Abstract
Previously a new method for ultrasound signal characterization using entropy Hf was reported, and it was demonstrated that in certain settings, further improvements in signal characterization could be obtained by generalizing to Renyi entropy-based signal characterization If(r) with values of r near 2 (specifically r=1.99) [M. S. Hughes et al., J. Acoust. Soc. Am. 125, 3141–3145 (2009)]. It was speculated that further improvements in sensitivity might be realized at the limit r→2. At that time, such investigation was not feasible due to excessive computational time required to calculate If(r) near this limit. In this paper, an asymptotic expression for the limiting behavior of If(r) as r→2 is derived and used to present results analogous to those obtained with If(1.99). Moreover, the limiting form If,∞ is computable directly from the experimentally measured waveform f(t) by an algorithm that is suitable for real-time calculation and implementation.
INTRODUCTION
In an earlier paper1 we reported on the application of Renyi entropy If(r), which is defined for all r<2 (r is roughly a reciprocal “temperature”), for the detection of changes in backscattered radio frequency (rf) ultrasound arising from the accumulation of targeted nanoparticles in the neovasculature in the insonified region of a tumor. That study was motivated by the observation that acoustic characterization of sparse collections of targeted perfluorocarbon nanoparticles presented challenges that might require the application of novel types of signal processing.2 We were able to show that signal processing based on a “moving window” Hf analysis [see Eq. 7] could detect accumulation of tissue-targeted nanoparticles 30 min following nanoparticle injection. The signal energy, defined as the sum of squares over the same moving window, was unable to distinguish measurements made at any time during the 1 h experiment (as was conventional B-mode imaging). Subsequently we determined that moving window If(r) analysis, with r=1.99, could distinguish the difference in backscatter measured at 0 and 15 min. Reduction in the accumulation time required to reach detectability from 30 to 15 min is clearly of significance: potentially reducing both patient discomfort and increasing clinical throughput. Moreover, although the computational effort to obtain the result precluded its clinical application with currently available equipment, the study raised the possibility of further sensitivity improvements by using values of r closer to the limiting value of 2, where If(r) approaches infinity. The purpose of the current study is to investigate the behavior of If(r) as r→2 by extracting its asymptotic form. While this involves use of the second derivatives of f(t) at its critical points, which can be expected to increase noise in the processing chain output, surprisingly the resulting signal processing scheme does not sacrifice sensitivity. Moreover, the operation count in this approach is lower than that used to produce the signal envelope, which currently is the standard for real-time ultrasonic imaging display, thus demonstrating its suitability for implementation in a real-time imaging system to facilitate detection of molecular epitopes associated with neovasculature in a growing tumor. As our technique is based on moving window analysis of digitized rf, which requires some sacrifice in spatial resolution, clinical implementation of entropy detection would probably follow the same approach currently employed in Doppler “imaging” systems, where the conventional B-mode image is color-coded according to the blood cell velocity to present a combined B-mode∕velocity image; similarly, a B-mode∕entropy image could be made as well.
APPROACH
All results in this study were obtained using the density function wf(y) of the continuous function y=f(t), assumed to underlie the sampled rf data. Subsequently, wf(y) was used to compute the entropy If(r). As described in previous studies wf(y) corresponds to the density functions used in statistical signal processing.1 In contradistinction to statistical signal processing, where f(t) is a random function, and often nowhere differentiable, we assume that the noise levels in our apparatus are low enough so that with sufficient signal averaging, noise may be eliminated, or at least reduced to a low enough level, that derivatives of f(t) may be accurately computed. From these derivatives the density function wf(y) may be computed,1 which then facilitates calculation of the quantities typically discussed in statistical signal processing (e.g., mean values, variances, and covariances).3, 4, 5 However, in that environment, the density function is usually assumed to be continuous, infinitely differentiable, and to approach zero at infinity. In our case wf(y) is not so well-behaved and has (integrable) singularities. While this renders calculation of the density function more difficult, applications of entropy imaging based on wf(y) have shown the cost to be justified in terms of increased sensitivity to subtle changes in scattering architecture that are often undetected by more conventional imaging.
We use the same conventions as in previous studies so that
| (1) |
where N is the number of laps [regions of monotonicity of f(t)], gk(y) is the inverse of f(t) in the kth-lap, and if y is not in the range of f(t) in the kth-lap, is taken to be 0.
We also assume that all experimental waveforms f(t) have a Taylor series expansion valid in the domain [0,1]. Then near a time tk such that f′(tk)=0,
| (2) |
where tk is a lap boundary. On the left side of this point Eq. 2 may be truncated to second order and inverted to obtain
| (3) |
with
| (4) |
The contribution to wf(y) from the right side of the lap boundary, from gk+1(y), is the same, so that the overall contribution to wf(y) coming from the time interval around tk is
| (5) |
for 0<f(tk)−y⪡1 for a maximum at f(tk) and 0<y−f(tk)⪡1 for a minimum. Thus, wf(y) has only a square root singularity (we have assumed that tk is interior to the interval [0,1]; if not, then the contributions to wf come from only the left or the right). If, additionally, f″(tk)=0, then the square root singularity in Eq. 4 will become a cube-root singularity, and so on, so that the density functions we consider will have only integrable algebraic singularities.
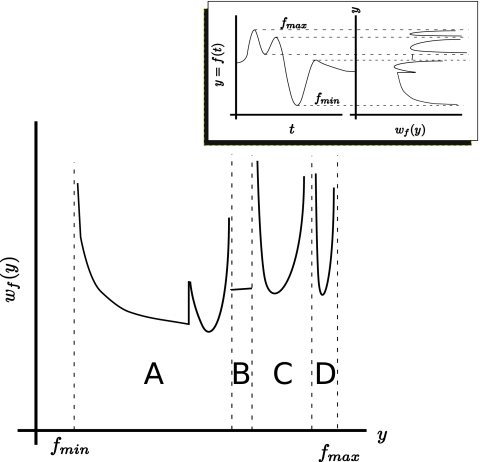
Figure 1 shows a typical waveform (inset) and its associated density function, which may be divided into four different regions, A–D, separated by singularities (dashed lines) corresponding to the critical points of f(t). In general, three types of behavior are possible in wf(y): continuous, finite jump discontinuity, and integrable singularity. In region A, there are two singularities and a finite jump discontinuity. In region B, wf(y) is continuous. In C and D, there are singularities at the region boundaries. This figure shows that the density functions possess significantly different attributes from those usually considered in statistical signal processing. To compare the current approach with that usually taken in discussions of “random variable theory,” we point out that a “random variable,” usually denoted X(t) [instead of our f(t)] is nothing more or less than a Lebesgue measurable function. In many applications of random variable theory, X(t) is everywhere continuous but nowhere differentiable (e.g., the Brownian motion), and various means are devised to estimate its probability density function. In the current investigation, it is not necessary to estimate wf(y) by these means since we may calculate it from f(t). However, as f(t) is assumed to be Lebesgue measurable, it is, in the strictly formal sense, also a random variable, and wf(y) is its probability density function. As we are investigating a subset of random variables, where it happens that the probability density function may be calculated from the random variable itself, we will carefully refrain from using this term, since it will only raise associations with the reader’s mind that are misleading in the current context.
Figure 1.
Plot of a typical density function wf(y) employed in our study. Inset shows a time-domain waveform f(t) with five critical points (left) and their relationship to the singularities of the associated density function wf(y) (right).
The mathematical characteristics of the singularities of wf(y) are important in order to guarantee the existence of the following integral on which we base our analysis of signals in this study:
| (6) |
which is known as the Renyi entropy.6 It is similar to the partition function in statistical mechanics with the parameter r playing the role of an “artificial” reciprocal temperature1, 7 (unrelated to the actual physical temperature in the scattering region); moreover, If(r)→−Hf, as r→1, using L’Hôpital’s rule, so that If is a generalization of Hf as follows:
| (7) |
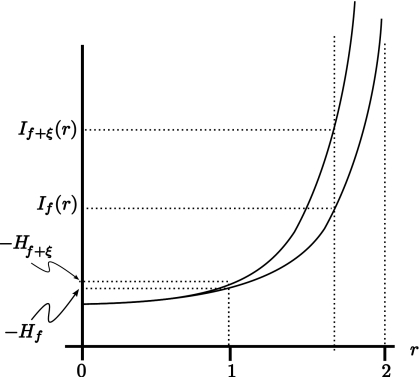
Previous studies have shown that this quantity can be more sensitive to subtle changes in scattering architecture than are more commonly used energy-based measures,2 with subsequent studies demonstrating further sensitivity improvements using If at the suitable value of r.1 For the density functions wf(y) encountered in our study, If(r) is undefined for r≥2, since as r→2−, the integral appearing in Eq. 6 will grow without bound due to the singularities in the density function wf(y) described in Eq. 5. The behavior as r→2 is dominated by contributions from these singularities, all of which correspond to critical points of f(t). This behavior is shown in Fig. 2. Moreover, as shown in the figure, it is possible that two slightly different functions, f(t) and f(t)+ξ(t), where ξ is small, may have entropies, Hf and Hf+ξ, that are close, as shown, but whose Renyi entropies, If(r) and If+ξ(r), diverge as r→2. Previous studies have shown that this can happen in practice.1 However, these results left open the possibility of further sensitivity gains. The purpose of the present study is to investigate the possibility of obtaining further sensitivity improvements by pushing toward this limit. As described in the Appendix0, the asymptotic form of If(r) as r→2 is given by
| (8) |
We will use this quantity to generate the images presented in the current study.
Figure 2.
Plots of If(r) and If+ξ(r) (left) showing that while If(1)=−Hf and If+ξ(1)=−Hf+ξ may be close, If(r) and If+ξ(r) diverge as r→2.
MATERIALS AND METHODS
Numerical computation of If,∞
Calculation of If,∞ via Eq. 8 is accomplished by fitting a cubic spline to the experimentally acquired data array using a well-known algorithm, which returns the second derivative of the cubic spline (in an array having the same length as the experimental data) and initializes data structures suitable for rapid computation of its first derivative.8 Subsequently, an array of corresponding first derivatives is computed and used to bracket the critical points of the spline (i.e., the zero crossings). Linear interpolation is then used to estimate the exact location of the bracketed zero crossings in order to obtain an algorithm suitable for real-time implementation in a medical imaging system. The total operation count is of order Nσ, where Nσ is the number of points processed, and is more than four orders of magnitude faster than the operation count 16 384Nσ required to compute If(r) used in our previous study.1 For comparison, we also note that the operation count required to produce the envelope of the same number of points (i.e., to produce a conventional B-mode image) would be of order Nσ log(Nσ) since computation of the envelope requires use of the fast Fourier transform; for the value of Nσ=512 used in our study below, this represents an increase in processing speed of roughly ninefold.
Simulations
The convergence properties, stability in the presence of noise, and effects of quantization error and sampling rate have been extensively evaluated using simulated data. Several types of waveforms have been investigated: Gaussian and parabolic waveforms, for which the exact value of If,∞ may be computed and linear combinations of exponentially damped sine waves that qualitatively resemble backscattered ultrasonic waveforms. Several carefully chosen example simulations illustrate guidelines for application of our algorithm in order to avoid potential artifacts produced by experimental factors.
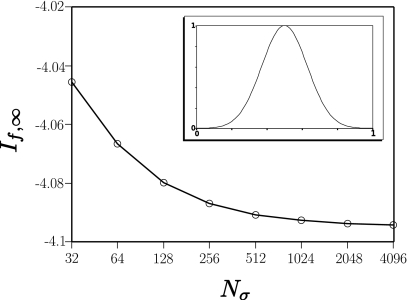
The first of these is Fig. 3, which shows a plot of If,∞ for a noise-free Gaussian pulse f(t)=e−30(t−0.5)2 for values of Nσ ranging from 32, 64, 128,…, 8192. Even at Nσ=32 the estimated value of If,∞ is within 1% of the exact value of log[1∕60]=−4.094. For moving window analysis of experimental data, Nσ is the length of the moving window. Choosing its length requires making trade-offs between sensitivity (smaller Nσ implying loss of sensitivity, but increased spatial resolution), noise level (smaller Nσ implying increased noise, but increased spatial resolution), and spatial resolution.
Figure 3.
Simulation for a noise-free Gaussian pulse showing the dependence of If,∞ on the number of sampled points Nσ.
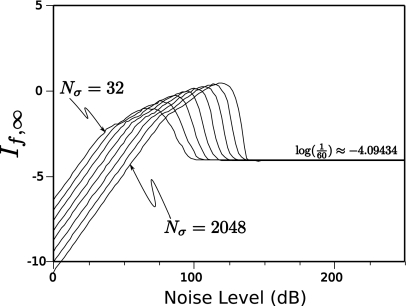
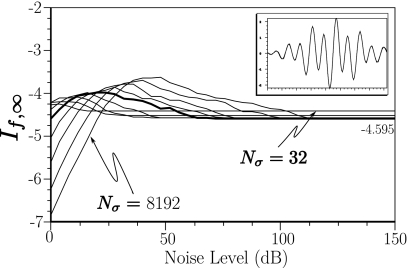
However, noise can have a significant effect on the calculation of If,∞. Figure 4 illustrates the impact of noise on the Gaussian pulse (f(t)=e−30(t−0.5)2) that was just discussed. As Nσ ranges from 32, 64, 128,…, 8192 and noise levels ranges from 0 to 150 dB, the calculated value of If,∞ can vary by over 100% of its actual value. Eventually, as Nσ increases and the noise level drops, our algorithm converges to a stable value. However, as the plots indicate, the noise requirements for a single peak function such as the Gaussian peak are quite stringent, being greater than 100 dB to obtain 10% accuracy.
Figure 4.
Simulation for a Gaussian pulse showing the dependence of If,∞ on the number of sampled points Nσ and noise level.
These requirements are less stringent if f(t) has several critical points. An example is shown in Fig. 5, which plots If,∞ for values of Nσ ranging from 32, 64, 128,…, 2048, and for noise levels ranging from 0 to 150 dB for the Gaussian modulated pulse f(t)=e−10(t−0.5)2 sin(20π(t−0.5))+0.7 sin(20π(t−0.5))+0.7 sin(10π(t−0.5)). As the plots indicate the noise requirements for a multipeak peak waveform f(t) are far less stringent with 87% accuracy being obtained at about 20 dB noise level for Nσ=512 (plotted using a heavier line in the plot family since these parameters match values used in the experimental portion of our study).
Figure 5.
Simulation for an unquantized Gaussian modulated pulse showing the dependence on the number of sampled points Nσ and noise level.
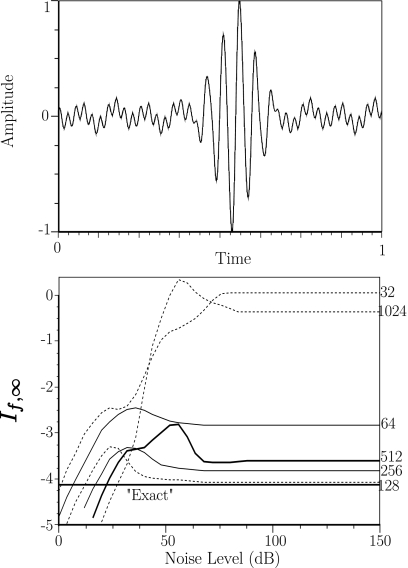
Figure 6 shows a plot of If,∞ for values of Nσ ranging from 32, 64, 128,…, 8192 for noise levels ranging from 0 to 150 dB for the simulated pulse f(t)=e−150(t−0.55)2 sin(40π(t−0.55))+0.7 sin(80π(t−0.55))+0.7 sin(20π(t−0.55))+0.03 sin(10π(t−0.55)). The “exact” answer is −4.149 073, found by running our algorithm with noise level set to zero, no quantization error, and Nσ=8192, and is also shown in the plot. The corresponding values of Nσ are indicated on the right side of the figure. For values of Nσ≤512 the error is less than 13%. We also note that for larger values of Nσ and lower levels of noise, our algorithm diverges with If,∞ becoming large and positive. This occurs only in quantized simulations and is the result of the long perfectly flat segments in the quantized data. This is an easily detected fault and, since the If,∞ images used in our experimental study had pixel values of approximately 7 bits∕symbol in magnitude on the regions used to estimate accumulation of targeted nanoparticles, can be ruled out as a possible artifact in our study.
Figure 6.
Top panel: the simulated backscatter signal described in the text. Bottom panel: plot showing the dependence of If,∞ on the number of sampled points Nσ and noise level at 8-bit quantization. The heavy black line labeled “Exact” is at 4.149, the limiting value of If,∞ obtained from our algorithm in the unquantized, noise-free case with Nσ=8192.
Nanoparticles for molecular imaging
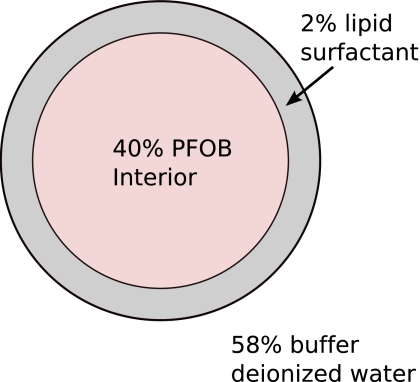
A cross-section of the spherical liquid nanoparticles used in our study is diagramed in Fig. 7. For in vivo imaging we formulated nanoparticles targeted to αvβ3-integrins of neovascularity in cancer by incorporating an “Arg-Gly-Asp” mimetic binding ligand into the lipid layer. Methods developed in our laboratories were used to prepare perfluorocarbon (perfluorooctylbromide, which remains in a liquid state at body temperature and at the acoustic pressures used in this study9) emulsions encapsulated by a lipid-surfactant monolayer.10, 11 The nominal sizes for each formulation were measured with a submicron particle analyzer (Malvern Zetasizer, Malvern Instruments). Particle diameter was measured at 200±30 nm.
Figure 7.
A cross-sectional diagram of the nanoparticles used in our study.
Animal model
The study was performed according to an approved animal protocol and in compliance with guidelines of the Washington University Institutional Animal Care and Use Committee.
The model used is the transgenic K14-HPV16 mouse in which the ears typically exhibit squamous metaplasia, a precancerous condition, associated with abundant neovasculature that expresses the αvβ3-integrin. Eight of these transgenic mice12, 13 were treated with 1.0 mg∕kg intravenous of either αvβ3-targeted nanoparticles (n=4) or untargeted nanoparticles (n=4) and imaged dynamically for 1 h using a research ultrasound imager (Vevo 660 40 MHz probe) modified to store digitized rf waveforms acquired at 0, 15, 30, and 60 min after injection of nanoparticles. In both targeted and untargeted cases, the mouse was placed on a heated platform maintained at 37 °C, and anesthesia was administered continuously with isoflurane gas (0.5%).
Ultrasonic data acquisition
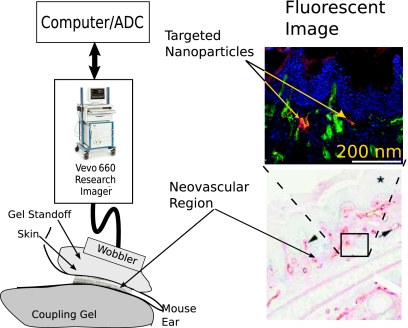
A diagram of our apparatus is shown in Fig. 8. rf data were acquired with a research ultrasound system (Vevo 660, Visualsonics, Toronto, Canada), with an analog port and a sync port to permit digitization. The tumor was imaged with a 40 MHz single element “wobbler” probe and the rf data corresponding to single frames were stored on a hard disk for later off-line analysis. The frames (acquired at a rate of 40 Hz) consisted of 384 lines of 4096 8-bit words acquired at a sampling rate of 500 MHz using a Gage CS82G digitizer card (connected to the analog-out and sync ports of the Vevo) in a controller PC. Each frame corresponds spatially to a region 0.8 cm wide and 0.3 cm deep.
Figure 8.
A diagram of the apparatus used to acquire rf data backscattered from K14-HPV16 transgenic mouse ears in vivo together with a histologically stained section of the ear indicating portions where αvβ3-targeted nanoparticles could adhere and a fluorescent image demonstrating presence of targeted nanoparticles.
The wobbler transducer used in this study is highly focused (3 mm in diameter) with a focal length of 6 mm and a theoretical spot size of 80×1100 μm (lateral beam width×depth of field at −6 dB), so that the imager is most sensitive to changes occurring in the region swept out by the focal zone as the transducer is “wobbled.” Accordingly, a gel standoff was used, as shown in Fig. 8, so that this region would contain the mouse ear.
A close-up view showing the placement of transducer, gel standoff, and mouse ear is shown in the bottom of the figure. Superposed on the diagram is a B-mode gray scale image (i.e., logarithm of the analytic signal magnitude). Labels indicate the location of skin (top of image insert), the structural cartilage in the middle of the ear, and a short distance below this, the echo from the skin at the bottom of the ear. Directly above this is an image of a histological specimen extracted from a K14-HPV16 transgenic mouse model that has been magnified 20 times to permit better assessment of the thickness and architecture of the sites where αvβ3-targeted nanoparticle might attach (red by β3 staining). Skin and tumor are both visible in the image. On either side of the cartilage (center band in image), extending to the dermal-epidermal junction, is the stroma. It is filled with neoangiogenic microvessels. These microvessels are also decorated with αvβ3 nanoparticles as indicated by the fluorescent image (labeled, in the upper right of the figure) of a bisected ear from an αvβ3-injected K14-HPV16 transgenic mouse. It is in this region that the αvβ3-targeted nanoparticles are expected to accumulate, as indicated by the presence of red β3 stain in the magnified image of a histological specimen also shown in the image.
Ultrasonic data processing
Each of the 384 rf lines in the data was first up-sampled from 4096 to 8192 points, using a cubic spline fit to the original data set in order to improve the stability of the thermodynamic receiver algorithms. A by-product of this “order Nσ” algorithm is simultaneous output of a corresponding array of second derivative values of the fit function.8 Next, a moving window analysis was performed on the second derivative data set, using Eq. 8 to compute If,∞, by moving a rectangular window (512 points long, 0.512 μs) in 0.064 μs steps (64 points), resulting in 121 window positions within the output data set. This produced an image for each time point in the experiment. The window length was chosen to match that used in previous studies;1, 2 it corresponds to the heavy black curve shown in Fig. 6. Analyses were also performed using window lengths of 256 (0.256 μs) and 128 points (0.256 μs). While they also produced statistically significant changes in If,∞ versus time, post-injection, the resulting If,∞ versus time curves were noisier, and required 1 h, post-injection, to exhibit statistically significant changes. As discussed previously, the optimum choice of window length requires trade-offs between sensitivity, noise level, and spatial resolution. In Sec. 4 we discuss the 512 point moving window length results since they correspond most closely to previous results, which were supported by independent histological results,1, 2 and produced images with sufficient spatial resolution to indentify relevant anatomical features in the mouse ear. Additionally, a major goal of this study was to assess the numerical stability of the algorithm, which is based on the second derivative of an experimentally measured data set, and thus contaminated by noise. Ordinarily, estimation of just the first derivative is difficult. However, in our application, the effects of noise might be mitigated by two factors: The second derivative is obtained from a global fit to the data, and it appears in the denominator of the expression for receiver output so that values having large error are likely to make small contributions to the sum appearing in Eq. 8.
Image processing
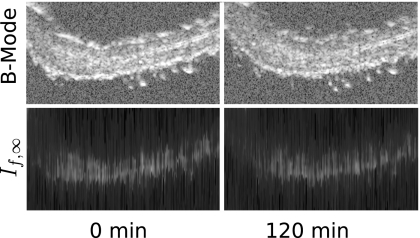
All rf data were processed off-line to reconstruct If,∞ images. Total analysis time using the new algorithm was less than 5 min on an eight core desktop computer [compared to the roughly week-long time required to execute the If(1.99) analysis on a cluster of just over 20 computers that was reported previously1]. A representative set of these images is shown in the bottom row of Fig. 9. For comparison, the top row shows conventional B-mode images, i.e., logarithm of the signal envelope. The left columns show images constructed from the rf data sets acquired 0 min after injection while the right column shows the images constructed from data acquired 120 min post-injection. The look-up-table of the entropy images have been inverted to produce a display in which pixels corresponding to tissue are brighter than surrounding pixels, in order to facilitate comparison with the conventional images. As expected, the conventional images exhibit higher spatial resolution compared with the If,∞ images, which employ a moving window in their construction. While the window is discernible in the entropy images, it is not a cause of major concern since our goal is automatic quantitative detection of changes in scattering architecture in the physical region represented by the image. As may be seen from the figure, the bright regions in If,∞ and conventional images are correlated. Moreover, as time passes from 0 to 120 min, both rows of images show a reduction in contrast between tissue of the ear (bright regions) and darker background (corresponding to the gel couplant). In the If,∞ images this corresponds to an increase in If,∞. As discussed below this effect is consistently observed in all If,∞ images from the group of K14-HPV16 transgenic mice injected with αvβ3-targeted nanoparticles, and it is not observed in If,∞ images from any of the control groups. As stated in previous publications, there are no statistically significant changes in signal energy images in any of the groups studied.2
Figure 9.
Top row: conventional B-mode images at 0 min (left), and after injection of αvβ3-targeted nanoparticles (right). Bottom row: corresponding If,∞ images.
Subsequently, a histogram of pixel values for the composite of the 0, 15, 30, and 60 min images was computed as described in previous papers.1, 2 Image segmentation of each type of image, at each time point in the experiment, was then performed automatically using its corresponding histogram according to the following threshold criterion: The lowest 7% of pixel values were classified as “targeted” tissue, while the remaining were classified as “untargeted” (histogram analysis was also performed using 90% and 87% thresholds, with 93% having the best statistical separation between time points). The mean value of pixels classified as targeted was computed at each time post-injection.
RESULTS AND DISCUSSION
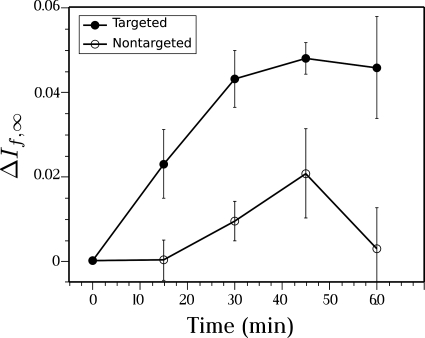
The results obtained after injection of targeted nanoparticles and nontargeted nanoparticles by If,∞ receiver are shown in Fig. 10. Both curves show the time evolution of the change (relative to 0 min) in mean value of receiver output in the enhanced regions of images obtained from the four animals in the targeted and the four animals in the nontargeted groups. Standard error bars are shown with each point. At 15 min the change in mean value of If,∞ is more than two standard errors from zero, implying statistical significance at the 95% level. There is no statistically significant change in image brightness for the nontargeted nanoparticles’ group. As the results show, the algorithm for computation of If,∞ is stable in the presence of experimental noise.
Figure 10.
If,∞ image enhancement, i.e., change relative to 0 min, obtained after injection of αvβ3-targeted nanoparticles (closed circles) and nontargeted nanoparticles (open circles) into four K14-HPV16 transgenic mice in each case.
The results presented in this paper extend earlier studies where it was shown that an entropy-based measure Hf was able to detect targeted nanoparticles in tumor neovasculature2 after 30 min of accumulation time. Subsequently, the time required to detect targeted nanoparticles was reduced to 15 min using a generalization of entropy If(r), with r=1.99, although the time required for signal analysis was greatly increased.1 In the current study based on If,∞, the analysis time has been reduced from days to minutes using an algorithm suitable for real-time implementation, while maintaining sensitivity that permits detection of nanoparticle accumulation at 15 min.
Real-time performance appears to have been purchased at the price of reduced statistical sensitivity, in view of prior observation that If(1.99) separated by over five standard errors from 0 at 15 min (Ref. 1) as compared to the two standard error separation obtained with the real-time receiver (see Fig. 10). It is possible that preprocessing of the data by bandpass filtering might improve the statistical performance of the algorithm without significant increase in computational overhead. This will be studied in a future report.
ACKNOWLEDGMENTS
This study was funded by NIH Grant Nos. EB002168, HL042950, and CO-27031, and NSF Grant No. DMS 0501079. The research was carried out at the Washington University Department of Mathematics and the School of Medicine.
APPENDIX: DERIVATION OF ASYMPTOTIC FORM
As described in Sec. 2, the limiting form of If(r) as r→2 will now be derived. The first step is to observe that the integral in Eq. 6 may split into two parts, one corresponding to the region where the function is clearly bounded and one corresponding to its singularities as shown in Fig. 11. Thus,
| (A1) |
where we have written Bk for the integral over the unshaded region between f(tk)+δk and f(tk+1)−δk+1 in Fig. 11. We observe that Bk is bounded as ϵ→0, while the integral appearing in Eq. A1 is not.
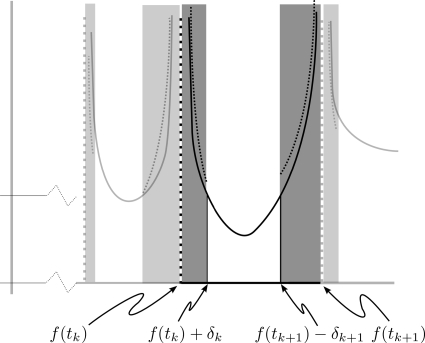
Figure 11.
An enlarged plot of a singularity of the density function wf(y)2−ϵ (solid curve) and Eq. 5 (dashed curves); quantities relevant for derivation of Eq. A12. As the shaded regions shrink the ratio between the dashed and solid curves approaches 1. The darker shading corresponds to the region discussed in the text.
Next, we consider the small interval of length δk near the singularity of wf(f(tk)) (shaded regions of Fig. 11). This is the singularity corresponding to the kth extrema of f(t): f(tk); also shown is the adjacent singularity corresponding to an extrema of f(t) at tk+1. The dashed lines in these regions represent the one over square root limiting form described in Eq. 5. By choosing δk small enough we may make the ratio of the solid and dashed curves arbitrarily close to 1. In other words, Eqs. 1, 5 tell us that in these shaded regions the following difference can be made as small as we like:
| (A2) |
where [assuming a minimum at f(tk), the argument for a maximum is similar]. Moreover, if a particular choice of δk yields the desired accuracy, i.e., makes the difference small enough, choosing a smaller value of δk will produce greater accuracy. Since the number of extrema in our time-domain function f(t) is finite, we pick the minimum δk, call it δ, yielding the desired accuracy in all of the shaded regions [i.e., at all singular points of wf(y)]. With this choice of δ Eq. A1 becomes
| (A3) |
and Eq. A2 becomes
| (A4) |
or
| (A5) |
where E>∣E(y)∣ may be chosen to be as small as we like by choosing small enough δ. As a result
| (A6) |
where, once again, may be made arbitrarily small, i.e., for every there exists some δ>0 such that for all y in between f(tk) and f(tk)+δ.
Combining Eqs. A3, A6 now yields
| (A7) |
The second integral above may be bounded by
| (A8) |
This inequality may be converted to an equality by replacing the factor with a smaller (positive) number. In general, this number will depend on the behavior of wf(y) near the singular point y=f(tk). For clarity, we denote this constant by . With this notation, Eq. A8 becomes
| (A9) |
where and hence may also be made as small as we wish by reducing δ. The common integral appearing in Eqs. A7, A8 may be computed as
| (A10) |
so that Eq. A7 becomes
| (A11) |
which we sum over all minima to obtain
| (A12) |
a sum of bounded and unbounded terms, whose unbounded term is computable directly from the experimentally accessible function f(t) using .
For the maximum we have the asymptotic term
| (A13) |
So that the contribution to Eq. 6 from all of the maxima becomes
| (A14) |
we now have a different expression for .
Adding the contributions for the maxima and minima we obtain
| (A15) |
Cross multiplying by ϵ
| (A16) |
taking the logarithm of both sides and letting ϵ→0 we have
| (A17) |
Now taking the limit δ→0 so that the we obtain
| (A18) |
This shows that as ϵ→0, the leading term in log∫wf(y)2−ϵdy always behaves like log 1∕ϵ, regardless of f(t); but the next term in the asymptotic expansion, the right-hand side of Eq. A18, does depend critically on f(t), and is the quantity we seek.
Multiplying both sides by 1∕(1−r)=1∕(1−2+ϵ)→−1 and then cancelling minus signs on both sides of the equation, we obtain
| (A19) |
For imaging applications, where offset removal and rescaling are typically performed when pixel values are assigned, we define the new quantity
| (A20) |
We will use this quantity to generate the images presented in Sec. 4.
References
- Hughes M. S., McCarthy J. E., Marsh J. N., Arbeit J. M., Neumann R. G., Fuhrhop R. W., Wallace K. D., Thomas T., Smith J., Agyem K., Znidersic D. R., Maurizi B. N., Baldwin S. L., Lanza G. M., and Wickline S. A., “Application of Renyi entropy to detect subtle changes in scattering architecture,” J. Acoust. Soc. Am. 125, 3141–3145 (2009). 10.1121/1.3097489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes M. S., McCarthy J. E., Marsh J. N., Arbeit J. M., Neumann R. G., Fuhrhop R. W., Wallace K. D., Znidersic D. R., Maurizi B. N., Baldwin S. L., Lanza G. M., and Wickline S. A., “Properties of an entropy-based signal receiver with an application to ultrasonic molecular imaging,” J. Acoust. Soc. Am. 121, 3542–3557 (2007). 10.1121/1.2722050 [DOI] [PubMed] [Google Scholar]
- Bucy R. S. and Joseph P. D., Filtering for Stochastic Processes With Applications to Guidance (Chelsea, New York, 1987). [Google Scholar]
- Wiener N., Extrapolation, Interpolation, and Smoothing of Stationary Time Series: With Engineering Applications (MIT, Cambridge, MA, 1949). [Google Scholar]
- Grenander U. and Rosenblatt M., Statistical Analysis of Stationary Time Series (Chelsea, New York, 1984). [Google Scholar]
- Cover T. M. and Thomas J. A., Elements of Information Theory (Wiley-Interscience, New York, 1991). [Google Scholar]
- Tolman R., The Principles of Statistical Mechanics (Dover, New York, 1979). [Google Scholar]
- Press W. H., Teukolsky S. A., Vetterling W. T., and Flannery B. P., Numerical Recipes in C, 2nd ed. (Cambridge University Press, Cambridge, 1992). [Google Scholar]
- Hughes M., Marsh J., Hall C., Fuhrhop R. W., Lacy E. K., Lanza G. M., and Wickline S. A., “Acoustic characterization in whole blood and plasma of site-targeted nanoparticle ultrasound contrast agent for molecular imaging,” J. Acoust. Soc. Am. 117, 964–72 (2005). 10.1121/1.1810251 [DOI] [PubMed] [Google Scholar]
- Flacke S., Fischer S., Scott M. J., Fuhrhop R. J., Allen J. S., McLean M., Winter P., Sicard G. A., Gaffney P. J., Wickline S. A., and Lanza G. M., “Novel MRI contrast agent for molecular imaging of fibrin implications for detecting vulnerable plaques,” Circulation 104, 1280–1285 (2001). 10.1161/hc3601.094303 [DOI] [PubMed] [Google Scholar]
- Lanza G. M., Wallace K. D., Scott M. J., Cacheris W. P., Abendschein D. R., Christy D. H., Sharkey A. M., Miller J. G., Gaffney P. J., and Wickline S. A., “A novel site-targeted ultrasonic contrast agent with broad biomedical application,” Circulation 94, 3334–3340 (1996). [DOI] [PubMed] [Google Scholar]
- Arbeit J. M., Riley R. R., Huey B., Porter C., Kelloff G., Lubet R., Ward J. M., and Pinkel D., “Chemoprevention of epidermal carcinogenesis in k14-hpv16 transgenic mice,” Cancer Res. 59, 3610–3620 (1999). [PubMed] [Google Scholar]
- Arbeit J. M., Mnger K., Howley P. M., and Hanahan D., “Progressive squamous epithelial neoplasia in k14-human papillomavirus type 16 transgenic mice,” J. Virol. 68, 4358–4368 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]