Abstract
Dicer, a ribonuclease essential for miRNA processing, is expressed abundantly in developing mouse cornea and lens. We studied the roles of Dicer and miRNAs in eye development by conditionally deleting the Dicer gene in the mouse lens and corneal epithelium. Adult Dicer conditional null (DicerCN) mice had severe microphthalmia with no discernible lens and a poorly stratified corneal epithelium. Targeted deletion of Dicer effectively inhibited miRNA processing in the developing lens at 12.5 day of embryogenesis (E12.5). Lens development initiated normally but underwent progressive dystrophy between E14.5 and E18.5. Microarray analysis revealed activation of P53 signaling in DicerCN lenses at E13.5, consistent with increased apoptosis and reduced cell proliferation between E12.5 and E14.5. Expression of Pax6 and other lens developmental transcription factors were not greatly affected between E12.5 and E14.5 but decreased as the lens degenerated. Our data indicated an indispensible role for Dicer and miRNAs in lens and corneal development.
Keywords: Dicer, microRNA, lens development
INTRODUCTION
The mammalian eye is a complex neurosensory organ. Development of a functional eye requires a series of interactions between different tissues (Lang, 2004; Cvekl and Duncan, 2007). Our interest has focused on the development of lens and cornea (Piatigorsky, 1998; Swamynathan et al., 2008). In brief, mouse eye development begins at 8.5 days of embryogenesis (E8.5) when the evaginating optic vesicle from the forebrain induces the lens placode which subsequently invaginates and separates from the surface ectoderm to form a lens vesicle. By E14.5, the anterior epithelial cells of the lens vesicle proliferate and form a cubical epithelium; the posterior cells of the lens vesicle stop dividing and differentiate into the primary non-nucleated fiber cells (Lang, 1999; Bhat, 2001). The ectodermal epithelial cells overlying the lens vesicle are destined to become the corneal epithelium (Zieske, 2004). At E12.5 the developing corneal epithelium becomes bilayered and neural crest cells start to migrate into the space between the developing lens and presumptive corneal epithelium, giving rise to the corneal endothelium and the stromal keratocytes. The corneal epithelium remains only 2–3 cell layers at postnatal day 10 (PN10). Stratification and maturation of the corneal epithelium coincides with eye opening two weeks after birth. By six weeks after birth, the corneal epithelium reaches 6–8 cell layers (Hay, 1979) and is composed of flattened cells at the surface and posterior columnar cells. At the molecular level, differentiation of the lens and cornea requires sequential activation of multiple transcription factors (e.g. Pax6, Prox1, and Pitx3 for lens development; Pax6, Hes1 and Klf4 for corneal development), as well as interactions between the transcription factors and signaling pathways from certain growth factors (e.g. Bmp and Fgf) (for reviews, see Ogino and Yasuda, 2000; Lang, 2004; Cvekl and Duncan, 2007).
microRNAs (miRNAs) are single-stranded, noncoding RNAs of 17–25 nucleotides. miRNA genes are transcribed by the RNA polymerase II (Pol II) as primary miRNAs (pri-miRNAs) (Lee et al., 2004). The pri-miRNAs are sequentially processed by two RNaseIII enzymes, Drosha and Dicer, to give rise to approximately 22 nucleotide-long double-stranded duplexes. The duplexes are incorporated into RISC (RNA-induced silencing complex), where the mature miRNA strands are directed to mRNA targets by sequence complementarity to the 3′-untranslated regions of the mRNAs (Lewis et al., 2005). miRNAs inhibit the expression of the mRNA targets in one of two ways, either by the cleavage and degradation of the mRNAs (Bagga et al., 2005; Lim et al., 2005), or inhibition of mRNA translation and expression (Gregory et al., 2005). Estimates indicate that miRNAs repress expression of more than 30% of the protein-coding genes at the posttranscriptional level in mammals. miRNAs can also lead to stimulation of mRNA translation (Pillai et al., 2005) and control of DNA transcription (Kim et al., 2008).
Hundreds of miRNA genes, mostly evolutionarily conserved, have been identified. Some are expressed in many tissues (cf let-7 family members), while others are expressed in a strict cell/tissue-specific pattern (cf muscle-specific miR-1 and -208) (Zhao et al., 2007). miRNA are key regulators in various aspects of tissue development and homeostasis (Kusenda et al., 2006; Zhao and Srivastava, 2007), including differentiation, apoptosis, proliferation, and the maintenance of cell and tissue identity during embryogenesis and adult life. miRNA expression has also been linked to disease. For example, aberrant expression of miR-21 has been associated with tumor initiation and progression (Shi et al., 2008). In particular, several miRNAs have been identified that are highly enriched in mouse cornea and lens (Ryan et al., 2006; Karali et al., 2007). Many of these corneal/lens miRNAs display spatial and temporal specificity, raising the question of how miRNAs function in cornea and lens development. Interestingly, a recent study has shown that suppression of lipid phosphatase SHIP2 expression by one of the corneal miRNAs, miR-205, can be antagonized by another corneal-enriched miRNA, miR-184 (Yu et al., 2008), suggesting complicated roles for miRNAs in regulation of ocular differentiation.
In the present investigation we investigated the general roles of miRNAs in corneal and lens development by focusing on Dicer, the essential ribonuclease for miRNA maturation. Our results reveal abundant expressions of Dicer and miRNAs during corneal and lens development. We created a conditional deletion of Dicer in the developing lens placode and presumptive corneal epithelium by using the Cre-LoxP technology (Ashery-Padan et al., 2000; Murchison et al., 2005). Our results reveal a critical role for Dicer in regulation of apoptosis and cell proliferation in the developing lens and cornea, and establish the importance of miRNAs expressed in the lens and cornea for proper development on the whole eye.
RESULTS
Dicer and miRNA Expression in the Developing Lens and Cornea
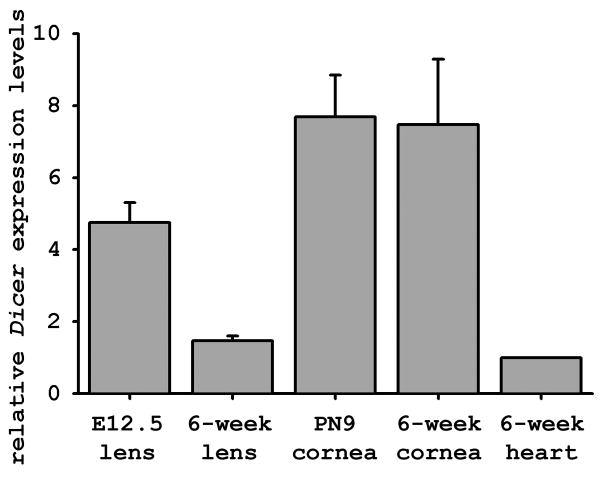
We first examined Dicer and miRNA expression during cornea and lens development in the mouse. As lens morphogenesis mainly occurs during embryonic development while cornea maturation is not complete until six weeks after birth, we examined Dicer expression in the developing mouse lens at E12.5 and developing mouse cornea at postnatal day (PN) 9; we also examined the expression in the mature lens and cornea of a 6-week-old wild type mouse. qRT-PCR analysis revealed that Dicer expression in the PN9 cornea was comparable to that in the mature cornea, which was 7 times higher than that in the mature heart (Chen et al., 2008; da Costa Martins et al., 2008) (Figure 1). By contrast, Dicer expression in the developing lens at E12.5 was about 3 times higher than in the mature lens of 6-week-old mice; Dicer expression in the mature lens was comparable to that in the mature heart. The decreased expression of Dicer transcripts in the mature lens reflects the non-nucleated, transcriptionally silent state of a majority of the mature lens fiber cells (Zelenka, 2004; Lovicu and McAvoy, 2005). In conclusion, the qRT-PCR assay demonstrated abundant Dicer expression in both the developing lenses and corneas.
Figure 1.
Expressions of Dicer during cornea and lens development of a wild type mouse. qRT-PCR was used to quantify Dicer mRNA levels in lenses and corneas at various developmental stages relative to the Dicer expression level in the 6-weeks-old mouse heart, which was arbitrarily set to 1. Data shown were obtained from three isolations of total RNA analyzed in triplicate. Error bars represent the standard deviation between experiments.
We next determined miRNA profiles in the wild type mouse cornea at PN9 before stratification of the corneal epithelium and at 6 weeks of age when the epithelium is fully stratified. Using Sanger miRBase Version 10.0, we analyzed 568 miRNAs across 4 biological replicates of the developing (PN9) and mature (6-week-old) mouse cornea. The microarray data have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE16209 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE16396). Abundant miRNAs were expressed in both the PN9 cornea and the mature 6-week-old cornea. Table 1A lists 10 miRNAs that gave the strongest hybridization signal in the 6-week mature cornea. In particular, miR-184 was highly enriched in both the developing and mature corneas and gave the highest hybridization signal of all the corneal miRNAs at both developmental stages. While Dicer expression levels were similar in the PN9 and 6-week-old cornea, the expression profiles of miRNAs were very different between the two developmental stages. 78 of the 568 (~13.7%) miRNAs showed differential expression between the PN9 and 6-week-old mouse corneas, with P values being <0.05. In particular, members of the miR-29 family, miR-29b and miR-29c, were up-regulated by more than 80-fold in the 6-week-old cornea. Table 1B lists the miRNAs showing the highest fold change between the two developmental stages. Abundant Dicer expression in the developing lens and cornea, as well as a dynamic change in miRNA expression during corneal maturation, suggests a critical role for Dicer and miRNAs in lens and cornea development.
Table 1.
miRNA expression profiles in mouse corneal development
| Table 1A. Most abundant miRNAs in 6-week-old mouse cornea | Table 1B. Differential expressed miRNAs between PN9 and 6-wk cornea |
|
|---|---|---|
| miRNA | fold change (6wk:PN9)1 | |
| mmu-miR-184 | mmu-miR-29b | 471.01 |
| mmu-miR-205 | mmu-miR-29c | 81.56 |
| mmu-miR-26a | mmu-miR-31* | 61.26 |
| mmu-miR-125b-5p | mmu-miR-141 | 38.29 |
| mmu-miR-23b | mmu-miR-31 | 36.85 |
| mmu-miR-23a | mmu-miR-434-3p | −32.10 |
| mmu-miR-709 | mmu-miR-101a | 31.58 |
| mmu-let-7a | mmu-miR-96 | 31.37 |
| mmu-miR-204 | mmu-miR-29a | 25.64 |
| mmu-let-7f | mmu-miR-382 | −22.22 |
fold change is base on expression in 6-wk cornea relative to that in PN9, with negative values indicating expression decrease in 6-wk cornea compared to PN9 cornea.
Conditional Knockout of Dicer in the Lens/Cornea Leads to Microphthalmia
To explore the role of Dicer and miRNAs in cornea and lens development, we generated a cornea/lens-specific deletion of the Dicer gene. We used an established DicerloxP/loxP mouse line in which the essential exons encoding the two RNaseIII domains are flanked by loxP sites (Murchison et al., 2005). DicerloxP/loxP mice were crossed with Le-Cre transgenic mice, where Cre recombinase and green fluorescent protein (GFP) are expressed via the Pax6 ectoderm enhancer (EE). Le-Cre is able to drive efficient deletion of target genes at the surface ectoderm-derived lens and cornea as early as E9.5 (Ashery-Padan et al., 2000). The resultant Dicer conditional null(DicerCN) mice were viable and fertile. However, as the Le-Cre also directs Dicer deletion in the pancreas (Ashery-Padan et al., 2000), the DicerCN mice have a short life span (less than 6 months), possibly due to low levels of blood insulin (0.1±0.05 ng/ml vs. 0.49±0.22 ng/ml in controls) and high levels of blood glucose (986±138 mg/dl vs. 151±15 mg/dl in controls).
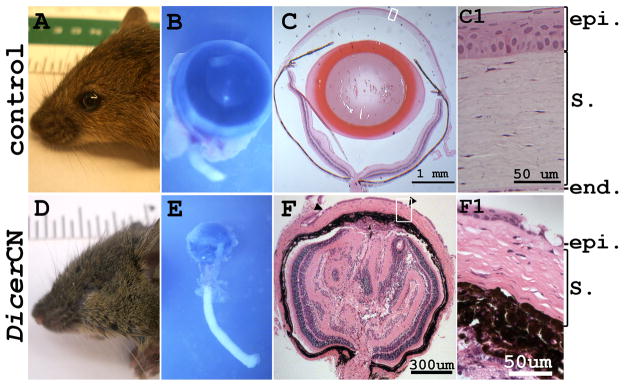
6-month-old DicerloxP/loxP mice without the Cre gene showed normal ocular morphology (Figure 2A) as did mice harboring the Cre transgene alone (Le-Cre; Dicer+/+) or Cre with the heterozygous floxed Dicer (Le-Cre; Dicer+/loxP) (data not shown). However, age-matched Le-Cre; DicerloxP/LoxP mice (DicerCN) were microphthalmic (Figure 2D). The eyelids were closed (not fused) and covered the entire eye. The beneath eyeballs were less than one tenth the size of the wild type control mice (Figure 2E). To study the morphology of the microphthalmic eyeballs, serial sagittal sections were cut through the whole eye. Figure 2F showed that a representative mid-plane section of DicerCN eyes without lens. The corneal epithelium was thinner and less stratified in the DicerCN mice (Figure 2F1), as compared to 6–8 layers of corneal epithelium in the age-matched controls (Figure 2C1). The basal layer of the DicerCN corneal epithelium appeared flattened in contrast to the columnar epithelium of the control cornea and large vacuoles were detected in the corneal epithelium of the DicerCN mice (arrowheads in Figure 2F). Although Dicer was deleted only in the developing corneal and lens epithelia, defects were observed throughout other ocular tissues. For example, the corneal stroma was thinner and the keratinocytes in the stroma had irregularly round- or star- shaped appearance (Figure 2F1). The iris and ciliary body were poorly developed. The iris without pupil opening is adherent to the cornea, the anterior chamber angle was absent, and the anterior chamber was flat. The inner retina was folded and disorganized. The sclera was poorly identifiable. Similar microphthalmic phenotypes have been observed in the PN3 and 3-month-old DicerCN mice (data not shown).
Figure 2.
Targeted deletion of Dicer in lens and cornea caused microphthalmia in DicerCN adult mice. The eyeballs were enucleated from a 6-month-old DicerCN mouse (B) and its age-matched wild type control (A). A typical mid-plane cross-section (C and F) showed lack of lens (F) in the conditional knockout. Close investigation of the eye structure showed that the corneal epithelium (F1) in the DicerCN mice was stratified into 3–4 layers while the epithelium in the control mice was stratified into 6–8 layers (C1). Abbreviations: epi: corneal epithelium; S.: corneal stroma; end: corneal endothelium.
Eye Development in DicerCN Mice
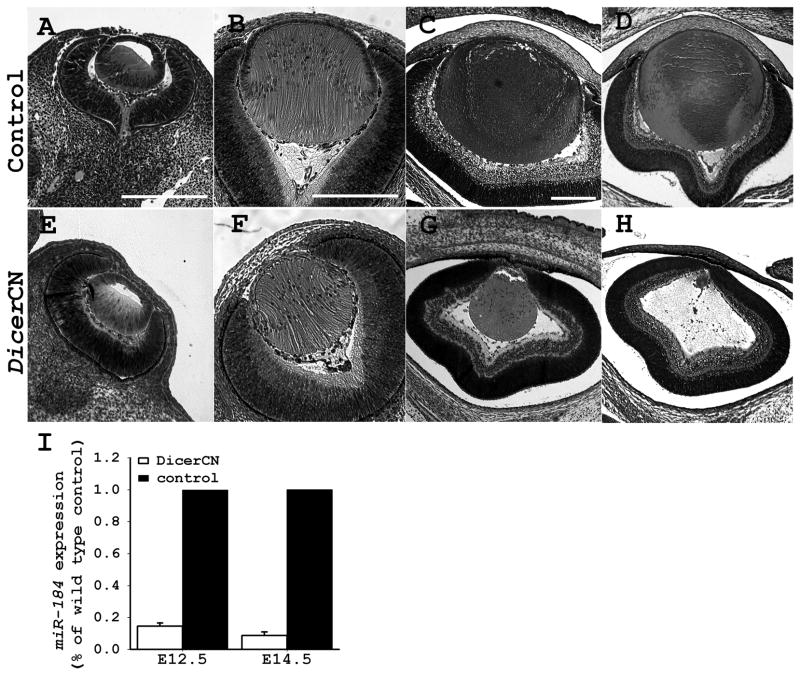
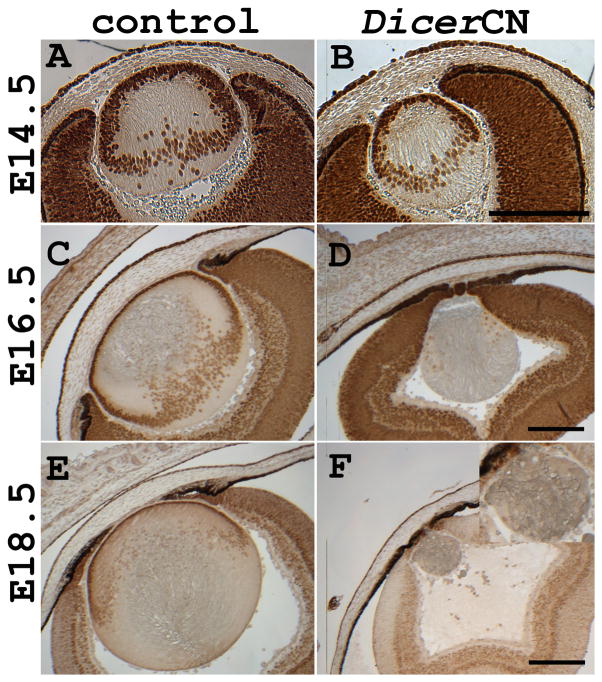
We next investigated whether the aphakia (lack of lens) of the adult DicerCN mice resulted from the failure to initiate lens specification, or from progressive lens dystrophy at later development. Inspection of the embryo revealed that the structure and morphology of the developing eye appeared normal at E12.5 in the DicerCN mice (Figure 3E). The lens vesicle pinched off from the surface ectoderm and the primary fiber cells started to form normally. The elongating lens primary fiber cells filled the lumen of the lens vesicle at E14.5 (Figure 3F). A distinct lens equatorial region was formed in DicerCN mice by E14.5, but the DicerCN lens appeared slightly smaller relative to the wild type lens and the lens epithelium contained fewer cells compared to the controls. There were no detectable histological abnormalities in cornea and retina at E14.5 in the DicerCN mice.
Figure 3.
Comparison of lens morphogenesis between DicerCN and control mice. (A–D): development of control lenses. (E–H): development of DicerCN lenses. (A and E): lenses at E12.5; (B and F): lenses at E14.5; (C and G): lenses at E16.5; (D and H): lenses at E18.5. Scale bar=200 um. (I). qRT-PCR analysis of the developing lenses showed that expression of miR-184 was significantly reduced in the DicerCN lenses at E12.5 and E14.5. Data shown were obtained from three isolations of total RNA analyzed in triplicate with each primer pair. Error bars represent the standard deviation between experiments.
By E16.5 the lens fiber cells were prominently dystrophic and the lens epithelium was not visible in the DicerCN mice (Figure 3G). Lens dystrophy continued at E18.5 (Figure 3H), leading to aphakia in the newborns (data not shown). The retinal pigmented epithelium at E18.5 extended to the anterior region of the eye and there was no discernible differentiation of ciliary body and iris (Figure 3H). In the control wild type embryos, protrusions and migration of the primitive eyelids proceeded normally and by E18.5, the eyes were completely covered with fused eyelids (data not shown). In contrast, migration of the primitive eyelids halted by E18.5 in DicerCN mice (Figure 3H) and eyelid fusion was never observed in the mutants during embryonic development (data not shown). Although Cre simultaneously deletes Dicer expression in the developing lens and cornea epithelium (Ashery-Padan et al., 2000), the morphology and integrity of the corneal epithelium appeared normal by E18.5, in contrast to the marked dystrophy of the lens at the same age.
We assessed Dicer inactivation and the consequent inhibition of miRNA maturation in the DicerCN mice during embryonic development. We performed Taqman® microRNA real-time PCR assays, which only detect the levels of mature miRNAs (Chen et al., 2005). Compared to the expression in controls, miR-184, a cornea/lens-specific miRNA (Ryan et al., 2006; Karali et al., 2007), was reduced by 85% in the DicerCN lens vesicles at E12.5, and more than 90% at E14.5 (Figure 3I). Significant reductions were also observed in expressions of other mature miRNAs as well as expression of functional Dicer transcripts at E12.5 (data not shown). Le-Cre transgenic mice start to express Cre recombinase in the prospective lens cells and corneal epithelial cells as early as E9.5 (Ashery-Padan et al., 2000; Miller et al., 2006). Therefore, the residual amount of functional miRNAs and Dicer mRNAs at E12.5 in the DicerCN lenses suggests a short delay between activation of Cre recombinase and effective ablations of all functional Dicer and mature miRNAs, a phenomenon that has also been observed in other tissues and cells where Dicer was inactivated (Andl et al., 2006; O’Rourke et al., 2007). This delay may arise from the long half-life of Dicer mRNA and/or miRNAs; alternatively, it may have resulted from incomplete Cre-mediated recombination.
Dicer Deletion Altered the Expression Profile of Protein-Coding Genes in Developing Lenses
In order to investigate the mechanism for the progressive lens dystrophy in DicerCN eyes, we compared expression profiles of protein-coding genes in wild type and DicerCN lenses at E13.5, at a time before gross morphological changes had occurred. Using the Affymetrix mouse gene 1.0 ST array, we analyzed gene expressions across five biological replicates of the wild type and mutant lenses at E13.5. The microarray data have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE16209 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE16209). Of the 28,853 identified transcripts, 1165 showed differential expression (accumulated FDR<0.05) between the wild type and the DicerCN lenses. 315 transcripts were up-regulated while 850 were down-regulated in the DicerCN lenses relative to the control lenses. Table 2 lists the top 10 most differentially expressed transcripts between the wild type and DicerCN lenses. Some of these differentially expressed genes may be direct targets for miRNAs, while expressions of others may be indirectly affected by the loss of miRNA expression. Using the terms from gene ontology, we studied the common characteristics among the differential expressed genes. Various subunits of the ribosomal complex (20 transcripts) were consistently down-regulated in the DicerCN lenses (data not shown), suggesting severely impaired machinery for protein biosynthesis in the absence of Dicer and miRNAs.
Table 2.
The most differentially expressed genes between the DicerCN and the control lenses at E13.5
| Unigene | Gene Description | Fold1 | Gene function |
|---|---|---|---|
| Mm.6253 | Crygs (crystallin, gamma S) | −10.24 | lens development |
| Mm.45494 | Gcg (glucagon) | −7.52 | insulin secretion |
| Mm.95780 | Lrtm1 (leucine-rich repeats and transmembrane domains 1) | −5.11 | --- |
| Mm.4258 | Ogn (osteoglycin) | −4.21 | --- |
| Mm.127156 | Pros1 (protein S, alpha) | 3.47 | blood coagulation |
| Mm.200608 | Clu (clusterin) | −3.46 | cell death |
| Mm.392646 | Btg2 (B-cell translocation gene 2) | 3.30 | transcription |
| Mm.260988 | Slc7a11 (solute carrier family 7, member 11) | −3.29 | amino acid transportation |
| Mm.4159 | Thbs1 (thrombospondin 1) | 3.22 | angiogenesis |
| Mm.405761 | Igfbp5 (insulin-like growth factor binding protein 5) | −3.20 | cell growth |
fold change is based on expressions in the DicerCN lenses relative to that in the control lenses, with negative value indicating expression decrease in the DicerCN compared to the control lenses.
Amongst the differentially expressed genes, there is a significant enrichment for the genes involved in apoptosis and cell death (Figure S1). In particular, many genes implicated in P53 signaling pathways, including Tp53inp1, Tnfrsf10A, Serpine2, and Tp53 itself, were up-regulated by at least 1.5-fold in the DicerCN lenses at E13.5 compared to that in the controls (Table 3). Many genes involved in cell cycle progression were also differentially expressed (Table 4), suggesting a de-regulation of cell cycle in DicerCN lenses at E13.5. Severe disruption of cell cycle control may lead to cessation of cell growth and induction of apoptosis in the DicerCN lenses as shown with other knockout mouse models (Clarke et al., 1992; Jacks et al., 1992; Lee et al., 1992; Nagahama et al., 2001).
Table 3.
differentially expressed genes affiliated with P53 signaling pathway
| Unigene | Gene Description | Fold1 |
|---|---|---|
| Mm.4159 | Thbs1 (thrombospondin 1) | 3.22 |
| Mm.253819 | Pik3r3 | 2.92 |
| Mm.393018 | Tp53inp1 (p53 inducible nuclear protein 1) | 2.79 |
| Mm.195663 | Cdkn1A (p21, Cip1) | 2.62 |
| Mm.3093 | Serpine2 (PAI-1) | 2.18 |
| Mm.101369 | Pik3cg | 1.96 |
| Mm.193430 | Tnfrsf10A (TrailR1) | 1.95 |
| Mm.273049 | Ccnd1 (Cyclin D1) | 1.79 |
| Mm.275071 | Jun | 1.63 |
| Mm.222 | Tp53 (tumor protein p53) | 1.49 |
| Mm.235194 | Akt3 | 1.33 |
| Mm.220289 | Apaf1 | 1.30 |
fold change is based on expressions in DicerCN lenses relative to that in the control lenses.
Table 4.
differentially expressed genes affecting cell cycle progression
| Unigene | Gene Description | Fold1 |
|---|---|---|
| Mm.222178 | Prkca (protein kinase C, alpha) | −2.18 |
| Mm.2444 | Myc | −2.01 |
| Mm.35605 | Cdh1 (E-cadherin)2 | −1.73 |
| Mm.168789 | Cdkn1c (p57, Kip2) | −1.37 |
| Mm.307932 | E2F2 (E2F transcription factor 2) | 1.37 |
| Mm.289662 | Ddx3x (DEAD/H box polypeptide 3, X-linked) | 1.37 |
| Mm.222 | Tp53 (tumor protein p53) | 1.49 |
| Mm.275071 | Jun | 1.63 |
| Mm.2314 | Prkcd (protein kinase C, delta) | 1.74 |
| Mm.273049 | Ccnd1 (cyclin D1) | 1.79 |
| Mm.193099 | FN1 (fibronectin 1) | 1.87 |
| Mm.195663 | Cdkn1a (p21, Cip1) | 2.62 |
fold change is based on expressions in the DicerCN lenses relative to that in the control lenses, with negative value indicating expression decrease in the DicerCN compared to the control lenses.
grey fonts mark the genes inhibiting while black fonts mark genes promoting cell cycle progression.
Gene set enrichment analysis also showed that a significant portion of the differentially expressed genes was involved in tissue specification and organ development (Figure S1 and Table S3). In particular, genes involved in epithelial cell differentiation, including Fzd1 and Cdh1, were differentially expressed in the DicerCN lenses. A few genes important for lens specification were down-regulated in the DicerCN lenses (Table 5). Lens crystallin genes, including βA2-, βA4-, γN-, and γS-crystallins, were down-regulated in the DicerCN lenses, with the expression of the lens-specific γS-crystallin (Sinha et al., 1998) down-regulated by more than 10-fold. It is noteworthy, however, that unlike β- and γ-crystallin gene expression, α-crystallin gene expression was not affected by the targeted deletion of Dicer. Also down-regulated in the DicerCN lenses were Fgfr2 and Cdh1 (E-Cadherin). Targeted deletion of these two genes in the lens is associated with impaired lens development and microphthalmia in adult eyes (Garcia et al., 2005; Pontoriero et al., 2008b).
Table 5.
Differential Expressed Genes Associated with Lens Development
| Unigene | Gene Description | Fold1 |
|---|---|---|
| Mm.6253 | Crygs (crystallin, gamma S) | −10.24 |
| Mm.63484 | Crygn (crystallin, gamma N) | −2.08 |
| Mm.35605 | Cdh1 (E-cadherin) | −1.73 |
| Mm.86656 | Cryba2 (crystallin, beta A2) | −1.7 |
| Mm.475710 | Fktn (fukutin) | −1.66 |
| Mm.16340 | Fgfr2 (fibroblast growth factor receptor 2) | −1.57 |
| Mm.1090 | Gpx1 (glutathione peroxidase 1) | −1.52 |
| Mm.221403 | Pdgfra | −1.5 |
| Mm.18213 | Tgfb2 (transforming growth factor, beta 2) | −1.42 |
| Mm.168789 | Cdkn1C (p57, Kip2) | −1.37 |
| Mm.40324 | Cryba4 (crystallin, beta A4) | −1.31 |
| Mm.272321 | Cited2 | −1.31 |
| Mm.268003 | Insr (insulin receptor) | 1.41 |
negative values indicate expression decrease in the DicerCN compared to the control lenses.
Decreased Cell Proliferation and Increased Cell Death in DicerCN Lens and Cornea
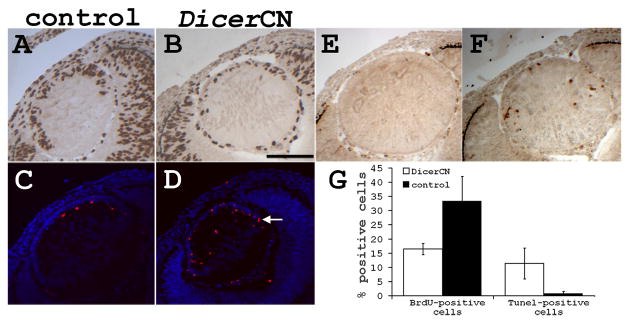
Inasmuch as the microarray data showed activation of apoptosis signaling pathway along with inhibition of cell growth pathway in the DicerCN lenses at E13.5, we investigated whether lens dystrophy in DicerCN mice was a result of aberrant cell proliferation and/or cell death. Proliferation of the lens epithelia was significantly reduced as early as E12.5 (Figure 4B). The BrdU labeling index of the control lens was about 33%, whereas 16% of the DicerCN lens cells were BrdU-positive (P=0.02; Figure 4G). By E14.5, lens proliferation was almost abolished in the lens epithelia of the DicerCN mice as judged by the BrdU incorporation, while the lens epithelia of the controls mice remained proliferative at the same embryonic stages (data not shown).
Figure 4.
DicerCN lenses had increased apoptosis and reduced proliferation at E12.5. (A, C, and E): control lenses. (B, D, and F): DicerCN lenses. (A–B): BrdU staining. (C–D): TUNEL staining (red) imposed on nuclear DAPI staining (purple). (E–F): active caspase-3 staining (red). Scale bar=150 um. White arrow marks the equatorial region of a developing lens. (G) Quantification of BrdU-positive cells and TUNEL-positive nuclei in control and DicerCN littermate lenses. At least three sections from each of the two control and two DicerCN embryos were counted.
TUNEL-staining revealed increased apoptosis in the mutant lens early during embryonic development. DicerCN lenses consistently showed a substantial increase of TUNEL-positive cells already at E12.5 (Figure 4D). Although the number of the total nuclei observed was comparable (p>0.1) in the control (277±91) and DicerCN (257±60) lenses at E12.5, 11% of the DicerCN lens nuclei were TUNEL-positive nuclei while only 0.7% of the control lenses nuclei contained TUNEL-positive nuclei (p<0.01; Figure 4G). Many of the TUNEL-positive cells were located in the epithelium of the DicerCN lenses, as well as in the equatorial region (arrows in Figure 4D). Consistent with increased apoptosis, active caspase-3 was detected in the DicerCN lens as early as E12.5 (Figure 4F), but not in the wild type lens (Figure 4E). As a result of the increased apoptosis and decreased cell proliferation, the number of total nuclei in the DicerCN lenses (207±31) was significantly reduced (p<0.02) as compared to that in the control lenses (380±41) by E14.5. The percentage of the TUNEL-positive lens cells was also higher in the E14.5 DicerCN lenses (8%) than in the control lenses (0.4%). The continued increase in apoptosis and cessation of cell proliferation at E14.5 accounted for the prominent lens atrophy observed between E14.5 to E18.5 in the DicerCN embryos.
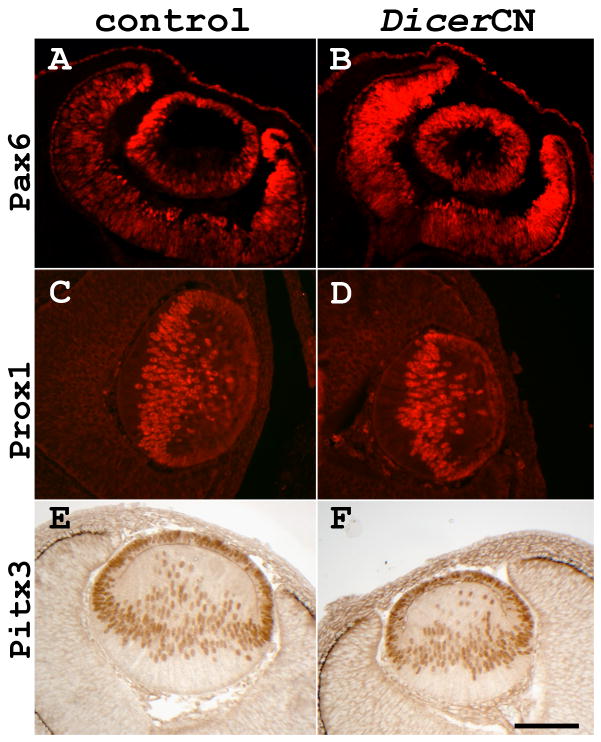
Expressions of Critical Developmental Transcription Factors in the DicerCN Mice
Microarray data suggested that mRNA levels of transcription factors critical for lens development, including Pax6, Prox1, Pitx3, and c-Maf, were not significantly changed in DicerCN lenses at E13.5 (data not shown). Also, based on immunostaining of the embryonic sections, Pax6, Prox1, and Pitx3 expressions were comparable in DicerCN and control lenses between E12.5 and E13.5 (Figure 5). There were overall fewer Pax6 positive cells in the E14.5 DicerCN lens (Figure 6B), consistent with the progression of apoptosis and lens dystrophy. Nonetheless, Pax6 staining in the remaining lens epithelial cells at E14.5 appeared similar to that in the wild type lens cells. Pax6 staining was gradually lost between E16.5 and E18.5 as the DicerCN lenses continued to degenerate (Figures 6D and 6F). In contrast to the loss of Pax6 expression in the lens by E18.5, Pax6 expression appeared similar in the corneal epithelium of the DicerCN and wild type mice at this stage.
Figure 5.
Immunostaining of critical transcription factors for lens specification. (A, C, and E): control lenses. (B, D, and F): DicerCN lenses. (A–B): anti-Pax6 staining at E12.5; (C–D): anti-Prox1 staining at E13.5; (E–F): anti-Pitx3 staining at E13.5. Scale bar=100 um
Figure 6.
Pax6 expression decreased at later developmental stages, coincident with lens dystrophy. Expression of Pax6 was examined using anti-Pax6 antibody at E14.5 (A–B), E16.5 (C–D), and E18.5 (E–F). (A, C, and E): control lenses; (B, D, and F): DicerCN lenses. Inset in Figure F shows the degenerative DicerCN lens at a higher magnification. Scale bar=200 um
DISCUSSION
The present results establish an indispensible role for Dicer in the development of the lens and cornea in the mouse. As Dicer primarily catalyzes miRNA maturation in mammalian species (Ambros et al., 2003; Lippman and Martienssen, 2004; Calabrese et al., 2007; Babiarz et al., 2008), our study implicates miRNAs in key aspects of lens and cornea development. Although Dicer was recently implicated in the biogenesis of endogenous siRNAs in mouse oocytes (Tam et al., 2008; Watanabe et al., 2008), sequencing of small RNA species from Dicer-null stem cells (Calabrese et al., 2007) has suggested that defects observed in Dicer deletion result from the loss of miRNAs rather than other small RNA species.
The progressive lens dystrophy in the DicerCN embryonic mice was accompanied by microphthalmia and supports the idea that the lens is a developmental organizer for the anterior segment of the eye (Coulombre, 1969; Beebe and Coats, 2000; Yamamoto and Jeffery, 2000; Kurita et al., 2003; Strickler et al., 2007). The severe microphthalmia in DicerCN mice resembled that in transgenic mice in which the lenses were ablated by lens-specific expression of a diphtheria toxin transgene (Zhang et al., 2007). The diphtheria toxin-induced microphthalmic eyes had a poorly differentiated ciliary body and iris, folded retina, and no significant anterior chamber. In contrast to the major abnormalities in the anterior regions of the DicerCN eyes, the retina of the DicerCN mice, although convoluted, was laminated and had differentiated cell layers in the newborns. We did observe, however, increased cell death in the DicerCN retina at E16.5 (data not shown) suggesting a role for lens on retinal cell survival, particularly the inner retina (Strickler et al., 2007).
The developing eyelids failed to migrate and fuse during embryogenesis, causing the defect of eye-open at birth (EOB) in the DicerCN mice. It has been shown that eyelid fusion has a protective role for the developing eye, particularly the cornea (Teramoto et al., 1988). Thus, failure of eyelid fusion may affect corneal maturation in the DicerCN mice. On the other hand, development of the corneal epithelium was less affected than that of the lens despite simultaneous deletion of the floxed Dicer in both tissues. The corneal epithelia of the adult DicerCN mice, however, were poorly stratified and displayed large vacuoles. At present, it is unclear whether the defect in stratification of the corneal epithelium is due to an autonomous role of Dicer and miRNA in corneal development or whether it is a secondary effect caused by lens dystrophy and/or eyelid defects.
Increased apoptosis in DicerCN lenses, along with decreased cell proliferation, resulted in massive lens dystrophy between E14.5 to E18.5. Microarray analysis of differential gene expression in the DicerCN lenses at E13.5 showed that many genes involved in the P53 pathway were up-regulated. Among them, Tp53inp1, Cdkn1a (p21Cip), Serpine2, and Tnfrsf10A are the key regulators of cell death and cell growth arrest (Okamura et al., 2001; Baetu and Hiscott, 2002; Jackson and Pereira-Smith, 2006; Kortlever and Bernards, 2006). Furthermore, phenotypes of the DicerCN lenses resembled the P53-dependent lens dystrophy in Creb2-deficient mice (Tanaka et al., 1998; Hettmann et al., 2000). In both cases, the initial stages of lens development including formation of the optic vesicle and primary lens fibers elongation occurred normally. However, at around E14.5 lens epithelial cells underwent massive apoptosis, causing aphakia in the newborns. Our data thus suggest that activation of the P53 signaling pathway may play an important role in lens dystrophy induced by Dicer and miRNA deletion. Indeed, recent studies have revealed involvement of multiple miRNAs in the P53 network, either as downstream effectors (He et al., 2007), or upstream regulators of P53 and/or its modifiers (Rane et al., 2009).
Lens development requires sequential activation of multiple transcription factors and growth factors signaling pathways. Our microarray and immunostaining studies suggest that, upon global removal of miRNAs, expression levels of lens transcription factors (Pax6, Prox1, Pitx3, Foxe3, Tcfap2a, and Sox2) known to direct lens development (Ogino and Yasuda, 2000; Cvekl and Duncan, 2007) were not significantly affected (either directly or indirectly) before lens dystrophy. The progressive lens dystrophy in DicerCN mice contrasts with the microphthalmic phenotypes caused by deletions of one of the aforementioned transcription factors. For example, defects in lens vesicle formation was detected as early as E10.5 when Pax6 (Ashery-Padan et al., 2000) was conditionally knocked out using the same Le-Cre transgenic line as used in the DicerCN mice. In addition, deletion of Foxe3 (Miller et al., 2006) or Tcfap2a (Miller et al., 2006; Pontoriero et al., 2008a) resulted in defective separation of the lens vesicle from the surface ectoderm between E9 and E10. Loss of Pax6 expression at later developmental stages was coincided with progressive lens dystrophy. It is possible that targeted Dicer deletion in the present study affected Pax6 expression at a later developmental stage due to relatively long half-lives of the involved mature miRNAs. Alternatively, our observations may suggest that loss of the critical transcription factors in the DicerCN lens was a consequence, rather than a cause, of lens dystrophy; and that regulation of lens development by miRNAs operates downstream from the sequential activation of lens-specific transcription factors.
Our microarray data raise the possibility that reduced expression of β- and γ-crystallin genes contributes to lens dystrophy in the DicerCN mice. γ-crystallins especially are lens-specific (Sinha et al., 1998) and γS-crystallin, which is highly enriched in lens fiber cells, has been associated with lens dystrophy and cataract (Sun et al., 2005). Fgfr2 expression is also down-regulated in the DicerCN lenses. FGF signaling has been implicated in lens induction (Faber et al., 2001) and the differentiation of lens fiber cells (Lang, 1999). Fgfr2 is required for lens fiber differentiation (Robinson, 2006). Fgfr2 conditional null lenses show increased apoptosis by E12.5, as do DicerCN lenses, and degenerate after birth (Garcia et al., 2005). The phenotypic similarities between the Dicer and Fgfr2 conditional knockout mice suggest that regulation of Fgfr2 levels in the developing lenses may be a pathway through which Dicer and miRNAs promote lens development.
Finally, although speculative, it may be of interest to consider the possibility that some miRNAs contribute to lens differentiation by suppressing expression of protein-coding genes that direct non-ocular cell fates. Itgb8 is an example of an up-regulated gene in DicerCN lenses that is implicated in vascular morphogenesis rather than lens differentiation (Zhu et al., 2002). Other up-regulated genes in DicerCN lenses involved in the development of various tissues are Eya1 (ear and kidney) (Xu et al., 1999), Klf-10 (bone morphogenesis) (Subramaniam et al., 2005; Hawse et al., 2008), Prkcd (immune tolerance) (Miyamoto et al., 2002) and Pik3cg (thymocyte development) (Sasaki et al., 2000). The functions of miR-133 in mesoderm development (Ivey et al., 2008) and of miR-196 in limb development (Hornstein et al., 2005) represent examples of this “fail-safe” mechanism of suppressing the expression of genes that are not appropriate for development of a specific tissue. Thus, we suggest that lens miRNAs may serve at a secondary level to limit the expression of genes that could interfere with proper lens development. Identification of miRNA target genes will be instrumental for further investigations of the roles for the many miRNAs that are dynamically expressed in the developing lens and cornea.
Experimental Procedures
Conditional disruption of Dicer
The Le-Cre mice in which the Pax6 P0 promoter drives the expression of Cre recombinase in the cornea, lens, and endocrine pancreas from E9.0 onward (Ashery-Padan et al., 2000) were mated to the mice that had LoxP sites inserted in the introns downstream of exon 21 and exon 23 of the Dicer gene (Murchison et al., 2005). The heterozygous progeny were crossed to obtain Le-Cre/−; DicerloxP/loxP (DicerCN) and DicerloxP/loxP (control)offsprings. Matings were set to ensure that only one copy of the transgene Cre was present, as homozygous Le-Cre can sometimes cause lens phenotype (Garcia et al., 2005). Genomic DNA isolated from tail clippings of these mice was assayed for the presence of the Dicer-LoxP (Murchison et al., 2005) and Le-Cre transgenes (Ashery-Padan et al., 2000) by PCR using specific primers. Mice studied here were on a mixed genetic background and maintained in accordance with the guidelines set forth by the Animal Care and Use Committee of the National Eye Institute, NIH.
Histology and Immunohistochemistry
Enucleated eyeballs or heads of the staged embryos were fixed in 10% buffered formalin, embedded in paraffin, and 5um sections were cut. For morphological studies, the slides were stained with hematoxylin and eosin (H&E; Poly Scientific, Bayshore, NY). Antibody staining was performed on deparaffinized sections after inactivation of the endogenous peroxidase activity with 0.3% H2O2 for 30 min. Antigen retrieval was performed in citrate buffer (pH 6.0) using a pressure cooker (Electron Microscopy Sciences, PA). For color development in immunohistochemistry, we used a biotinylated secondary antibody from the Vectastain Elite ABC Kit (Vector Laboratories, Burlingame, CA), followed by incubation with diaminobenzidine (DAB; Vector Laboratories). For immuno-detection in immunofluorescence, we applied on the sections Alexa-fluor-568-labeled anti-rabbit antibody (Invitrogen, Carlsbad, CA) following primary antibody incubation. For cell proliferation studies on the embryonic eyes, pregnant female mice were injected intraperitoneally with 100 μg 5-bromo-2′-deoxyuridine(BrdU) per gram of body weight and sacrificed 4–6 hours later. Heads of the staged embryos were fixed, embedded in paraffin and sectioned. Deparaffinized sections were incubated with a monoclonal anti-BrdU antibody, followed by a color reaction using a Vectastain Elite ABC kit as described above. Apoptosis was analyzed via in situ labeling of DNA fragments using an ApopTag Red In Situ kit (Millipore, Billirica, MA)according to the manufacturer’s instructions. Sections were then counterstained either with DAPI (4′, 6′-diamidino-2-phenylindole) using Prolong Gold anti-fade reagent (Invitrogen, Carlsbad, CA) or hematoxylin.
For the quantification of BrdU-positive cells and TUNEL-positive nuclei at E12.5 and E14.5 lenses, at least three sections from each of the two control and two DicerCN embryos were counted. Percentages of the positive cells were calculated as the number of BrdU (or TUNEL)-positive cells divided by total numbers of nuclei for each section.
The following antibodies were used in this study: rat anti-BrdU (1:200; Catalog #: OBT0030G, Accurate Chemical and Scientific, Westbury, NY); rabbit anti-Pax6 (1:5000; Catalog #: PRB-278P, Covance, Vienna, VA); rabbit anti-Prox1 (1:10000; Catalog #: PRB-238C, Chemicon, Temecula, CA); rabbit anti-active caspase-3 (1:200; Catalog #: 9661, Cell Signaling Technology, Danvers, MA); rabbit anti-Pitx3 (1:200; Catalog #: 38-2850, Invitrogen, Carlsbad, CA).
Isolation of total RNA, real time qRT-PCR, and microarray analyses
Intact lenses were dissected from five control and five DicerCN mice at E13.5 and immediately snap-frozen in liquid nitrogen. The pair of lenses from each mouse was combined for isolation of total RNAs so that five independent samples for each group (control versus DicerCN lenses) were submitted for gene expression analysis. Microarray analysis was performed at Expression Analysis (Durham, NC). After confirming the RNA quality with bioanalyzer, 100 ng total RNAs per sample were labeled and hybridized with mouse gene 1.0 ST arrays from Affymetrix (Santa Clara, CA). The microarray data have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE16209 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE16209). Expression profiling of the E13.5 control and DicerCN lenses was compared by a Permutation Analysis for Differential Expression (PADE). The False Discovery Rate (FDR) was calculated for each potential differentially expressed transcript. Calculation of fold change was based on the control group relative to the DicerCN group, with a negative value indicating expression reduction in the DicerCN group relative to the controls. Differentially expressed transcripts were selected according to the following criteria: (1) the accumulated FDR was less than 0.05 (maximal p-value for the selected transcripts was less than 0.01), and (2) the absolute fold change of the average expression signal from the two groups was greater than 1.3 between the control and DicerCN. Data were analyzed through the use of Ingenuity Pathways Analysis (Ingenuity® Systems, www.ingenuity.com, Ingenuity Systems Inc, Redwood City, CA).
Corneas were dissected from C57BL/6 mice sacrificed at PN9 and 6 weeks (adult) after birth. Total RNAs were isolated from ten 6-week and sixteen PN9 whole corneas. Cornea dissection and RNA extraction for both the 6-week and PN9 corneas were repeated four times to collect altogether eight RNA samples (four adult cornea RNAs and four PN9 cornea RNAs). The samples were sent to LC Sciences LLC (Houston, TX) for miRNA microarray assay. All probe sequences in the array were based on Sanger miRBase Release 10.0. The signals were presented after background subtraction, normalization and detection evaluation. Statistic significance was set by P-value < 0.05. The microarray data have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE16209 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE16396).
miRNA expression levels were examined using the TaqMan® MicroRNA Assay Kit (Applied Biosystems, Foster City, CA). The kit uses gene-specific stem-loop reverse transcription primers and TaqMan probes to detect only mature miRNA transcripts in a two-step qRT-PCR assay. Either U6 snRNA or 18S rRNA was used as an internal control for the normalization in the Taqman microRNA assay. There were no significant differences in the outcome when using different internal controls. Dicer mRNA levels were examined via SYBR Green real time qRT-PCR using either GAPDH or 18S rRNA as the internal control. The primer pair for Dicer used in the PCR step is as follows: 5′-AAG GGC AGA GCG CAA GTC AGT CA-3′ and 5′-ACA CAC GCC TCC TAC CAC TAC AAC AC-3′. The two primers reside at exon 22 and exon 23, respectively, which are flanked by the loxP sites in the DicerloxP/loxP mice (Murchison et al., 2005). Thus, conditional excision of the fragment between the loxP sites by the Cre recombinase removed the template for the primer pair in the Dicer mRNAs, and therefore no product is predicted in the qRT-PCR assays.
Supplementary Material
Acknowledgments
This work was supported by the intramural research program of NEI, NIH.
We are indebted to Dr. R. Ashery-Padan of Tel Aviv University for sharing Le-Cre transgenic mice with us and to Dr. G. Hannon of Cold Spring Harbor Laboratory for providing us with Dicer conditional knockout mice. We thank Dr. Oksana Gavrilova at National Institute of Diabetes and Digestive and Kidney Diseases for the assistance in testing the blood insulin and glucose levels. We thank the NEI Histology Core, particularly Dr. Chi-Chao Chen, for help in histological studies. We are grateful to the NEI Transgenic Animals and Genome Manipulation Section for their technical support; to Dr. Janine Davis for discussions and critical comments on the manuscript.
References
- Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr Biol. 2003;13:807–818. doi: 10.1016/s0960-9822(03)00287-2. [DOI] [PubMed] [Google Scholar]
- Andl T, Murchison EP, Liu F, Zhang Y, Yunta-Gonzalez M, Tobias JW, Andl CD, Seykora JT, Hannon GJ, Millar SE. The miRNA-processing enzyme dicer is essential for the morphogenesis and maintenance of hair follicles. Curr Biol. 2006;16:1041–1049. doi: 10.1016/j.cub.2006.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashery-Padan R, Marquardt T, Zhou X, Gruss P. Pax6 activity in the lens primordium is required for lens formation and for correct placement of a single retina in the eye. Genes Dev. 2000;14:2701–2711. doi: 10.1101/gad.184000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baetu TM, Hiscott J. On the TRAIL to apoptosis. Cytokine Growth Factor Rev. 2002;13:199–207. doi: 10.1016/s1359-6101(02)00006-0. [DOI] [PubMed] [Google Scholar]
- Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- Beebe DC, Coats JM. The lens organizes the anterior segment: specification of neural crest cell differentiation in the avian eye. Dev Biol. 2000;220:424–431. doi: 10.1006/dbio.2000.9638. [DOI] [PubMed] [Google Scholar]
- Bhat SP. The ocular lens epithelium. Biosci Rep. 2001;21:537–563. doi: 10.1023/a:1017952128502. [DOI] [PubMed] [Google Scholar]
- Calabrese JM, Seila AC, Yeo GW, Sharp PA. RNA sequence analysis defines Dicer’s role in mouse embryonic stem cells. Proc Natl Acad Sci U S A. 2007;104:18097–18102. doi: 10.1073/pnas.0709193104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JF, Murchison EP, Tang R, Callis TE, Tatsuguchi M, Deng Z, Rojas M, Hammond SM, Schneider MD, Selzman CH, Meissner G, Patterson C, Hannon GJ, Wang DZ. Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure. Proc Natl Acad Sci U S A. 2008;105:2111–2116. doi: 10.1073/pnas.0710228105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke AR, Maandag ER, van Roon M, van der Lugt NM, van der Valk M, Hooper ML, Berns A, te Riele H. Requirement for a functional Rb-1 gene in murine development. Nature. 1992;359:328–330. doi: 10.1038/359328a0. [DOI] [PubMed] [Google Scholar]
- Coulombre AJ. Regulation of ocular morphogenesis. Invest Ophthalmol. 1969;8:25–31. [PubMed] [Google Scholar]
- Cvekl A, Duncan MK. Genetic and epigenetic mechanisms of gene regulation during lens development. Prog Retin Eye Res. 2007;26:555–597. doi: 10.1016/j.preteyeres.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Costa Martins PA, Bourajjaj M, Gladka M, Kortland M, van Oort RJ, Pinto YM, Molkentin JD, De Windt LJ. Conditional dicer gene deletion in the postnatal myocardium provokes spontaneous cardiac remodeling. Circulation. 2008;118:1567–1576. doi: 10.1161/CIRCULATIONAHA.108.769984. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber SC, Dimanlig P, Makarenkova HP, Shirke S, Ko K, Lang RA. Fgf receptor signaling plays a role in lens induction. Development. 2001;128:4425–4438. doi: 10.1242/dev.128.22.4425. [DOI] [PubMed] [Google Scholar]
- Garcia CM, Yu K, Zhao H, Ashery-Padan R, Ornitz DM, Robinson ML, Beebe DC. Signaling through FGF receptor-2 is required for lens cell survival and for withdrawal from the cell cycle during lens fiber cell differentiation. Dev Dyn. 2005;233:516–527. doi: 10.1002/dvdy.20356. [DOI] [PubMed] [Google Scholar]
- Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- Hawse JR, Iwaniec UT, Bensamoun SF, Monroe DG, Peters KD, Ilharreborde B, Rajamannan NM, Oursler MJ, Turner RT, Spelsberg TC, Subramaniam M. TIEG-null mice display an osteopenic gender-specific phenotype. Bone. 2008;42:1025–1031. doi: 10.1016/j.bone.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay ED. Development of the vertebrate cornea. Int Rev Cytol. 1979;63:263–322. doi: 10.1016/s0074-7696(08)61760-x. [DOI] [PubMed] [Google Scholar]
- He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettmann T, Barton K, Leiden JM. Microphthalmia due to p53-mediated apoptosis of anterior lens epithelial cells in mice lacking the CREB-2 transcription factor. Dev Biol. 2000;222:110–123. doi: 10.1006/dbio.2000.9699. [DOI] [PubMed] [Google Scholar]
- Hornstein E, Mansfield JH, Yekta S, Hu JK, Harfe BD, McManus MT, Baskerville S, Bartel DP, Tabin CJ. The microRNA miR-196 acts upstream of Hoxb8 and Shh in limb development. Nature. 2005;438:671–674. doi: 10.1038/nature04138. [DOI] [PubMed] [Google Scholar]
- Ivey KN, Muth A, Arnold J, King FW, Yeh RF, Fish JE, Hsiao EC, Schwartz RJ, Conklin BR, Bernstein HS, Srivastava D. MicroRNA regulation of cell lineages in mouse and human embryonic stem cells. Cell Stem Cell. 2008;2:219–229. doi: 10.1016/j.stem.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T, Fazeli A, Schmitt EM, Bronson RT, Goodell MA, Weinberg RA. Effects of an Rb mutation in the mouse. Nature. 1992;359:295–300. doi: 10.1038/359295a0. [DOI] [PubMed] [Google Scholar]
- Jackson JG, Pereira-Smith OM. p53 is preferentially recruited to the promoters of growth arrest genes p21 and GADD45 during replicative senescence of normal human fibroblasts. Cancer Res. 2006;66:8356–8360. doi: 10.1158/0008-5472.CAN-06-1752. [DOI] [PubMed] [Google Scholar]
- Karali M, Peluso I, Marigo V, Banfi S. Identification and characterization of microRNAs expressed in the mouse eye. Invest Ophthalmol Vis Sci. 2007;48:509–515. doi: 10.1167/iovs.06-0866. [DOI] [PubMed] [Google Scholar]
- Kim DH, Saetrom P, Snove O, Jr, Rossi JJ. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci U S A. 2008;105:16230–16235. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortlever RM, Bernards R. Senescence, wound healing and cancer: the PAI-1 connection. Cell Cycle. 2006;5:2697–2703. doi: 10.4161/cc.5.23.3510. [DOI] [PubMed] [Google Scholar]
- Kurita R, Sagara H, Aoki Y, Link BA, Arai K, Watanabe S. Suppression of lens growth by alphaA-crystallin promoter-driven expression of diphtheria toxin results in disruption of retinal cell organization in zebrafish. Dev Biol. 2003;255:113–127. doi: 10.1016/s0012-1606(02)00079-9. [DOI] [PubMed] [Google Scholar]
- Kusenda B, Mraz M, Mayer J, Pospisilova S. MicroRNA biogenesis, functionality and cancer relevance. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2006;150:205–215. doi: 10.5507/bp.2006.029. [DOI] [PubMed] [Google Scholar]
- Lang RA. Which factors stimulate lens fiber cell differentiation in vivo? Invest Ophthalmol Vis Sci. 1999;40:3075–3078. [PubMed] [Google Scholar]
- Lang RA. Pathways regulating lens induction in the mouse. Int J Dev Biol. 2004;48:783–791. doi: 10.1387/ijdb.041903rl. [DOI] [PubMed] [Google Scholar]
- Lee EY, Chang CY, Hu N, Wang YC, Lai CC, Herrup K, Lee WH, Bradley A. Mice deficient for Rb are nonviable and show defects in neurogenesis and haematopoiesis. Nature. 1992;359:288–294. doi: 10.1038/359288a0. [DOI] [PubMed] [Google Scholar]
- Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. Embo J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Lippman Z, Martienssen R. The role of RNA interference in heterochromatic silencing. Nature. 2004;431:364–370. doi: 10.1038/nature02875. [DOI] [PubMed] [Google Scholar]
- Lovicu FJ, McAvoy JW. Growth factor regulation of lens development. Dev Biol. 2005;280:1–14. doi: 10.1016/j.ydbio.2005.01.020. [DOI] [PubMed] [Google Scholar]
- Miller LA, Smith AN, Taketo MM, Lang RA. Optic cup and facial patterning defects in ocular ectoderm beta-catenin gain-of-function mice. BMC Dev Biol. 2006;6:14. doi: 10.1186/1471-213X-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto A, Nakayama K, Imaki H, Hirose S, Jiang Y, Abe M, Tsukiyama T, Nagahama H, Ohno S, Hatakeyama S, Nakayama KI. Increased proliferation of B cells and auto-immunity in mice lacking protein kinase Cdelta. Nature. 2002;416:865–869. doi: 10.1038/416865a. [DOI] [PubMed] [Google Scholar]
- Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci U S A. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagahama H, Hatakeyama S, Nakayama K, Nagata M, Tomita K, Nakayama K. Spatial and temporal expression patterns of the cyclin-dependent kinase (CDK) inhibitors p27Kip1 and p57Kip2 during mouse development. Anat Embryol (Berl) 2001;203:77–87. doi: 10.1007/s004290000146. [DOI] [PubMed] [Google Scholar]
- O’Rourke JR, Georges SA, Seay HR, Tapscott SJ, McManus MT, Goldhamer DJ, Swanson MS, Harfe BD. Essential role for Dicer during skeletal muscle development. Dev Biol. 2007;311:359–368. doi: 10.1016/j.ydbio.2007.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino H, Yasuda K. Sequential activation of transcription factors in lens induction. Dev Growth Differ. 2000;42:437–448. doi: 10.1046/j.1440-169x.2000.00532.x. [DOI] [PubMed] [Google Scholar]
- Okamura S, Arakawa H, Tanaka T, Nakanishi H, Ng CC, Taya Y, Monden M, Nakamura Y. p53DINP1, a p53-inducible gene, regulates p53-dependent apoptosis. Mol Cell. 2001;8:85–94. doi: 10.1016/s1097-2765(01)00284-2. [DOI] [PubMed] [Google Scholar]
- Piatigorsky J. Gene sharing in lens and cornea: facts and implications. Prog Retin Eye Res. 1998;17:145–174. doi: 10.1016/s1350-9462(97)00004-9. [DOI] [PubMed] [Google Scholar]
- Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- Pontoriero GF, Deschamps P, Ashery-Padan R, Wong R, Yang Y, Zavadil J, Cvekl A, Sullivan S, Williams T, West-Mays JA. Cell autonomous roles for AP-2alpha in lens vesicle separation and maintenance of the lens epithelial cell phenotype. Dev Dyn. 2008a;237:602–617. doi: 10.1002/dvdy.21445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontoriero GF, Smith AN, Miller LA, Radice GL, West-Mays JA, Lang RA. Cooperative roles for E-cadherin and N-cadherin during lens vesicle separation and lens epithelial cell survival. Dev Biol. 2008b doi: 10.1016/j.ydbio.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rane S, He M, Sayed D, Vashistha H, Malhotra A, Sadoshima J, Vatner DE, Vatner SF, Abdellatif M. Downregulation of MiR-199a Derepresses Hypoxia-Inducible Factor-1{alpha} and Sirtuin 1 and Recapitulates Hypoxia Preconditioning in Cardiac Myocytes. Circ Res. 2009 doi: 10.1161/CIRCRESAHA.108.193102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson ML. An essential role for FGF receptor signaling in lens development. Semin Cell Dev Biol. 2006;17:726–740. doi: 10.1016/j.semcdb.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan DG, Oliveira-Fernandes M, Lavker RM. MicroRNAs of the mammalian eye display distinct and overlapping tissue specificity. Mol Vis. 2006;12:1175–1184. [PubMed] [Google Scholar]
- Sasaki T, Irie-Sasaki J, Jones RG, Oliveira-dos-Santos AJ, Stanford WL, Bolon B, Wakeham A, Itie A, Bouchard D, Kozieradzki I, Joza N, Mak TW, Ohashi PS, Suzuki A, Penninger JM. Function of PI3Kgamma in thymocyte development, T cell activation, and neutrophil migration. Science. 2000;287:1040–1046. doi: 10.1126/science.287.5455.1040. [DOI] [PubMed] [Google Scholar]
- Shi XB, Tepper CG, deVere White RW. Cancerous miRNAs and their regulation. Cell Cycle. 2008;7:1529–1538. doi: 10.4161/cc.7.11.5977. [DOI] [PubMed] [Google Scholar]
- Sinha D, Esumi N, Jaworski C, Kozak CA, Pierce E, Wistow G. Cloning and mapping the mouse Crygs gene and non-lens expression of [gamma]S-crystallin. Mol Vis. 1998;4:8. [PubMed] [Google Scholar]
- Strickler AG, Yamamoto Y, Jeffery WR. The lens controls cell survival in the retina: Evidence from the blind cavefish Astyanax. Dev Biol. 2007;311:512–523. doi: 10.1016/j.ydbio.2007.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam M, Gorny G, Johnsen SA, Monroe DG, Evans GL, Fraser DG, Rickard DJ, Rasmussen K, van Deursen JM, Turner RT, Oursler MJ, Spelsberg TC. TIEG1 null mouse-derived osteoblasts are defective in mineralization and in support of osteoclast differentiation in vitro. Mol Cell Biol. 2005;25:1191–1199. doi: 10.1128/MCB.25.3.1191-1199.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Ma Z, Li Y, Liu B, Li Z, Ding X, Gao Y, Ma W, Tang X, Li X, Shen Y. Gamma-S crystallin gene (CRYGS) mutation causes dominant progressive cortical cataract in humans. J Med Genet. 2005;42:706–710. doi: 10.1136/jmg.2004.028274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swamynathan SK, Davis J, Piatigorsky J. Identification of candidate Klf4 target genes reveals the molecular basis of the diverse regulatory roles of Klf4 in the mouse cornea. Invest Ophthalmol Vis Sci. 2008;49:3360–3370. doi: 10.1167/iovs.08-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, Hannon GJ. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Tsujimura T, Takeda K, Sugihara A, Maekawa A, Terada N, Yoshida N, Akira S. Targeted disruption of ATF4 discloses its essential role in the formation of eye lens fibres. Genes Cells. 1998;3:801–810. doi: 10.1046/j.1365-2443.1998.00230.x. [DOI] [PubMed] [Google Scholar]
- Teramoto S, Fujii S, Yoshida A, Shirasu Y. Morphological and genetic characteristics of the open-eyelid mutant spontaneously occurring in NC-strain mice. Jikken Dobutsu. 1988;37:455–462. doi: 10.1538/expanim1978.37.4_455. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, Surani MA, Sakaki Y, Sasaki H. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- Xu PX, Adams J, Peters H, Brown MC, Heaney S, Maas R. Eya1-deficient mice lack ears and kidneys and show abnormal apoptosis of organ primordia. Nat Genet. 1999;23:113–117. doi: 10.1038/12722. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Jeffery WR. Central role for the lens in cave fish eye degeneration. Science. 2000;289:631–633. doi: 10.1126/science.289.5479.631. [DOI] [PubMed] [Google Scholar]
- Yu J, Ryan DG, Getsios S, Oliveira-Fernandes M, Fatima A, Lavker RM. MicroRNA-184 antagonizes microRNA-205 to maintain SHIP2 levels in epithelia. Proc Natl Acad Sci U S A. 2008;105:19300–19305. doi: 10.1073/pnas.0803992105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelenka PS. Regulation of cell adhesion and migration in lens development. Int J Dev Biol. 2004;48:857–865. doi: 10.1387/ijdb.041871pz. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Overbeek PA, Govindarajan V. Perinatal ablation of the mouse lens causes multiple anterior chamber defects. Mol Vis. 2007;13:2289–2300. [PubMed] [Google Scholar]
- Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Srivastava D. A developmental view of microRNA function. Trends Biochem Sci. 2007;32:189–197. doi: 10.1016/j.tibs.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Zhu J, Motejlek K, Wang D, Zang K, Schmidt A, Reichardt LF. beta8 integrins are required for vascular morphogenesis in mouse embryos. Development. 2002;129:2891–2903. doi: 10.1242/dev.129.12.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zieske JD. Corneal development associated with eyelid opening. Int J Dev Biol. 2004;48:903–911. doi: 10.1387/ijdb.041860jz. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.