Abstract
Candida albicans is a major fungal systemic pathogen in humans. Genetic manipulation of C. albicans is unwieldy. We report here a strategy that is useful and successful for large-scale genetic manipulation of C. albicans genes of interest: use of the UAU1 cassette on a Tn7 transposon. Streamlined yet admittedly flawed disruption techniques, such as the one described here, may prove vital to uncovering the genetic basis of fungal virulence.
Keywords: UAU1, large-scale gene disruption, mutants
1. Introduction
Until recently, Candida albicans was considered a genetically intractable organism for several reasons. Firstly, its genome was not sequenced. Secondly, it is a diploid that lacks a complete sexual cycle, thus making null mutants requires two successive transformations. Lastly, C. albicans transformations are not very efficient. From the time that the first C. albicans gene was disrupted in 1993 [1] and up until November 2002, less than 150 mutants had been defined out of greater that 7,000 ORFs in the haploid complement of its genome. Now in this postgenomic era, we have easily accessible online databases containing the DNA sequences for our favorite C. albicans ORFs (See The Candida Genome Database at http://www.candidagenome.org/; the Stanford Candida albicans Sequence Assembly at http://www-sequence.stanford.edu/group/candida/search.html; and the Pasteur Institute CandidaDB at http://genolist.pasteur.fr/CandidaDB/). The accessibility of the fully sequenced genome should aid in the first difficult part of a large-scale gene disruption procedure – deciding on a group of genes to disrupt.
Once a group of genes has been chosen, the next hard part is the actual large-scale genetic manipulation. We describe here a procedure that we find useful and successful for the rapid production of homozygous null mutants through just a single transformation. This disruption procedure involves the use of the UAU1 cassette (Figure 1) [2]. The UAU1 cassette consists of a functional C. albicans ARG4 gene flanked by nonfunctional URA3 deletion derivatives (ura3Δ3’-ARG4-ura3Δ5’), thus conferring an Arg+ Ura− phenotype after transformation when the cassette has inserted into the first allele. At this point, the heterozygous UAU1 insertion can become homozygous through a mitotic gene conversion event. Then, by selecting for the Arg+ Ura+ phenotype, we select for those segregants that have undergone a mitotic gene conversion followed by a looping out of the ARG4 gene between the URA3 segments in one of the alleles (the URA3 segments share ~500 bp of overlapping homology for recombination), thus yielding an intact URA3 gene. Homozygous Arg+ Ura+ isolates can then be genotyped using colony PCR for the absence of a wild-type allele, and the presence of the insertion allele. Typically, after screening all of the Arg+ Ura+ isolates by PCR, two types of genotypes are observed. The first type consists of the intended homozygote disruption mutants, and the second consists of allelic triplications, where a copy of the wildtype allele is present in addition to the two disrupted alleles. It is unknown how this triplication event occurs, but one could fathom that it may occur via a translocation, a tandem duplication, or non-disjunction.
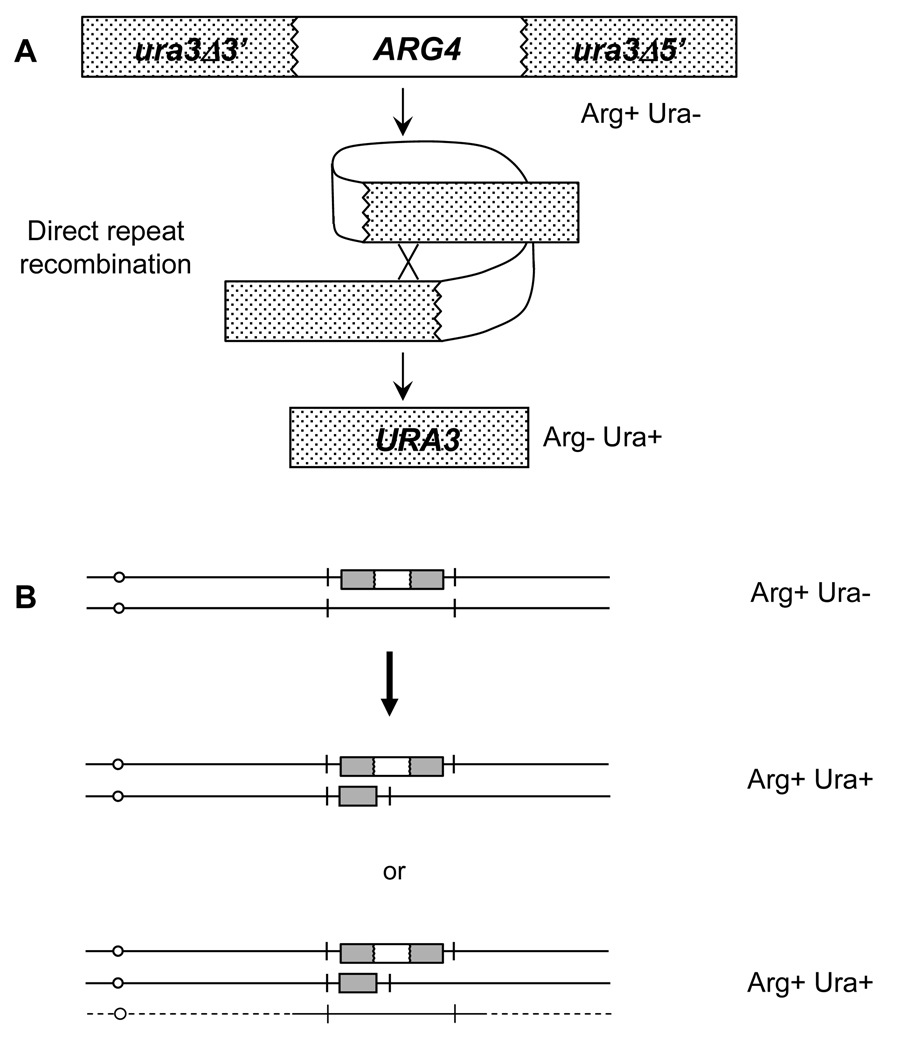
Figure 1.
Genetic properties of the UAU1 cassette. (A) Conversion of UAU1 to URA3. Recombination excises the ARG4 gene and results in an Arg− Ura+ phenotype. (B) Outcome of double-disruption selection with the UAU1 cassette yields the intended homozygote disruption mutant and/or a triplication segregant.
This triplication byproduct of using the UAU1 cassette to disrupt genes has its advantages. Since deletion of essential genes, by definition, results in lethality, failure to obtain null deletion mutants of a gene has led to the assumption that said gene is essential. This may not always be the case; it is possible that the gene may not be essential, but simply difficult to delete due to such things as a highly repetitive sequence, or the presence of closely related genes. The UAU1 cassette is an excellent test to identify potential essential genes. We deduce that a UAU1 insertion in nonessential genes produces both homozygous mutants and triplication segregants, whereas in essential genes, the UAU1 cassette yields only triplication segregants [2]. Since essential genes are putative drug targets, the UAU1 cassette may prove beneficial in this area. Thus, the UAU1 cassette is useful for both the rapid and systematic disruption of genes, as well as for the identification of essential genes.
2. Materials
2.1 Reagents for Preparation of C. albicans Genomic DNA
YPD (10 g yeast extract, 20 g peptone, 20 g glucose, to 1000 mL with dH20) liquid media. Supplement YPD medium with 80 µg/µL uridine if using a Ura− C. albicans strain (see Note 1)
TENTS (1% SDS, 2% Triton X-100, 0.1 M NaCl in 1×TE). Filter sterilize.
Acid washed beads (sterilize by autoclaving).
Phenol/chloroform/isoamyl alcohol (25:24:1, v/v).
Ethanol.
1× Tris-EDTA (1×TE), pH 7.4
10 mg/mL RNase.
10 M NH4OAc.
2.2 PCR Reagents
1× Tris-EDTA (1×TE), pH 7.4.
10× PCR buffer (100 mM Tris-HCl, pH 8.3, 500 mM KCl, 15 mM MgCl2, 0.01% gelatin).
25 mM MgCl2.
5 mM dNTPs.
Taq DNA polymerase.
10 µM forward ORF primer.
10 µM reverse ORF primer.
2.3 Ligation Reagents
pGEMT-Easy vector (Promega).
T4 DNA ligase (Promega).
2× ligation buffer (Promega).
2.4 Materials for E. coli Transformation, Plasmid Extraction/Purification, and Cloning Confirmation
2.5 Reagents for GPS Transposon Mutagenesis
Donor plasmid pDDB166 (Figure 2).
Target plasmid pGEMT-Easy-ORF.
10× GPS Buffer (New England Biolabs).
TnsABC* Transposase (New England Biolabs).
Start Solution (New England Biolabs).
PI-SceI (New England Biolabs).
10× PI-SceI Buffer (New England Biolabs).
100× BSA (New England Biolabs).
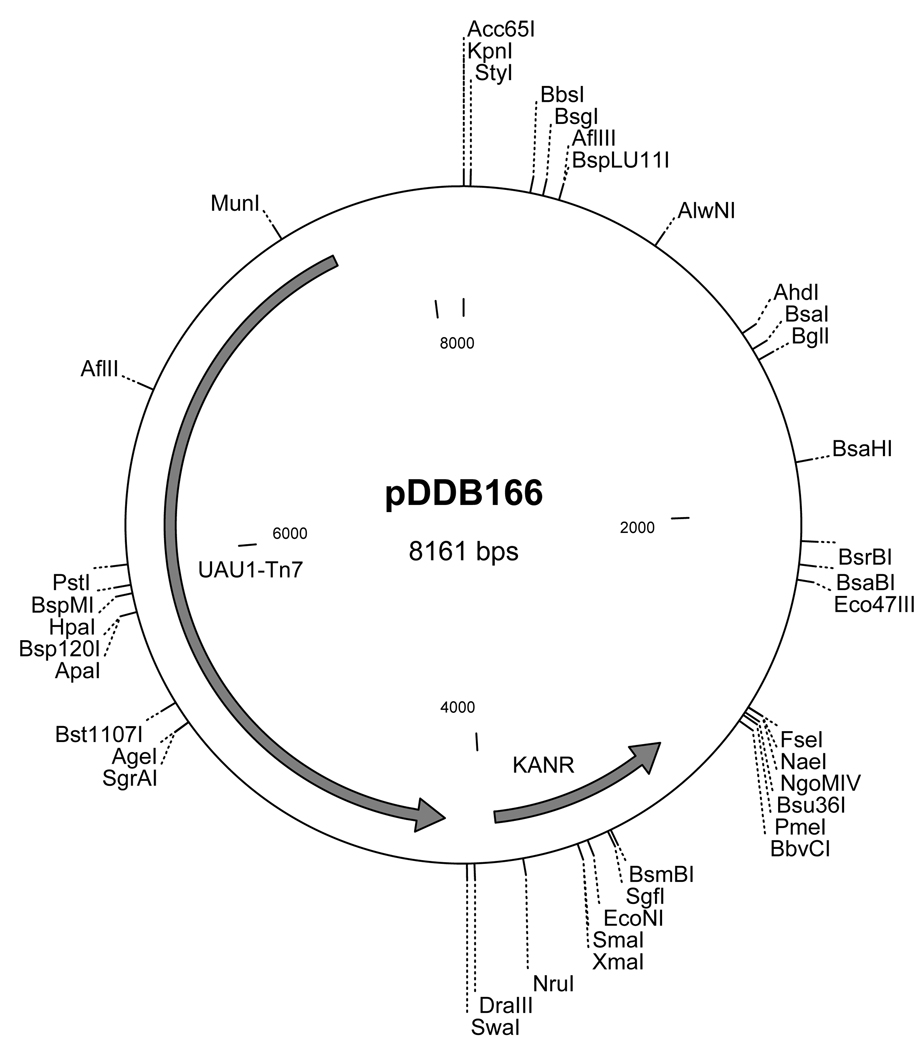
Figure 2.
Donor plasmid pDDB166 containing the UAU1 cassette on a Tn7 transposon. Plasmid pDDB166 confers kanamycin resistance.
2.6 Reagents for Transformation of Mutagenesis Reaction into E. coli, Extraction of Mutagenized Plasmid DNA, and Confirmation of Mutagenesis into ORF of interest
Chemically competent E. coli.
LB+Amp+Kan (100 µg/mL and 50 µg/mL, respectively) plates.
LB liquid media.
LB+Amp+Kan (100 µg/mL and 50 µg/mL, respectively) liquid media.
Montage Plasmid Miniprep96 kit (Millipore) (see Note 7).
Restriction enzyme NotI (New England Biolabs) (see Note 11).
10× NE Buffer 3 (New England Biolabs).
100× BSA (New England Biolabs).
Restriction enzyme PmeI (New England Biolabs).
10× NE Buffer 2 (New England Biolabs).
2.7 Reagents for Digestion of Plasmids for Transformation, and Transformation into C. albicans
C. albicans strain BWP17 [3].
YPD+Uri (80 µg/µL) plates.
YPD+Uri (80 µg/µL) liquid media.
Extracted plasmid DNA containing Tn7-UAU1 in middle of ORF.
Restriction enzyme NotI (New England Biolabs) (see Note 11).
10× NE Buffer 3 (New England Biolabs).
100× BSA (New England Biolabs).
LATE (0.1M LiOAc in 1× TE buffer).
Calf thymus DNA (9.1 mg/mL) (Sigma).
PLATE (8 mL 50% PEG + 1 mL 10× TE + 1 mL 1 M LiOAc).
YPD plates.
SC-Arg plates.
SC-Arg-Ura plates.
2.8 Reagents for Confirmation of Homozygous Insertion Mutants
10 µM Arg4-detect primer (sequence GGAATTGATCAATTATCTTTTGAAC) (see Note 17).
10 µM forward ORF primer.
10 µM forward ORF primer.
10× PCR buffer (100 mM Tris-HCl, pH 8.3, 500 mM KCl, 15 mM MgCl2, 0.01% gelatin).
25 mM MgCl2.
5 mM dNTPs.
Taq DNA polymerase.
3. Methods
The methods to follow describe in detail a large-scale C. albicans disruption procedure in a step-by-step format. We begin with a description of the preparation of C. albicans genomic DNA for use in a PCR reaction to amplify your ORFs of interest. We follow with in depth instructions on large-scale PCR protocols, and cloning of the ORFs into a suitable plasmid vector. We then describe a transposon mutagenesis protocol using the UAU1 cassette on a Tn7 transposon into the plasmid vector containing the ORFs of interest, followed by the final selection of C. albicans homozygous disruption mutants.
3.1 Preparation of C. albicans Genomic DNA
Inoculate a 5 mL culture of YPD or YPD+Uri (see Note 1) liquid media with C. albicans reference strain (see Note 3). Grow the culture overnight with agitation at 30°C.
Spin down the entire 5 mL culture at low speed, and aspirate supernatant.
Add 500 µL TENTS, and resuspend.
Transfer the resuspended mixture to a fresh eppendorf tube containing 200 µL of sterile acid washed beads.
Add 500 µL phenol/choloroform/isoamylalcohol.
Vortex for 2 min.
Spin down tubes at 14,000 rpm at 4°C for 10 min.
Transfer aqueous phase to a new eppendorf tube.
Add 1 mL 100% ethanol.
Place at −20°C for at least 1 h.
Spin down tubes at 14,000 rpm at 4°C for 15 min.
Aspirate supernatant, and resuspend in 200 µL 1×TE.
Add 1 µL 10 mg/mL RNase.
Place at room temperature for 30 min.
Add 40 µL 10 M NH4OAc.
Add 500 µL 100% ethanol, and mix by inversion.
Place at −20°C for 30 min.
Spin down tubes at 14,000 rpm for 5 min. Decant supernatant.
Add 1 mL 70% Ethanol to pellet, and immediately decant.
Place tubes open in a speed vacuum for 5–10 min or until dry.
Resuspend gently in 50 µL 1×TE (see Note 2).
3.2 PCR of ORFs of Interest
Primers are designed for amplification of ORFs of interest from the start codon to the stop codon using 20 bp oligonucleotides. Primers are diluted to 100 µM in 1× TE, and further diluted to a working stock of 10 µM in dH20.
PCR is performed off of a diluted genomic DNA template (prepared in Method 3.1) isolated from a C. albicans reference strain containing the ORFs of interest for amplification (see Note 3).
Dilute genomic DNA template for amplification by PCR. If genomic DNA was isolated as in Method 3.1, a 1:1000 dilution in dH20 is recommended for the PCR reaction.
- An example of a typical large-scale PCR reaction for one sample is listed below (see Note 4). Mix well.
10× PCR buffer 5 µL 25 mM MgCl2 1 µL 5 mM dNTPs 2 µL Genomic DNA template (1/1000) 1 µL 10 µM forward ORF primer 2 µL 10 µM reverse ORF primer 2 µL Taq DNA polymerase 0.2 µL dH20 36.8 µL Total Volume 50 µL - An example of a typical large-scale PCR program for amplification of most C. albicans ORFs is listed below (see Note 5).
Step 1 94°C for 3 min Step 2 94°C for 1 min Step 3 50°C for 1 min Step 4 72°C for 4 min Step 5 35 times to Step 2 Step 6 72°C for 10 min Step 7 4°C/End Run 5 µL of PCR product on an agarose gel containing ethidium bromide. Typically, a single band of the correct size for the ORF is present. No purification of the PCR sample is necessary for the ligation reaction to follow (see Note 6).
3.3 Ligation of PCR Products into Plasmid Vector pGEMT-Easy
- Ligate PCR product into pGEMT-Easy as detailed below.
2× ligation buffer 7.5 µL T4 DNA ligase 1 µL pGEMT-Easy vector 1 µL PCR product insert 3 µL dH20 2.5 µL Total Volume 15 µL Mix gently.
Allow reaction to sit for 2–4 h at room temperature or overnight at 4°C.
3.4 Transformation of Ligation Reaction into E. coli, Extraction of Plasmid DNA, and Cloning Confirmation
Thaw chemically competent E. coli on ice.
Add entire 15 µL ligation reaction to appropriate volume of thawed chemically competent E. coli.
Allow mixture to sit on ice for 10 min.
Heat shock mixture at 42°C for 45 s.
Place on ice for an additional 3 min.
Plate directly on LB+Amp+X-Gal plates for selection of the ORF inserted into the pGEMT-Easy vector. Grow overnight at 37°C.
Select 8 white colonies, and inoculate separately into 2 mL LB+Amp liquid media.
Grow overnight at 37°C with agitation.
Spin down cultures and extract plasmid DNA using the Montage Plasmid Miniprep96 kit (see Note 7).
- Digest a portion of extracted plasmid DNA in order to determine if the ORF of interest has inserted correctly into the pGEMT-Easy vector. An example digestion is listed below using the restriction enzyme NotI (see Note 8).
Extracted plasmid DNA 3 µL 10× NE Buffer 3 2 µL 100× BSA 0.2 µL NotI 0.2 µL dH2O 14.6 µL Total Volume 20 µL Mix gently.
Allow the reaction to digest for 2 h at 37°C.
Run 10 µL of digestion reaction on an agarose gel containing ethidium bromide. Using NotI releases a ~3 kb band for the pGEMT-Easy vector backbone. The gel should thus resolve a ~3 kb band representing the vector, as well as a band the expected size for the inserted ORF of interest (see Note 9).
3.5 GPS Transposon Mutagenesis
The following steps require use of the plasmid pDDB166 as the donor plasmid containing the UAU1 cassette on a Tn7 transposon (Figure 2). Plasmid pDDB166 was constructed by destroying the ScaI site of pAED98 [4] so that it no longer confers resistance to ampicillin, but retains resistance to kanamycin. The target plasmid for the mutagenesis reaction is the pGEMT-Easy plasmid containing the inserted ORF of interest (or pGEMT-Easy-ORF) for disruption. Donor plasmid pDDB166 is kanamycin resistant and target plasmid pGEMT-Easy-ORF is ampicillin resistant. When mutagenesis into the target plasmid is successful, clones will be both ampicillin and kanamycin resistant. The mutagenesis reaction described below is modified from the GPS-Mutagenesis System instruction manual (New England Biolabs).
Dilute donor plasmid pDDB166 and target plasmid pGEMT-Easy-ORF to a concentration of 20 ng/µL (see Note 10).
- Add the following reagents for the GPS mutagenesis reaction:
10× GPS Buffer 2 µL Donor pDDB166 (20 ng/εL) 1 µL Target pGEMT-Easy-ORF (20 ng/εL) 4 µL dH20 11 µL Total Volume 18 µL Mix gently.
Add 1 µL TnsABC* Transposase and mix gently.
Place the reaction at 37°C for 15 min.
Add 1 µL Start Solution, and mix gently.
Place the reaction at 37°C for 1 h.
Heat inactivate the transposase at 75°C for 10 min.
- To destroy the pDDB166 donor backbone, add the following reagents to the reaction:
10× PI-SceI Buffer 5 µL 100× BSA 0.5 µL PI-SceI 6 µL dH20 18.5 µL Total Volume 30 µL Place the reaction at 37°C for 2–3 h.
Place the reaction at 75°C for 10 min.
3.6 Transformation of Mutagenesis Reaction into E. coli, Extraction of Mutagenized Plasmid DNA, and Confirmation of Mutagenesis into ORF of interest
Thaw chemically competent E. coli on ice.
Add 10 µL of undiluted mutagenesis reaction to appropriate volume of thawed chemically competent E. coli.
Allow mixture to sit on ice for 10 min.
Heat shock mixture at 42°C for 45 s.
Place on ice for an additional 5 min.
Add 1 mL LB, and incubate for 1 h at 37°C with agitation for outgrowth.
Spin down cells at low speed for 1 min, decant supernatant, and resuspend cell pellet in 100 µL dH2O.
Plate on LB+Amp+Kan plates. Grow overnight at 37°C.
Select 8 white colonies, and inoculate separately into 2 mL LB+Amp+Kan liquid media.
Grow overnight at 37°C with agitation.
Spin down cultures and extract plasmid DNA using the Montage Plasmid Miniprep96 kit (see Note 7).
- Digest a portion of the extracted plasmid DNA in order to determine if the Tn7-UAU1 cassette has inserted into the ORF of interest in the target plasmid. An example digestion is listed below using the restriction enzyme NotI (see Note 11).
Extracted plasmid DNA 3 µL 10× NE Buffer 3 2 µL 100× BSA 0.2 µL NotI 0.2 µL dH2O 14.6 µL Total Volume 20 µL Mix gently.
Allow the reaction to digest for 2 h at 37°C.
Run 10 µL of digestion reaction on an agarose gel containing ethidium bromide.
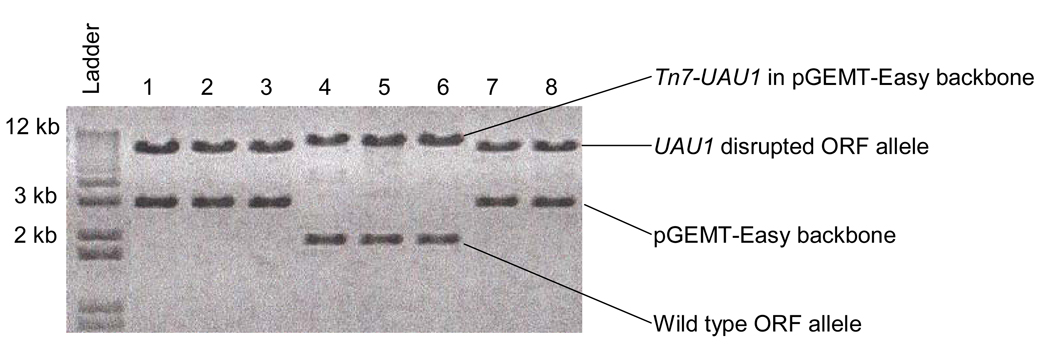
Two scenarios are detectable on a gel when the Tn7-UAU1 cassette is inserted into the pGEMT-Easy-ORF target plasmid, and digested with NotI (see Note 12). The first possibility is insertion of the Tn7-UAU1 cassette into the pGEMT-Easy backbone of the pGEMT-Easy-ORF plasmid. The second possibility is insertion of the Tn7-UAU1 cassette into the ORF of the pGEMT-Easy-ORF plasmid. The latter is the desired scenario. Figure 3 is an example mutagenesis of an ORF of size 1800 bp. Clones 1, 2, 3, 7, and 8 are examples of the desired scenario where the Tn7-UAU1 cassette has inserted successfully into the 1800 bp ORF (Figure 3, upper band for clones 1–3, and 7–8), leaving behind the pGEMT-Easy backbone of ~3 kb (Figure 3, lower ~3 kb band for clones 1–3, and 7–8) upon NotI digestion. Clones 4, 5, and 6 are examples of where the Tn7-UAU1 cassette has inserted into the pGEMT-Easy backbone of the pGEMT-Easy-ORF plasmid; not into the ORF (Figure 3, upper band for clones 4–6), leaving behind the ORF of 1800 bp (Figure 3, lower 1800 bp band for clones 4–6) upon NotI digestion.
- Digest a portion of the desired scenario extracted plasmid DNA, where the Tn7-UAU1 cassette has inserted successfully into the ORF of interest, in order to approximate where the Tn7-UAU1 cassette has inserted into the ORF. An example digestion is listed below using the restriction enzymes NotI and PmeI; a PmeI site is present on one end on the Tn7-UAU1 cassette (Figure 2).
Extracted plasmid DNA containing Tn7-UAU1 in ORF 3 µL 10× NE Buffer 2 2 µL 100× BSA 0.2 µL NotI 0.2 µL PmeI 0.2 µL dH2O 14.4 µL Total Volume 20 µL Mix gently.
Allow the reaction to digest for 2 h at 37°C.
Run 10 µL of digestion reaction on an agarose gel containing ethidium bromide.
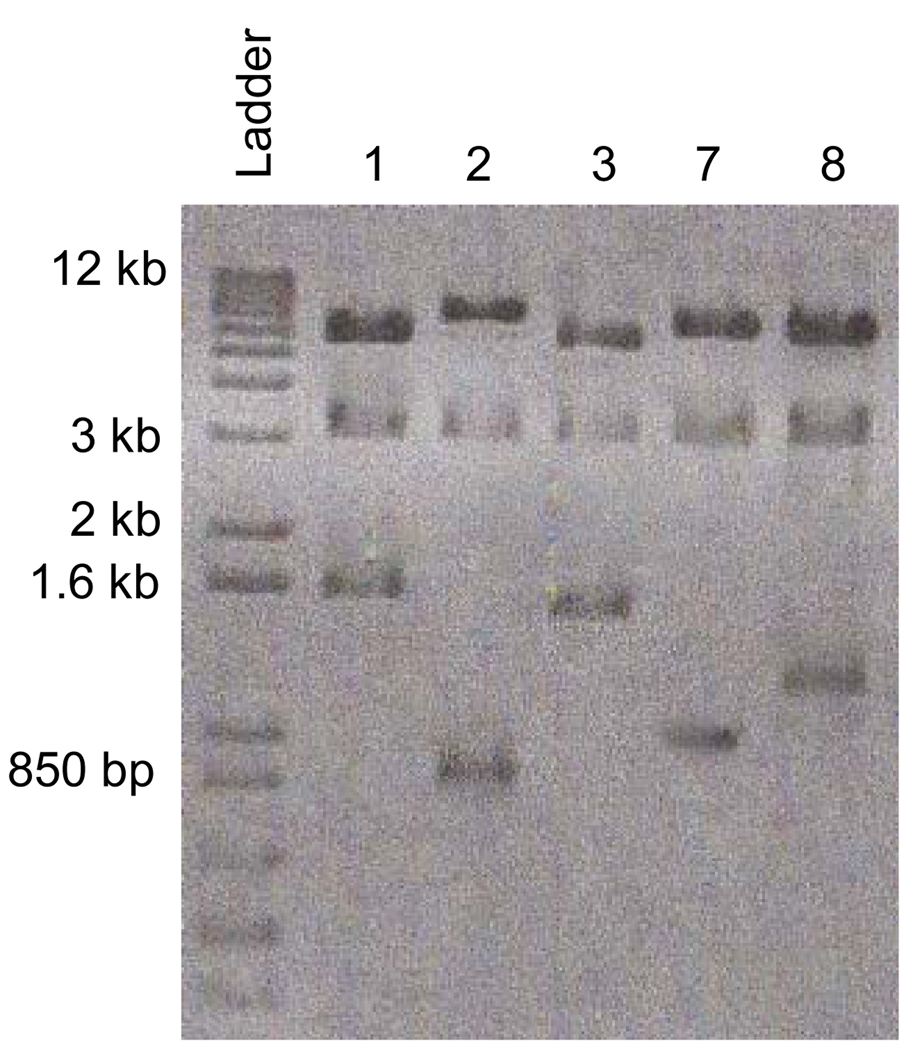
Since the PmeI site is on one end of the Tn7-UAU1 cassette, digestion with PmeI and NotI for the clones where the Tn7-UAU1 cassette has inserted into the ORF, will produce 3 detectable bands on an agarose gel (Figure 4). We will focus on the smallest band because this band is the size from one end of the Tn7-UAU1 cassette to the closest adjacent end of the ORF (see Note 13). Figure 4 is an example of how we confirmed where the Tn7-UAU1 cassette inserted into the 1800 bp ORF from Figure 3 for clones 1, 2, 3, 7, and 8. It is evident from the mutagenesis reaction that the Tn7-UAU1 cassette hopped into different regions of the ORF (Figure 4). For the sake of simplicity, we will go further will clones 2 and 7 because the Tn7-UAU1 cassette inserted approximately into the middle of the 1800 bp ORF as the smallest band for digested clones 2 and 7 is ~900 bp and ~980 bp, respectively (Figure 4).
Figure 3.
Mutagenesis of an ORF of size 1800 bp. Clones 1, 2, 3, 7, and 8 are examples of where the Tn7-UAU1 cassette has inserted successfully into the 1800 bp ORF, leaving behind the pGEMT-Easy backbone of ~3 kb upon NotI digestion. Clones 4, 5, and 6 are examples of where the Tn7-UAU1 cassette has inserted into the pGEMT-Easy backbone of the pGEMT-Easy-ORF plasmid; not into the ORF, leaving behind the ORF of 1800 bp upon NotI digestion.
Figure 4.
Confirmation of location in which the Tn7-UAU1 cassette has inserted into the 1800 bp ORF for clones 1, 2, 3, 7, and 8. Clones 2 and 7 contain the Tn7-UAU1 cassette inserted approximately into the middle of the 1800 bp ORF as the smallest band for digested clones 2 and 7 is ~900 bp and ~980 bp, respectively.
3.7 Digestion of Plasmids Containing the Tn7-UAU1 Cassette in the Middle of the ORF and Transformation into C. albicans
Streak out C. albicans strain BWP17 [3] for singles on a YPD+Uri plate. Grow at 30°C for 2 days.
Culture a single colony in 5 mL YPD+Uri liquid media. Grow overnight at 30°C with agitation.
The following day determine the OD600 of the overnight culture.
Dilute the sample to OD600 = 0.2 in 50 mL YPD+Uri liquid media.
Incubate the diluted culture at 30°C until the culture reaches an OD600 of ~0.8 (see Note 14).
- During this incubation period, digest 10 µL of each extracted plasmid containing the Tn7-UAU1 cassette inserted into the middle of the ORF of interest with a restriction enzyme that releases the ORF containing the Tn7-UAU1 cassette from the pGEMT-Easy vector backbone. An example digestion is listed below using the restriction enzyme NotI.
Extracted plasmid DNA containing Tn7-UAU1 in middle of ORF 10 µL 10× NE Buffer 3 2 µL 100× BSA 0.2 µL NotI 0.5 µL dH2O 7.3 µL Total Volume 20 µL Mix gently.
Allow the reaction to digest at 37°C until the C. albicans culture is ready for transformation (~3–4 h).
When the C. albicans culture has reached an OD600 of ~0.8, pour the culture into a 50 mL conical tube, and spin at low speed (~5,000 rpm) for 5 min.
Discard the supernatant, and wash the cell pellet by gently resuspending it in 5 mL sterile dH2O (do not vortex).
Spin at low speed for 5 min, and discard the supernatant.
Resuspend the cell pellet in 500 µL of LATE.
- To set up C. albicans transformation reactions, add for each transformation:
LATE cell suspension 100 µL Calf thymus DNA 10 µL Digest from Step 6 20 µL Mix gently.
Incubate for 30 min at 30°C.
Add 700 µL of freshly made PLATE, and incubate overnight at 30°C (see Note 15).
Heat shock the cell mixture at 44°C for 20 min.
Spin cells down for 30 s at low speed, and aspirate the supernatant.
Wash the cell pellet by resuspending in 1 mL YPD+Uri.
Spin cells down for 30 s at low speed, and decant the supernatant.
Resuspend the cells gently in 100 µL of YPD+Uri, and plate on Beefy SC-Arg plates.
Grow for 2 days at 30°C.
Pick 12 colonies from each transformation plate, streak for singles on SC-Arg plates, and grow for 2 days at 30°C. These transformants should be heterozygous mutants.
Patch a single colony from each of the 12 heterozygote mutants (streaked for singles) onto YPD plates (see Note 16). Grow for 2 days at 30°C.
Replica plate the YPD patches onto SC-Arg-Ura plates.
Grow for 4–5 days at 30°C. Some of these transformants should be homozygous mutants.
Pick one colony from each quarter, and streak for singles on SC-Arg-Ura plates. Grow for 2 days at 30°C.
3.8 Confirmation of Homozygous Insertion Mutants
-
28Colony PCR directly off of a single colony using 3 primers. An example of a typical large-scale colony PCR reaction for one sample is listed below.
10× PCR buffer 5 µL 25 mM MgCl2 3 µL 5 mM dNTPs 2 µL 10 µM Arg4-detect primer 1 µL 10 µM forward ORF primer 1 µL 10 µM reverse ORF primer 1 µL dH20 37 µL Total Volume 50 µL -
29
Add a single colony to the PCR mixture.
-
30
Before adding the tubes to the PCR block, start the program and pause it at the first step at 94°C. When the block has reached 94°C, add the tubes containing the PCR mixture with the colony. Allow the tubes to boil in the thermocycler (paused at 94°C) for 8 min.
-
31
Add 0.5 µL Taq DNA polymerase to each tube.
-
32Unpause the program and let it run to completion. The colony PCR program is listed below.
Step 1 94°C for 2 min Step 2 94°C for 45 s Step 3 50°C for 45 s Step 4 72°C for 4 min Step 5 35 times to Step 2 Step 6 72°C for 12 min Step 7 4°C/End -
33
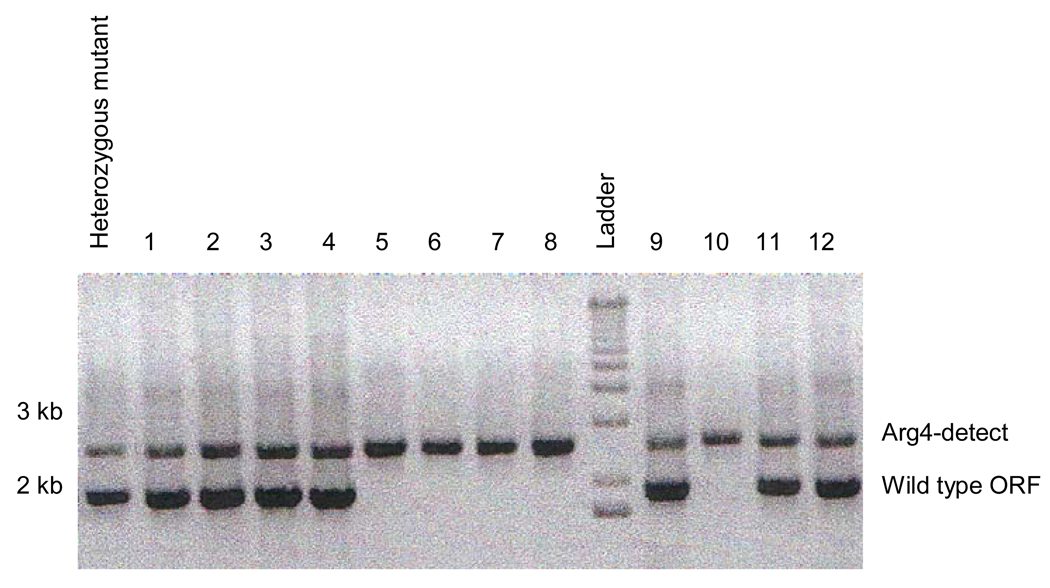
Run 10 µL of the colony PCR on an agarose gel containing ethidium bromide. Figure 5 is an example of a colony PCR from a mutagenesis of an ORF of size 1800 bp. The heterozygote mutant (far left lane) for the corresponding putative homozygote mutants (colonies 1–12) contains the Arg4-detect band (upper band) and the lower 1800 bp band corresponding to the size of the ORF (Figure 5). PCR off of the putative homozygous mutant colonies 1–12, shows two different scenarios. PCR off of colonies 1–4, 9, and 11–12 resemble the banding pattern of the heterozygote mutant with the Arg4-detect band (upper band) and the lower 1800 bp band corresponding to the size of the ORF (Figure 5); these colonies are not homozygous mutants, but correspond to triplication derivatives. PCR off of colonies 5–8, and 10 contain just the Arg4-detect band (upper band) (Figure 5); these colonies are homozygous mutants.
Figure 5.
Colony PCR from a mutagenesis of an ORF of size 1800 bp. The heterozygote mutant (far left lane) for the corresponding putative homozygote mutants (colonies 1–12) contains the Arg4-detect band (upper band) and the lower 1800 bp band corresponding to the size of the wild type ORF. PCR off of the putative homozygous mutant colonies 1–12, shows two different scenarios. PCR off of colonies 1–4, 9, and 11–12 resemble the banding pattern of the heterozygote mutant with the Arg4-detect band (upper band) and the lower 1800 bp band corresponding to the size of the wild type ORF; these colonies are triplication derivatives. PCR off of colonies 5–8, and 10 contain just the Arg4-detect band (upper band); these colonies are homozygous mutants.
Acknowledgments
This work was supported by NIH grants R01 AI067703 and T32 DK007786.
Footnotes
C. albicans strains that are Ura− (with the URA3 gene disrupted) require supplementation with uridine at 80 mg/l because disruption of URA3 blocks the ability to synthesize uridine. Supplementation with uracil, which is typical for S. cerevisiae media recipes, is not adequate for growth: C. albicans may lack the ability to take uracil up efficiently or to employ the uracil salvage pathway.
Genomic DNA is extremely delicate. To avoid shearing, genomic DNA should never be vortexed.
We typically extract genomic DNA from reference strains BWP17 (Arg− Ura-His−) [3], DAY185 (Arg+ Ura+ His+) [5], or DAY286 (Arg+ Ura+ His−) [4].
This is an example PCR reaction that works for the amplification of most C. albicans ORFs. Harder to amplify ORFs may require other additions.
This is a generalized PCR program that works for most C. albicans ORFs less than 4 kb in length. Larger ORFs may require longer extension times.
It is unnecessary to clean up the PCR sample for direct ligation into plasmid vector pGEMT-Easy as long as a single band of the correct size is present. In fact, we find that purification of the sample using the Qiagen PCR purification and/or gel extraction kits significantly reduces the DNA concentration of the sample as well as the ligation efficiency, and should only be used if multiple bands are present.
Plasmid DNA can be extracted and purified using several methods or several commercially available kits depending on the number of samples. For a small number of samples, the standard alkaline lysis or boiling mini-preparation methods are acceptable. For a medium number of samples, the QIAprep Spin Miniprep kit (Qiagen) is ideal. For a large number of samples, we recommend the Montage Plasmid Miniprep96 kit (Millipore) because it allows for the rapid extraction and purification of plasmid DNA in 96-well plates for large-scale procedures.
Several restriction enzymes could be used to confirm the presence of the ORF insert into the pGEMT-Easy vector. We use NotI here because it is a rare cutter, and is not present in most C. albicans ORFs. Thus, it is most useful for a large-scale procedure. However, if NotI is present in your ORF of interest, several other convenient restriction enzymes are present in the pGEMT-Easy vector (See Promega pGEMT-Easy manual for details).
Certain ORFs are the same size or close in size to the ~3 kb pGEMT-Easy vector backbone that is released when the ORF-containing plasmid is digested with NotI. If this is the case, other restriction enzymes should be considered for use instead of NotI that cut at different sites within the vector backbone, thus altering the size of the released vector backbone fragment.
The mutagenesis reaction is extremely sensitive to the ratio of donor to target plasmid concentration, and thus concentrations of plasmids should be exact.
Several restriction enzymes could be used to confirm the presence of the Tn7-UAU1 cassette into the ORF of interest in the target plasmid. If NotI is present in the ORF of interest, PvuII is another useful alternative.
The insertion of the Tn7-UAU1 cassette into the ORF of interest versus the target plasmid backbone is dependent on the size of the ORF. The chance of the transposon hopping into the ORF is based on probability. Larger ORFs have a higher likelihood of being hopped into versus smaller ORFs. For smaller ORFs, it is not a bad idea to screen more clones.
We chose to continue with clones 2 and 7 in this example because the Tn7-UAU1 cassette inserted approximately into the middle of the 1800 bp ORF. For a large-scale disruption procedure, we recommend picking clones with middle insertions because this allows for much flanking homology for transformation intoC. albicans. More homology ensures that the disrupted ORF will more successfully homologously recombine to the native locus of said ORF in C. albicans. Other insertion sites may also be of interest for particular ORFs. If so, we recommend sequencing from one transposon junction using Primer S (New England Biolabs), followed by BLASTN analysis against The Candida Genome Database at http://www.candidagenome.org/.
The doubling time of C. albicans is ~1.5 h.
PEG is cytotoxic, thus incubating for greater than 16 h significantly reduces the transformation efficiency.
We recommend dividing a YPD plate into quarters, and thickly patching the single colony onto one quarter of the YPD plate. Due to the fact that the gene conversion event transferring the UAU1 cassette to the other allele occurs very infrequently, a thick patch of cells is necessary.
Arg4-detect primer is a reverse primer on one end of ARG4.
References
- 1.Fonzi WA, Irwin MY. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Enloe B, Diamond A, Mitchell AP. A single-transformation gene function test in diploid Candida albicans. J Bacteriol. 2000;182:5730–5736. doi: 10.1128/jb.182.20.5730-5736.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilson RB, Davis D, Mitchell AP. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J Bacteriol. 1999;181:1868–1874. doi: 10.1128/jb.181.6.1868-1874.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davis DA, Bruno VM, Loza L, Filler SG, Mitchell AP. Candida albicans Mds3p, a conserved regulator of pH responses and virulence identified through insertional mutagenesis. Genetics. 2002;162:1573–1581. doi: 10.1093/genetics/162.4.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis D, Wilson RB, Mitchell AP. RIM101-dependent and-independent pathways govern pH responses in Candida albicans. Mol Cell Biol. 2000;20:971–978. doi: 10.1128/mcb.20.3.971-978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]