Abstract
ZntB is the distant homolog of CorA Mg2+ transporter within the metal ion transporter superfamily. It was early reported that the ZntB from Salmonella typhimurium facilitated efflux of Zn2+ and Cd2+, but not Mg2+. Here, we report the 1.90 Å crystal structure of the intracellular domain of ZntB from Vibrio parahemolyticus. The domain forms a funnel-shaped homopentamer that is similar to the full-length CorA from Thermatoga maritima, but differs from two previously reported dimeric structures of truncated CorA intracellular domains. However, no Zn2+ or Cd2+ binding sites were identified in the high-resolution structure. Instead, 25 well-defined Cl− ions were observed and some of these binding sites are highly conserved within the ZntB family. Continuum electrostatics calculations suggest that the central pore of the funnel is highly attractive for cations, especially divalents. The presence of the bound Cl− ions increases the stability of cations along the pore suggesting they could be important in enhancing cation transport.
Keywords: ZntB, CorA, transporter, structure, pentamer, electrostatic, Cl− ions
Introduction
The metal ion transporter (MIT) superfamily is composed of a large and diverse group of essential membrane proteins found in all three kingdoms of life involved in metal ion transport (uptake or efflux) across membranes. Among MITs, the Mg2+ transporters of the CorA family, primarily responsible for Mg2+ uptake in most prokaryotes (and some eukaryotes), have been thus far the most extensively characterized. The three recently determined full-length Thermatoga maritima CorA (Tm-CorA) lower resolution structures (2.9, 3.7, and 3.9 Å) have revealed a striking homopentamer with two transmembrane α-helical spanners per monomer and a large funnel-shaped intracellular assembly.1–3 However, two structures of cytoplasmic domains of Tm-CorA (1.85 Å)1 and Archaeoglobus fulgidus CorA (Af-CorA) (2.9 Å)3 showed dimeric arrangements of the CorA subunits. The observed conformational differences between the full-length Tm-CorA pentamer and the Af-CorA intracellular domain dimer were used to propose a close/open CorA translocation cycle model.3 Two conserved metal binding sites were found on the interface between monomers in the intracellular domain in both the CorA pentamers and dimers. Analysis of metal binding sites suggested a possible gating mechanism linking the pore to the intracellular concentration of Mg2+.1 Other divalent cations, such as Co2+ and Ca2+ can also be accommodated by CorA,2,3 but it is not clear how binding the metal ions can influence transport through the pore. In one study, a putative Cl− anion was found near the metal-binding sites and was suggested to be important in stabilizing the conformation of the N-terminal subdomain.2
Zinc is the second most abundant transition metal in biological systems and is essential for many cellular processes4,5 though the concentration of free zinc in the cell is low.6 Zinc excess is highly toxic and the range of tolerable concentrations is narrow.7 Therefore, multiple zinc specific transport systems maintain homeostasis in the cell. In eubacteria, zinc uptake is facilitated by the ZnuABC8 and ZupT9 transport systems and zinc efflux is maintained by the ZntA,10 ZitB,11 and ZntB12 efflux systems.
ZntB proteins are sequence homologs of the CorA-type Mg2+ transporters.13 These proteins belong to the 2-TM-GxN clad with the signature sequence G[I,V]N.13 The ZntB protein from Salmonella typhimurium was early reported as a zinc transporter, facilitating Zn2+ (and Cd2+) ions efflux across the membrane, but not transporting Mg2+ ions.12 The ZntB transporter homologs are exclusively identified in the protobacteria of the α-, β-, and γ-subgroups with distant relatives found in plants.13
The ZntB protein from Vibrio parahemolyticus (Vp-ZntB), a major diarrheagenic Vibrio species, was identified but has not been functionally characterized.14 It shows 30% primary sequence identity with S. typhimurium ZntB (St-ZntB) and 17% with Tm-CorA. The crystal structure of the Vp-ZntB cytoplasmic domain determined at 1.90 Å resolution reveals a funnel-shaped homopentamer, similar to the full-length Tm-CorA structures. The high-resolution funnel structure enables us to examine the transporter in more detail and identify 25 bound Cl− ions that seem to promote divalent metal ion transport.
Results
The structures of Vp-ZntB monomer and pentamer
A Vp-ZntB cytoplasmic domain monomer is composed of two closely associated subdomains, a N-terminal α/β/α sandwich subdomain (residues 1–125) and a C-terminal coiled-coil subdomain (residues 126–203) [Fig. 1(A)]. The α/β/α subdomain consists of a twisted central β-sheet with seven mainly anti-parallel β-strands (β2↑β1↓β3↑β7↑β6↓β5↑β4↓). On one side, the β-sheet is flanked by two anti-parallel α-helices connected between the β3 and β4. On the other side, the β-sheet interacts with the α3 helix and the N-terminal part of α4 and the C-terminal region of the α6 helix from the coiled-coil subdomain. The coiled-coil subdomain is composed of three long anti-parallel α-helices, α4, α5, and α6 (the stalk helix in Tm-CorA). The links between them are all well structured. A coiled-coil structure is characterized by heptad sequence repeats (abcdefg)n, in which the residues at positions 1 (a) and 4 (d) are mostly hydrophobic and form core-like inter-helical interactions15,16 [highlighted in Fig. 1(B)].
Figure 1.
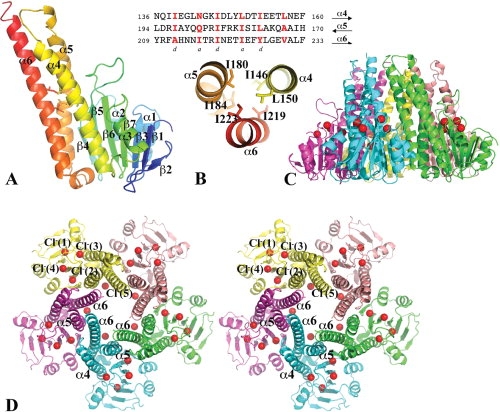
The structure of the Vp-ZntB cytoplasmic domain. (A) A ribbon drawing of the Vp-ZntB cytoplasmic domain monomer structure with the secondary structure in rainbow colors from N- (blue) to C-terminus (red). The subunit can be divided into an N-terminal α/β/α sandwich domain on the right side and a C-terminal coiled-coil like domain on the left side. Six residues [see Fig. 1(B)] contributing to two layers of inter-helical interactions are drawn in stick form. (B) The heptad sequence repeats in the coiled-coil forming helices, α4, α5, and α6. The residues at positions a and d are mostly hydrophobic and form core inter-helical interactions, only two layers of interactions are shown. (C) Ribbon drawings of Vp-ZntB cytoplasmic domain pentamer in the side view. (D) The stereo view of the pentamer from the top. Five monomers are colored differently. The bound Cl− anions, which form a ring, are drawn as red balls. The large opening at the funnel bottom is formed with five α5 helices and five α6 helices. The top small opening is formed by five α6 helices. The second half of the α6 helix interacts with two neighboring α6 helices and one neighboring α6 helix. This interaction seems critical to the formation of the pentamer. Figures 1, 2, and 3 were prepared using the program Pymol (http://www.PyMOL.org).
Five Vp-ZntB cytoplasmic domain monomers form a funnel-shaped assembly with approximate five-fold symmetry [Fig. 1(C,D)]. The coiled-coil subdomains exclusively contribute to the formation of the pentamer. The large opening of the funnel is formed by five α5 and five α6 stalk helices while the small opening is formed by the C-terminal portions of five α6 stalk helices. Across the monomer–monomer interface, there are numerous interactions, including salt-bridges (E221-R63, E221-R185, and E236-K175), hydrogen bonds (H56-N214, H207-D70, N214-R63, N214-S65, and T222-R182), and hydrophobic interactions involving residues I69, I171, I178, F111, F225, and L232 (see Fig. 2). The total buried surface at the monomer–monomer interface is ∼2100 Å2, a number that is larger than a “typical” protein interface [average surface area is 1600 (±400) Å2].17 The Gibbs free energy ΔG resulted from the formation of the interface is −8.6 kcal/mol.18 These data indicate that the subunit interface is stable and the ZntB cytoplasmic domain pentameric assembly can form in the absence of transmembrane spanners.
Figure 2.
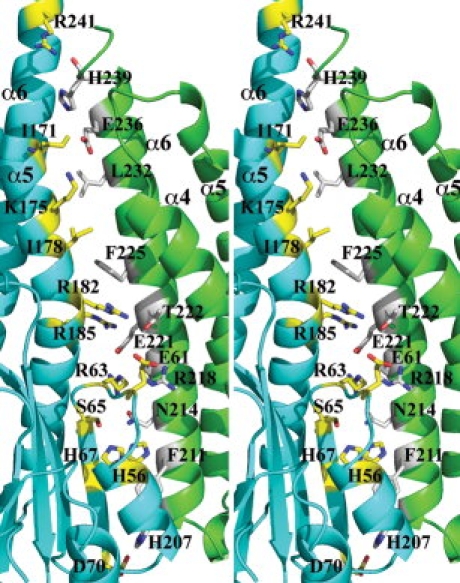
Interactions across the monomer–monomer interface. A stereo ribbon drawing of one monomer–monomer interface. The residues involved in the interaction across the interface are drawn in sticks and colored differently according to monomers. Salt bridges include K175-E236, R63-E221, and D70-H207. Hydrogen bonds include R182-T22 and S65-N214. There is also water molecules involved in hydrogen bonds with protein residues. The residues R185 and F225 may form a π-cation interaction.
Ion-binding sites
In the anomalous difference Fourier maps calculated from selenium and zinc edge data, besides the 35 peaks (corresponding to the Se atoms) in the anomalous difference map calculated from selenium absorption edge peak data there were no other significant peaks. X-ray fluorescence measurements of Vp-ZntB crystals also indicated the absence of zinc or other common transition metal atoms (Co, Mn, and Cd) in the Vp-ZntB structure.
However, during solvent model building, five electron density peaks (initially assigned as water molecules) per monomer showed significant positive densities in Fo−Fc Fourier difference map (2.5−4.5 σ) in all equivalent positions of the five monomers [Figs. 1(D) and 3]. The arrangement of their neighboring protein atoms indicated that they were Cl− ions (protein was crystallized in the presence 0.2M MgCl2).19 Cl− ions were then assigned to these sites and refined with full occupancies. Their refined B-factors were similar to that of their neighboring protein atoms. To further verify the presence of Cl− ions in the ZntB structure, anomalous diffraction data were collected at an energy that allows direct detection of anomalous diffraction originating from certain types of atoms such as sulfur, phosphorus, chlorine, or transition metals present in the protein structure. At 7 keV, the anomalous scattering factor δf′ of a chlorine atom is equivalent to 0.913 electron and gives a stronger signal than a sulfur atom with δf′ equivalent to 0.729 electron (http://henke.lbl.gov/optical_constants/asf.html). Therefore, by combining the anomalous data with their distinct protein binding chemistry,19 the Cl− ions can be unambiguously identified in the structure. The anomalous difference map [Fig. 3(F)] calculated from the data collected at 7 keV clearly illustrates the anomalous scatterers in the Vp-ZntB structure. They include seven Se atoms from SeMet residues, the S atom of Cys33 (not shown in the figure) and the five Cl− ions per Vp-ZntB monomer. Within Cl− ions, the anomalous peaks of Cl−(1) and Cl−(2) are the highest, ranging from 6.1 σ to 8.5 σ. The peaks of Cl−(3) ions are within 4.7 σ–7.0 σ. The five peaks of Cl−(4) ions have a wider range in height and they are 2.9 σ, 3.5 σ, 5.9 σ, 6.1 σ, and 6.8 σ, respectively. The Cl−(5) ions are relatively weak scatterers and their five peaks are in the height of 3.0 σ, 3.2 σ, 3.6 σ, 5.0 σ, and 5.7 σ. No bound Mg2+ was observed in the structure though crystallization buffer included 0.2M MgCl2. In fact, no metal ion was identified in the structure. However, this doesn't exclude the possibility that Mg2+ ion could bind to the transmembrane domain.
Figure 3.
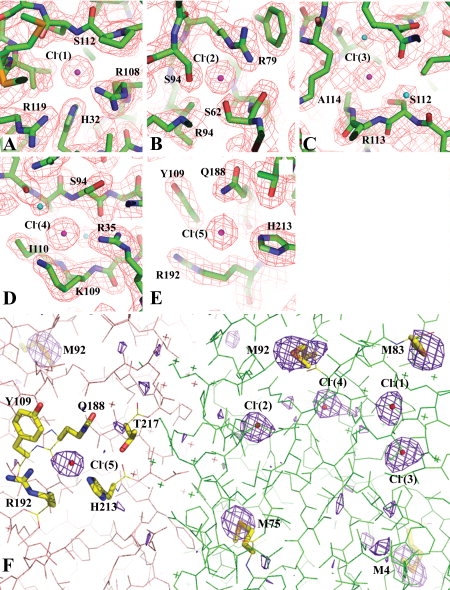
The Cl− binding sites and Cl− anomalous diffration. The Cl− binding sites and anomalous difference maps obtained using data collected at 7 keV. (A–E) The five Cl− ions and their interactions with neighboring protein residues. The omitted 2Fo–Fc Fourier difference maps in all these figures were contoured at a 1.3 σ level. These maps were calculated using a 1.9 Å data set (Table I). (F) The anomalous difference map contoured at 3 σ level calculated at 2.5 Å using data collected at 7 keV. The map shows the anomalous scatterers including five chloride ions (in the format of small red balls) and several selenium atoms from SeMet residues (in the stick format). The sulfur atom of Cys33 also shows a similar anomalous peak (out of the view in this figure). Two Vp-ZntB monomers in the background are drawn in two different colors. The “X” represents water molecules.
The low energy anomalous diffraction data conclusively confirm the presence of Cl− ions in the ZntB structure. Interestingly, the Cl− ions are not located within monomer–monomer interfaces [Fig. 1(C,D)]. The first four are largely associated with the α/β/α subdomain. The Cl−(1) [Fig. 3(A)] is in a pocket between the β-sheet and the α3 helix of the α/β/α subdomain. It interacts directly with H32 (NE2 atom, ∼3.07 Å) from β3, R108 (NH1, ∼3.26 Å) from β7 and the amide group of S112 (∼3.25 Å) from the β7_α3 loop. Both H32 and R108 are highly conserved in the ZntB transporters. It also forms a water-bridged interaction with R119 (NH1) from the α3 helix. The Cl−(2) [Fig. 3(B)] is in a shallow pocket between the β-sheet and the α2_β4 loop. It directly interacts with highly conserved R79 (NE, ∼2.86 Å) from β5, S94 (OG, ∼3.01 Å) from β6 and S62 (OG, ∼2.95 Å) from the α2_β4 loop. Both S62 and S94 also form hydrogen bonds to the neighboring R96. The Cl−(3) [Fig. 3(C)] is uniquely located at the N-terminal end of the α3 helix, interacting with the two amides from R113 (∼3.31 Å) and A114 (∼3.23 Å), respectively. It also bonds to S112 (OG, ∼3.02 Å). The Cl−(3) is positioned to interact with the amino (positive) end of the α3 helix dipole. The Cl−(4) [Fig. 3(D)] interacts with R35 (NH1, ∼3.45 Å) from the end of β3, and the two amides, one from S94 (∼3.24 Å) at the beginning of β6 and the other from I110 at the end of β7. It also interacts with the amide or the side chain of K109. Finally, the Cl−(5) [Fig. 3(E)] is located on the internal wall of the transporter funnel. It is the only Cl− ion identified in the structure to be associated with the coiled-coil subdomain. The Cl−(5) binds to H213 (NE2, ∼3.13 Å) from the α8 stalk helix and Q188 (NE2, ∼3.24 Å) from α5. It also interacts with nearby R192 from the α5.
Structural alignment of Vp-ZntB and Tm-CorA
The intracellular domains of Tm-CorA and Vp-ZntB are structurally similar both at the level of monomer and pentameric assembly. Nevertheless, the monomer and pentamer structures cannot be simply superimposed because the relative orientation the α/β/α and the coiled-coil subdomains are different. Separately, Vp-ZntB and Tm-CorA subdomains align well, the α/β/α subdomains can be superimposed with an RMSD value of 2.26 Å (for Cα of 111 residues aligned out 130 with sequence identity of 10.8%). Similarly, the two coiled-coil subdomains can be aligned with an RMSD value of 2.78 Å (for Cα of 106 residues aligned out 119 with a sequence identity of 10.4%). The change in orientation between the two subdomains seems to result in a larger funnel opening in Vp-ZntB [Fig. 4(B)].
Figure 4.
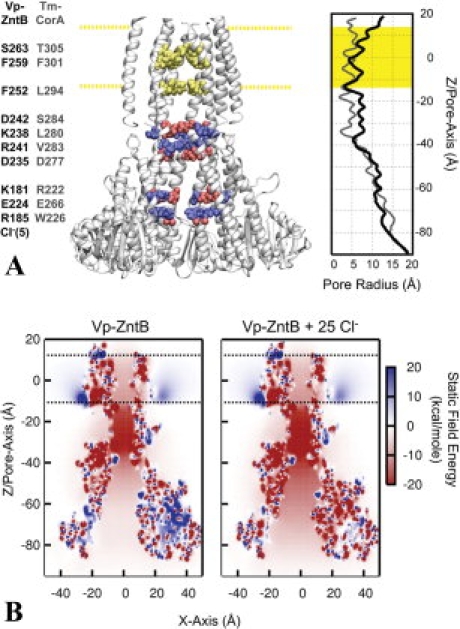
The architecture and static field potential of full-length ZntB. (A) Full-length ZntB, comprised of the crystallographic cytoplasmic domain with a homology modeled transmembrane domain based on Tm-CorA, is shown with one of the monomers removed for a clearer view of the pore. Cytoplasmic residues that contribute significantly to the electrostatic energy of an ion along the central pore are highlighted blue for positively charged residues (R185, K181, K238, and R241) and red for negative [E224, D235, D242, and Cl−(5)]. Residues in the transmembrane domain forming constriction sites are highlighted in yellow (S263, F259, and F252). The corresponding residues of Tm-CorA are listed adjacent to the residues of Vp-ZntB. The membrane is positioned between −12.5 Å < Z < 12.5 Å, as represented by the highlighted yellow regions. The radius profile is shown on the right hand side for the Vp-ZntB and Tm-CorA. (B) Static field potential energy maps of ZntB with and without Cl− ions. The energy of a positive charge in the electrostatic potential produced by the protein charge distribution is shown for every position along the XZ plane of the system. To accentuate the bare static field created by the protein charges and bound Cl− ions, no counter-ion screening was considered inside the pore (i.e., κTM-pore/cyto-pore = 0).
Full-length Vp-ZntB modeling and pore radius profile
Though the overall sequence identity between CorA and ZntB transporters is low (17%), their C-terminal transmembrane regions show significantly higher sequence similarity (55% with 29% of the residues being identical) (Supporting Information Fig. S1). On the basis of the structural and sequence alignment of Vp-ZntB and Tm-CorA (Supporting Information Fig. S1), a full-length Vp-ZntB model was built using the 2.9 Å structure of Tm-CorA2 [Fig. 4(A)].
Internal radius profiles were calculated for both Vp-ZntB and Tm-CorA [Fig. 4(A)]. In general, the Tm-CorA pore is too constricted to allow for ion permeation, which is in line with the supposition that the structure is in a closed state.1 On the other hand, the Vp-ZntB full-length model is more open with only a few regions reducing the pore to <5 Å [Fig. 4(A)]. The first constriction site occurs just below the membrane into the cytosol and is formed by a group of charged residues including D235, K238, R241, and D242. Tm-CorA is similarly narrow in this area but contains mostly neutral residues at the corresponding sites. The other two constriction sites are in the transmembrane domain and are created by residues S263, F259, and F252 [Fig. 4(A)].
Electrostatic properties of the pore
Static field potentials for Vp-ZntB were calculated along a cross-section of Vp-ZntB [Fig. 4(B)]. The pore of Vp-ZntB shows predominantly negative potentials throughout including the funnel-like entrance into the cytoplasm. The addition of the 25 Cl− anions significantly increases the negative potential of the Vp-ZntB and neutralizes most of the positive surface patches [Fig. 4(B)].
To further investigate the energetics involved in the ion transport process, the electrostatic free energy of transferring a cation from bulk and into the pore, ΔΔGint, was calculated along the central axis (see Fig. 5). This energy includes reaction field effects in addition to the static field, and also introduces some ionic screening inside the pore to estimate these interactions under multi-ion conditions. In these calculations, only the cytoplasmic results are reported as the modeled transmembrane region is too narrow to allow for an adequate interpretation of the electrostatics results.20 The energy profiles demonstrate four distinct features: (a) an initial stabilization along the entrance of the pore at Z = −80 Å, (b) a potential barrier around Z = −60 Å, (c) a small plateau region around Z = −40 Å, and (d) a deep electrostatic well at Z = −35 Å. The calculations reveal that the affinity of a positive ion inside the cytoplasmic entrance of the pore depends strongly on the presence of the Cl− ions detected in the ZntB structure. Including these ions in the calculations completely removes the barrier in the region (b) and makes the entire vestibule attractive for cations (see Fig. 5). Further calculations were performed with ions of charge +1 or +2, revealing that the pore is more attractive for divalent cations than monovalent. The barrier at Z = −60 Å is largely contributed by the sum of the effects from residues K181, R185, and E224 (Supporting Information Fig. S2). The Cl−(5) is located at the inner surface of the funnel [Figs. 1(D) and 3(E)] near residue R192 and close to R185, thereby counteracting the positive charge contributed by the basic residues in this neighborhood. A mixture of charged residues are responsible for the strong affinity well located at Z = −35 Å, which corresponds to the constriction site located below the membrane in Figure 4(A). The important residues are D235, K238, R241, and D242 with the strongest contributor being D242.
Figure 5.

Electrostatic free energy of cations along the cytoplasmic pore of Vp-ZntB. The four different locations along the pore are described in the text. The different curves correspond to monovalent (Q = +1) or divalent (Q = +2) pore ions and the presence of Cl− in the protein structure (x-ray Cl−). Ionic screening corresponding to 150 mM salt was included inside the wide cytoplasmic pore.
Discussion
The structure of the Vp-ZntB cytoplasmic domain indicates that these transporters can assemble pentamers in the absence of the transmembrane spanners; therefore, parts of the determinants of pentamer formation may lie within cytoplasmic domains.
Similar to the full-length Tm-CorA structures, the coiled-coil subdomain of Vp-ZntB exclusively contributes to the formation of the funnel-shaped pentamer. Across the monomer–monomer interface there are numerous interactions, including salt-bridges, direct and water mediated hydrogen bonds, and hydrophobic contacts (see Fig. 2). Interestingly, none of the residues involved in the monomer–monomer interactions is universally conserved in CorA and ZntB across different species. Although there seem to be no signature interactions across the monomer–monomer interface, the large buried molecular surface resulted from one pair of monomer–monomer contact (∼2100 Å2) and resulted favorable Gibbs free energy ΔG (−8.6 kcal/mol) indicate that the intracellular domain is sufficient to assemble a pentamer. In the full-length CorA structure, the total surface area buried due to monomer–monomer contact is about 4400 Å2 including 2720 Å2 from the intracellular region (1–282aa). The contribution of the soluble domain to Gibbs free energy ΔG from the monomer–monomer interaction in CorA is −6.7 kcal/mol, about one fourth of the total energy change due to the formation of monomer–monomer interface. This suggests that the intracellular domain of CorA-like transporters holds a rather significant fraction of determinants for pentamer formation.
As discussed earlier, the α6 stalk helix is the primary contributor of the monomer–monomer interaction along the entire length of the funnel. The helix also contributes to the formation of the coiled-coil subdomain of the intracellular domain [Fig. 1(B)]. Any truncation at the middle of stalk helix α6 will undoubtedly disrupt the stability of the coiled-coil subdomain and result in an incorrect folding of the intracellular domain. An improperly folded intracellular domain could alternate the monomer–monomer interaction and consequently their packing. The reported CorA intracellular domain dimers are likely to be artifacts of extensive truncation of stalk helix α6, in which the coiled-coil subdomain was dissociated.1,3 A Vp-ZntB construct truncated at N220, corresponding to the truncated Tm-CorA (1–266),1 would have dramatically reduced the monomer–monomer interface (a loss of ∼1000 Å2 buried area) and is not expected to form a pentamer.
In Tm-CorA structures, two divalent metal (Mg2+, Co2+, or Ca2+) binding sites and a putative Cl− binding site were located at the monomer–monomer interface between N-terminal α/β/α domains. These metal binding-sites were believed to be mostly occupied by Mg2+ ions in the CorA structure. They seem to be one of the hallmarks of the Mg2+ transporters subgroup A. However, the residues involved in the metal bindings are not conserved in the CorA subgroup B,21 nor in the ZntB subfamily (Supporting Information Fig. S1). It is unknown if there are any other potential Mg2+ binding-sites in the CorA subgroup B. The bound Mg2+ was proposed to increase the stability of the pentameric assembly in CorA, at least for CorA subgroup A. In the high-resolution structure of the Vp-ZntB, no structurally ordered Mg2+, Zn2+, and other relevant divalent metal ions were identified. Instead, 25 well ordered Cl− anions were found bound to the protein and positioned to neutralize local positive charges, forming a Cl− ring in the middle of a cytoplasmic pentamer. The ZntB chloride ions binding sites show characteristic halide anion binding environment and distances as described previously for chloride and bromide ions.19 Chloride ion shows preferences for nitrogen (His, Arg, Asn) but also binds to oxygen atoms. The Vp-ZntB molecule is quite acidic with a calculated pI of 5.24, but it prefers to bind chloride over magnesium ions. Moreover, incorporation of 25 Cl− anions would drop the protein pI by ∼1 unit and create strong negative electrostatic potential on the molecular surface and along the central pore as demonstrated by the electrostatics calculations, thereby making it more attractive for positively charged metal ions. Indeed as it was shown for other metal channels, electrostatic forces may play an important role in metal transport.22–24
The cation selectivity of CorA and ZntB was attributed to the difference in their signature motifs (CorA:YGMNFxxMPEL, ZntB: GxxG[I,V]NxGGxP).12,25 The motif is located between two transmembrane α-helices on the outer surface of the membrane and it tends to be disordered in the crystal structures.1–3 The mechanism of the ion selectivity for ZntB as a cation exporter has never been clear and it is not obvious why the metal ion needs to cross the entire pore to be selected for passage. While possible Mg2+ gating mechanisms through the transmembrane were discussed in the Tm-CorA structures,1–3 N314 residue at the external entrance to the pore and a pair of hydrophobic rings formed by M291 and L294, respectively, seemed to play important roles in selecting ion size and possible ion dehydration/rehydration steps. The equivalent residue of N314 of Tm-CorA is identical in Vp-ZntB (N272) while M291/L294 of Tm-CorA are replaced by S249/F252. D242 of Vp-ZntB, corresponding to S284 of Tm-CorA, is located just before the narrowest pore segment [Fig. 4(A)]. The side chains of the five D242 point to the center of the channel, forming an “aspartate ring” just like that formed by the D277 residues of Tm-CorA. The corresponding residue of the D277 of Tm-CorA in Vp-ZntB is the highly conserved D235 in ZntB family [Fig. 4(A)]. The side chains of five D235 also point into the channel but they are farther apart as compared with D277 in Tm-CorA, forming a second but wider “aspartate ring.” Since the “aspartate ring” is believed to be important in Mg2+ movement by regulating ion selection or dehydration,2,26 the presence of similar “aspartate rings” in Vp-ZntB suggests similar mechanisms in supporting metal ion movement in CorA and ZntB.
The negative electrostatic surface potential of the cytoplasmic domain pentamer of Vp-ZntB (75 formal negative charges), enhanced by the binding of 25 chloride ions is likely to increase the local concentrations of metal ions inside the intracellular funnel, favoring the outward direction of flux. In addition, there is a process of charge selectivity that occurs inside the large cytoplasmic domain. The ability for a protein environment to favor divalents over monovalents involves a balancing act between different terms of the electrostatic free energy. This is given by ΔΔGint(Q) = (1/2)AQ2 + BQ + C where Q is the ion charge, A is the reaction field created by the ion in the protein dielectric environment, B is the static field created by the protein charges, and C is the effect of the ion dielectric on the protein charges.27 For a pore to demonstrate a general preference for divalents, ΔΔGint(+2)−ΔΔGint(+1) < 0 must be satisfied such that (3/2)A + B < 0. Since A is always positive for a single ion, this implies that the static field must be negative and greater than 150% of the reaction field. The funnel of Vp-ZntB creates a wide aqueous pore (small A) that is very negatively charged (large B) that provides this electrostatic preference for the stabilization of divalents over monovalent ions.
The lack of well-defined functional data of ZntB transporters limits our comparison of CorA and ZntB from their structural properties to their functions such as cation selectivity. However, the high-resolution Vp-ZntB structure suggests that ionic properties independent of the protein may be important in conferring selectivity. For instance, Mg2+ has a 1000 fold slower water substitution rate than Zn2+ or Cd2+.28 If the transport of metal ions is limited by this rate, then this, along with potential preferable interactions with the proteins, could contribute a transport mechanism selective for different divalent cations. Therefore, the binding of anions such as chloride and other effectors could potentially represent a regulatory mechanism. We speculate that the VP-ZntB transporter functions by combining the great attractor for divalent metal ions with the ion selectivity filter to passively maintain the homeostasis of the metal ions it transports. The protein provides great metal “ion attractors” on the cytoplasmic site and filter out ions that can pass through the selectivity filter most likely composed of the region just below the membrane and transmembrane domain. The transportation of metal ions can be further enhanced by binding chloride ions that can significantly lower the electrostatic barrier across the pore. Nevertheless, further experiments are required to verify the cation selectivity of ZntB transporter.
Materials and Methods
Expression and crystallization
The gene of cytoplasmic domain of Vp-ZntB (residues 1–249) was cloned in the pMCSG729 and overexpressed in E. coli BL21 (DE3). The cells were grown at 37°C in seleno-methionine (SeMet) containing enriched M9 medium under conditions known to inhibit methionine biosynthesis.30 The SeMet labeled Vp-ZntB protein was purified using Ni-affinity chromatography. The final protein buffer includes 20 mM HEPES pH 8.0, 250 mM NaCl, and 2 mM dithiothreitol (DTT). The protein (1 mM) was screened for crystallization conditions using the sitting drop vapor diffusing technique at 4°C. Crystals appeared under the conditions including 0.2M MgCl2, 0.1M Tris pH 8.5, 30% PEG 400 (Supporting Information SM1 and SM2).
Structural determination
The crystals of SeMet labeled Vp-ZntB were flash-frozen in liquid nitrogen. X-ray diffraction data were collected at 100 K at the 19ID beamline of the Structural Biology Center at the Advanced Photon Source at Argonne National Laboratory. Multi-wavelength anomalous diffraction (MAD) data sets were collected at the wavelengths near selenium and zinc absorption peaks from a single protein crystal (Table I). To verify the presence of chlorine ions, additional data set with multiple paths was collected at low energy (7 keV) (Table I). All diffraction data were processed and the structure was determined by using the programs integrated within the HKL-3000 suite.31 Further model buildings and refinement were performed using the programs COOT32 and REFMAC33 (Table I), respectively (Supporting Information SM3 and SM4).
Table I.
Crystallographic Statistics
| Data collection | Se absorption edge | Zn absorption edge | Low energy |
|---|---|---|---|
| Space group | P21 | P21 | P21 |
| Unit cell (Å, °) | a = 64.67, b = 130.18, c = 80.05, β = 103.4 | a = 64.39, b = 130.21, c = 79.39, β = 103.4 | a = 64.60, b = 130.31, c = 79.92, β = 103.4 |
| MW Da (residue) | 29021.6a (249) | ||
| Mol (AU) | 5 | ||
| SeMet (AU) | 30 | ||
| Wavelength (Å) | 0.9793 (peak), 0.9795 (infl.) | 1.2823 (peak) | 1.7712 |
| Resolution (Å) | 44.2–2.10 | 30.8–1.90 | 77.85–2.50 |
| Number of unique reflections | 73,213b | 97,781b | 42,472 |
| Redundancy | 3.3 (1.7)c | 6.0 (3.1)d | 17.1 (9.1)e |
| Completeness (%) | 96.7 (78.9)c | 98.1 (81.8)d | 95.1 (77.2)e |
| Rmerge (%) | 10.2 (48.4)c | 10.4 (84.5)d | 7.9 (33.3)e |
| I/σ (I) | 17.8 (1.8)c | 23.2 (1.2)d | 65.1 (8.8) |
| Phasing | |||
| RCullis (Anomalous) (%) | 69 (Peak), 82 (Inflection) | ||
| Figure of merit (%) | 26.2 | ||
| Refinement | |||
| Resolution | 44.2–2.10 | 30.8–1.90 | 77.85–2.50 |
| Reflections (work/test) | 69,412/3675 | 92,732/4875 | 40,309/2298 |
| Rcrystal/Rfree (%) | 17.95/24.53 | 0.187/0.223 | 0.178/0.235 |
| RMS deviation from ideal geometry Bond length (Å)/angle (°) | 0.017/1.65 | 0.012/1.284 | 0.013/1.423 |
| No. of atoms (Protein/HETATM) | 10,044/695 | ||
| Mean B-value (Åb) (Mainchain/Sidechain) | 32.14/34.47 | ||
| Ramachandran plot statistic (%) | |||
| Residues in most favored regions | 92.8 | ||
| In additional allowed regions | 6.7 | ||
| In generously allowed regions | 0.5 | ||
| In disallowed region |
Not including cloning artifact.
Including Bijvoet pairs.
Last resolution bin, 2.10–2.15 Å.
Last resolution bin, 1.90–1.95 Å.
Last resolution bin, 2.50–2.57 Å.
Full-length Vp-ZntB modeling
The stalk helices from one full-length Vp-ZntB monomer and one Tm-CorA intracellular domain monomer were structurally aligned (Supporting Information Fig. S1). The transmembrane domain of Vp-ZntB was built following the trace of the Tm-CorA transmembrane domain and their sequence alignment in this region. A full-length Vp-ZntB monomer was then used to generate a full-length pentamer based on the intracellular domain pentameric assembly. The pentamer was then subjected to a refinement of idealization.34
Electrostatic calculation
All calculations were performed using CHARMM version c33a2.35 The pore of Vp-ZntB was aligned along the Z-axis with the transmembrane helices centered along Z = 0. For the calculation of the radius, a dummy sphere was positioned along the pore and the radius was iteratively increased to determine the minimum van der Waals contact distance. This radius was then used to define a 1 Å thick grid from which the volume of free space was estimated. By assuming the pore as a series of cylindrical segments of cross-sectional area A, the effective pore radius was calculated by r2 = (A/π).36 For the continuum electrostatics calculations, finite difference solutions of the Poisson-Boltzmann equation were determined using the PBEQ module of CHARMM.35 The solvent dielectric was defined as ɛsolvent = 80 and the protein dielectric as ɛprotein = 2 using the reentrant surface with water probe radius 1.4 Å. The membrane was established between Z = −12.5 Å and Z = 12.5 Å with ɛmembrane = 2, and the transmembrane pore was defined by cutting out a cylinder with a radius of 6.5 Å and ɛcylinder = 80. Ionic screening corresponding to 150 mM salt concentration was considered at all points outside the transmembrane region, except for the static field potential map calculations where there was no ionic screening defined within the interior of the cytoplasmic pore. All protein charges were kept in their typical protonation state. The finite difference calculations were performed with a grid of 299 × 305 × 307 with initial cell size of 1.0 Å followed by a focused grid of 0.5 Å. The electrostatic interaction free energy profiles were calculated by performing three separate calculations ΔΔGint = ΔGion+protein−ΔGprotein−ΔGion.
Acknowledgments
The authors thank the members of Structural Biology Center at Argonne National Laboratory for their help with data collection at the 19ID and 19BM beamlines, and Lindsey Butler for help in preparation of this manuscript. The coordinates of the ZntB cytoplasmic domain pentamer have been deposited in Protein Data Bank (PDB) under the accession code 3CK6.
References
- 1.Lunin VV, Dobrovetsky E, Khutoreskaya G, Zhang R, Joachimiak A, Doyle DA, Bochkarev A, Maguire ME, Edwards AM, Koth CM. Crystal structure of the CorA Mg2+ transporter. Nature. 2006;440:833–837. doi: 10.1038/nature04642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eshaghi S, Niegowski D, Kohl A, Martinez Molina D, Lesley SA, Nordlund P. Crystal structure of a divalent metal ion transporter CorA at 2.9 angstrom resolution. Science. 2006;313:354–357. doi: 10.1126/science.1127121. [DOI] [PubMed] [Google Scholar]
- 3.Payandeh J, Pai EF. A structural basis for Mg2+ homeostasis and the CorA translocation cycle. EMBO J. 2006;25:3762–3773. doi: 10.1038/sj.emboj.7601269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vallee BL, Falchuk KH. The biochemical basis of zinc physiology. Physiol Rev. 1993;73:79–118. doi: 10.1152/physrev.1993.73.1.79. [DOI] [PubMed] [Google Scholar]
- 5.Berg JM, Shi Y. The galvanization of biology: a growing appreciation for the roles of zinc. Science. 1996;271:1081–1085. doi: 10.1126/science.271.5252.1081. [DOI] [PubMed] [Google Scholar]
- 6.Cousins RJ. Absorption, transport, and hepatic metabolism of copper and zinc: special reference to metallothionein and ceruloplasmin. Physiol Rev. 1985;65:238–309. doi: 10.1152/physrev.1985.65.2.238. [DOI] [PubMed] [Google Scholar]
- 7.Outten CE, Tobin DA, Penner-Hahn JE, O'Halloran TV. Characterization of the metal receptor sites in Escherichia coli Zur, an ultrasensitive zinc(II) metalloregulatory protein. Biochemistry. 2001;40:10417–10423. doi: 10.1021/bi0155448. [DOI] [PubMed] [Google Scholar]
- 8.Patzer SI, Hantke K. The ZnuABC high-affinity zinc uptake system and its regulator Zur in Escherichia coli. Mol Microbiol. 1998;28:1199–1210. doi: 10.1046/j.1365-2958.1998.00883.x. [DOI] [PubMed] [Google Scholar]
- 9.Grass G, Wong MD, Rosen BP, Smith RL, Rensing C. ZupT is a Zn(II) uptake system in Escherichia coli. J Bacteriol. 2002;184:864–866. doi: 10.1128/JB.184.3.864-866.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beard SJ, Hashim R, Membrillo-Hernâandez J, Hughes MN, Poole RK. Zinc(II) tolerance in Escherichia coli K-12: evidence that the zntA gene (o732) encodes a cation transport ATPase. Mol Microbiol. 1997;25:883–891. doi: 10.1111/j.1365-2958.1997.mmi518.x. [DOI] [PubMed] [Google Scholar]
- 11.Grass G, Fan B, Rosen BP, Franke S, Nies DH, Rensing C. ZitB (YbgR), a member of the cation diffusion facilitator family, is an additional zinc transporter in Escherichia coli. J Bacteriol. 2001;183:4664–4667. doi: 10.1128/JB.183.15.4664-4667.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Worlock AJ, Smith RL. ZntB is a novel Zn2+ transporter in Salmonella enterica serovar Typhimurium. J Bacteriol. 2002;184:4369–4373. doi: 10.1128/JB.184.16.4369-4373.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knoop V, Groth-Malonek M, Gebert M, Eifler K, Weyand K. Transport of magnesium and other divalent cations: evolution of the 2-TM-GxN proteins in the MIT superfamily. Mol genet genomics. 2005;274:205–216. doi: 10.1007/s00438-005-0011-x. [DOI] [PubMed] [Google Scholar]
- 14.Makino K, Oshima K, Kurokawa K, Yokoyama K, Uda T, Tagomori K, Iijima Y, Najima M, Nakano M, Yamashita A, Kubota Y, Kimura S, Yasunaga T, Honda T, Shinagawa H, Hattori M, Iida T. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V cholerae. Lancet. 2003;361:743–749. doi: 10.1016/S0140-6736(03)12659-1. [DOI] [PubMed] [Google Scholar]
- 15.Lumb KJ, Kim PS. A buried polar interaction imparts structural uniqueness in a designed heterodimeric coiled coil. Biochemistry. 1995;34:8642–8648. doi: 10.1021/bi00027a013. [DOI] [PubMed] [Google Scholar]
- 16.Harbury PB, Zhang T, Kim PS, Alber T. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science. 1993;262:1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- 17.Lo Conte L, Chothia C, Janin J. The atomic structure of protein-protein recognition sites. J Mol Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 18.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 19.Dauter Z, Dauter M, de La Fortelle E, Bricogne G, Sheldrick GM. Can anomalous signal of sulfur become a tool for solving protein crystal structures? J Mol Biol. 1999;289:83–92. doi: 10.1006/jmbi.1999.2743. [DOI] [PubMed] [Google Scholar]
- 20.Allen TW, Andersen OS, Roux B. On the importance of atomic fluctuations, protein flexibility, and solvent in ion permeation. J Gen Physiol. 2004;124:679–690. doi: 10.1085/jgp.200409111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niegowski D, Eshaghi S. The CorA family: structure and function revisited. Cell Mol Life Sci. 2007;448:613–616. doi: 10.1007/s00018-007-7174-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Niu X, Brelidze TI, Magleby KL. Ring of negative charge in BK channels facilitates block by intracellular Mg2+ and polyamines through electrostatics. J Gen Physiol. 2006;128:185–202. doi: 10.1085/jgp.200609493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Long SB, Campbell EB, Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 24.Kloda A, Ghazi A, Martinac B. C-terminal charged cluster of MscL, RKKEE, functions as a pH sensor. Biophys J. 2006;90:1992–1998. doi: 10.1529/biophysj.105.075481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kehres DG, Lawyer CH, Maguire ME. The CorA magnesium transporter gene family. Microb Comp Genomics. 1998;3:151–169. doi: 10.1089/omi.1.1998.3.151. [DOI] [PubMed] [Google Scholar]
- 26.Koronakis V, Sharff A, Koronakis E, Luisi B, Hughes C. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature. 2000;405:914–919. doi: 10.1038/35016007. [DOI] [PubMed] [Google Scholar]
- 27.Roux B, MacKinnon R. The cavity and pore helices in the KcsA K+ channel: electrostatic stabilization of monovalent cations. Science. 1999;285:100–102. doi: 10.1126/science.285.5424.100. [DOI] [PubMed] [Google Scholar]
- 28.Hille B. Ion channels of excitable membrane. 3rd ed. Sunderland, MA: Sinauer Associates, Inc; 2001. [Google Scholar]
- 29.Stols L, Gu M, Dieckman L, Raffen R, Collart FR, Donnelly MI. A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expr Purif. 2002;25:8–15. doi: 10.1006/prep.2001.1603. [DOI] [PubMed] [Google Scholar]
- 30.Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J Mol Biol. 1993;229:105–124. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- 31.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution–from diffraction images to an initial model in minutes. Acta crystallogr D Biol Crystallogr. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 32.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 33.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 34.CCP4. The CCP4 suite: programs for protein crystallography. Acta crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 35.MacKerell JAD, Brooks B, Brooks ICL, Nilsson L, Roux B, Won Y, Karplus M. Encyclopedia of Computational Chemistry. Chichester, U.K.: John Wiley and Sons Inc.,; 1998. CHARMM: the energy function and its parameterization with an overview of the program; pp. 271–277. [Google Scholar]
- 36.Jogini V, Roux B. Electrostatics of the intracellular vestibule of K+ channels. J Mol Biol. 2005;354:272–288. doi: 10.1016/j.jmb.2005.09.031. [DOI] [PubMed] [Google Scholar]


