Abstract
The structure of recombinant domain III of the envelope protein (rED3) of yellow fever virus (YFV), containing the major neutralization site, was determined using NMR spectroscopy. The amino acid sequence and structure of the YFV-rED3 shows differences from ED3s of other mosquito-borne flaviviruses; in particular, the partially surface-exposed BC loop where methionine-304 and valine-324 were identified as being critical for the structure of the loop. Variations in the structure and surface chemistry of ED3 between flaviviruses affect neutralization sites and may affect host cell receptor interactions and play a role in the observed variations in viral pathogenesis and tissue tropism.
INTRODUCTION
Yellow fever virus (YFV) is an arthropod-borne virus belonging to the family Flaviviridae, genus Flavivirus. Most flaviviruses are typically transmitted by either mosquitoes or ticks, and include major human pathogens such as YFV, dengue virus (DENV types 1–4), West Nile virus (WNV), Japanese encephalitis virus (JEV), and tick-borne encephalitis virus (TBEV). Yellow fever is an acute viral disease that causes hemorrhagic fever and jaundice. The virus is transmitted between humans by the Aedes aegypti mosquito and about 200,000 cases are reported annually, including 30,000 deaths. Because no treatment or cure exists for yellow fever, there is great interest in developing strategies to control the disease. Unlike other mosquito-borne flaviviruses, YFV has a tropism for the liver and causes a viscerotropic disease whereas many other mosquito-borne flaviviruses have a tropism for the brain, or in the case of the DEN viruses they target cells of reticuloendothelial origin.
The YFV genome is an 11kb single-stranded positive-sense RNA genome coding for a polyprotein, which is post- and co-translationally processed into three structural proteins and seven non-structural proteins. The largest of the structural proteins, the envelope (E) protein, is the major component of the virion surface. It is the primary immunogen and plays a central role in receptor binding and membrane fusion (Heinz and Allison, 2003). The structure of the ectodomain (the soluble N-terminal portion, consisting of 395 residues) of the E protein of TBEV was determined by x-ray crystallography (Rey et al., 1995). Based on this structure, three distinct structural domains, domains I, II and III, have been identified in the ectodomain. This structure has been confirmed by x-ray crystallographic studies of other flaviviruses, including DENV1 (Nayak et al., 2009), DENV2 (Modis et al., 2003), DENV3 (Modis et al., 2005) and WNV (Kanai et al., 2006; Nybakken et al., 2006). Domains I and II lie parallel to the virion surface in the mature, pre-fusion form. They contain the fusion peptide and the hinge region, both involved in the low-pH induced conformational change observed upon fusion and entry into the cell, and the N-linked glycosylation site(s) (Rey et al., 1995). Domain III (ED3) is involved in receptor binding and contains epitopes critical for type-specific neutralization of the virus (i.e., those neutralization epitopes that distinguish each flavivirus, e.g. YFV from DENV2) (Chu et al., 2005; Crill and Roehrig, 2001). The major neutralization epitopes of WNV (Beasley and Barrett, 2002; Nybakken et al., 2005; Sanchez et al., 2005), YFV (Ryman et al., 1998), DENV2 (Hiramatsu et al., 1996; Roehrig et al., 1998; Gromowski and Barrett, 2007; Sukupolvi-Petty et al., 2007), TBEV (Mandl et al., 1989; Holzmann et al., 1997) and JEV (Cecilia and Gould, 1991; Wu and Chen, 2001; Lin and Wu, 2003; Wu et al., 1997, 2003, 2004; Goncalvez et al., 2008) have all been mapped to ED3.
Cryoelectron microscopic reconstructions of several flaviviruses indicate that the E protein is arranged as dimers parallel to the virion surface, such that ED3 projects slightly above the viral surface (Kuhn et al., 2002; Mukhopadhyay et al., 2003). Interactions between five ED3 subunits at the virion 5-fold axes of symmetry form pores on the virion surface where cell receptors may bind. NMR-derived solution structures of the JEV (Wu et al., 2003), WNV (Volk et al., 2004), Omsk hemorrhagic fever virus (OHFV (Volk et al., 2006)), Langat virus (LGTV (Mukherjee et al., 2006)) and DENV4 (Volk et al., 2007b) rED3 illustrate an overall similar structural fold for this domain of these flaviviruses, with specific differences between those viruses transmitted by mosquito or tick vectors. In this study we have solved the solution structure of rED3 of wild-type strain Asibi of YFV and demonstrate that it is markedly different to ED3 of other mosquito-borne flaviviruses that have been solved.
RESULTS
Quality of the NMR structure
The 20 final structures of YFV-rED3 in the ensemble (Fig 1A and B) had low molecular and restraint energy penalties. The structure presented here is well defined, as shown by the r.m.s.d. values and restraint violations listed in Table 1. The final structures, determined in an automated fashion, had 13 ± 2 distance violations over 0.3 Å, 3 ± 1 violation over 0.5 Å and 2 ± 1 dihedral angle violations over 10°, and no dihedral angle violations over 20° (Table 1). Thus, 99.9% of the NMR-derived restraints fit the structures determined. Most of the violations occur in four or fewer of the twenty structures, although six are nearly always violated. The violations occur because the NOE interactions and cut-off distances were set in an automated fashion into distance spins based on crosspeak volumes, disregarding confounding effects such as amide proton exchange rates, equivalent geminal methyl groups, ambiguous NOE assignments and differing spin-spin relaxation rates. The r.m.s.d. on the distance restraint error was 0.019 ± 0.071 Å, and the r.m.s.d. on dihedral angle error was 0.54 ± 1.51°. The structural ensemble has an average pairwise backbone atom r.m.s.d of 1.38 ± 0.44 Å and an average pairwise heavy atom r.m.s.d of 1.59 ± 0.41 Å. The program PROCHECK was used to analyze the quality of the final ensemble. Analysis of the non-glycine, non-proline residues indicated that 98.7% of these residues are in the two most favored regions of a Ramachandran plot. Specifically, 81.8% of the residues are in the most favored regions, 16.9% are in the additionally allowed regions, 1.1% are in the generously allowed regions and 0.2% are in the disallowed regions.
Figure 1.
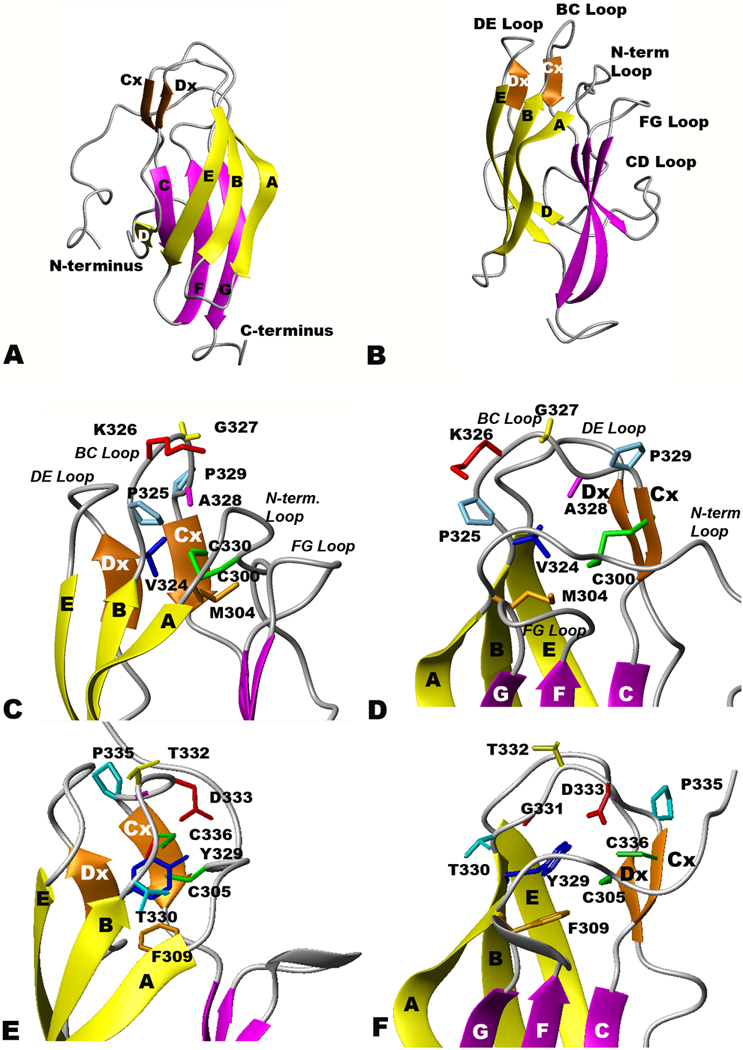
Ribbon diagrams of rED3 of YFV and WNV. Beta sheets 1–3 are colored yellow, orange and magenta, respectively, and the disulfide bridge between C300 and C330 is colored green. (A and B) Two orthogonal views of the NMR-derived YF rED3 backbone atom structures (C and D) Surface loop structure of YF rED3, and (E and F)WN rED3 (Volk et al., 2004).
Table 1.
Summary of NMR structure constraints and statistics
| Total Restraints | 1833 |
| NOE restraints | 1278 |
| Intra-residue | 609 |
| Sequential | 319 |
| Medium range | 59 |
| Long rang | 291 |
| Talos phi/psi dihedral restraints | 170 |
| Omega dihedral restraints | 111 |
| Chirality restraints | 274 |
| Structural Statistics | |
| NOE violations > 0.5 Å | 3 ± 1 |
| NOE violations > 0.3 Å | 13 ± 2 |
| Dihedral angle violation > 20° | 0 |
| Dihedral angle violation > 10° | 2 ± 1 |
| r.m.s.d from ideal geometry | |
| Bond lengths (Å) | 0.013 |
| Bond angles (°) | 2.1 |
| Restraint error r.m.s.d. | |
| Distance restraints (Å) | 0.019 ± 0.071 |
| Dihedral restraints (Å) | 0.54 ± 1.51 |
| Atomic Pairwise r.m.s.d. | |
| Backbone atoms | 1.38 ± 0.44 |
| All heavy atoms | 1.59 ± 0.41 |
| Ramachandran statistics | |
| Most favored regions (%) | 81.8 |
| Additionally allowed regions (%) | 16.9 |
| Generously allowed regions (%) | 1.1 |
| Disallowed regions (%) | 0.2 |
Structural details of the NMR ensemble
The overall structure ensemble of YFV ED3 determined by NMR (Figure 1A and B; Table 2, all amino acid numbers in the text refer to amino acid number in the specific viral E protein being discussed, unless otherwise defined) is similar to that reported for the ED3 of other flaviviruses, including DENV1 (Nayak et al., 2009), DENV2 (Modis et al., 2003), DENV3 (Modis et al., 2005), DENV4 (Volk et al., 2007b), JEV (Wu et al., 2003), LGTV (Mukherjee et al., 2006), OHFV (Volk et al., 2006), TBEV (Rey et al., 1995), and WNV (Volk et al., 2004). The YFV-rED3 structure has nine β-strands in three β-sheets arranged in an IgG-like β-barrel configuration. The first β-sheet (yellow) contains β-strands from Ser305 to Asp312, β-strand B from Val318 to Lys323, β-strand D, from Ile348 to Leu349, and β-strand E, from Glu362 to Asn368. The second β-sheet (orange) is formed by only two short β-strands, Cx and Dx, encompassing residues Cys330-Lys331 and Ile355-Ala356, respectively. The last β-sheet (magenta) is comprised of β-strand C from Val334-Ala337, β-strand F from Gly372-Val378 and β-strand G from Leu385-Lys391. Both the overall global fold and the secondary structures of YFV-rED3 are grossly similar to the structures reported for mosquito-borne DENV1, DENV2, DENV3, DENV4, JEV and WNV rED3, although small differences in the lengths of beta sheets do exist. However, the major difference between the YFV-ED3 structure and other flavivirus structures is found at the surface-exposed loops; particularly in the BC loop. This difference is directly related to the addition of Pro325 in YFV (found in no other flavivirus; see Fig 1C and D), and the presence of relatively small, non-aromatic residues at positions Met304 and Val324 of YFV ED3 (Fig. 1C and D and Table 2) compared to other mosquito borne flaviviruses (see WNV ED3 in Fig 1E and F).
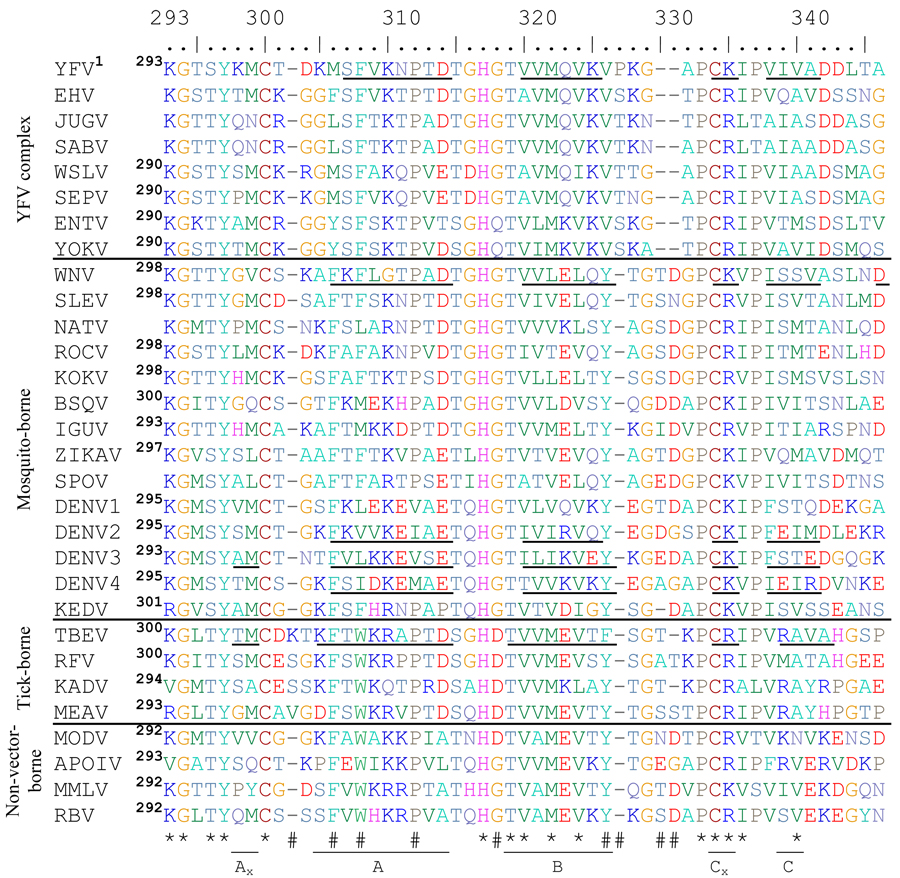
Table 2.
Alignment of flavivirus ED3 proteins from mosquito-, tick- and non-vector-borne flaviviruses. Asterisks (*) indicate highly conserved residues and (#) hashes indicate residues with group-specific differences. Beta strands are indicated with underlined residues and are labeled underneath using TBEV(Rey et al., 1995) nomenclature. The number of the first amino acid of ED3 for each flavivirus is shown in superscript to the right of the name of each flavivirus. Those without available full length E genes sequences are left blank. Biochemically similar amino acids are the same color to allow easier understanding of the alignment.
 |
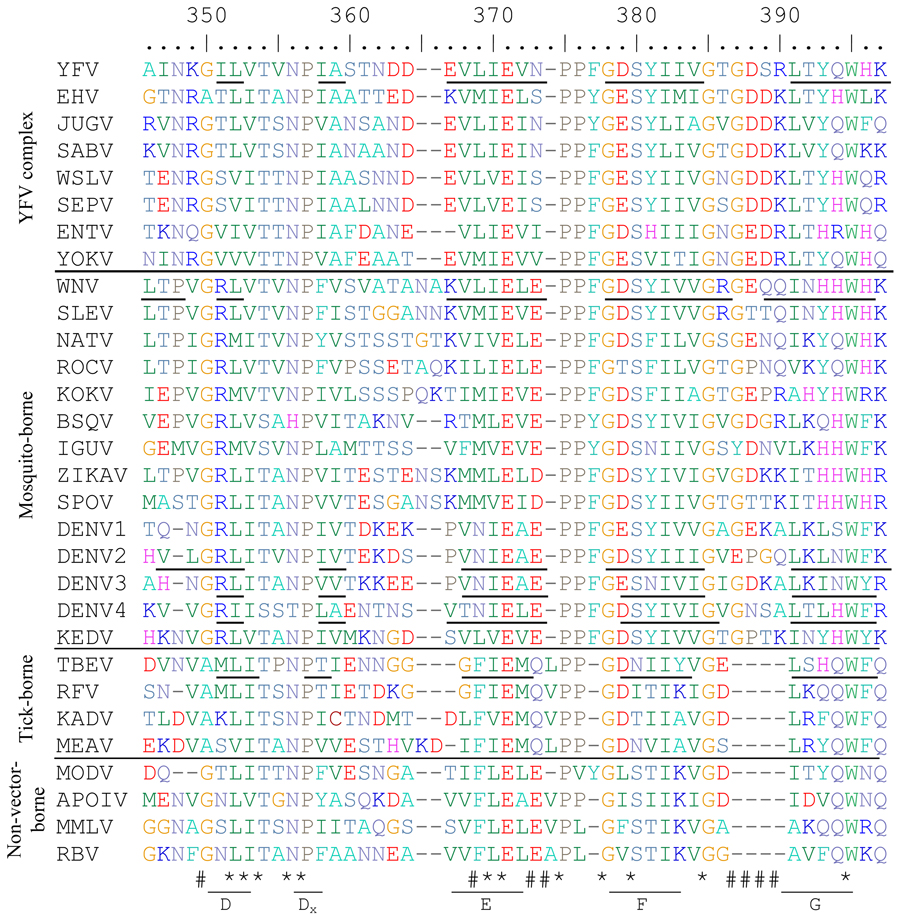 |
ICTV abbreviations used are as follows: Yellow fever virus=YFV, Edge Hill virus=EHV, Jugra virus=JUGV, Saboya virus=SABV, Wesselsbron virus=WSLV, Sepik virus=SEPV, Entebbe bat virus=ENTV, Yokose virus=YOKV, West Nile virus=WNV, St Louis encephalitis virus=SLEV, Ntaya virus=NATV, Rocio virus=ROCV, Kokobera virus=KOKV, Bussuquara virus=BSQV, Iguape virus=IGUV, Zika virus=ZIKAV, Spondweni virus=SPOV, dengue virus=DENV, Kedougou virus=KEDV, tick-borne encephalitis virus=TBEV, Royal Farm virus=RFV, Kadam virus=KADV, Meaban virus=MEAV, Modoc virus=MODV, Apoi virus=APOIV, Montana myotis leukoencephalitis virus=MMLV, Rio Bravo virus=RBV
The residues comprising the flavivirus BC loops differ significantly in mosquito and tick vectors and between flavivirus complexes (Table 2). All of the mosquito-borne flaviviruses, excluding the YFV complex, contain a conserved tyrosine immediately before the BC loop (amino acid position 329 for WNV in Table 2 and Fig 1E and F), which has been shown to be essential for viability of WNV (Zhang, S and Beasley, DWC, unpublished data) and presumably plays a role in stabilizing the ED3 protein fold while some of the tick-borne viruses have a phenylalanine substitution in place of the tyrosine. The phenylalanine at amino acid position 305 in the alignment (equivalent to F309 in WNV and M304 in YFV), which is packed closely with the tyrosine at position 329 in the WNV-ED3 structure, is also conserved in these viruses. In contrast, YFV-rED3, as well as other YFV complex viruses (Wesslesbron [WSLV], Sepik [SEPV], Saboya [SABV], Jugra [JUGV], Edge Hill [EHV], Yokose [YOKV] and Entebbe Bat viruses [ENTV]), contain a methionine at position 304 and a valine at position 324. Immediately following the valine at 324, the BC loop of YFV both starts and ends with a proline (amino acids 325 and 329) whereas all other flaviviruses have BC loops ending with a proline only. The proline present at position 325 in the YFV E protein removes the need for a tyrosine or phenylalanine at position 324 by forcing the BC loop to start turning towards the next beta strand. The smaller sizes of Met304, relative to a phenylalanine, and Val324, relative to either a tyrosine or a phenylalanine, allows the length of the BC loop to be smaller in YFV and related viruses compared to the other mosquito-borne flaviviruses.
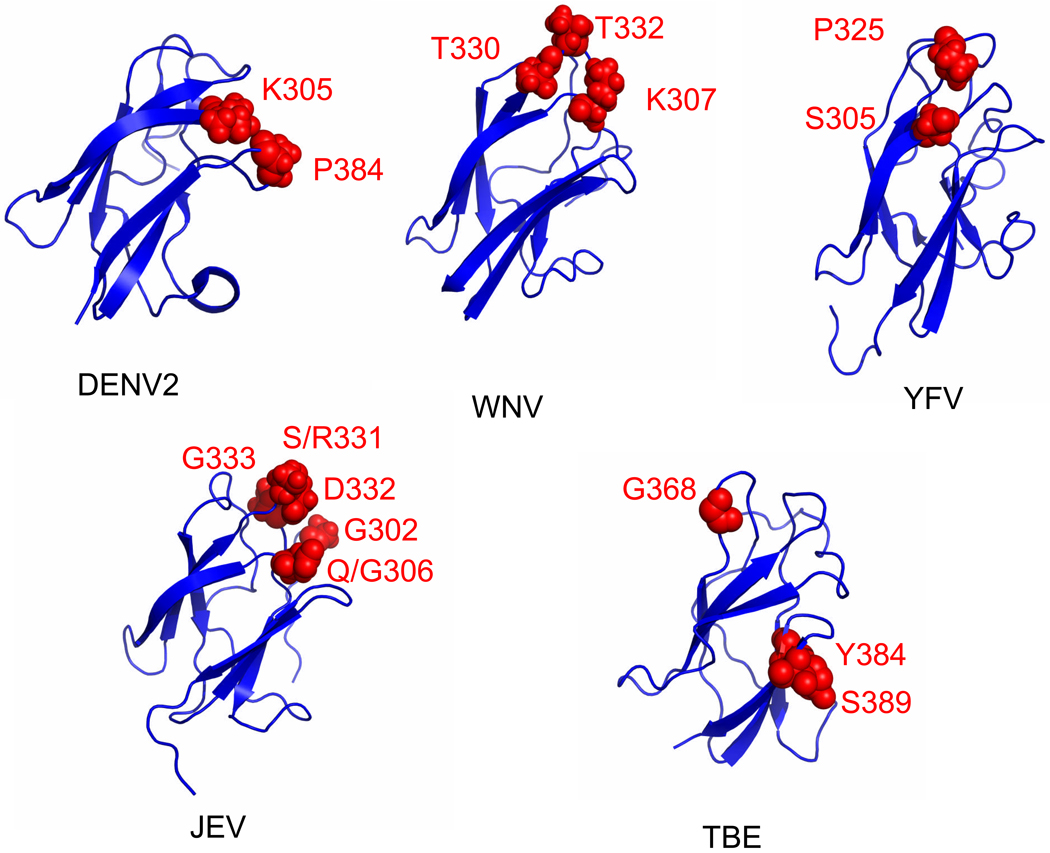
These differences in loops are unique in the YFV complex viruses and would be predicted to contribute to differences in antigenicity and differences in the individual amino acids that constitute the major type-specific neutralization epitopes on different flaviviruses. In particular, the major neutralization epitope in YFV involves the serine at residue 305 and proline at residue 325 (Ryman et al., 1998), while it is the lysine at residue 307, the threonine at residue 330 and threonine at residue 332 for WNV (Beasley and Barrett, 2002; Nybakken et al., 2005); the lysine at residue 305 and proline at residue 384 for DENV2 (Hiramatsu et al., 1996; Gromowski and Barrett, 2007; Sukupolvi-Petty et al., 2007); the glycine at residue 302, glutamine or glycine at residue 306, serine or arginine at residue 331, aspartic acid at residue 332, and glycine at residue 333 for JEV (Cecilia and Gould, 1991; Wu and Chen, 2001; Lin and Wu, 2003; Goncalvez et al., 2008), and the glycine at residue 368, tyrosine at residue 384, and serine at residue 389 for TBEV (Mandl et al., 1989; Holzmann et al., 1991). Based on this information, Figure 2 shows that the location of the type-specific epitopes associated with neutralization for WNV, JEV, TBEV, DENV2 and YFV viruses are not in identical locations on ED3. (i.e., YFV: 305 and 325 [B–C loop], DENV2: 305 and 284 [F–G loop], WNV: 310 and 332 [B–C loop], JEV: 302, 306, 331, 332 and 333 [B–C loop]), TBEV: 384 and 389 Thus, different surface exposed loops on ED3 of different flaviviruses are important for neutralizing epitopes.
Figure 2.
Neutralizing epitopes on ED3 of yellow fever (YF), dengue-2 (DEN2). Japanese encephalitis (JE), tick-borne encephalitis (TBE) and West Nile (WN) viruses. Red dots identify amino acids recognized by type-specific monoclonal antibodies.
DISCUSSION
Identification of structural and/or amino acid differences in ED3 has revealed differences in the critical type-specific neutralization epitopes that is leading to a greater understanding of how each flavivirus is distinguished immunologically. The newly determined structure of the ED3 of YFV, representing a major flavivirus serocomplex not previously subjected to detailed structural analysis, was compared with the structure of the ED3s of other flaviviruses.
The structure of the ED3 of YFV differs from the structures of other mosquito-borne flaviviruses; in particular the surface exposed loops, especially the BC loop, are different (see Fig. 1C, D, E and F). In YFV, the BC loop is one amino acid shorter than in the mosquito-borne and non-vector-borne viruses, but the same length as most tick-borne viruses. Although the function of the BC loop is unknown, it contains the major neutralization determinant for YFV (residue 325), WNV (residue 332; Beasley and Barrett, 2002; Nybakken et al., 2005) and JEV (residue 333; Wu and Chen, 2001). The structures in Figure 1C, D, E and F suggest that not all flavivirus type-specific neutralization epitopes are in analogous positions, which supports the hypothesis that the function of the BC loop may be different for at least YFV, WNV and JEV. In addition, a nearby loop, the FG loop, located on the same surface of ED3, has been shown to be the major neutralization determinant for TBEV and DENV2 (see Figure 2) and is involved in vector-specific receptor binding of DENV2 (Hung et al., 2004). Like the BC loop, the FG loop is longer in most mosquito-borne viruses than the tick-borne viruses. The differences in this loop, in combination with other variations in surface chemistry, most likely contribute to the diversity in antigenicity, and possibly receptor binding and host specificity and tissue tropism.
The overall structure of the ED3s of most mosquito-borne flaviviruses, including DENV1 (Nayak et al., 2009), DENV2 (Modis et al., 2003), DENV4 (Volk et al., 2007b), WNV (Volk et al., 2004) and JEV (Wu et al., 2003) are very similar, and comparison of the amino acid sequences reveal several motifs unique to these virus complexes. The same is true of the tick-borne viruses such as TBEV (Rey et al., 1995), LGTV (Mukherjee et al., 2006) and OHFV (Volk et al., 2006) (see Table 2). In contrast, the structure of the YFV ED3 has several unique differences when compared with other mosquito-borne flaviviruses; in particular, the surface exposed BC loop is shorter in YFV than any other mosquito-borne virus. These differences are reflected in the amino acid sequence of this region, and due to a high level of similarity of the amino acid sequence of ED3 between members of the YFV complex (see Table 2), these structural differences can be expected to occur in other members of the YFV complex.
METHODS
Protein Expression and Purification
Uniformly 15N,13C-labeled human YFV-rED3 protein (Asibi strain) encompassing residues (Ser288-Lys398) was expressed using the pET-15b vector (Novagen), with an added Methionine residue on the N-terminus but lacking the N-terminal His-tag sequence encoded in that plasmid. The cells were lysed using the native lysis buffer and centrifuged to obtain the YFV-rED3 protein along with the crude cell debris in the pellet. The pellet is then dissolved in denaturing lysis buffer containing 6 M Guanidine Hydrochloride to solubilize the protein. The insoluble cell debris is removed by centrifugation. The Guanidine HCl in the supernatant was then removed by dialysis (6 × 1/2000 dilution). The expressed protein was filtered through an Amicon centrifugal filter concentrator with a 50 kDa molecular weight cut-off to remove proteins with higher molecular weight. Size Exclusion Chromatography was performed using Sephadex G-75 beads to further purify the protein. Centricon concentrators with a 3 kDa cut-off membrane were used for the final concentration step and to remove low-molecular weight impurities, as well as to exchange the material into the final NMR buffer.
NMR Spectroscopy and the Generation of NMR Restraints
The NMR samples contained 0.1–0.4 mM protein in 50 mM deuterated Tris (pH 5.8), 50 mM NaCl, 1mM NaN3 in 90% H2O and 10% D2O. NMR experiments were performed at 25 °C on Varian Inova 750 MHz (UTMB) or 600 MHz (with cold probe, Rice University) spectrometers with triple resonance probes. The 13C and 15N dimensions were referenced indirectly using frequency ratios. Sequence-specific backbone assignments were obtained using the 2D 1H, 15N-HSQC, 3D HNCACB, 3D CBCA(CO)NH and 3D HNCO experiments as described previously (Volk et al., 2007a). Non-aromatic side chain assignments were obtained using the HCCH-TOCSY, TOCSY-[1H,15N]-HSQC, H(C)CH-TOCSY, H(CCO)NH and C(CO)NH experiments as described previously (Volk et al., 2007a). Aromatic proton assignments were obtained from the (HB)CB(CGCD)HD and (HB)CB(CGCDCE)HE experiments. A NOESY-[1H,15N]-HSQC experiment provided several missing side chain assignments as described previously (Volk et al., 2007a). Stereo-specific assignments for some of the side-chain protons were obtained after initial rounds of structure calculations using ambiguous restraints. The NMR spectra were processed in VNMRJ (Varian Inc.) or Felix2000 (Felix,Inc.) software. SANE was used to facilitate the assignment of the 15N-edited or 13C-edited NOE cross peaks and for the generation of restraints as described previously (Volk et al., 2007a). Chemical shifts, distance cutoffs and contribution cutoffs were used within the program. The NMR restraints were separated into four bins, based on the NOESY cross-peak volumes from which they were derived, with upper distance limits of 2.5, 3.5, 4.5 and 6.0 Å for all NOE data. The 1833 NOE-based restraints (see Table 1) consist of 609 intra-residue, 319 sequential, 59 medium-range and 291 long-range distance restraints. TALOS was used to derive 170 phi/psi dihedral angle restraints based on the chemical shifts of the amino acids. Additional angular restraints for the omega angles and correct chiralities were generated within AMBER6.0.
Molecular Dynamics Calculations
One hundred random structures were generated by annealing the protein at 700 K, obtaining the coordinates every 5 ps and minimizing the structures obtained. The structures were then subjected to r-MD using dihedral angle restraints (Table 1) followed by the application of all restraints at 300 K. Finally, the structures were energy minimized for 2,000 steps. Twenty structures with the lowest restraint penalties were then chosen for the structural ensemble. The SANDER module within AMBER6.0 (Case et al., 1999) was used for all NMR structure calculations and MIDAS (Ferrin et al., 1988) and MOLMOL (Koradi, Billeter, and Wuthrich, 1996) were used to visualize the structures. Coordinates for the ensemble of NMR structures of YFV-rED3 have been deposited with the Protein Data Bank (PDB ID 2JQM) and the chemical shifts have been deposited with the BMRB (Volk et al., 2007a) accession code 15034).
ACKNOWLEDGEMENTS
This work was supported by the Pediatric Dengue Vaccine Initiative, the Centers for Disease Control (U90CCU618754), the NIAID (U01 AI054827), and the Welch Foundation (H1296). We thank Sean Moran for assistance with the Rice University Varian Inova 600 MHz NMR spectrometer whose Carbon-Enhanced HCN Cold Probe was obtained through the SPRING (Strategic Partnership for Research in Nanotechnology Grant) AFRL (AFOSR) FA9550-04-1-0328.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The atomic coordinates (2JQM) and NMR restraints (2JV6) for YFV-rED3 were deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/). NMR chemical shifts (15034) have been deposited at the BioMagResBank, University of Wisconsin-Madison (http://www.bmrb.wisc.edu/). The sequence of the WSLV has been deposited with Genbank (accession number EU707555).
References
- Beasley DW, Barrett AD. Identification of neutralizing epitopes within structural domain III of the West Nile virus envelope protein. J Virol. 2002;76(24):13097–13100. doi: 10.1128/JVI.76.24.13097-13100.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billoir F, de Chesse R, Tolou H, de Micco P, Gould EA, de Lamballerie X. Phylogeny of the genus flavivirus using complete coding sequences of arthropod-borne viruses and viruses with no known vector. J Gen Virol. 2000;81(Pt 3):781–790. doi: 10.1099/0022-1317-81-3-781. [DOI] [PubMed] [Google Scholar]
- AMBER6.0. San Francisco, CA: [Google Scholar]
- Cecilia D, Gould EA. Nucleotide changes responsible for loss of neuroinvasiveness in Japanese encephalitis virus neutralization-resistant mutants. Virology. 2001;181(1):70–77. doi: 10.1016/0042-6822(91)90471-m. [DOI] [PubMed] [Google Scholar]
- Chu JJ, Rajamanonmani R, Li J, Bhuvanakantham R, Lescar J, Ng ML. Inhibition of West Nile virus entry by using a recombinant domain III from the envelope glycoprotein. J Gen Virol. 2005;86(Pt 2):405–412. doi: 10.1099/vir.0.80411-0. [DOI] [PubMed] [Google Scholar]
- Crill WD, Roehrig JT. Monoclonal antibodies that bind to domain III of dengue virus E glycoprotein are the most efficient blockers of virus adsorption to Vero cells. J Virol. 2001;75(16):7769–7773. doi: 10.1128/JVI.75.16.7769-7773.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrin TE, Huang CC, Jarvis LE, Langridge R. The MIDAS display system. Journal of Molecular Graphics. 1988;6(1):13–27. [Google Scholar]
- Gaunt MW, Sall AA, de Lamballerie X, Falconar AK, Dzhivanian TI, Gould EA. Phylogenetic relationships of flaviviruses correlate with their epidemiology, disease association and biogeography. J Gen Virol. 2001;82(Pt 8):1867–1876. doi: 10.1099/0022-1317-82-8-1867. [DOI] [PubMed] [Google Scholar]
- Goncalvez AP, Chien CH, Tubthong K, Gorshkova I, Roll C, Donau O, Schuck P, Yoksan S, Wang SD, Purcell RH, Lai CJ. Humanized monoclonal antibodies derived from chimpanzee Fabs protect against Japanese encephalitis virus in vitro and in vivo. J Virol. 2008;82(14):7009–7021. doi: 10.1128/JVI.00291-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromowski GD, Barrett ADT. Characterization of an Antigenic Site that contains a Dominant, Type-Specific, Neutralization Determinant on the Envelope Protein Domain III (ED3) of Dengue 2 virus. Virology. 2007;366:349–360. doi: 10.1016/j.virol.2007.05.042. [DOI] [PubMed] [Google Scholar]
- Heinz FX, Allison SL. Flavivirus structure and membrane fusion. Adv Virus Res. 2003;59:63–97. doi: 10.1016/s0065-3527(03)59003-0. [DOI] [PubMed] [Google Scholar]
- Hiramatsu K, Tadano M, Men R, Lai CJ. Mutational analysis of a neutralization epitope on the dengue type 2 virus (DEN2) envelope protein: monoclonal antibody resistant DEN2/DEN4 chimeras exhibit reduced mouse neurovirulence. Virology. 1996;224(2):437–445. doi: 10.1006/viro.1996.0550. [DOI] [PubMed] [Google Scholar]
- Holzmann H, Stiasny K, Ecker M, Kunz C, Heinz FX. Characterization of monoclonal antibody-escape mutants of tick-borne encephalitis virus with reduced neuroinvasiveness in mice. J Gen Virol. 1997;78(Pt 1):31–37. doi: 10.1099/0022-1317-78-1-31. [DOI] [PubMed] [Google Scholar]
- Hung JJ, Hsieh MT, Young MJ, Kao CL, King CC, Chang W. An external loop region of domain III of dengue virus type 2 envelope protein is involved in serotype-specific binding to mosquito but not mammalian cells. J Virol. 2004;78(1):378–388. doi: 10.1128/JVI.78.1.378-388.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai R, Kar K, Anthony K, Gould LH, Ledizet M, Fikrig E, Marasco WA, Koski RA, Modis Y. Crystal structure of west nile virus envelope glycoprotein reveals viral surface epitopes. J Virol. 2006;80(22):11000–11008. doi: 10.1128/JVI.01735-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph. 1996;14(1):51–55. 29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- Kuhn RJ, Zhang W, Rossmann MG, Pletnev SV, Corver J, Lenches E, Jones CT, Mukhopadhyay S, Chipman PR, Strauss EG, Baker TS, Strauss JH. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell. 2002;108(5):717–725. doi: 10.1016/s0092-8674(02)00660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuno G, Chang GJ. Biological transmission of arboviruses: reexamination of and new insights into components, mechanisms, and unique traits as well as their evolutionary trends. Clin Microbiol Rev. 2005;18(4):608–637. doi: 10.1128/CMR.18.4.608-637.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuno G, Chang GJ, Tsuchiya KR, Karabatsos N, Cropp CB. Phylogeny of the genus Flavivirus. J Virol. 1998;72(1):73–83. doi: 10.1128/jvi.72.1.73-83.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CW, Wu SC. A functional epitope determinant on domain III of the Japanese encephalitis virus envelope protein interacted with neutralizing-antibody combining sites. J Virol. 2003;77(4):2600–2606. doi: 10.1128/JVI.77.4.2600-2606.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandl CW, Guirakhoo F, Holzmann H, Heinz FX, Kunz C. Antigenic structure of the flavivirus envelope protein E at the molecular level, using tick-borne encephalitis virus as a model. J Virol. 1989;63(2):564–571. doi: 10.1128/jvi.63.2.564-571.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modis Y, Ogata S, Clements D, Harrison SC. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc Natl Acad Sci U S A. 2003;100(12):6986–6991. doi: 10.1073/pnas.0832193100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modis Y, Ogata S, Clements D, Harrison SC. Variable surface epitopes in the crystal structure of dengue virus type 3 envelope glycoprotein. J Virol. 2005;79(2):1223–1231. doi: 10.1128/JVI.79.2.1223-1231.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee M, Dutta K, White MA, Cowburn D, Fox RO. NMR solution structure and backbone dynamics of domain III of the E protein of tick-borne Langat flavivirus suggests a potential site for molecular recognition. Protein Sci. 2006;15(6):1342–1355. doi: 10.1110/ps.051844006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S, Kim BS, Chipman PR, Rossmann MG, Kuhn RJ. Structure of West Nile virus. Science. 2003;302(5643):248. doi: 10.1126/science.1089316. [DOI] [PubMed] [Google Scholar]
- Nayak V, Dessau M, Kucera K, Anthony K, Ledizet M, Modis Y. Crystal structure of dengue virus type 1 envelope protein in the postfusion conformation and its implications for membrane fusion. J Virol. 2009;83(9):4338–4344. doi: 10.1128/JVI.02574-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nybakken GE, Nelson CA, Chen BR, Diamond MS, Fremont DH. Crystal structure of the West Nile virus envelope glycoprotein. J Virol. 2006;80(23):11467–11474. doi: 10.1128/JVI.01125-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nybakken GE, Oliphant T, Johnson S, Burke S, Diamond MS, Fremont DH. Structural basis of West Nile virus neutralization by a therapeutic antibody. Nature. 2005;437(7059):764–769. doi: 10.1038/nature03956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey FA, Heinz FX, Mandl C, Kunz C, Harrison SC. The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature. 1995;375(6529):291–298. doi: 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- Roehrig JT, Bolin RA, Kelly RG. Monoclonal antibody mapping of the envelope glycoprotein of the dengue 2 virus. Virology. 1988;246(2):317–328. doi: 10.1006/viro.1998.9200. Roehrig JT, Bolin RA, Kelly RG. [DOI] [PubMed] [Google Scholar]
- Ryman KD, Ledger TN, Campbell GA, Watowich SJ, Barrett AD. Mutation in a 17D-204 vaccine substrain-specific envelope protein epitope alters the pathogenesis of yellow fever virus in mice. Virology. 1998;244(1):59–65. doi: 10.1006/viro.1998.9057. [DOI] [PubMed] [Google Scholar]
- Sánchez MD, Pierson TC, McAllister D, Hanna SL, Puffer BA, Valentine LE, Murtadha MM, Hoxie JA, Doms RW. Characterization of neutralizing antibodies to West Nile virus. Virology. 2005;336(1):70–82. doi: 10.1016/j.virol.2005.02.020. [DOI] [PubMed] [Google Scholar]
- Sukupolvi-Petty S, Austin SK, Purtha WE, Oliphant T, Nybakken GE, Schlesinger JJ, Roehrig JT, Gromowski GD, Barrett AD, Fremont DH, Diamond MS. Type- and subcomplex-specific neutralizing antibodies against domain III of dengue virus type 2 envelope protein recognized adjacent epitopes. J Virol. 2007;81:12816–12826. doi: 10.1128/JVI.00432-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk DE, Beasley DW, Kallick DA, Holbrook MR, Barrett AD, Gorenstein DG. Solution structure and antibody binding studies of the envelope protein domain III from the New York strain of West Nile virus. J Biol Chem. 2004;279(37):38755–38761. doi: 10.1074/jbc.M402385200. [DOI] [PubMed] [Google Scholar]
- Volk DE, Chavez L, Beasley DW, Barrett AD, Holbrook MR, Gorenstein DG. Structure of the envelope protein domain III of Omsk hemorrhagic fever virus. Virology. 2006;351(1):188–195. doi: 10.1016/j.virol.2006.03.030. [DOI] [PubMed] [Google Scholar]
- Volk DE, Gandham SHA, May FJ, Anderson A, Barrett AD, Gorenstein DG. NMR Assignments of the Yellow Fever Virus Envelope Protein Domain III. Biomol. NMR Assign. 2007a;1:49–50. doi: 10.1007/s12104-007-9000-9. [DOI] [PubMed] [Google Scholar]
- Volk DE, Lee YC, Li X, Thiviyanathan V, Gromowski GD, Li L, Lamb AR, Beasley DW, Barrett AD, Gorenstein DG. Solution structure of the envelope protein domain III of dengue-4 virus. Virology. 2007b;364(1):147–154. doi: 10.1016/j.virol.2007.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SC, Lian WC, Hsu LC, Liau MY. Japanese encephalitis virus antigenic variants with characteristic differences in neutralization resistance and mouse virulence. Virus Res. 51(2):173–181. doi: 10.1016/s0168-1702(97)00098-1. [DOI] [PubMed] [Google Scholar]
- Wu SC, Lin CW. Neutralizing peptide ligands selected from phage-displayed libraries mimic the conformational epitope on domain III of the Japanese encephalitis virus envelope protein. Virus Res. 2001;76(1):59–69. doi: 10.1016/s0168-1702(01)00246-5. [DOI] [PubMed] [Google Scholar]
- Wu KP, Wu CW, Tsao YP, Kuo TW, Lou YC, Lin CW, Wu SC, Cheng JW. Structural basis of a flavivirus recognized by its neutralizing antibody: solution structure of the domain III of the Japanese encephalitis virus envelope protein. J Biol Chem. 2003;278(46):46007–46013. doi: 10.1074/jbc.M307776200. [DOI] [PubMed] [Google Scholar]
- Wu SC, Lin YJ, Chou JW, Lin CW. Construction and characterization of a Fab recombinant protein for Japanese encephalitis virus neutralization. Vaccine. 2004;23(2):163–171. doi: 10.1016/j.vaccine.2004.05.016. [DOI] [PubMed] [Google Scholar]