed GalNAc-modified DBP because the genetic variation of human DBP creates a natural site-directed mutagenesis study in which T420 is eliminated to create the Gc2 allele product. In the 32 cases of Gc2 allele product examined in this study, none were found to carry the single GalNAc residue observed on many Gc1 allele products-even when Gc2 protein was found paired in heterozygous fashion with Gc1 protein carrying the residual GalNAc residue. (Notably, no Gc2 allele products were found to carry the trisaccharide either.) These data suggest that the GalNAc monosaccharide observed is attached to T420-the same site of NeuNAc-Gal-GalNAc-Thr glycosylation suggested in previous studies by other investigators.13,14,18 With regard to glycosylation at T418, previous work of ours using tandem mass spectrometry of DBP peptides has confirmed trisaccharide glycosylation of T420 and has gone on to show that a disaccharide glycosylated form of DBP appears, in some Gc1-derived protein molecules, to have the carbohydrate attached at T418.8 Thus, glycosylation of Gc1 allele products at T418 may be involved with formation of active GcMAF and may serve as a way out of the apparent contradictions between the data presented here and conclusions of molecular structure drawn from activity studies.1–3,15,17 Due to its extremely low natural abundance, it is unlikely that true GcMAF from any allele product would be detected by MSIA without isolation from the most abundant forms of DBP.
Additionally, it may be that the existence of the observed GalNAc-modified DBP (which is too abundant in cancer patients to be GcMAF) solves the contradictory reports of Kanan et al.19 and Yamamoto et al.5–7,17,20 The former found noninducible generation of GalNAc-only modified DBP via lectin-based immunoassay which, when believed to be GcMAF, is in conflict with the activity based studies of Yamamoto et al. which suggest that a mediator of inflammation such as lysophosphatidylcholine must be present to induce production of GcMAF. The GalNAc monosaccharide-modified DBP observed in this report would produce positive results in a lectin-based immunoassay performed on samples tested in the absence of a mediator of inflammation, showing itself to be “noninducible.” Yet, as described earlier, this “noninducible” species is far too abundant in cancer patients to be active GcMAF.
Methods
Electrospray ionization-based mass spectrometric immunoassay (ESI-MSIA) was employed to ascertain the relative degree of trisaccharidic glycosylation of serum or plasma DBP in 56 human cancer patients (nine breast cancer, 11 colorectal cancer, nine prostate cancer, and 27 pancreatic cancer patients) and 29 healthy humans.
MSIA is a high throughput-amenable analytical technique in which a native protein of interest is purified by immunoprecipitation then analyzed intact by mass spectrometry. Since its inception in 1995 by R.W. Nelson et al.,21 it has been applied to study a variety of proteins and peptides,22–36 and has served as the foundation for the nascent field of Population Proteomics.37–42 Roughly, MSIA can be envisioned as ultra high resolution, semiquantitative Western blotting in which protein variants are thoroughly resolved and their molecular masses determined to within less than 2 Da.
Materials
Polyclonal rabbit anti-human DBP (GC-Globulin) antibodies (Cat. No. A0021) were obtained from DAKO (Carpinteria, CA). According to the manufacturer's specifications, this antibody is for in vitro diagnostic use and is intended for the determination of DBP in gel immunoprecipitation techniques and for phenotyping of DBP by immunofixation. Premixed MES-buffered saline powder packets were from Pierce (Rockford, IL). Isolation of DBP from plasma was carried out with proprietary MSIA pipette tips from Intrinsic Bioprobes (Tempe, AZ) derivatized with the DBP antibodies via 1,1′-carbonyldiimidazole (CDI) chemistry as previously described.33,43 Serum samples from 29 cancer patients (nine breast cancer, 11 colorectal cancer, nine prostate cancer) of African-American, Caucasian, and Hispanic origin, along with age, gender, and race-matched controls were purchased from ProMedDx (Norton, MA). Twenty seven plasma samples from pancreatic cancer patients were generously donated by Dr. Douglas Lake of the School of Life Sciences at Arizona State University. All cancer patients were diagnosed by qualified physicians. Protein Captrap catridges for LC/MS were obtained from Michrom Bioresources (Auburn, CA). Premade 10 mM Hepes-buffered saline (HBS) was bought from Biacore (Piscataway, NJ). All other chemicals were obtained from Sigma-Aldrich (St. Louis, MO).
Sample preparation for the analysis of intact DBP by MSIA ESI-TOF-MS
With the aid of a Beckman Multimek 96-channel automated pipetter, MSIA pipette tips that had been preactivated with CDI33,43 were derivatized with polyclonal DBP antibodies by repetitively flowing (aspirating and dispensing 750 times) 50 μL volumes of antibody solution (150 μL/well; 0.05 g/L in 0.1M MES buffered saline, pH 4.7) through the tips. Antibody-linked tips were stored in HBS at 4°C until the day of use at which time they were prerinsed (400 μL/well; 150 μL aspirate and dispense cycles; 10 cycles) with HBS then used to extract DBP from individual samples (25 μL of human serum or plasma diluted with 75 μL of HBS) at room temperature (100 μL/well; 50 μL aspirate and dispense cycles; 750 cycles). Pipette tips were then ejected from the robot and allowed to sit in their respective plasma samples at room temperature until they were individually (manually) washed (by drawing from a fresh reservoir of liquid and dispensing to waste) and eluted as follows: Five cycles of 200 μL of HBS, five cycles of 200 μL distilled water, five cycles 200 μL of 2M ammonium acetate/acetonitrile (3:1 v/v), ten cycles of 200 μL of distilled water. Elution was accomplished by briefly air-drying the pipette frits then drawing 5 μL of a mixture of 100% formic acid/acetonitrile/distilled water (9/5/1 v/v/v), mixing over the pipette affinity capture frit for 20–30 seconds, and dispensing into a 96-conical well polypropylene autosampler tray. Frits were then washed with an additional 5.5 μL distilled water which was used to dilute the eluted sample. Six microliters was injected into the LC-TOF-MS well within 10 min of elution to avoid protein formylation or other potential modifications from the elution solvent.
ESI-TOF-MS
A trap-and-elute form of sample concentration/solvent exchange rather than traditional LC was used for these analyses. Six-microliter samples were injected by a Spark Holland Endurance autosampler in microliter pick-up mode and loaded by an Eksigent nanoLC*1D at 10 μL/min (90/10 water/acetonitrile containing 0.1% formic acid, Solvent A) onto a protein captrap (polymeric/reversed phase sorbent, Michrom Bioresources, Auburn, CA) configured for unidirectional flow on a 6-port divert valve. After 2 min, the divert valve position was automatically toggled and flow rate over the protein captrap cartridge changed to 1 μL/min solvent A (running directly to the ESI inlet) which was immediately ramped over 8 min of a linear gradient from 10 to 90% solvent B (100% acetonitrile). By 10.2 min, the analysis was completed and the flow rate back to 100% solvent A.
DBP and its variants eluted between 5.5 and 7.5 min into a Bruker MicrOTOF-Q (Q-TOF) mass spectrometer operating in positive ion, TOF-only mode, acquiring spectra in the m/z range of 50–3000. ESI settings for the Agilent G1385A capillary nebulizer ion source were as follows: End Plate Offset −500 V, Capillary −4500 V, Nebulizer nitrogen 2 Bar, Dry Gas nitrogen 3.0 L/min at 225°C. Data were acquired in profile mode at a digitizer sampling rate of 2 GHz. Spectra rate control was by summation at 1 Hz.
Data analysis for intact DBP
Approximately 1 min of recorded spectra were averaged across the chromatographic peak apex of DBP elution. The ESI charge-state envelope was deconvoluted with Bruker DataAnalysis v3.4 software to a mass range of 1000 Da on either side of any identified peak. Deconvoluted spectra were baseline subtracted and all peaks were integrated. Tabulated mass spectral peak areas were exported to a spreadsheet for further calculation and determination of the peak areas of interest relative to all other forms of DBP present in the mass spectrum.
Conclusions
Trisaccharide glycosylated-DBP circulates in abundant quantities (comparable to that observed in healthy individuals) in the blood of breast, colorectal, pancreatic, and prostate cancer patients, making it unlikely that it is a depleted GcMAF precursor in cancer patients. Alternative hypotheses regarding the molecular and/or structural origins of GcMAF must be considered to explain the relative inability of cancer patient serum to activate macrophages.
Acknowledgments
Thanks to Dr. Douglas Lake of the School of Life Sciences at Arizona State University for his generous donation of pancreatic cancer patient plasma samples. Also, thanks to Jim Tassano for helpful discussions.
Glossary
Abbreviations:
- CDI
1,1′-carbonyldiimidazole
- DBP
vitamin D binding protein
- ESI-MSIA
electrospray ionization-based mass spectrometric immunoassay
- Gal
galactose
- GalNAc
N-acetylgalactosamine
- GcMAF
Gc macrophage activating factor
- HBS
hepes-buffered saline
- MES
2-(N-morpholino)ethanesulfonic acid
- nagalase
α-N-acetylgalactosaminidase
- NeuNAc
N-acetylneuraminic acid.
References
- 1.Yamamoto N, Suyama H, Nakazato H, Yamamoto N, Koga Y. Immunotherapy of metastatic colorectal cancer with vitamin D-binding protein-derived macrophage-activating factor. Gc MAF Cancer Immunol Immunother. 2008;57:1007–1016. doi: 10.1007/s00262-007-0431-z. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 2.Yamamoto N, Suyama H, Yamamoto N. Immunotherapy for prostate cancer with Gc protein-derived macrophage-activating factor. Gc MAF Transl Oncol. 2008;1:65–72. doi: 10.1593/tlo.08106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamamoto N, Suyama H, Yamamoto N, Ushijima N. Immunotherapy of metastatic breast cancer patients with vitamin D-binding protein-derived macrophage activating factor (GcMAF) Int J Cancer. 2008;122:461–467. doi: 10.1002/ijc.23107. [DOI] [PubMed] [Google Scholar]
- 4.Yamamoto N. Structural definition of a potent macrophage activating factor derived from vitamin D3-binding protein with adjuvant activity for antibody production. Mol Immunol. 1996;33:1157–1164. doi: 10.1016/s0161-5890(96)00081-8. [DOI] [PubMed] [Google Scholar]
- 5.Yamamoto N, Homma S. Vitamin D3 binding protein (group-specific component) is a precursor for the macrophage-activating signal factor from lysophosphatidylcholine-treated lymphocytes. Proc Natl Acad Sci USA. 1991;88:8539–8543. doi: 10.1073/pnas.88.19.8539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamamoto N, Homma S, Millman I. Identification of the serum factor required for in vitro activation of macrophages. Role of vitamin D3-binding protein (group specific component, Gc) in lysophospholipid activation of mouse peritoneal macrophages. J Immunol. 1991;147:273–280. [PubMed] [Google Scholar]
- 7.Yamamoto N, Kumashiro R. Conversion of vitamin D3 binding protein (group-specific component) to a macrophage activating factor by the stepwise action of beta-galactosidase of B cells and sialidase of T cells. J Immunol. 1993;151:2794–2802. [PubMed] [Google Scholar]
- 8.Borges CR, Jarvis JW, Oran PE, Nelson RW. Population studies of vitamin d binding protein microheterogeneity by mass spectrometry lead to characterization of its genotype-dependent o-glycosylation patterns. J Proteome Res. 2008;7:4143–4153. doi: 10.1021/pr8002936. [DOI] [PubMed] [Google Scholar]
- 9.Borges CR, Jarvis JW, Oran PE, Rogers SP, Nelson RW. Population studies of intact Vitamin D binding protein by affinity capture ESI-TOF-MS. J Biomol Tech. 2008;19:167–176. [PMC free article] [PubMed] [Google Scholar]
- 10.Coppenhaver DH, Sollenne NP, Bowman BH. Post-translational heterogeneity of the human vitamin D-binding protein (group-specific component) Arch Biochem Biophys. 1983;226:218–223. doi: 10.1016/0003-9861(83)90287-4. [DOI] [PubMed] [Google Scholar]
- 11.Svasti J, Bowman BH. Human group-specific component. Changes in electrophoretic mobility resulting from vitamin D binding and from neuraminidase digestion. J Biol Chem. 1978;253:4188–4194. [PubMed] [Google Scholar]
- 12.Svasti J, Kurosky A, Bennett A, Bowman BH. Molecular basis for the three major forms of human serum vitamin D binding protein (group-specific component) Biochemistry. 1979;18:1611–1617. doi: 10.1021/bi00575a036. [DOI] [PubMed] [Google Scholar]
- 13.Viau M, Constans J, Debray H, Montreuil J. Isolation and characterization of the O-glycan chain of the human vitamin-D binding protein. Biochem Biophys Res Commun. 1983;117:324–331. doi: 10.1016/0006-291x(83)91579-6. [DOI] [PubMed] [Google Scholar]
- 14.Speeckaert M, Huang G, Delanghe JR, Taes YE. Biological and clinical aspects of the vitamin D binding protein (Gc-globulin) and its polymorphism. Clin Chim Acta. 2006;372:33–42. doi: 10.1016/j.cca.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 15.Yamamoto N, Naraparaju VR, Urade M. Prognostic utility of serum alpha-N-acetylgalactosaminidase and immunosuppression resulted from deglycosylation of serum Gc protein in oral cancer patients. Cancer Res. 1997;57:295–299. [PubMed] [Google Scholar]
- 16.Korbelik M, Naraparaju VR, Yamamoto N. The value of serum alpha-N-acetylgalactosaminidase measurement for the assessment of tumour response to radio- and photodynamic therapy. Br J Cancer. 1998;77:1009–1014. doi: 10.1038/bjc.1998.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamamoto N, Naraparaju VR, Asbell SO. Deglycosylation of serum vitamin D3-binding protein leads to immunosuppression in cancer patients. Cancer Res. 1996;56:2827–2831. [PubMed] [Google Scholar]
- 18.Arnaud J, Constans J. Affinity differences for vitamin D metabolites associated with the genetic isoforms of the human serum carrier protein (DBP) Hum Genet. 1993;92:183–188. doi: 10.1007/BF00219689. [DOI] [PubMed] [Google Scholar]
- 19.Kanan RM, Cook DB, Datta HK. Lectin immunoassay for macrophage-activating factor (Gc-MAF) produced by deglycosylation of Gc-globulevidence for noninducible generation of Gc-MAF. Clin Chem. 2000;46:412–414. [PubMed] [Google Scholar]
- 20.Yamamoto N, Naraparaju VR, Orchard PJ. Defective lymphocyte glycosidases in the macrophage activation cascade of juvenile osteopetrosis. Blood. 1996;88:1473–1478. [PubMed] [Google Scholar]
- 21.Nelson RW, Krone JR, Bieber AL, Williams P. Mass spectrometric immunoassay. Anal Chem. 1995;67:1153–1158. doi: 10.1021/ac00103a003. [DOI] [PubMed] [Google Scholar]
- 22.Kiernan UA, Addobbati R, Nedelkov D, Nelson RW. Quantitative multiplexed C-reactive protein mass spectrometric immunoassay. J Proteome Res. 2006;5:1682–1687. doi: 10.1021/pr0601133. [DOI] [PubMed] [Google Scholar]
- 23.Kiernan UA, Nedelkov D, Nelson RW. Multiplexed mass spectrometric immunoassay in biomarker research: a novel approach to the determination of a myocardial infarct. J Proteome Res. 2006;5:2928–2934. doi: 10.1021/pr060062+. [DOI] [PubMed] [Google Scholar]
- 24.Kiernan UA, Nedelkov D, Niederkofler EE, Tubbs KA, Nelson RW. High-throughput affinity mass spectrometry. Methods Mol Biol. 2006;328:141–150. doi: 10.1385/1-59745-026-X:141. [DOI] [PubMed] [Google Scholar]
- 25.Kiernan UA, Nedelkov D, Tubbs KA, Niederkofler EE, Nelson RW. Proteomic characterization of novel serum amyloid P component variants from human plasma and urine. Proteomics. 2004;4:1825–1829. doi: 10.1002/pmic.200300690. [DOI] [PubMed] [Google Scholar]
- 26.Kiernan UA, Nedelkov D, Tubbs KA, Niederkofler EE, Nelson RW. Selected expression profiling of full-length proteins and their variants in human plasma. Clin Proteomics J. 2004;1:7–16. [Google Scholar]
- 27.Kiernan UA, Tubbs KA, Gruber K, Nedelkov D, Niederkofler EE, Williams P, Nelson RW. High-throughput protein characterization using mass spectrometric immunoassay. Anal Biochem. 2002;301:49–56. doi: 10.1006/abio.2001.5478. [DOI] [PubMed] [Google Scholar]
- 28.Kiernan UA, Tubbs KA, Nedelkov D, Niederkofler EE, McConnell E, Nelson RW. Comparative urine protein phenotyping using mass spectrometric immunoassay. J Proteome Res. 2003;2:191–197. doi: 10.1021/pr025574c. [DOI] [PubMed] [Google Scholar]
- 29.Kiernan UA, Tubbs KA, Nedelkov D, Niederkofler EE, Nelson RW. Comparative phenotypic analyses of human plasma and urinary retinol binding protein using mass spectrometric immunoassay. Biochem Biophys Res Commun. 2002;297:401–405. doi: 10.1016/s0006-291x(02)02212-x. [DOI] [PubMed] [Google Scholar]
- 30.Kiernan UA, Tubbs KA, Nedelkov D, Niederkofler EE, Nelson RW. Detection of novel truncated forms of human serum amyloid A protein in human plasma. FEBS Lett. 2003;537:166–170. doi: 10.1016/s0014-5793(03)00097-8. [DOI] [PubMed] [Google Scholar]
- 31.Niederkofler EE, Kiernan UA, O'Rear J, Menon S, Saghir S, Protter AA, Nelson RW, Schellenberger U. Detection of endogenous B-type natriuretic peptide at very low concentrations in patients with heart failure. Vol. 1. Circulation: Heart Failure; 2008. pp. 258–264. [DOI] [PubMed] [Google Scholar]
- 32.Niederkofler EE, Tubbs KA, Gruber K, Nedelkov D, Kiernan UA, Williams P, Nelson RW. Determination of beta-2 microglobulin levels in plasma using a high-throughput mass spectrometric immunoassay system. Anal Chem. 2001;73:3294–3299. doi: 10.1021/ac010143j. [DOI] [PubMed] [Google Scholar]
- 33.Niederkofler EE, Tubbs KA, Kiernan UA, Nedelkov D, Nelson RW. Novel mass spectrometric immunoassays for the rapid structural characterization of plasma apolipoproteins. J Lipid Res. 2003;44:630–639. doi: 10.1194/jlr.D200034-JLR200. [DOI] [PubMed] [Google Scholar]
- 34.Tubbs KA, Kiernan UA, Niederkofler EE, Nedelkov D, Bieber AL, Nelson RW. High-throughput MS-based protein phenotyping: application to haptoglobin. Proteomics. 2005;5:5002–5007. doi: 10.1002/pmic.200500176. [DOI] [PubMed] [Google Scholar]
- 35.Tubbs KA, Kiernan UA, Niederkofler EE, Nedelkov D, Bieber AL, Nelson RW. Development of recombinant-based mass spectrometric immunoassay with application to resistin expression profiling. Anal Chem. 2006;78:3271–3276. doi: 10.1021/ac060013g. [DOI] [PubMed] [Google Scholar]
- 36.Tubbs KA, Nedelkov D, Nelson RW. Detection and quantification of beta-2-microglobulin using mass spectrometric immunoassay. Anal Biochem. 2001;289:26–35. doi: 10.1006/abio.2000.4921. [DOI] [PubMed] [Google Scholar]
- 37.Nedelkov D. Population proteomics: addressing protein diversity in humans. Expert Rev Proteomics. 2005;2:315–324. doi: 10.1586/14789450.2.3.315. [DOI] [PubMed] [Google Scholar]
- 38.Nedelkov D. Population proteomics: investigation of protein diversity in human populations. Proteomics. 2008;8:779–786. doi: 10.1002/pmic.200700501. [DOI] [PubMed] [Google Scholar]
- 39.Nedelkov D, Kiernan UA, Niederkofler EE, Tubbs KA, Nelson RW. Investigating diversity in human plasma proteins. Proc Natl Acad Sci USA. 2005;102:10852–10857. doi: 10.1073/pnas.0500426102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nedelkov D, Kiernan UA, Niederkofler EE, Tubbs KA, Nelson RW. Population proteomics: the concept, attributes, and potential for cancer biomarker research. Mol Cell Proteomics. 2006;5:1811–1818. doi: 10.1074/mcp.R600006-MCP200. [DOI] [PubMed] [Google Scholar]
- 41.Nedelkov D, Phillips DA, Tubbs KA, Nelson RW. Investigation of human protein variants and their frequency in the general population. Mol Cell Proteomics. 2007;6:1183–1187. doi: 10.1074/mcp.M700023-MCP200. [DOI] [PubMed] [Google Scholar]
- 42.Nedelkov D, Tubbs KA, Niederkofler EE, Kiernan UA, Nelson RW. High-throughput comprehensive analysis of human plasma proteins: a step toward population proteomics. Anal Chem. 2004;76:1733–1737. doi: 10.1021/ac035105+. [DOI] [PubMed] [Google Scholar]
- 43.Hermanson GT, Krishna Mallia A, Smith PK. Immobilized affinity ligand techniques. San Diego, CA: Academic Press; 1992. [Google Scholar]


