Summary
CD69 is a type II C-type lectin involved in lymphocyte migration and cytokine secretion. CD69 expression represents one of the earliest available indicators of leukocyte activation and its rapid induction occurs through transcriptional activation. In this study we examined the molecular mechanism underlying mouse CD69 gene transcription in vivo in T and B cells. Analysis of the 45kb region upstream of the CD69 gene revealed evolutionary conservation at the promoter and at four non-coding sequences (CNS) that were called CNS1, CNS2, CNS3 and CNS4. These regions were found to be hypersensitive sites in DNase I digestion experiments and chromatin immunoprecipitation assays showed specific epigenetic modifications. CNS2 and CNS4 displayed constitutive and inducible enhancer activity in transient transfection assays in T cells. Using a transgenic approach to test CNS function, we found that the CD69 promoter conferred developmentally regulated expression during positive selection of thymocytes but could not support regulated expression in mature lymphocytes. Inclusion of CNS1 and CNS2 caused suppression of CD69 expression whereas further addition of CNS3 and CNS4 supported developmental-stage and lineage-specific regulation in T cells but not in B cells. We concluded CNS1-4 are important cis-regulatory elements that interact both positively and negatively with the CD69 promoter and that differentially contribute to CD69 expression in T and B cells.
Introduction
Leukocyte activation involves coordinated changes in the expression of key genes involved in the inflammatory cascade and leukocyte migration to promote effective immune responses against diverse pathogens and malignant cells. Recently, several reports using in vivo animal models have highlighted the role of the CD69 membrane molecule in both cytokine gene regulation and cell migration upon leukocyte activation. CD69 has been shown to be involved in the inhibition of lymphocyte egress from lymphoid organs in response to INFαβ through a mechanism that involves downregulation of S1P1 receptors (1). Overexpression of CD69 in transgenic mice supports a role in thymocyte migration (2, 3). CD69 deficient mice display enhanced resistance to MHC class Ĩ tumor growth and increased susceptibility to collagen induced arthritis and Listeria M. infection, associated with increased cellular recruitment and altered cytokine production and apoptosis (4, 5 and manuscript submitted for publication). Moreover, in vivo blocking of CD69 with monoclonal antibodies resulted in exacerbated autoimmune and antitumor responses (6). Recently, it has been demonstrated that CD69+CD4+CD25− cells represent a new subset of regulatory T cells involved in tumor-induced immunosuppression (7). Although CD69 is expressed during lymphocyte development (8), positive and negative selection of thymocytes is normal in CD69 deficient mice and only minor alterations in the pre-B cell compartment have been detected (9).
The CD69 gene is located in the natural killer complex (NKC) on mouse chromosome 6 and human chromosome 12. This complex includes a variety of genes encoding C-type lectins with diverse expression patterns and functions in the immune system (10). CD69 is expressed on the surface of activated leukocytes through a mechanism that involves ras and raf activation and calcium release (11, 12). A variety of agents, including anti-CD3 antibodies, TNFα, INFαβ, polyI:C or phorbol esters, can up-regulate CD69 in vitro. Transcripts are detected as early as 30 min after T cell stimulation and cell surface protein is observed 3h later (13). CD69 transcription, however, is transient and returns to an “off” state at later times (14). Transient transfection experiments showed that both the mouse and human CD69 promoters can direct reporter transcription in cells stimulated with PMA plus ionomycin (14, 15). Cis-elements contributing to this inducibility were mapped to the proximal promoter region and these elements were shown to interact with transcription factors Erg-1, Erg-3, ATF-3/CREB and AP-1 upon stimulation (16). Basal CD69 transcription was also detected by transient transfection of mouse and human promoter constructs and was attributed to the −78 to +16 region of the human CD69 gene. Interestingly, the transcription factor Sp1 was shown to constitutively bind to this region. In another study, an NFκB motif at position −223 of the human CD69 promoter was shown to be required for transcriptional induction of CD69 in response to TNFα (17).
Transient transfection assays do not account for potential influences of the chromatin environment, which may require the action of distal enhancers, silencers and insulators for efficient gene expression, and may require DNA methylation and histone modifications to regulate access to transcription factors. Epigenetic regulation has been shown to be critical for inducible expression of a variety of immune system genes, including IL-4, INFγ, INFβ and IL-12 (18–21). Therefore, the goal of this study was to define the epigenetic changes and cis-acting elements important for regulated CD69 gene expression in lymphocytes in vivo. We searched for distal regulatory elements using cross-species sequence comparison, DNase I hypersensitivity mapping and transcriptional activity analysis in transient transfection assays and we then used a transgenic approach to test the functional significance of candidate cis-regulatory elements in vivo. Our results indicate that unusual as well as common transcriptional regulatory mechanisms control expression of the CD69 gene in different lymphocyte populations.
Materials and Methods
Comparative genomic analysis
Sequence comparison of mouse, human and dog CD69 was performed using VISTA Browser from the Lawrence Berkeley National Laboratory and available at http://pipeline.lbl.gov/cgi-bin/gateway2.
DNase I Hypersensitivity assay
Rag2−/−x Tcrb transgenic mice (RXβ) and hemaglutinin (HA)-TCR transgenic mice were described previously (22, 23). Red blood cells were lysed in 0.15M NH4Cl, 1mM KHCO3, 1mM Na2-EDTA, pH 7.4 for 3min at 4°C and subsequently washed in 30ml of cold PBS. Thymocytes (107/ml) were permeabilized with 0.067mg/ml lysolecithicin in buffer C (0.15M Sucrose, 80mM KCl, 30mM Hepes pH7.4, 5mM MgCl2, 5m CaCl2) for 5 minutes. DNase I was added at a final concentration of 0, 4, 8, 12, 16, 20, 24, 28, or 32 units/ml for 10min on ice. Reactions were stopped by the addition of EDTA, SDS and Proteinase K to final concentrations of 10mM, 0.4% (w/vol) and 0.4mg/ml, respectively, and were incubated overnight at 37°C. DNA was purified by phenol, phenol:chloroform and chloroform extractions and ethanol precipitation, taking care not to shear the genomic DNA. Purified DNA (10μg) was incubated overnight at 37°C with an excess of EcoRI restriction enzyme. Digests were separated by 0.7% (wt/vol) agarose gel electrophoresis and were analysed by Southern blot with 32P-labeled DNA probes (primers to generate probes, Supplementary Table 1).
Reporter constructs and transgenic mice
hCD2 reporter constructs were generated by modification of the mCD8 reporter vector (24). To generate construct 1 (fig. 4B), the promoter of mCD69 (−645/+1) was PCR amplified (Expand High Fidelity PCR System, Roche) and cloned into pstBlue vector (Vector Acceptor Kit, Novagen). Restriction sites introduced during PCR amplification were used to excise the mCD69 promoter from this plasmid with ApaI and ClaI and ligate it to the ApaI/ClaI digested hCD2 construct. To ensure that we incorporated conserved sequences and DNase I hypersensitive sites, primers for genomic regions encompassing CNS1 (233bp), CNS2 (451bp), CNS3 (439bp) and CNS4 (181bp) were designed (primer sequences, Supplementary table 1). PCR products for CNS1-4 fragments were 807bp, 932bp, 600bp and 707bp in length respectively. To generate construct 2, CNS1 and CNS2 were first combined into pstBlue cloning vector. CNS2/CNS1 fragment was then excised from this construct using ApaI and XbaI and inserted into XbaI/ApaI sites upstream of the mCD69 promoter in construct 1. Similarly, to make construct 3, CNS3 and CNS4 were combined into pstBlue, digested with XbaI and inserted into the XbaI site upstream of CNS2 in construct 2. Importantly, NotI restriction sites were flanking all three hCD2 reporter constructs. Construct 1 had a size of 3.7 kb and was separated from vector backbone using ApaI/NotI digestion. Construct 2 was 5.4 kb and was separated with XbaI/NotI digestion and construct 3, with a size of 6.7 kb, with NotI digestion. Purified DNA was then microinjected into fertilized eggs. Founders were identified by PCR and copy number was determined by quantitative real-time PCR. Input DNA was determined using primers ExonIIIF and ExonIIIR (primers sequences, supplementary table 1). Handling of mice and experimental procedures were in accordance with institutional requirements for animal care and use.
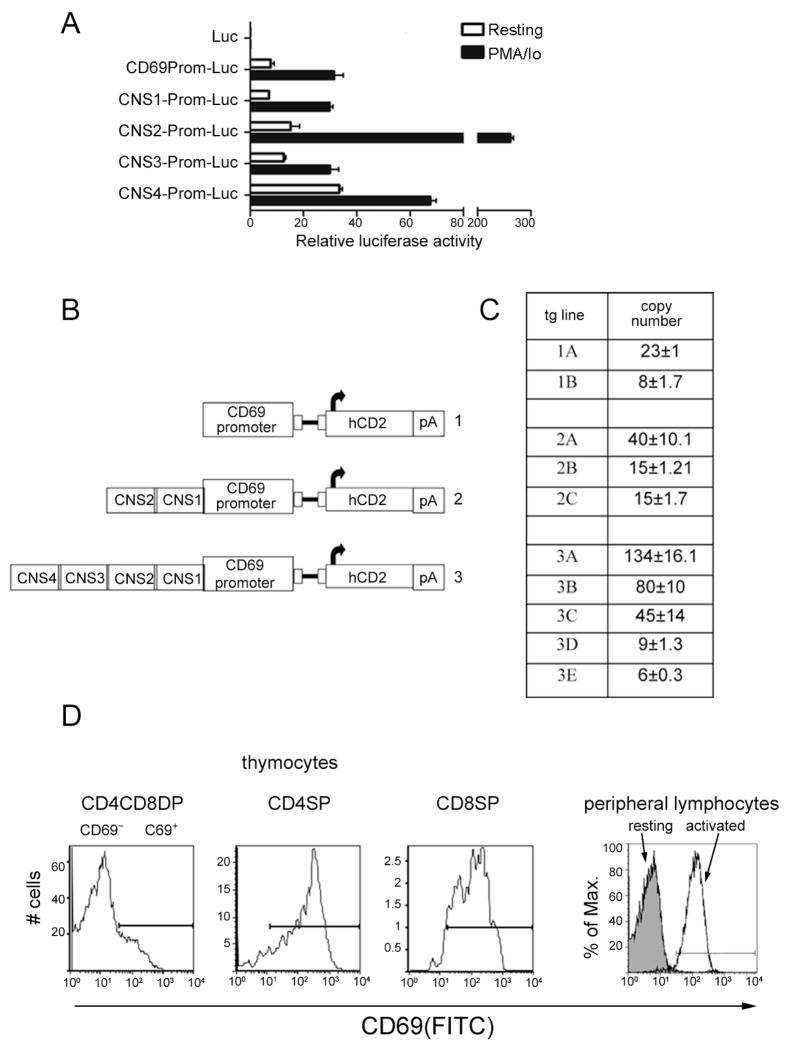
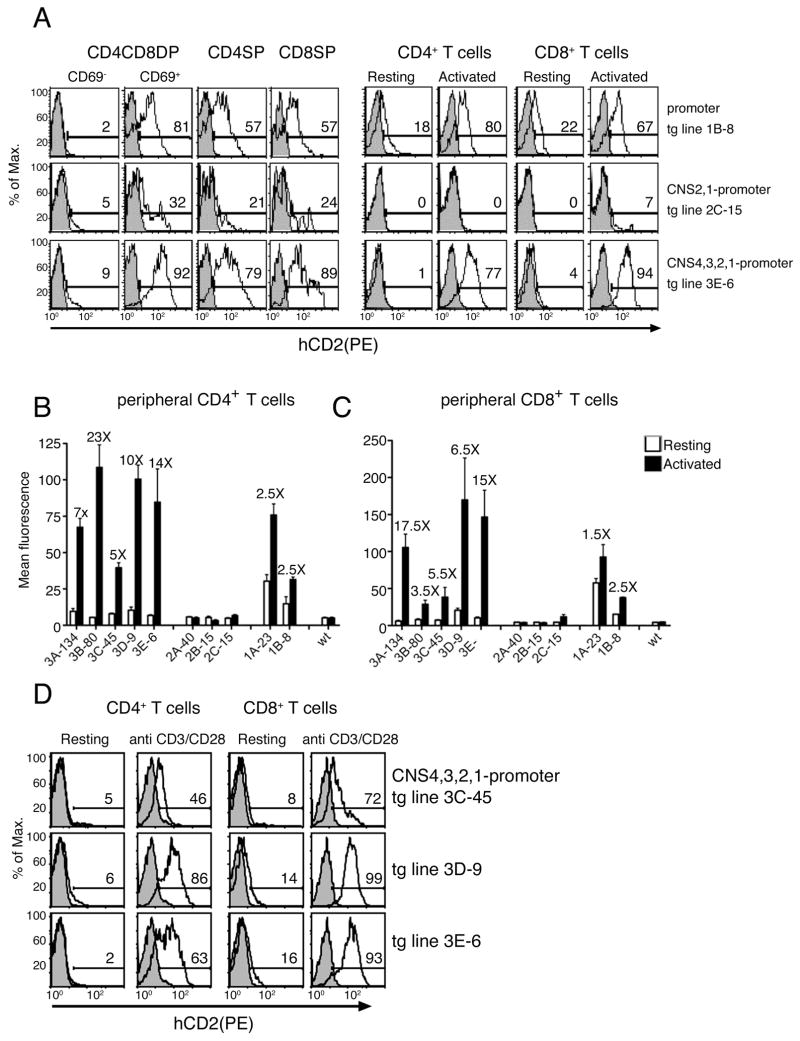
Figure 4. Characterization of CNS function in transfected cells and transgenic mice.
(A) Luciferase activity of reporter constructs in unstimulated or stimulated Jurkat cells. Results are expressed as the mean ±SEM of duplicate transfections and are representative of three experiments. (B) Representation of hCD2 reporter constructs used to generate transgenic mice. (C) Copy numbers of transgenic lines were determined by real time PCR. Values for construct 1 represent the average amplification using hCD2 primers. Values for construct 2 represent the average amplification using CNS1 and hCD2 primers, and values for construct 3 represent the average amplification using CNS4, CNS3, CNS1 and hCD2 primers. (D) Flow cytometric analysis of CD69 expression in thymus and resting and activated peripheral lymphocytes, indicating gating for CD69+ and CD69− populations.
Purification of cell populations and cell culture
To enrich for T cell populations, lymph node cell suspensions were passed through a nylon wool column (Polysciences) following the manufacturer’s instructions. T cell purity, determined by flow cytometry, was >85%. Splenocytes of TCRβ−/−TCRδ−/− mice were used to obtain B cells (>95% B220+ by flow cytometry).
Prior to chromatin immunoprecipitation analysis of peripheral lymphocyte populations, cells were stimulated in vitro with 10ng/ml PMA (Sigma Biochemical) and 0.5μM Calcium Ionophore (Sigma) for 6h at 37°C. CD69 expression was then analyzed by flow cytometry.
For hCD2 analysis in peripheral lymphocytes, splenocytes of wild-type and transgenic mice were activated by incubation with 10ng/ml PMA and 1μM Io or 5μg/ml plate-bound anti-CD3 (clone 145-2C11, eBiosciences) and anti-CD28 (clone 37.51, eBiosciences) monoclonal antibodies overnight at 37°C. In experiments using PMA plus ionomycin, data was collected from experiments in which activated lymphocytes were >90% CD69+.
Poly (I:C) treatment
Mice were injected intraperitoneally (i.p.) with 500μg of Poly (I:C) (Sigma). After 18h of treatment, spleens were obtained from animals and lymphocyte suspensions were prepared for FACS analysis.
Chromatin immunoprecipitation
For analysis of thymocytes, immunoprecipitations were performed on purified mononucleosomes as described previously (25). Briefly, thymocytes were lysed and nuclei were treated with microccocal nuclease to produce a partial chromatin digest. After removal of linker histone H1, chromatin was fractionated on a sucrose gradient. For analysis of peripheral lymphocytes, immunoprecipitations were performed on paraformaldehyde-crosslinked chromatin prepared as described previously (26). Sonication was used to obtain DNA fragments ranging from 300 to 500 bp. With either approach, anti-diacetylated H3, anti-dimethylated H3 K4 and control rabbit-IgG antibodies (Upstate Biotechnology) were used for immunoprecipitation and bound and input fractions were quantified using SYBR green real-time PCR (Roche). Analysis of the constitutively active carbamoyl transferase dihydrorotase (CAD) gene was used to normalize values of different samples. Primer sequences are provided in supplementary Table 1.
Flow cytometry
Cell suspensions from spleen, thymus and bone marrow were stained with FITC-CD69 (BD Bioscience), PE-hCD2 (Caltag), PerCP-CD4 (BD Bioscience), APC-CD8 (BD Bioscience), FITC-CD24 (HSA, eBiosciences), PE-Cy5 IgM (eBioscience) or APC-B220 (eBioscience) antibodies. Data was collected on a FACSCalibur or FACSCanto (BD Bioscience) and was analyzed using Flow Jo software.
Luciferase reporter constructs and cell transfections
For the firefly luciferase vector we used the pXPG vector (27). Primers used to amplify fragments were the same as for the hCD2 reporter constructs with modifications in the restriction sites (PromF contained a XhoI restriction site and other primers a BamHI restriction site). The CD69 promoter was digested with XhoI and BamHI and ligated to XhoI/BamHI digested pXPG-Luc vector. CNS1-4 fragments were digested with BamHI and ligated to BamHI digested CD69 prom-pXPG-Luc vector. Construct sequences were confirmed by restriction enzyme digestion and sequence analysis.
For transient transfection assays, a total of 5×105 Jurkat cells were plated into a 24-well plate and transfected with 1μg of specific firefly luciferase test plasmid, 20ng of pRL-TK renilla luciferase control plasmid and 4μl of Superfect (Qiagen) according to manufacturer’s instructions. Twenty-four hours after transfection, cells were stimulated with 10ng/ml PMA and 1μM Io or 5μg/ml plate bound anti-CD3 (clone OKT3, eBiosciences) and anti-CD28 (clone CD28.2, eBiosciences) monoclonal antibodies or were mock incubated. A luciferase assay was performed twenty-four hours later using the Dual-Luciferase Reporter Assay System (Promega). Transfections were performed in duplicate and values were normalized to Renilla luciferase activity.
Results
Conservation of the CD69 locus
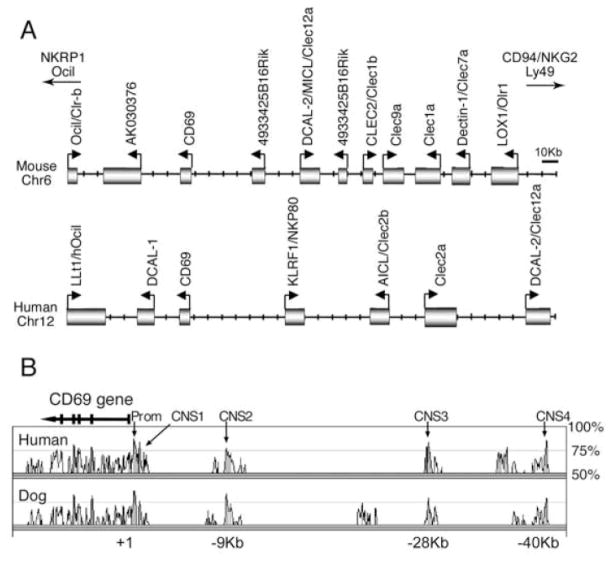
CD69 is a type II transmembrane C-type lectin encoded in the NK complex on mouse chr6 and human chr12 (Fig. 1A). The gene spans approximately 7.5 kb and contains 5 exons. The first two exons encode the cytoplasmic and transmembrane domains and exons III, IV and V encode the extracellular portions of the molecule. The defined murine CD69 promoter (−656 to +1 relative to the transcription start site) is the only cis-acting element known to regulate CD69 expression (13). We wanted to identify other potential cis-acting sequences involved in CD69 gene regulation. As cross-species genome analysis has been useful for this purpose (28), we compared mouse, human and dog genomic sequences by the means of VISTA Browser (29). Using the default parameters for defining a conserved non-coding sequence (CNS) element (70% identity over 100bp length), four elements upstream of the CD69 gene were identified (Fig. 1B). CNS1was upstream of and contiguous to the promoter and CNS2, CNS3 and CNS4 were 9 kb, 28 kb and 40 kb away from the main site of transcription initiation, respectively. Marked conservation was also observed at the promoter. As there are substantial differences in genomic organization of the human and mouse NK complexes upstream of CD69 (Fig. 1A), we hypothesized that the four CNSs may regulate CD69 rather than neighbouring gene expression.
Figure 1. Genomic organization and conservation of the CD69 locus.
(A) Genomic organization of the human and mouse CD69 gene and neighbouring genes in the natural killer complex. The diagram is drawn to scale according to the most recent gene annotations (July 2007 for mice and March 2006 for human) at the University of California Santa Cruz web site (http://genome.ucsc.edu). Boxes are genes and arrows indicated transcriptional initiation sites and orientation. (B) VISTA Browser diagram identifying conserved noncoding sequences upstream of the CD69 gene. The mouse sequence is shown on the X axis and percentage of similarity to the human and dog genomes on the Y axis. Non-coding sequences of at least 100-bp long with more than 70% sequence identity are shown are indicated with arrows.
Accessibility of the CD69 locus
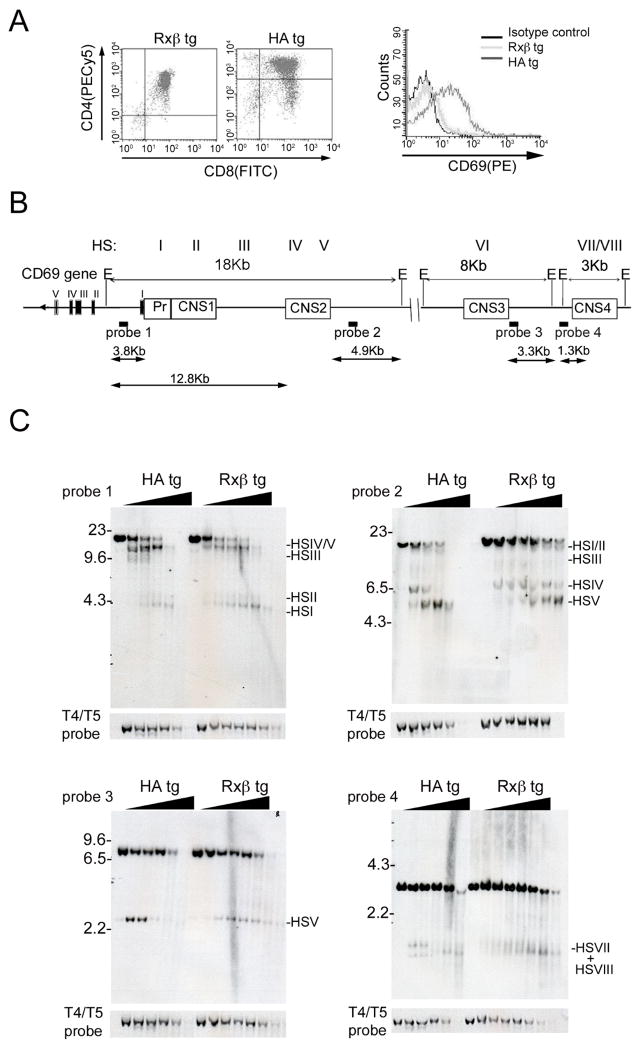
Chromatin regulatory regions are typically characterized by DNase I hypersensitive sites (HSs), that reflect nuclear factor binding and disrupted nucleosome organization. We conducted DNase I HS assays and southern blots to determine whether CNS1-4 represented DNase HSs. CD69 is upregulated by thymocyte positive selection and different TCR transgenic mouse strains express characteristic quantities of CD69 based on both the selecting background and the strength of TCR-MHC:peptide interactions (30). We used thymocytes from B10.D2 H-2Kd mice expressing a transgenic TCR specific for a hemagglutinin peptide presented by H-2Kd (HA) to isolate thymocytes with high CD69 expression and thymocytes from Rag2−/−mice that express a Tcrb transgene (Rxβ) to isolate thymocytes with low CD69 expression (Fig. 2A). Total thymocytes from these strains were treated with varying concentrations of DNase I and genomic DNA was extracted, digested with EcoRI, and analyzed by southern blot (Fig. 2B). A site within the inactive trypsinogen gene was used as an internal control to compare chromatin digestion in different samples. Probes 1 and 2, which hybridized to different ends of the same genomic EcoRI fragment (Fig. 2B), detected five distinct HSs (Fig. 2C, upper panels). HS I and II corresponded to the CD69 promoter and CNS1, respectively, whereas HSIV and HSV mapped to CNS2. HSIII was evident as a cluster of weak HSs that was only detected in Rxβ and that mapped to a less conserved sequence between CNS1 and CNS2. HSVI, corresponding to CNS3, was detected using probe 3 (Fig. 2C, lower left panel). Probe 4 revealed weak HSs VII and VIII that corresponded to CNS4 (Fig. 2C, lower right panel). Hence these experiments identified eight distinct DNase I hypersensitive sites (HSs) mapping predominantly to the defined CNSs. CD69+ and CD69− cells presented similar patterns of DNase I sensitivity.
Figure 2. Conserved noncoding sequences are constitutive hypesensitive sites.
(A) Flow cytometry analysis showing CD69 expression in Rxβ and HA transgenic thymi. (B) Map of the CD69 locus showing locations of CNSs and the genomic fragments expected from EcoRI digestion. Probes used for southern blot are shown and HSs are indicated. (C) Representative DNase I analyses using the indicated probes. Total thymocytes from Rxβ and HA thymocytes were treated with increasing amounts of DNase I. EcoRI digested DNA was examined by Southern Blot. Size markers and relevant bands are denoted.
CD69 locus histone modifications
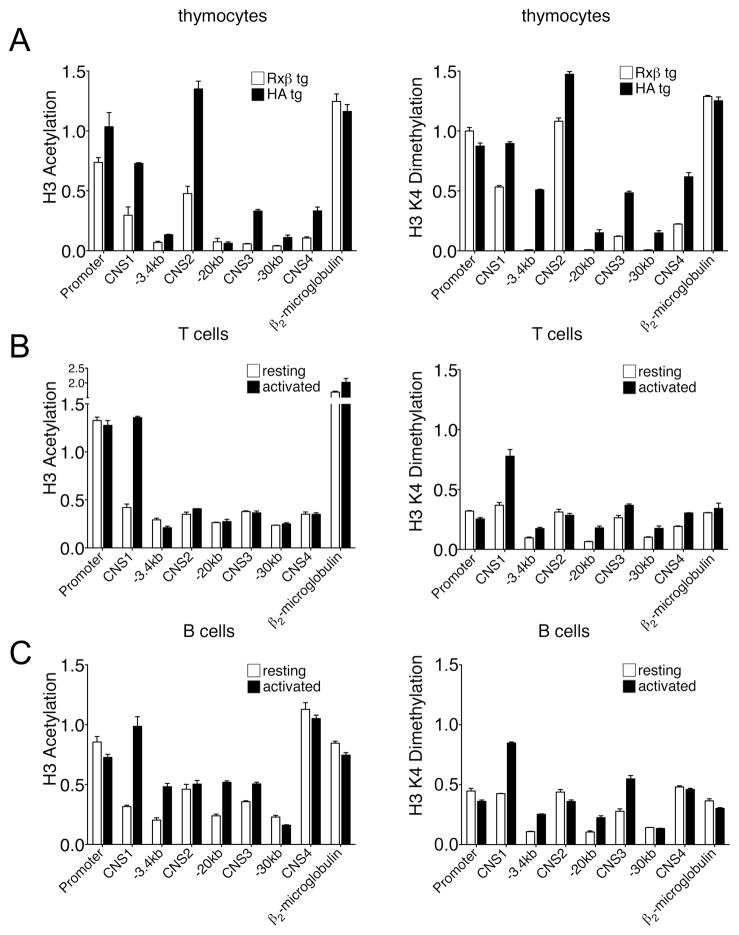
One mechanism by which chromatin structure participates in the regulation of gene expression, is through the modification of histone tails (31). Histone H3 acetylation and lysine 4 dimethylation are associated with active and poised chromatin, respectively(32). We assessed their levels by chromatin immunoprecipitation at conserved genomic regions (CD69 proximal promoter, CNS1, CNS2, CNS3, and CNS4) as well as at several nonconserved regions (−3.4 kb, −20 kb and −30 kb). The β2-microglobulin gene was used as positive control.
Mononucleosomes were prepared from Rxβ and HA tg thymocytes and were then immunoprecipitated with anti-acetylated histone H3, anti-dimethylated histone H3 K4 or control IgG antibodies. Coprecipitated DNA was purified and subjected to real time PCR to quantify the recovery of regions of interest (Fig. 3A). Results are expressed as the ratio of immunoprecipitated and input DNA normalized to the constitutively active control gene CAD. The promoter demonstrated high levels of H3 acetylation and H3 K4 dimethylation in both CD69− Rxβ thymocytes and CD69+ HA tg thymocytes. CD69− Rxβ thymocytes demonstrated moderate acetylation at CNS1 and CNS2, and very low levels at CNS3 and CNS4. CD69+ HA tg thymocytes displayed increases in H3 acetylation at all of these sites. A similar overall pattern was observed for H3 K4 dimethylation, with the exception that this modification was only modestly increased at CNS2 in CD69+ cells. With one exception (H3 K4 dimethyation at position −3.4 kb in CD69+ thymocytes), the two modifications were very low at all nonconserved sites tested.
Figure 3. Epigenetic profile of the CD69 locus in T and B cells.
Chromatin immunoprecipitation with antisera against acetylated H3 and dimethylated H3 K4. (A) ChIP performed on purified mononucleosomes obtained from total thymocytes of Rxβ and HA transgenic mice. (B) ChIP performed on sonicated chromatin from purified T cells either unstimulated or stimulated with 10ng/ml PMA plus 0.5μM Io for 6h. Purity as determined by flow cytometry was greater than 85%. (C) ChIP performed on sonicated chromatin from B cells isolated from TCRδ−/−β−/− mice, either unstimulated or stimulated with 10ng/ml PMA plus 0.5μM Io for 6h. Flow cytometry analysis showed that more than 95% of spleen cells were B220+. Bars represents the abundance of indicated DNA sequences in immunoprecipitated samples and are expressed as the ratio of immunoprecipitated DNA and input DNA normalized to the abundance of the constitutively active control CAD gene. The data are representative of two experiments and are expressed as the mean ± SEM of triplicate PCRs.
To investigate CD69 locus chromatin structure in peripheral lymphocytes, lymph node T cells and splenic B cells were treated in vitro with PMA and ionomycin for 6h to upregulated CD69 expression or were left untreated. Chromatin was then cross-linked with paraformaldehyde, fragmented by sonication, and immunoprecipitated as above. As in thymocytes, acetylation of H3 and dimethylation of H3 K4 were enriched at the promoter in resting and activated T and B cells (Figs. 3B,C). Also as in thymocytes, H3 acetylation and H3 K4 dimethylation increased substantially at CNS1 upon activation of both peripheral T and B cells (Figs. 3B, C). However, unlike in thymocytes, we detected only very low H3 acetylation at CNS2, CNS3 and CNS4 in both resting and activated peripheral T cells. Moreover H3 K4 dimethylation at these sites, although mildly elevated in resting T cells, was not inducible. B cells displayed increased amounts of both modifications at CNS2, CNS3 and CNS4, but only CNS3 showed inducible modification. Moreover, in striking contrast to thymocytes, H3 acetylation and H3 K4 dimethylation were constitutively high at CNS4 in B cells.
The above experiments revealed three important aspects of CD69 gene chromatin structure. First, the CD69 promoter is constitutively associated with active chromatin modifications in all three cell types. Second, CNS1 becomes hyperacetylated and hypermethylated upon CD69 induction in all three cell types. Third, histone modifications at CNS2, CNS3 and CNS4 undergoes dynamic changes during T cell development but are differentially modified in peripheral T and B cells. This suggests the possibility of distinct mechanisms of CD69 gene regulation in the various cell types.
Analysis of promoter and CNS1-4 function
The correlation between DNA conservation and the presence of HSs and positive histone marks at the promoter and CNS1-4 prompted us to test their regulatory properties. We prepared luciferase reporter constructs under the control of the CD69 promoter linked to CNS fragments (Fig. 4A). Plasmids were transiently transfected into Jurkat cells, cultured for 24hr, and then stimulated or not with PMA/Io for 24hr more before cells extracts were harvested for luciferase activity. Stimulation conferred a 4-fold increase in the activity of the CD69 promoter. Inducible activity of the CD69 promoter, however, was greatly enhanced in the presence of CNS2. This fragment conferred a 2-fold increase in basal activity and 20-fold increase over basal activity under conditions of stimulation. CNS4 conferred an approximately 5-fold increase in basal transcription activity and a further 2-fold increase under conditions of stimulation. In contrast, reporter activity was not altered when CNS1 or CNS3 was linked to the CD69 promoter. Luciferase activity of these constructs was also determined upon stimulation with immobilized antibodies against CD3 and CD28 (Supplementary Fig. 1). CNS2 was found to confer inducibility to this stimulus, although the magnitude (3-fold) was lower than with PMA/Io. These results provide initial evidence that CNS2 and CNS4 may be enhancers elements for CD69 gene transcription.
To assess their in vivo functional properties, transgenic mice were generated using a hCD2 expression construct as a reporter. Transgenic constructs containing the mCD69 promoter alone upstream of hCD2 (construct 1) or together with all four CNS (construct 3) were generated (Fig. 4B). Due to the strong enhancer activity observed for CNS2 element in luciferase assays, transgenic mice containing CNS1 and CNS2 upstream of the promoter (construct 3) were also generated. Two transgenic lines were obtained for construct 1, three for construct 2 and five for construct 3 (Fig. 4C). Copy numbers were determined by quantitative real time PCR (Fig. 4C).
We assessed the magnitude and fidelity of hCD2 reporter expression on gated CD69+ and CD69− thymocytes and on purified resting or activated mature lymphocyte populations by flow cytometry (Fig. 4D). Tg lines 1A-23 and 1B-8 (containing 23 and 8 copies of the transgene, respectively) that contained only the CD69 promoter, expressed hCD2 in both thymocytes and peripheral lymphocyte populations (Fig. 5A and supplementary Figs. 2A, 3A). hCD2 expression in DP and SP thymocyte populations was strictly correlated with endogenous CD69 expression, suggesting that the promoter was sufficient to confer specificity to CD69 expression during positive selection (Fig. 5A). However, aberrant expression of hCD2 was observed in resting peripheral T (Fig. 5A) and B (Fig. 6A) lymphocytes, suggesting the need for additional elements.
Figure 5. Reporter expression in thymocytes and peripheral T cells of transgenic mice.
(A) Cytometric analysis of hCD2 expression in transgenic lines 1B-8, 2C-15 and 3E-6 from thymocytes (CD69+ and CD69− CD4CD8DP, CD4SP and CD8SP) and splenocytes (resting or activated with PMA/Io, CD4+ and CD8+) (open histograms). Analyses of control non-transgenic mice are also shown (shaded histograms). (B) and (C) Mean fluorescence intensity of the hCD2 expression in resting and activated peripheral CD4+T cells and CD8+ T cells. Values reflect the mean ± SEM of 5 to 10 mice. For each transgenic line, fold-increase in MFI in response to activation is indicated above the bars. (D) Cytometric analysis of hCD2 expresion in transgenic lines 3C-45, 3D-9 and 3E-6 activated or not with plate bound anti CD3/CD28 antibodies. Analyses of control non-transgenic mice are also shown (shaded histograms).
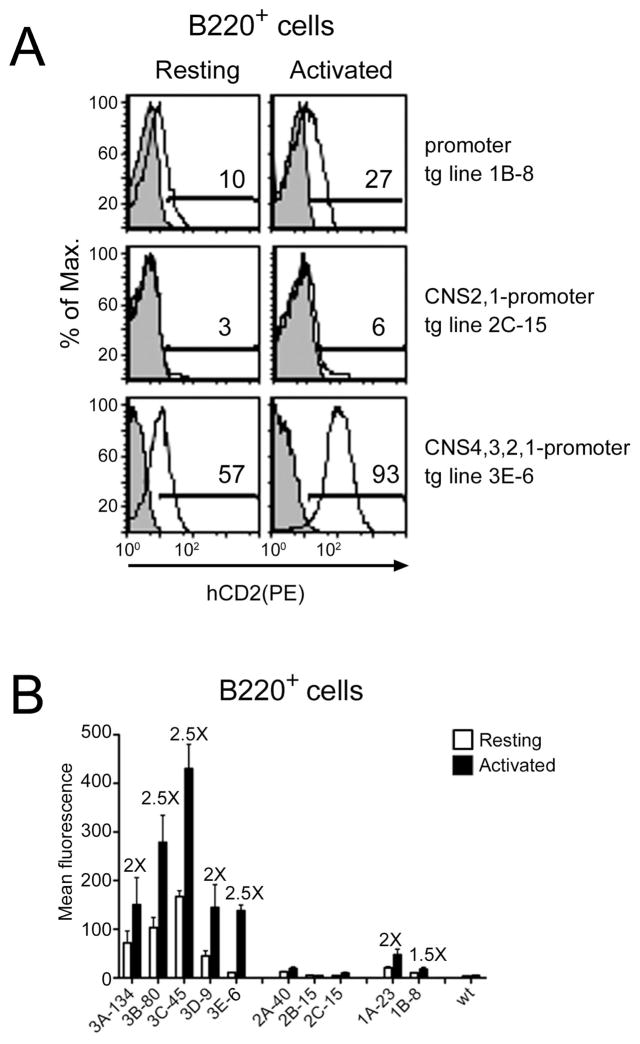
Figure 6. Reporter expression in resting and activated peripheral B cells of transgenic mice.
(A) Cytometric analysis of hCD2 expression in transgenic lines 1B-8, 2C-15 from resting and activated with PMA/Io B220+ splenocytes (open histograms). Analyses of control non-transgenic mice are also shown (shaded histograms). (B) Mean fluorescence intensity of the hCD2 expression in resting and activated peripheral B220+ cells. Values reflect the mean ± SEM of 5 to 10 mice. For each transgenic line, fold-increase in MFI in response to activation is indicated above the bars
We tested for inducibility of the mCD69 promoter by overnight activation of isolated peripheral T and B lymphocytes with PMA and ionomycin. Because even resting cells expressed hCD2, we used the mean fluorescence intensity (MFI) of the whole population to measure changes in hCD2 expression. Inducibility was calculated as the ratio of the MFI for the hCD2 staining between activated and resting cells. We detected 1.5- to 2.5-fold hCD2 inducibility in activated T cells (Figs. 5B, 5C) and 1.5- to 2-fold inducibility in activated B cells (Figs. 6B), suggesting that promoter elements support some inducibility in vivo.
Unexpectedly, inclusion of CNS1 and CNS2 upstream of the CD69 promoter (tg lines 2A-40, 2B-15 and 2C-15) suppressed hCD2 expression in both thymocytes and peripheral T and B cell populations (Figs. 5A, 6A and supplementary Figs. 2B, 3B). hCD2 expression was substantially reduced in CD69+ DP and SP thymocytes, and was essentially eliminated in resting and activated peripheral CD4+ and CD8+ T cells (Fig. 5A). Expression was also eliminated in resting and activated peripheral B cells in two of the three tg lines (Fig. 6A supplementary figure 3B). These data suggest that CD69 gene expression in lymphocytes is regulated by silencer elements mapping to CNS1 or CNS2.
In contrast to the results obtained with construct 2, transgenic lines carrying construct 3, including the CD69 promoter and all four CNSs (tg lines 3A-134, 3B-80, 3C-45, 3D-9 and 3E-6) revealed a recovery of hCD2 expression (Figs.5A, 6A and supplementary Figs. 2C, 3C). Expression of the hCD2 reporter correlated well with CD69 expression in all thymocyte populations, ranging from 92% to 22% in DP thymocytes and 89% to 15% in CD4SP and CD8SP thymocytes (supplementary Fig. 2C). Within the SP compartments, hCD2 expression also correlated with mCD69 expression, since downregulation of CD69 and HSA in more mature SP thymocytes was associated with downregulation of reporter expression (Supplementary Fig. 4). Notably, high copy tg lines 3C-45, 3B-80 and 3A-134, but not low copy lines 3E-6 and 3D-9, displayed variegated hCD2 expression indicative of genomic position effects. In the low copy lines hCD2 expression faithfully mimicked mCD69 expression, and expression levels were greater than those observed with construct 1, containing the promoter alone (compare tg line 1B-8 and 3E-6 in Fig. 5A).
Analysis of construct 3 expression in mature CD4+ and CD8+ T cells stimulated with PMA/Io revealed hCD2 expression to be restricted to activated cells in four of five tg lines (3E-6, 3C-45, 3B-80 and 3A-134) with percentages ranging from 99% to 10% (supplementary Fig. 2C). However, hCD2 expression was inducible in all five lines (range 23-fold to 3.5-fold) and inducibility was substantially higher than for tg lines containing construct 1 with the promoter alone. Tg line 3E-6, with the lowest copy number, was the most tightly regulated, with negligible hCD2 expression in resting CD4+ and CD8+ T cells, and abundant hCD2 expression in the vast majority of activated CD4+ and CD8+ T cells (Figs. 5A). Inducibility was 14-fold and 15-fold in CD4+ and CD8+ T cells, respectively (Figs. 5B, 5C). As in thymocytes, expression was highly variegated in the three tg lines with the highest copy numbers (3C-45, 3B-80 and 3A-134) (supplementary Fig. 2C).
Expression of construct 3 in peripheral T cells was also inducible following direct engagement of TCR with anti-CD3 plus anti-CD28 antibodies (Fig. 5D). Moreover, the kinetics hCD2 expression paralleled mCD69 expression, since both were detected 2h after stimulation, were maximally detected at 12–24h and expression was reduced thereafter (supplementary Figs. 5A, B). Since CD69 expression can be induced in vivo by infectious agents that cause in the production of type I interferons, we also analyzed hCD2 induction following in vivo administration of poly I:C (supplementary Fig. 6). The hCD2 expression was induced after poly I:C treatment indicating that CNSs can also respond to stimuli distinct from the TCR pathway. These results suggest that the combination of CNS1-4 contributes to regulated CD69 expression in both thymocytes and peripheral T cells under a variety of activation conditions.
We further examined the expression of hCD2 in B cells. The combination of CNS1-4 clearly enhanced hCD2 expression as compared to the promoter alone (Figs. 6A and supplementary Fig. 3C). However, hCD2 expression was not concordant with mCD69 expression, since substantial numbers of resting B cells were hCD2+ in all five transgenic lines (range 22% to 79%)(Supplementary Fig. 2C). This may reflect the constitutive and abundant H3 acetylation and H3 K4 dimethylation detected at CNS4 in B cells (Fig. 3C). Stimulation with PMA and ionomycin resulted in 2–2.5-fold inducibility in the various lines (Fig. 6B). However, the failure to appropriately suppress expression in resting B cells suggests that there are different requirements for regulated CD69 gene expression in B and T cells.
As dysregulated expression of hCD2 was observed in mature B cells, we evaluated whether CNSs and the promoter may have an effect on hCD2 expression at different stages of B cell development (supplementary Fig. 7). Results indicate that hCD2 expression was already present in B220intIgM+ immature B cells in transgenic lines containing the promoter alone or in combination with CNS1-4 elements and suggests that specific signals received at this stage of development induce the disregulated expression of hCD2 that is also observed in mature B cells.
Collectively, these experiments indicate that the CD69 promoter has strong activity and by itself can faithfully direct CD69 transcription in developing thymocytes. CNS1 and CNS2 appear to repress promoter activity whereas CNS3 and CNS4 appear to counteract this repression, resulting in more tightly regulated expression in peripheral T lymphocytes and enhanced expression in both thymocytes and peripheral T lymphocytes. The same elements are insufficient to faithfully direct CD69 expression in the B cell lineage.
Discussion
In this study we have provided insights into the mechanism that regulate CD69 gene expression in vivo by the identification of new cis-regulatory elements and analysis of their chromatin structure and function. We identified four conserved noncoding sequences upstream of the CD69 promoter. Chromatin and functional analyses indicated that the CD69 promoter adopts an open chromatin conformation and can direct reporter gene transcription in transgenic mice. However, the reporter, unlike mCD69, was expressed in unstimulated peripheral lymphocytes, suggesting that other elements must participate to achieve appropriate specificity. We detected several DNase I hypersensitive sites that mapped to conserved noncoding sequences (CNSs) and that displayed epigenetic profiles that were distinct in T and B cells and dynamic during T cell development. Analysis of CNS function in transient transfection assays revealed constitutive and inducible enhancer activity for both CNS2 and CNS4, but no apparent activity for CNS1 and CNS3. However, we found that transgenic mice bearing CNS1 and CNS2 reduced reporter expression in T and B cells and that this inhibition was only overcome by inclusion of CNS3 and CNS4. The combination of all four CNSs allowed for high level reporter expression that was appropriately regulated in T cells but not B cells.
DNA sequence comparison has become a useful tool to identify specific remote elements that participate in the regulation of gene expression (19, 33, 34). We observed discrete islands of conservation in a 45 kb region upstream of the CD69 gene and found them to correspond to sites displaying hypersensitivity to DNase I digestion. It is noteworthy that these HSs were equally sensitive to DNase I digestion in CD69− and CD69+ lymphocytes. CD69 is one of the earliest antigens expressed upon activation and, therefore, it is possible that these regions are occupied by proteins even before activation. In accord with our results, the first genome-wide map of DNase I HSs in human CD4+ T cells identified HSs lying within the CD69 promoter, CNS2, CNS3 and CNS4 in the human CD69 locus (35).
Previous work showed that the CD69 promoter can support rapid induction of reporter gene expression in transient transfection assays. Here we found that the CD69 promoter is always associated with an “active” chromatin configuration, even in situations where CD69 is not expressed. The promoter displayed high level histone H3 acetylation and H3 K4 dimethylation in all thymocyte and peripheral lymphocyte populations. These two modifications have been shown to mark transcriptional competence of the IL-4 and IL-2 loci, respectively, in T cells (36, 37) and may be important to maintain transcriptional competence of the CD69 gene as well. Interestingly, genome-wide analysis of chromatin modifications in unstimulated human CD4+ T cells demonstrated enrichment of both histone H3 K4 trimethylation and RNA polymerase II at the promoter, exon I and intron I of mCD69 (34). These data suggest that, as for other genes that require rapid induction, the CD69 promoter may constitutively harbor a promoter-paused RNA polymerase II (38, 39). CNS1 is the distal region of the C69 promoter and one notable result is that this region showed a peak of permissive histone marks that correlated with CD69 expression in all cell populations analysed. However, the promoter alone showed inducible activity in both transient and transgenic reporters, and at least in the case of transient assays, CNS1 had no influence on basal or inducible promoter activity. This suggests that inducible histone modifications at this site may depend on other elements.
Tissue- and developmental stage-specific gene expression depends on both activators and silencers. A well-studied case is that of CD4, in which a silencer represses the CD4 promoter and enhancer in DN and CD8 SP thymocytes (40, 41). Similarly, regulated IL-4 transcription requires the action of a 3′ silencer element that can suppress reporter gene expression in transgenic mice (18) and whose germline deletion results in the aberrant expression of IL-4 in Th1 cells (34). Moreover, there are examples of bifunctional elements, which function as both enhancers and silencers in a developmental-stage specific way (42, 43). Our results from transgenic mice indicates that CNS1, CNS2, or the combination of the two elements can play a repressive role in CD69 gene expression in thymocytes, as well as stimulated T and B cells. Nevertheless, CNS2 displayed strong, inducible enhancer activity when tested in isolation by transient transfection into Jurkat. Notably, the detected enhancer activity is consistent with the inducible histone H3 acetylation detected at this site in thymocytes. We suggest that CNS2 is an inducible thymocyte enhancer whose activity in the context of chromatin requires the activity of other CD69 regulatory elements. Remarkably, inclusion of CNS3 and CNS4 in transgenic reporter constructs led to both a recovery of reporter gene expression and tightly regulated reporter gene expression in thymocytes and peripheral T cells. Based on its inducible enhancer activity in Jurkat and inducible histone acetylation in thymocytes, CNS4 appears to function as a thymocyte enhancer as well. Thus we suggest that CNS4 may not only function as an enhancer in the context of the endogenous CD69 locus, but may, perhaps in conjunction with CNS3, function as an anti-silencer that switches CNS2 from silencer to enhancer activity. We propose that all four CNSs may converge to interact physically and functionally in the form of an active chromatin hub, as initially described for the b-globin locus (44). Perhaps consistent with this, a previous study suggested that CD69 may be regulated by the architectural protein SATB1(45).
We note that our conclusions about the activities of CNS1-4 in vivo assume that these elements function normally in the context of our relatively compact transgenic reporter. Prior studies have validated this approach in other systems (18, 46). However, we cannot rule out that our transgenic constructs lack relevant elements from DNA segments that normally separate the CNSs, or cannot adopt important three dimensional chromatin configurations that are important for physiological regulation. To further study mechanisms of CD69 regulation in vivo we will analyze mice transgenic for a bacterial artificial chromosome containing the mCD69 locus.
As compared to thymocytes, the mechanism of induction of CD69 gene expression in mature T cells appears distinct, as only CNS1 displayed inducible H3 histone acetylation and H3 K4 dimethylation in the latter cell population. This was true despite the fact that, as for thymocytes, CNS2-4 were needed for tightly regulated, inducible expression in peripheral T cells of transgenic mice. We suggest that CNS2-4 activity during T cell development may establish a specific chromatin context that is required for proper regulation of the CD69 locus in mature T cells.
Although the combination of all four CNSs was capable of promoting high level and tightly regulated reporter gene expression in thymocytes and peripheral T lymphocyte populations, it did not do so in all transgenic lines. One explanation for this is that additional elements may be required to reconstitute a CD69 locus control region that can consistently overcome chromosomal position effects. However, we note that hCD2 expression was both low and variegated in those transgenic lines containing particularly high copy reporter arrays. Thus it is possible that RNA interference may cause transgene silencing in these lines(47).
The unexpected finding that transgenic mice with all CNSs mis-expressed the hCD2 reporter in unstimulated B cells indicates that CD69 gene transcription is differentially controlled in B and T cells. Thus it seems likely that the construct lacked an element required to repress the expression in unstimulated B cells. Further evidence for differential regulation in T and B cells was obtained from the epigenetic profile of CD69, which revealed distinct patterns of histone modifications at CNS2, CNS3 and CNS4 between T and B cells. In this regard, IL-2 transcription is thought to be regulated by different mechanisms in CD4 and CD8 lymphocytes and IL-4 and IL-13 transcription is reduced in Th2 cells but not in mast cells in mice deficient for the CNS1 region (48, 49). Interestingly, transgenic mice bearing a 30kb genomic fragment containing Ly49A, another gene in the NK complex that encodes a C-type lectin, also showed aberrant expression in B cells. Perhaps similar mechanisms are used to suppress CD69 and Ly49A expression in B cells (50).
Based on our findings, we propose that the CD69 promoter and upstream elements display an accessible chromatin structure prior to CD69 transcription to support rapid gene induction upon stimulation. Gene induction requires the combined activity of multiple upstream CNSs, two of which display classical enhancer activity in T cells. The interdependence of these CNSs may also allow for physiological repression of CD69 expression by disruption CNS3 and CNS4 interactions with CNS1 and CNS2. Finally, an as yet uncharacterized B cell specific element may be required to suppress CNS activity in unstimulated B cells. The above results indicate CD69 gene regulation is complex and likely differs in different cell types. Future studies are required to elucidate the nature of CD69 regulatory elements and the mechanism through which they regulate CD69 expression.
Supplementary Material
Acknowledgments
We thank Dr. Wilfried Ellmeier (Viena, Austria) for providing the hCD2 containing vector used in this study. We are grateful to Dr. Yiping Yang (Durham, USA) and his lab for kindly providing HA transgenic mice. We also thank the transgenic mouse facility at Duke and Zanchun Huang and Sheila Ortega for maintaining the mouse colonies.
Abbreviations
- CNS
conserved non coding region
- Tg
transgenic
- ChIP
chromatin immunoprecipitation
- DP
double positive
- SP
single positive
- WT
wild type
- MFI
mean fluorescence intensity
Footnotes
This work was supported by grants (SAF2005-04876 and SAF2007-64310) from the Ministerio de Educación, Ciencia y Tecnología. B. Vázquez and T. Laguna were supported by predoctoral fellowships from the Spanish Ministry of Education and Science.
References
- 1.Shiow LR, Rosen DB, Brdickova N, Xu Y, An J, Lanier LL, Cyster JG, Matloubian M. CD69 acts downstream of interferon-alpha/beta to inhibit S1P1 and lymphocyte egress from lymphoid organs. Nature. 2006;440:540–544. doi: 10.1038/nature04606. [DOI] [PubMed] [Google Scholar]
- 2.Feng C, Woodside KJ, Vance BA, El-Khoury D, Canelles M, Lee J, Gress R, Fowlkes BJ, Shores EW, Love PE. A potential role for CD69 in thymocyte emigration. Int Immunol. 2002;14:535–544. doi: 10.1093/intimm/dxf020. [DOI] [PubMed] [Google Scholar]
- 3.Nakayama T, Kasprowicz DJ, Yamashita M, Schubert LA, Gillard G, Kimura M, Didierlaurent A, Koseki H, Ziegler SF. The generation of mature, single-positive thymocytes in vivo is dysregulated by CD69 blockade or overexpression. J Immunol. 2002;168:87–94. doi: 10.4049/jimmunol.168.1.87. [DOI] [PubMed] [Google Scholar]
- 4.Esplugues E, Sancho D, Vega-Ramos J, Martinez C, Syrbe U, Hamann A, Engel P, Sanchez-Madrid F, Lauzurica P. Enhanced antitumor immunity in mice deficient in CD69. J Exp Med. 2003;197:1093–1106. doi: 10.1084/jem.20021337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sancho D, Gomez M, Viedma F, Esplugues E, Gordon-Alonso M, Garcia-Lopez MA, de la Fuente H, Martinez AC, Lauzurica P, Sanchez-Madrid F. CD69 downregulates autoimmune reactivity through active transforming growth factor-beta production in collagen-induced arthritis. J Clin Invest. 2003;112:872–882. doi: 10.1172/JCI19112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esplugues E, Vega-Ramos J, Cartoixa D, Vazquez BN, Salaet I, Engel P, Lauzurica P. Induction of tumor NK-cell immunity by anti-CD69 antibody therapy. Blood. 2005;105:4399–4406. doi: 10.1182/blood-2004-10-3854. [DOI] [PubMed] [Google Scholar]
- 7.Han Y, Guo Q, Zhang M, Chen Z, Cao X. CD69+ CD4+ CD25− T cells, a new subset of regulatory T cells, suppress T cell proliferation through membrane-bound TGF-beta1. J Immunol. 2009;182:111–120. doi: 10.4049/jimmunol.182.1.111. [DOI] [PubMed] [Google Scholar]
- 8.Yamashita I, Nagata T, Tada T, Nakayama T. CD69 cell surface expression identifies developing thymocytes which audition for T cell antigen receptor-mediated positive selection. Int Immunol. 1993;5:1139–1150. doi: 10.1093/intimm/5.9.1139. [DOI] [PubMed] [Google Scholar]
- 9.Lauzurica P, Sancho D, Torres M, Albella B, Marazuela M, Merino T, Bueren JA, Martinez AC, Sanchez-Madrid F. Phenotypic and functional characteristics of hematopoietic cell lineages in CD69-deficient mice. Blood. 2000;95:2312–2320. [PubMed] [Google Scholar]
- 10.Kelley J, Walter L, Trowsdale J. Comparative genomics of natural killer cell receptor gene clusters. PLoS Genet. 2005;1:129–139. doi: 10.1371/journal.pgen.0010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D’Ambrosio D, Cantrell DA, Frati L, Santoni A, Testi R. Involvement of p21ras activation in T cell CD69 expression. Eur J Immunol. 1994;24:616–620. doi: 10.1002/eji.1830240319. [DOI] [PubMed] [Google Scholar]
- 12.Taylor-Fishwick DA, Siegel JN. Raf-1 provides a dominant but not exclusive signal for the induction of CD69 expression on T cells. Eur J Immunol. 1995;25:3215–3221. doi: 10.1002/eji.1830251203. [DOI] [PubMed] [Google Scholar]
- 13.Ziegler SF, Levin SD, Johnson L, Copeland NG, Gilbert DJ, Jenkins NA, Baker E, Sutherland GR, Feldhaus AL, Ramsdell F. The mouse CD69 gene. Structure, expression, and mapping to the NK gene complex. J Immunol. 1994;152:1228–1236. [PubMed] [Google Scholar]
- 14.Lopez-Cabrera M, Santis AG, Fernandez-Ruiz E, Blacher R, Esch F, Sanchez-Mateos P, Sanchez-Madrid F. Molecular cloning, expression, and chromosomal localization of the human earliest lymphocyte activation antigen AIM/CD69, a new member of the C-type animal lectin superfamily of signal-transmitting receptors. J Exp Med. 1993;178:537–547. doi: 10.1084/jem.178.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ziegler SF, Ramsdell F, Hjerrild KA, Armitage RJ, Grabstein KH, Hennen KB, Farrah T, Fanslow WC, Shevach EM, Alderson MR. Molecular characterization of the early activation antigen CD69: a type II membrane glycoprotein related to a family of natural killer cell activation antigens. Eur J Immunol. 1993;23:1643–1648. doi: 10.1002/eji.1830230737. [DOI] [PubMed] [Google Scholar]
- 16.Castellanos Mdel C, Lopez-Giral S, Lopez-Cabrera M, de Landazuri MO. Multiple cis-acting elements regulate the expression of the early T cell activation antigen CD69. Eur J Immunol. 2002;32:3108–3117. doi: 10.1002/1521-4141(200211)32:11<3108::AID-IMMU3108>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 17.Lopez-Cabrera M, Munoz E, Blazquez MV, Ursa MA, Santis AG, Sanchez-Madrid F. Transcriptional regulation of the gene encoding the human C-type lectin leukocyte receptor AIM/CD69 and functional characterization of its tumor necrosis factor-alpha-responsive elements. J Biol Chem. 1995;270:21545–21551. doi: 10.1074/jbc.270.37.21545. [DOI] [PubMed] [Google Scholar]
- 18.Lee GR, Fields PE, Flavell RA. Regulation of IL-4 gene expression by distal regulatory elements and GATA-3 at the chromatin level. Immunity. 2001;14:447–459. doi: 10.1016/s1074-7613(01)00125-x. [DOI] [PubMed] [Google Scholar]
- 19.Hatton RD, Harrington LE, Luther RJ, Wakefield T, Janowski KM, Oliver JR, Lallone RL, Murphy KM, Weaver CT. A distal conserved sequence element controls Ifng gene expression by T cells and NK cells. Immunity. 2006;25:717–729. doi: 10.1016/j.immuni.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 20.Weinmann AS, Plevy SE, Smale ST. Rapid and selective remodeling of a positioned nucleosome during the induction of IL-12 p40 transcription. Immunity. 1999;11:665–675. doi: 10.1016/s1074-7613(00)80141-7. [DOI] [PubMed] [Google Scholar]
- 21.Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell. 2000;103:667–678. doi: 10.1016/s0092-8674(00)00169-0. [DOI] [PubMed] [Google Scholar]
- 22.Shinkai Y, Koyasu S, Nakayama K, Murphy KM, Loh DY, Reinherz EL, Alt FW. Restoration of T cell development in RAG-2-deficient mice by functional TCR transgenes. Science. 1993;259:822–825. doi: 10.1126/science.8430336. [DOI] [PubMed] [Google Scholar]
- 23.Morgan DJ, Liblau R, Scott B, Fleck S, McDevitt HO, Sarvetnick N, Lo D, Sherman LA. CD8(+) T cell-mediated spontaneous diabetes in neonatal mice. J Immunol. 1996;157:978–983. [PubMed] [Google Scholar]
- 24.Ellmeier W, Sunshine MJ, Losos K, Hatam F, Littman DR. An enhancer that directs lineage-specific expression of CD8 in positively selected thymocytes and mature T cells. Immunity. 1997;7:537–547. doi: 10.1016/s1074-7613(00)80375-1. [DOI] [PubMed] [Google Scholar]
- 25.McMurry MT, Krangel MS. A role for histone acetylation in the developmental regulation of VDJ recombination. Science. 2000;287:495–498. doi: 10.1126/science.287.5452.495. [DOI] [PubMed] [Google Scholar]
- 26.Carabana J, Ortigoza E, Krangel MS. Regulation of the murine Ddelta2 promoter by upstream stimulatory factor 1, Runx1, and c-Myb. J Immunol. 2005;174:4144–4152. doi: 10.4049/jimmunol.174.7.4144. [DOI] [PubMed] [Google Scholar]
- 27.Bert AG, Burrows J, Osborne CS, Cockerill PN. Generation of an improved luciferase reporter gene plasmid that employs a novel mechanism for high-copy replication. Plasmid. 2000;44:173–182. doi: 10.1006/plas.2000.1474. [DOI] [PubMed] [Google Scholar]
- 28.Loots GG, Locksley RM, Blankespoor CM, Wang ZE, Miller W, Rubin EM, Frazer KA. Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science. 2000;288:136–140. doi: 10.1126/science.288.5463.136. [DOI] [PubMed] [Google Scholar]
- 29.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Puls KL, Hogquist KA, Reilly N, Wright MD. CD53, a thymocyte selection marker whose induction requires a lower affinity TCR-MHC interaction than CD69, but is up-regulated with slower kinetics. Int Immunol. 2002;14:249–258. doi: 10.1093/intimm/14.3.249. [DOI] [PubMed] [Google Scholar]
- 31.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 32.Schneider R, Bannister AJ, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat Cell Biol. 2004;6:73–77. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- 33.Shnyreva M, Weaver WM, Blanchette M, Taylor SL, Tompa M, Fitzpatrick DR, Wilson CB. Evolutionarily conserved sequence elements that positively regulate IFN-gamma expression in T cells. Proc Natl Acad Sci U S A. 2004;101:12622–12627. doi: 10.1073/pnas.0400849101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ansel KM, Greenwald RJ, Agarwal S, Bassing CH, Monticelli S, Interlandi J, Djuretic IM, Lee DU, Sharpe AH, Alt FW, Rao A. Deletion of a conserved Il4 silencer impairs T helper type 1-mediated immunity. Nat Immunol. 2004;5:1251–1259. doi: 10.1038/ni1135. [DOI] [PubMed] [Google Scholar]
- 35.Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE. High-Resolution Mapping and Characterization of Open Chromatin across the Genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adachi S, Rothenberg EV. Cell-type-specific epigenetic marking of the IL2 gene at a distal cis-regulatory region in competent, nontranscribing T-cells. Nucleic Acids Res. 2005;33:3200–3210. doi: 10.1093/nar/gki637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Avni O, Lee D, Macian F, Szabo SJ, Glimcher LH, Rao A. T(H) cell differentiation is accompanied by dynamic changes in histone acetylation of cytokine genes. Nat Immunol. 2002;3:643–651. doi: 10.1038/ni808. [DOI] [PubMed] [Google Scholar]
- 38.Cui K, Tailor P, Liu H, Chen X, Ozato K, Zhao K. The chromatin-remodeling BAF complex mediates cellular antiviral activities by promoter priming. Mol Cell Biol. 2004;24:4476–4486. doi: 10.1128/MCB.24.10.4476-4486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zou YR, Sunshine MJ, Taniuchi I, Hatam F, Killeen N, Littman DR. Epigenetic silencing of CD4 in T cells committed to the cytotoxic lineage. Nat Genet. 2001;29:332–336. doi: 10.1038/ng750. [DOI] [PubMed] [Google Scholar]
- 41.Sawada S, Scarborough JD, Killeen N, Littman DR. A lineage-specific transcriptional silencer regulates CD4 gene expression during T lymphocyte development. Cell. 1994;77:917–929. doi: 10.1016/0092-8674(94)90140-6. [DOI] [PubMed] [Google Scholar]
- 42.Bilic I, Koesters C, Unger B, Sekimata M, Hertweck A, Maschek R, Wilson CB, Ellmeier W. Negative regulation of CD8 expression via Cd8 enhancer-mediated recruitment of the zinc finger protein MAZR. Nat Immunol. 2006;7:392–400. doi: 10.1038/ni1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He X, Park K, Wang H, He X, Zhang Y, Hua X, Li Y, Kappes DJ. CD4-CD8 lineage commitment is regulated by a silencer element at the ThPOK transcription-factor locus. Immunity. 2008;28:346–358. doi: 10.1016/j.immuni.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 44.Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 45.Alvarez JD, Yasui DH, Niida H, Joh T, Loh DY, Kohwi-Shigematsu T. The MAR-binding protein SATB1 orchestrates temporal and spatial expression of multiple genes during T-cell development. Genes Dev. 2000;14:521–535. [PMC free article] [PubMed] [Google Scholar]
- 46.Decker T, Pasca di Magliano M, McManus S, Sun Q, Bonifer C, Tagoh H, Busslinger M. Stepwise activation of enhancer and promoter regions of the B cell commitment gene Pax5 in early lymphopoiesis. Immunity. 2009;30:508–520. doi: 10.1016/j.immuni.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 47.Pal-Bhadra M, Bhadra U, Birchler JA. RNAi related mechanisms affect both transcriptional and posttranscriptional transgene silencing in Drosophila. Mol Cell. 2002;9:315–327. doi: 10.1016/s1097-2765(02)00440-9. [DOI] [PubMed] [Google Scholar]
- 48.Yui MA, Hernandez-Hoyos G, Rothenberg EV. A new regulatory region of the IL-2 locus that confers position-independent transgene expression. J Immunol. 2001;166:1730–1739. doi: 10.4049/jimmunol.166.3.1730. [DOI] [PubMed] [Google Scholar]
- 49.Mohrs M, Blankespoor CM, Wang ZE, Loots GG, Afzal V, Hadeiba H, Shinkai K, Rubin EM, Locksley RM. Deletion of a coordinate regulator of type 2 cytokine expression in mice. Nat Immunol. 2001;2:842–847. doi: 10.1038/ni0901-842. [DOI] [PubMed] [Google Scholar]
- 50.Tanamachi DM, Moniot DC, Cado D, Liu SD, Hsia JK, Raulet DH. Genomic Ly49A transgenes: basis of variegated Ly49A gene expression and identification of a critical regulatory element. J Immunol. 2004;172:1074–1082. doi: 10.4049/jimmunol.172.2.1074. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.