Abstract
Cardiovascular diseases impose enormous social and economic burdens on both individual citizens and on society as a whole. Clinical indicators such as high blood pressure, blood cholesterol and obesity have had some utility in identifying those who are at increased risk of cardiovascular events. However, there remains an urgent need for sensitive and specific indicators, preferably acquired through minimally invasive means, to help stratify patients for more personalized health care. As such, there has been a steadily growing interest in searching for ‘omic’ biomarkers of cardiovascular diseases. Historically, the transition of cardiac biomarker discovery to implementation has been a lengthy and somewhat unregulated process. Recent technological advancements, as well as concurrent efforts by regulatory agencies such as the Food and Drug Administration (United States) and Health Canada to establish policies and guidelines in the ‘omic’ arena, have helped propel the discovery and validation of biomarkers forward. The present paper provides perspective on current strategies in the bio-marker development pathway, as well as the potential limitations associated with each step from discovery to clinical uptake. Canadian biomarker studies now underway illustrate the possibilities for assessment of risk, diagnosis, prognosis and response to therapy, and for the drug discovery process.
Keywords: Biomarker, Cardiovascular, Genomics
Abstract
Les maladies cardiovasculaires imposent un fardeau social et économique énorme à la fois sur les citoyens touchés et sur l’ensemble de la société. Des indicateurs cliniques comme l’hypertension, le cholestérol sanguin et l’obésité ont une certaine utilité pour dépister les personnes plus vulnérables à des événements cardiovasculaires. Il est toutefois urgent de déterminer des indicateurs sensibles et spécifiques, acquis de préférence par des moyens très peu effractifs, afin de contribuer à stratifier les patients de manière qu’ils reçoivent des soins plus personnalisés. À cet égard, on s’intéresse de plus en plus à la recherche des biomarqueurs « omiques » des maladies cardiovasculaires. Par le passé, la transition de la découverte des biomarqueurs cardiaques à leur implantation s’est révélée un processus fastidieux et plutôt non réglementé. Les récentes avancées technologiques, de même que les efforts concomitants d’organismes de réglementation comme la Food and Drug Administration des États-Unis et Santé Canada afin d’établir des politiques et des lignes directrices dans le domaine « omique », ont contribué à accélérer les découvertes et la validation des biomarqueurs. Le présent article donne une perspective des stratégies actuelles sur la voie du développement des biomarqueurs ainsi que les restrictions potentielles liées à chaque étape entre la découverte et la mise en œuvre clinique. Les études canadiennes en cours sur les biomarqueurs démontrent les possibilités d’évaluation du risque, de diagnostic, de pronostic et de réponse aux traitements, ainsi que le processus de découverte des médicaments.
Globally, the number of people at excess cardiovascular risk over their lifetimes continues to rise. As such, cardiovascular disease remains a major economic and social burden, with immense implications for public health and economic well-being. Based on the last survey conducted by Statistics Canada (2004; released in 2006), heart disease and stroke are still responsible for more deaths and disability in Canadians than any other diseases, and cost the Canadian economy over $18 billion per year in direct and indirect health care costs (1–3).
Several known but, unfortunately, all-too-commonly coexistent factors (ie, dyslipidemia, systemic inflammation, high blood pressure, tobacco use, overweight, diabetes and obesity) have all been associated with increased risk of cardiovascular events. However, due to the myriad of phenotypes with which patients may present, either during or as a result of the influence of factors before the overt heart failure stage, there is a growing interest in novel cardiovascular biomarkers or biomarker sets to better stratify patients for more personalized care. For example, heart failure, the leading cause of human morbidity and mortality worldwide, can result from various causes such as ischemia, infection, immunity, toxicity, metabolic abnormalities or genetic factors. A singular factor or a combination of factors can trigger aberrant oxidative, immune, inflammatory, reparative or remodelling events that can lead to structural and functional impairment of the heart. However, despite the range of ‘initiators’ involved, the fundamental mechanisms are often shared regardless of the etiology of heart disease. That said, ‘gold’ lies in the particular biological processes that underlie uniqueness of risk and pathology in cardiovascular disease.
Thus, to better manage patients from various disease groups – those at risk, at early stage of myopathy or vascular occlusive disease, having progressive myocardial disease, or near end-stage heart failure – it is crucial to identify biomarkers of precise and accurate value. Specifically, through biomarker discovery, we can strengthen our current knowledge of classical and marginal phenotypes, and one day change the way we define the various categories and stages of heart muscle disease. From determination of the likelihood of the patient to experience cardiovascular events to reliably diagnosing, monitoring or providing prognostic guidance related to the severity and progression of diseases, novel molecular biomarkers hold immense potential for good. They provide the opportunity to improve the intervention, management and, ultimately, the lives of cardiac patients. However, many questions need to be answered and numerous obstacles need to be overcome in the biomarker development process. How do we derive or discover new biomarkers or biomarker sets? How do we test their ‘validity’? When are they good enough for clinical use?
BIOMARKERS – CHARACTERISTICS AND HISTORY
A biomarker, as defined by the Food and Drug Administration (FDA) of the United States, is any “characteristic that is objectively measured and evaluated as an indicator of normal biological processes, pathogenic processes, or pharmacologic responses to a therapeutic intervention” (4). Similarly, according to Health Canada, a biomarker is described as any “measurable characteristic that is an indicator of normal biologic processes, pathogenic processes, and/or response to therapeutic or other interventions” (5). It is interesting to note that, by the current definitions of biomarkers, it has been argued that one can approach the biomarker discovery process in an existential fashion. Such a perspective means that, to a significant extent, we do not necessarily need to know the detailed identity, function or biological relationship of a given biomarker or biomarker set for it to be reproducibly and objectively useful and clinically valuable (TJ Triche, personal communication).
Historically, cardiac biomarkers have been identified, sometimes serendipitously, through targeted studies of physiological processes. Many biomarkers discovered decades ago in the pre-‘omics’ era (Table 1) are still being used in medical practice. From aspartate transaminase to C-reactive protein and creatine kinase, brain natriuretic peptide (BNP) and cardiac troponins, the road to establishing the particular utility of biomarkers in clinical practice has been a time-consuming and an often iterative process.
TABLE 1.
Historical perspective on cardiac biomarkers
| Year | Examples of cardiac biomarkers | References |
|---|---|---|
| 1954 | First known marker of acute myocardial infarction (AMI), aspartate transaminase, was reported | Karmen et al, 1955 (38) |
| 1956 | Blood levels of C-reactive protein, an acute-phase protein discovered in the 1930s, were found to be generally higher in heart failure subjects | Tillet et al, 1930 (39); Elster et al, 1956 (40) |
| 1977 | Creatine phosphokinase was shown to be potentially useful in diagnosis of AMI in 1963. In 1977, creatine kinase MB isoenzyme (CK-MB) was reported to correlate with angiographic estimates of myocardial infarct size; several conditions, in addition to AMI, were reported to cause serum CK-MB elevation | Sorensen, 1963 (41); Rogers et al, 1977 (42); Tsung, 1981 (43) |
| 1981 | Atrial natriuretic peptide, first observed as granules in atrial cardiocytes in guinea pigs in the 1950s by Kisch, then described as a hormone produced by the heart in rats, was later (in 1986) found to be elevated in congestive heart failure in humans | Kisch, 1956 (44); de Bold et al, 1981 (45); Burnett et al, 1986 (46) |
| 1987 | Radioimmunoassay for cardiac-specific troponin I (part of the troponin complex discovered in the 1960s by Ebashi et al) was described as a potential diagnostic tool for AMI. CK-MB, C-reactive protein and troponin I were later re-evaluated as biomarkers of cardiac muscle damage in humans in an independent study | Ebashi et al, 1965 (47); Cummins et al, 1987 (48,49) |
| 1990 | Brain natriuretic peptide elevation was also found in patients with congestive heart failure. Increased plasma level of brain natriuretic peptide was later (in 1993) reported in AMI patients | Mukoyama et al, 1990 (50); Morita et al, 1993 (51) |
| Present | Currently, there are over 360,000 peer-reviewed biomarker reports (not limited to cardiac biomarkers). Of those, only several hundred have been Food and Drug Administration (USA)- approved or -cleared, including perhaps several dozen genomic or genetic markers | Mansfield, 2008 (52) |
BIOMARKER DEVELOPMENT: AN OVERVIEW
In general, the biomarker development process can be broken down into three stages: discovery, internal validation and external validation.
Biomarker discovery
With the recent explosion of high-performance ‘omic’ technologies – genomics, proteomics and metabolomics, among others – the rate at which biomarker candidates are being discovered is now faster than ever. During the discovery phase, single or multiple platforms are used to identify potential candidate biomarkers in a given patient population, typically from a small geographical area (ie, a single institutional catchment). This quest involves selection of patients with clear clinical phenotypes, and standardized operating protocol-driven collection and processing of single or multiple timepoint samples for analysis. Given that biological variability can lead to substantial differences in body fluid and tissue composition as well as associated biomarker measurements, basic data such as sex, age, hormonal status, race, ethnic background, disease history and severity of underlying conditions should be carefully considered during patient selection procedures for biomarker discovery and development efforts (6).
In the case of multiplatform ‘omic’ biomarker discovery, data within each platform are analyzed using suitable, tiered statistical methods to identify differentially expressed genes, proteins or metabolites in normal subjects versus patients in cohorts, from baseline status throughout the course of illness, addressing patients with different stages of disease. Biomarker candidates may be identified via single timepoint analysis or derived serially over time. In addition to the statistical approaches, data-and literature-mining techniques must also be applied to find potential candidates that can later be validated. Bioinformatical tools are also applied in parallel to other strategies to help deduce potential functional or pathway associations among candidate biomarkers. Once candidate biomarkers from each platform have been identified, combinatorial analyses may be carried out for further assessment and refinement. Which specific combination of genomic, proteomic or metabolomic biomarkers, independent from or in conjunction with known clinical variables, provides the best tool to stratify patients of interest?
It is not always practical to pursue validation of all candidate bio-markers identified during the discovery stage. Thus, it is important from both time and cost point-of-views to establish parameters for scientifically rational, statistically sound, evidence-based selection or rejection of biomarker candidates (7–9). Only the ‘best’ candidates should move forward into the validation stage. The decision to move a candidate biomarker forward is not only dependent on its statistical or bioinformatical significance, but also largely based on its potential to contribute cost-effectively to disease management or prevention (10).
Biomarker validation
Biomarker validation is currently a lengthy and complex process. Not surprisingly, with the availability of ‘omic’ platforms, candidate bio-marker discovery now commonly outruns the rate at which the candidates are being validated. This situation has created a bottleneck in the biomarker development and translation process (11,12). To help address this issue, the FDA has led the way by creating initial guidelines, such as Guidance for Industry: Bioanalytical Method Validation, published in 2001 (13). The FDA also launched the Critical Path Initiative in 2004 in an effort to improve the biomarker development process (14). Further, the FDA has helped to introduce the concept of classes of biomarkers based on their validity in Guidance for Industry: Pharmacogenomic Data Submission, published in 2005 (15). The FDA classifies (pharmacogenomic) biomarkers as exploratory or valid (16). Valid biomarkers are further classified as ‘probable’ or ‘known’, depending on the level of confidence they attain during the validation process (Table 2) (16).
TABLE 2.
Definitions of known valid and probable valid biomarkers
| Biomarker validity | Definition |
|---|---|
| Known | A biomarker that is measured in an analytical test system with well-established performance characteristics and for which there is widespread agreement in the medical or scientific community about the physiological, toxicological, pharmacological or clinical significance of the results |
| Probable | A biomarker that is measured in an analytical test system with well-established performance characteristics and for which there is a scientific framework or body of evidence that appears to elucidate the physiological, toxicological, pharmacological or clinical significance of the test results |
Data from reference 16
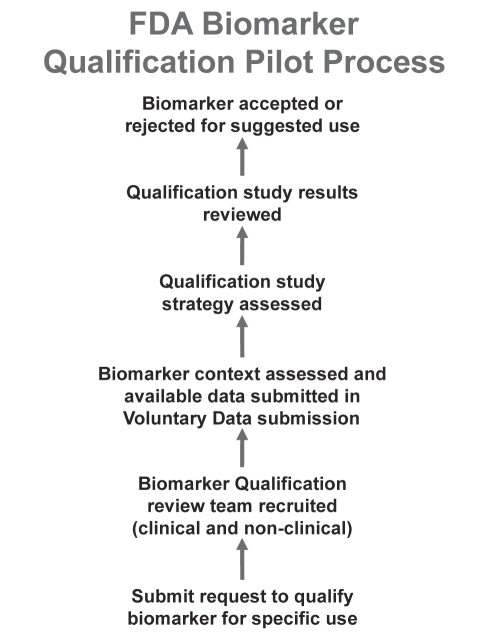
The term ‘validation’, particularly in the arena of biomarker research, is a broad concept that can be used to describe anything from analytical and technical validation to the biological characteristics of the biomarkers identified (17). As such, the FDA has adopted the term ‘qualification’ to describe the assessment of a biomarker for its utility in clinical process. In other words, it is the evidentiary process by which data are provided to link biomarkers with biology, and to show that each biomarker identified is qualified for application in a specific context (18). The pilot structure of this qualification process is shown in Figure 1.
Figure 1).
United States Food and Drug Administration (FDA) biomarker qualification pilot process. Figure adapted from reference 16
Despite the availability of guidelines such as those described above, there remains a lack of sufficient guidance on what validation process(es) are recommended or required to transition from exploratory to valid biomarkers, or from probable valid to known valid bio-markers (19,20).
The lack of universal validation guidelines is partially attributable to the diverse nature of biomarker research and development (20–22). In fact, questions repeatedly asked during the biomarker development process ultimately determine the validation strategy: What is the context of the application? Is the biomarker going to be predictive, diagnostic or prognostic? How ‘good’ does the biomarker need to be? Is it likely to replace the current gold standard, or to augment or supplement it?
In essence, the foundational thrust of validation efforts is to ensure that the biomarkers, or their assays, are ‘reliable for intended use’– a principle commonly referred to as ‘fit-for-purpose’ (23,24). Validation of a biomarker or biomarker set is an evolutionary and integrative process. Ideally, both analytical or method validation, determined by assay performance characteristics, and clinical validation (qualification), showing an evidentiary link of the biomarker with a biological process or clinical end point, should be carried out continuously and concurrently (22,25). The key parameters that serve as focal points of method validation include accuracy, precision, sensitivity, specificity, reproducibility and stability. Whenever possible, assay and analytical sensitivity and specificity, as well as quality control and quality assurance, should be established early in biomarker development. This approach ensures the robustness of the analytical methods used in clinical phases of validation or qualification, and the integrity of the evidence collected.
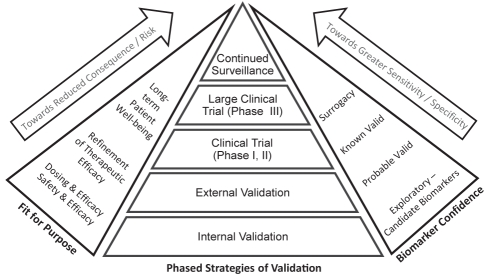
As mentioned earlier, the fit-for-purpose strategy is context specific, and the validation criteria are dependent on the intended use of the biomarker. For instance, a biomarker used to include or exclude patients from treatment likely requires a different level of sensitivity and specificity than one used to determine drug dosing, toxicity or safety. A biomarker intended as a population screen must be very sensitive and extremely specific (26). This framework is further described in Figure 2, which breaks down biomarker validation into five major phased strategies – the internal validation (within the initial cohort; often done to evaluate the sensitivity and specificity of the selected candidate biomarker), the external validation (same type of platform and statistical or informatical analyses as internal validation but in a second, independent external cohort), the initial (phases 1 and 2) clinical trials, followed by larger clinical trials and continued surveillance. As shown in Figure 2, exploratory biomarkers may need to go through both internal validation and external validation to reach the ‘probable’ valid status. Subsequently, biomarkers may be put through prospective clinical trials to become ‘known’ valid. In a few instances, the biomarker may demonstrate enough sensitivity and specificity as it moves through the different phases of validation and clinical trials to eventually reach surrogacy, considered to be the ‘holy grail’ of biomarker development. This latter status is a lofty perch only achieved by a few biomarkers.
Figure 2).
Fit-for-purpose biomarker validation. Biomarker development and validation is driven by its intended use or fit-for-purpose. The principle of fit-for-purpose strategy is that biomarkers with false-positive or -negative indications pertaining to high patient consequences and risks require various phases of validation. There are five major validation phases: internal validation, external validation, clinical trials (phase I and II; checking for safety and efficacy), large clinical trials (phase III) and continued surveillance. Sensitivity and specificity correlate with the intended purpose of the bio-marker. The level of biomarker confidence that a candidate biomarker can attain depends on the phase of validation that has been reached. In ideal cases, biomarkers may reach the point of surrogacy and replace known clinical end points. However, this designation requires agreement with regulatory authorities, as well as long-term data and evidence for safety and efficacy, because the consequences of a flawed surrogate biomarker are high
POTENTIAL PITFALLS
Issues that may come up during the biomarker discovery stage (ie, limited availability of quality clinical specimens and clear phenotypes) can also occur during the validation phases described above. The lack of available samples or patient enrolment can ultimately translate to a decrease in biomarker confidence, as there may be insufficient long-term data, and in certain contexts, insufficiently robust evidence of safety and efficacy. Collaboration between academia and industry partners will likely make the biomarker development process more efficient and economically feasible than working independently (27). Unfortunately, even during collaborations, there may be inadequate data-sharing due to proprietary interests. Examples of potential pitfalls are summarized in Table 3. Similar to the idea that biomarker development is dependent on the intended use of the biomarker, the consequence of a flawed biomarker is also, in fact, heavily influenced by the context of application. This is one reason why surrogate biomarkers, for example, are difficult to develop and yet may be associated with either great rewards or comparably large risks.
TABLE 3.
Potential pitfalls in biomarker development
| Patient variability – interindividual and intraindividual |
| Limited availability of samples |
| Lack of method standards, quality assurance, quality control or standard operating protocol-driven sample collection and processing |
| Intellectual property protection – lack of collaboration |
| Inadequate sharing of data between academia and industry partners |
| Lack of clear regulatory guidance |
| Insufficient long-term data |
| Insufficient evidence of safety and efficacy |
As noted, surrogate end point biomarkers have the potential to play essential roles in pharmaceutical and therapeutic developments in coming years. Regulatory agencies in both Europe and North America have implemented strategies for streamlining drug development in response to plunging rates of drug discovery. Biomarkers play a central role in the efforts to tackle the current crisis facing the pharmaceutical industry, which has recently witnessed a 20-year low in the introduction of new chemical entities (28). Current costs of bringing a drug to market have skyrocketed in the past decade, in large part due to the overwhelming rate of clinical trial failure. The resulting pressure for successful compounds to reap a profit has led to intense marketing by pharmaceutical companies, in addition to increased pressure on regulatory bodies to ensure patient safety in the face of market forces encouraging high uptake rates. The need for effective surrogate bio-markers is clear; the potential to evaluate a predictor in place of clinical measurement of disease would allow for shorter clinical trials, more cost-effective drug discovery and more rapid implementation of beneficial therapies. But in spite of the immense potential for surrogate markers in correcting the ‘pipeline problems’ of pharmaceutical development (29), it is essential that these biomarkers are suitable, as cautionary tales of inappropriate surrogate marker application abound. A parable of illustrative value relates to using cardiac arrhythmias as a surrogate biomarker for survival following myocardial infarction (MI). The identification of ventricular premature depolarizations following MI was established as being associated with increased risk of arrhythmic death compared with patients without such arrhythmia. Consequently, antiarrhythmic drugs, including flecainide and encainide, were implemented in clinical practice in an effort to improve survival post-MI. The antiarrhythmogenic activity of these drugs was monitored by echocardiogram, and while both compounds suppressed ventricular arrhythmias, the Cardiac Arrhythmia Suppression Trial (CAST) demonstrated that both drugs increased arrhythmic death (30). In this instance, the use of cardiac arrhythmia as a surrogate end point for survival was dramatically inadequate, and preliminary data from CAST were so overwhelming as to result in an early discontinuation of the treatment arms of the trial.
The use of pharmacogenomic biomarkers is increasing as high throughput genomic technologies become more accessible. Such markers have utility both in the drug development pathway for patient responder identification and in clinical practice for dose optimization. The identification of single nucleotide polymorphisms (SNPs) in genes involved in the metabolism of warfarin is of particular note, because this has led to successful commercialization of a test that has emerged as having value in the clinic. Warfarin is the most widely prescribed anticoagulant in North America, and while efficacious as a pharmaceutical intervention, it harbours a significant risk of serious hemorrhage that is compounded by the sizable range in effective daily dose among patients: the dose range extends from 0.5 mg/day to 60 mg/day, and it has a narrow therapeutic window (31). Warfarin exists as a racemic mixture of R-and S-enantiomers, each of which is metabolized by different enzymes in the cytochrome c complex. The R-enantiomer is the more potent of the two, and as such, SNPs in CYP2C9, the enzyme that metabolizes it, are of significance in its pharmacodynamics. The downstream epoxide-reductase VKOR in the vitamin K-dependent clotting cascade also harbors SNPs that affect the efficacy of warfarin in anticoagulation (31,32). Genotypes CYP2C9 and VKOR play into the phenotype of drug metabolism, and by harnessing advances in genomic technology, clinical genotyping tools such as the Verigene Warfarin Metabolism Nucleic Acid test (Nanosphere Inc, USA) have come to market, demonstrating the applicability of these tools in personalized medical care. Indeed, the Verigene test was the first FDA-approved genetic test for warfarin sensitivity and represents a significant step forward (33).
Perhaps the most significant cardiovascular biomarker to be successfully implemented in the clinic in recent years is BNP. As one of four natriuretic peptides regulating blood pressure, electrolyte balance and fluid volume, BNP acts as an antagonist to the renin-angiotensin-aldosterone system, and is released in response to hemodynamic stress. A complex relationship between BNP and heart function exists: BNP is released by atrial myocytes acting to reduce hemodynamic strain. BNP levels, including active and inactive hormone, are elevated in the blood in the setting of heart failure. The measurement of blood BNP and its inactive pro-hormone, N-terminal-proBNP, increases the accuracy of heart failure diagnosis, when combined with traditional clinical observations such as from physical examination, history and chest x-ray. The negative predictive value of this biomarker is 98% (34), and its applicability is appropriate given the clinical context of heart failure; the ability to exclude heart failure in a patient is indispensable. As a result, the implementation of BNP as a tool in clinical practice has resulted in reduced time from patient diagnosis to discharge; prevented unnecessary admission to hospital; reduced costs of conventional management; and reduced adverse events, mortality and patient hospital days (35).
The promise of improvements in clinical care and accelerated drug development offered by successful biomarkers, juxtaposed with the catastrophic consequences of their failures, brings into clear focus the need for well-designed and rigorously executed assessment for bio-marker identification, validation, qualification and implementation.
BIOMARKERS IN TRANSPLANTATION STUDY
Vital organ failure is a leading cause of disability and premature death worldwide. Organ transplantation is now a common, and often the only available, procedure for patients with end-stage organ failure. Transplantation restores the life and health of more than 40,000 patients per year worldwide. However, long-term allograft (the transplanted organ) survival has improved only marginally since the early 1980s. Immune and inflammatory insults are the major causes of acute rejection, an early immune attack on transplanted allografts characterized by cellular and antibody-mediated injury; and chronic rejection, a persistent immune attack characterized not only by visible injury, but also reparative healing, with mesenchymal cell proliferation, tissue remodelling and fibrosis. The rejection can be controlled with drugs that suppress the immune system, but they must be administered carefully. Too much immunosuppressive therapy can cause complications due to the immune system’s inability to fight diseases, including infection, cancer, diabetes, heart disease and kidney failure. Certain therapies contribute to post-transplant lymphoproliferative syndromes. Transplant failure and treatment complications consume enormous health care resources.
A major difficulty in the management of transplant recipients is the lack of prognostic assays capable of predicting graft injury. Current rejection diagnostic approaches still have room for improvement in terms of their sensitivities and specificities. Tissue biopsies, the current gold standard, are invasive, expensive, subject to sampling error and subjectively interpreted, and they incur patient risk. The objectives of the Biomarkers in Transplantation initiative are to identify effective and widely applicable biomarkers in heart, kidney and liver transplants using high-performance genomics and proteomics that diagnose acute and chronic rejection, predict rejection or immune accommodation of solid organ allografts, and forecast the response to therapies that individual transplant recipients receive. Implementation of these biomarkers into the clinic will eliminate the need for biopsy and can help personalize the immunosuppressive treatment of transplant patients. This will mean a minimum of 50% reduction of health care costs associated with transplant patient monitoring and care.
The strategy employed by the Biomarkers in Transplantation team is to collect serial biopsy, blood and urine samples, from pre-transplant through three years post-transplant, from subjects who received heart, kidney or liver transplants in Vancouver, British Columbia, and who consented to enroll in the study. Selected samples are analyzed using transcriptomic (Affymetrix Human Genome U133 Plus 2.0 chips [USA]), proteomic (iTRAQ MALDI TOF-TOF [36]) and metabolomic (Nuclear Magnetic Resonance Spectroscopy [37]) platforms. The biomarker classifier panels developed are validated internally in samples collected within the same site but from a separate cohort than the one used in the classifier development. The biomarker panels will be validated externally using Canada-wide, multisite transplant subject samples. This latter external qualification phase will be run under the umbrella of the Centre of Excellence for the Prevention of Organ Failure (PROOF), a Centre of Excellence for Commercialization and Research initiative.
THE PROOF CENTRE
Organ failure of hearts, lungs and kidneys is epidemic. Preventing organ failure is an ethical and economic imperative. The PROOF Centre links research with industry, health and policy. Through commercialization partnerships, the PROOF Centre delivers personalized and preventive tools and products to patients in our health care system at risk for or suffering from heart, lung, and kidney failure.
The PROOF Centre has several goals. The first goal is to use expertise and passion to help Canadians avoid preventable organ failure. Second, the PROOF Centre aims to attract, retain and nurture young international researchers and commercialization entrepreneurs. Third, the PROOF Centre will create a first class incubator infrastructure that will sustain itself in the future. Finally, the PROOF Centre will ensure that the best ideas are brought to bear on the commercialization and translational pipeline to benefit Canadians. The PROOF Centre’s vision is to reduce economic, social and personal costs imposed by the epidemic of heart, lung and kidney failure, while harnessing innovation to create value from new predictive, diagnostic and prognostic biomarker tools. The PROOF Centre’s mission is to ensure the timely translation of solution-oriented discoveries that move away from drug treatment-only strategies toward prevention and effective early detection of heart, lung and kidney failure. The PROOF Centre will help Canadians by speeding up health care improvements. Through the PROOF Centre and related efforts, Canadians can be world leaders in the prevention of organ failure due to chronic disease.
FUTURE OUTLOOK AND OPPORTUNITIES
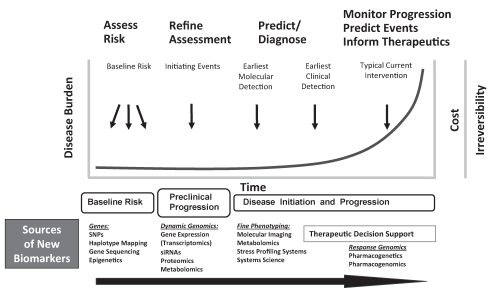
Much of current biomarker development efforts are concentrated toward the right end of the curve in Figure 3, aimed at identifying noninvasive ways of monitoring progression and diagnosing clinical events in relation to current or new therapeutic agents. While interventions during later stages of disease development can still provide significant impact on patient management, the cost of medical care can be quite high (eg, cardiac assist device implantation) and the pathological condition may have already become irreversible. Canada can contribute to the ‘omic’ biomarker field by moving upstream and identifying biomarkers that can diagnose or predict the disease when it is not as advanced (Figure 3). An even bigger contribution can come from the discovery of biomarkers used in assessing baseline risk and initiating events of organ disease. In Figure 3, different sources of bio-markers used during the evolving timecourse of disease development are shown. It will take an integrative and staged approach in the assessment of biomarker sets of different types to move toward more personalized management of organ failure and its prediction. The coming few years will see many breakthroughs in this regard.
Figure 3).
Different uses and sources of novel biomarkers pertinent to the course of risk and disease development (adapted from PP Liu). siRNA small interfering RNA; SNP Single nucleotide polymorphism
Footnotes
FINANCIAL SUPPORT: Heart & Stroke Foundation, Canadian Institutes of Health Research, Genome Canada, Genome British Columbia, Networks of Centres of Excellence Centres of Excellence for Commercialization and Research, IBM, Novartis, Pfizer and IO Informatics.
CONFLICTS OF INTEREST: The authors have no conflicts of interest to declare.
REFERENCES
- 1.Heart & Stroke Foundation of BC & Yukon Statistics 2008. <www.heartandstroke.bc.ca/site/c.kpIPKXOyFmG/b.3644453/k.3454/Statistics.htm> (Version current 2008)
- 2.Public Health Agency of Canada Economic burden of illness in Canada 1998. <www.phac-aspc.gc.ca/publicat/ebic-femc98/> (Version current April 25, 2009).
- 3.Causes of death, 2004. Released 2006. For provincial and national mortality tables, go to <http://www.heartandstroke.on.ca/site/c.pvI3IeNWJwE/b.3581729/k.359A/Statistics.htm> and click on the Statistics Canada link. (Version current at May 11, 2009).
- 4.Biomarkers Definitions Working Group Biomarkers and surrogate endpoints: Preferred definitions and conceptual framework. Clin Pharmacol Ther. 2001;69:89–95. doi: 10.1067/mcp.2001.113989. [DOI] [PubMed] [Google Scholar]
- 5.Health Canada Adoption of ICH Guidance: Guidance Document Definitions for Genomic Biomarkers, Pharmacogenomics, Pharmacogenetics, Genomic Data and Sample Coding Categories, ICH Topic E15 2008. <www.hc-sc.gc.ca/dhp-mps/alt_formats/hpfb-dgpsa/pdf/prodpharma/e15-eng.pdf> (Version current April 25, 2009).
- 6.Moore RE, Kirwan J, Doherty MK, Whitfield PD. Biomarker discovery in animal health and disease: The application of post-genomic technologies. Biomark Insights. 2007;2:185–96. [PMC free article] [PubMed] [Google Scholar]
- 7.Hunt SM, Thomas MR, Sebastian LT, et al. Optimal replication and the importance of experimental design for gel-based quantitative proteomics. J Proteome Res. 2005;4:809–19. doi: 10.1021/pr049758y. [DOI] [PubMed] [Google Scholar]
- 8.Lee JW, Figeys D, Vasilescu J. Biomarker assay translation from discovery to clinical studies in cancer drug development: Quantification of emerging protein biomarkers. Adv Cancer Res. 2007;96:269–98. doi: 10.1016/S0065-230X(06)96010-2. [DOI] [PubMed] [Google Scholar]
- 9.Listgarten J, Emili A. Statistical and computational methods for comparative proteomic profiling using liquid chromatography-tandem mass spectrometry. Mol Cell Proteomics. 2005;4:419–34. doi: 10.1074/mcp.R500005-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Bast RC, Jr, Lilja H, Urban N, et al. Translational crossroads for biomarkers. Clin Cancer Res. 2005;11:6103–8. doi: 10.1158/1078-0432.CCR-04-2213. [DOI] [PubMed] [Google Scholar]
- 11.Baker M. In biomarkers we trust? Nat Biotechnol. 2005;23:297–304. doi: 10.1038/nbt0305-297. [DOI] [PubMed] [Google Scholar]
- 12.Benowitz S. Biomarker boom slowed by validation concerns. J Natl Cancer Inst. 2004;96:1356–7. doi: 10.1093/jnci/96.18.1356. [DOI] [PubMed] [Google Scholar]
- 13.FDA Guidance for Industry: Bioanalytical Method Validation 2001. <www.fda.gov/CDER/GUIDANCE/4252fnl.pdf> (Version current April 25, 2009).
- 14.FDA Innovation or Stagnation: Challenge and Opportunity on the Critical Path to New Medical Products 2004. <www.fda.gov/oc/initiatives/criticalpath/whitepaper.pdf> (Version current April 25, 2009).
- 15.FDA Guidance for Industry: Pharmacogenomic Data Submissions 2005. <www.fda.gov/cber/gdlns/pharmdtasub.pdf> (Version current April 25, 2009).
- 16.Goodsaid F, Frueh F. Biomarker qualification pilot process at the US Food and Drug Administration. AAPS J. 2007;9:E105–108. doi: 10.1208/aapsj0901010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonassi S, Neri M, Puntoni R. Validation of biomarkers as early predictors of disease. Mutat Res. 2001:480–481. 349–58. doi: 10.1016/s0027-5107(01)00194-4. [DOI] [PubMed] [Google Scholar]
- 18.Goodsaid F.Impact of the biomarker qualification project on drug developmentCritical Markers of Disease Symposium 2008. <www.cmod.org/slide_presentations/resources_slides_2008.html> (Version current April 25, 2009).
- 19.Goodsaid F, Frueh F. Process map proposal for the validation of genomic biomarkers. Pharmacogenomics. 2006;7:773–82. doi: 10.2217/14622416.7.5.773. [DOI] [PubMed] [Google Scholar]
- 20.Lee JW, et al. Method validation and measurement of biomarkers in nonclinical and clinical samples in drug development: A conference report. Pharm Res. 2005;22:499–511. doi: 10.1007/s11095-005-2495-9. [DOI] [PubMed] [Google Scholar]
- 21.Colburn WA. Biomarkers in drug discovery and development: From target identification through drug marketing. J Clin Pharmacol. 2003;43:329–41. doi: 10.1177/0091270003252480. [DOI] [PubMed] [Google Scholar]
- 22.Wagner JA, Williams SA, Webster CJ. Biomarkers and surrogate end points for fit-for-purpose development and regulatory evaluation of new drugs. Clin Pharmacol Ther. 2007;81:104–7. doi: 10.1038/sj.clpt.6100017. [DOI] [PubMed] [Google Scholar]
- 23.Ilyin SE, Belkowski SM, Plata-Salaman CR. Biomarker discovery and validation: Technologies and integrative approaches. Trends Biotechnol. 2004;22:411–6. doi: 10.1016/j.tibtech.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 24.Lee JW, et al. Fit-for-purpose method development and validation for successful biomarker measurement. Pharmaceut Res. 2006;23:312–28. doi: 10.1007/s11095-005-9045-3. [DOI] [PubMed] [Google Scholar]
- 25.Wagner JA. Overview of biomarkers and surrogate endpoints in drug development. Dis Markers. 2002;18:41–6. doi: 10.1155/2002/929274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Normolle D, Ruffin Iv MY, Brenner D. Design of early validation trials of biomarkers. Cancer Inform. 2005;1:25–31. [PMC free article] [PubMed] [Google Scholar]
- 27.McCormick T, Martin K, Hehenberger M. The evolving role of biomarkers: Focusing on patients from research to clinical practice, in IBM (Imaging) Biomarker Summit III. Nice; France: 2007. [Google Scholar]
- 28.Woodcock J, Woosley R. The FDA critical path initiative and its influence on new drug development. Ann Rev Med. 2008;59:1–12. doi: 10.1146/annurev.med.59.090506.155819. [DOI] [PubMed] [Google Scholar]
- 29.Krishna R, Herman G, Wagner JA. Accelerating drug development using biomarkers: A case study with sitagliptin, a novel DPP4 inhibitor for type 2 diabetes. AAPS J. 2008;10:401–9. doi: 10.1208/s12248-008-9041-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pratt CM, Moye LA. The cardiac arrhythmia suppression trial. Casting suppression in a different light. Circulation. 1995;91:245–7. doi: 10.1161/01.cir.91.1.245. [DOI] [PubMed] [Google Scholar]
- 31.Hirsh J, et al. American Heart Association/American College of Cardiology Foundation guide to warfarin therapy. J Am Coll Cardiol. 2003;41:1633–52. doi: 10.1016/s0735-1097(03)00416-9. [DOI] [PubMed] [Google Scholar]
- 32.Herman D, et al. Influence of CYP2C9 polymorphisms, demographic factors and concomitant drug therapy on warfarin metabolism and maintenance dose. Pharmacogenomics J. 2005;5:193–202. doi: 10.1038/sj.tpj.6500308. [DOI] [PubMed] [Google Scholar]
- 33.FDA FDA clears genetic lab test for warfarin sensitivity 2007. <www.fda.gov/bbs/topics/NEWS/2007/NEW01701.html> (Version current April 25, 2009).
- 34.Anderson KM. Clinical uses of brain natriuretic peptide in diagnosing and managing heart failure. J Am Acad Nurse Pract. 2008;20:305–10. doi: 10.1111/j.1745-7599.2008.00327.x. [DOI] [PubMed] [Google Scholar]
- 35.Godkar D, et al. B-type natriuretic peptide (BNP) and proBNP: Role of emerging markers to guide therapy and determine prognosis in cardiovascular disorders. Am J Ther. 2008;15:150–6. doi: 10.1097/MJT.0b013e31815af96f. [DOI] [PubMed] [Google Scholar]
- 36.Zieske LR. A perspective on the use of iTRAQ reagent technology for protein complex and profiling studies. J Exp Bot. 2006;57:1501–8. doi: 10.1093/jxb/erj168. [DOI] [PubMed] [Google Scholar]
- 37.Wishart DS. Metabolomics: A complementary tool in renal transplantation. Contrib Nephrol. 2008;160:76–87. doi: 10.1159/000125935. [DOI] [PubMed] [Google Scholar]
- 38.Karmen A, Wroblewski F, Ladue JS. Transaminase activity in human blood. J Clin Invest. 1955;34:126–31. doi: 10.1172/JCI103055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tillett WS, Francis T., Jr Serological reactions in pneumonia with a nonprotein somatic fraction of pneumococcus. J Exp Med. 1930;52:561–85. doi: 10.1084/jem.52.4.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elster SK, Braunwald E, Wood HF. A study of C-reactive protein in the serum of patients with congestive heart failure. Am Heart J. 1956;51:533–41. doi: 10.1016/0002-8703(56)90099-0. [DOI] [PubMed] [Google Scholar]
- 41.Sorensen NS. Creatine phosphokinase in the diagnosis of myocardial infarction. Acta Med Scand. 1963;174:725–34. [PubMed] [Google Scholar]
- 42.Rogers WJ, McDaniel HG, Smith LR, Mantle JA, Russel RO, Jr, Rackley CE. Correlation of angiographic estimates of myocardial infarct size and accumulated release of creatine kinase MB isoenzyme in man. Circulation. 1977;56:199–205. doi: 10.1161/01.cir.56.2.199. [DOI] [PubMed] [Google Scholar]
- 43.Tsung SH. Several conditions causing elevation of serum CK-MB and CK-BB. Am J Clin Pathol. 1981;75:711–5. doi: 10.1093/ajcp/75.5.711. [DOI] [PubMed] [Google Scholar]
- 44.Kisch B. Electron microscopy of the atrium of the heart. I. Guinea pig. Exp Med Surg. 1956;14:99–112. [PubMed] [Google Scholar]
- 45.de Bold AJ, Borenstein HB, Veress AT, Sonnenberg H. A rapid and potent natriuretic response to intravenous injection of atrial myocardial extract in rats. Life Sci. 1981;28:89–94. doi: 10.1016/0024-3205(81)90370-2. [DOI] [PubMed] [Google Scholar]
- 46.Burnett JC, Jr, Kao PC, Hu DC, et al. Atrial natriuretic peptide elevation in congestive heart failure in the human. Science. 1986;231:1145–7. doi: 10.1126/science.2935937. [DOI] [PubMed] [Google Scholar]
- 47.Ebashi S. Third component participating in the superprecipitation of “natural actomyosin”. Nature. 1963;200:1010. doi: 10.1038/2001010a0. [DOI] [PubMed] [Google Scholar]
- 48.Cummins B, Auckland ML, Cummins P. Cardiac-specific troponin-I radioimmunoassay in the diagnosis of acute myocardial infarction. Am Heart J. 1987;113:1333–44. doi: 10.1016/0002-8703(87)90645-4. [DOI] [PubMed] [Google Scholar]
- 49.Cummins P, Young A, Auckland ML, Michie CA, Stone PC, Shepstone BJ. Comparison of serum cardiac specific troponin-I with creatine kinase, creatine kinase-MB isoenzyme, tropomyosin, myoglobin and C-reactive protein release in marathon runners: Cardiac or skeletal muscle trauma? Eur J Clin Invest. 1987;17:317–24. doi: 10.1111/j.1365-2362.1987.tb02194.x. [DOI] [PubMed] [Google Scholar]
- 50.Mukoyama M, Nakao K, Saito Y, et al. Increased human brain natriuretic peptide in congestive heart failure. N Engl J Med. 1990;323:757–8. doi: 10.1056/NEJM199009133231114. [DOI] [PubMed] [Google Scholar]
- 51.Morita E, Yasue H, Yoshimura M, et al. Increased plasma levels of brain natriuretic peptide in patients with acute myocardial infarction. Circulation. 1993;88:82–91. doi: 10.1161/01.cir.88.1.82. [DOI] [PubMed] [Google Scholar]
- 52.Mansfield E.International Partnership for Critical Markers of Disease<www.cmod.org> (Version current at May 12, 2009).