Abstract
Spinal muscular atrophy is the second most common fatal childhood disorder. Core clinical features include muscle weakness caused by degenerating lower motor neurons and a high incidence of bone fractures and hypercalcemia. Fractures further compromise quality of life by progression of joint contractures or additional loss of motor function. Recent observations suggest that bone disease in spinal muscular atrophy may not be attributed entirely to lower motor neuron degeneration. The presence of the spinal muscular atrophy disease-determining survival motor neuron gene (SMN), SMN expression, and differential splicing in bone-resorbing osteoclasts was recently discovered. Its ubiquitous expression and the differential expression of splice variants suggest that SMN has specific roles in bone cell function. SMN protein also interacts with osteoclast stimulatory factor. Mouse models of human spinal muscular atrophy disease suggest a potential role of SMN protein in skeletal development. Dual energy x-ray absorptiometry analysis demonstrated a substantial decrease in total bone area and poorly developed caudal vertebra in the mouse model. These mice also had pelvic bone fractures. Studies delineating SMN signaling mechanisms and gene transcription in a cell-specific manner will provide important molecular insights into the pathogenesis of bone disease in children with spinal muscular atrophy. Moreover, understanding bone remodeling in spinal muscular atrophy may lead to novel therapeutic approaches to enhance skeletal health and quality of life. This article reviews the skeletal complications associated with spinal muscular atrophy and describes a functional role for SMN protein in osteoclast development and bone resorption activity.
Keywords: spinal muscular atrophy, bone fractures, SMN protein, osteoclasts
Spinal muscular atrophy is the second most common fatal autosomal recessive childhood disorder. It occurs at a frequency of 1 case per 8000 newborns. The disease is characterized by degeneration of lower motor neurons.1 Spinal muscular atrophy is clinically classified on the bases of age at onset and clinical course into type 1 (severe), type 2 (intermediate), and type 3 (mild) forms, with heterogeneity of disease.2 Type 1 spinal muscular atrophy manifests the most severely, with generalized muscle weakness and hypotonia from early infancy, usually leading to death within the first 2 years. Children with type 2 spinal muscular atrophy cannot walk or stand unaided and survive beyond 4 years. Patients with type 3 spinal muscular atrophy have proximal muscle weakness leading to gait abnormality, with onset in childhood. All 3 subtypes of spinal muscular atrophy demonstrate linkage to the survival motor neuron protein (SMN)-encoding gene region on chromosome 5q13.3
SMN is a 38-kDa protein ubiquitously expressed in human tissues and highly conserved in diverse species. In healthy humans, the gene encoding SMN protein is present as telomeric (SMN1) and centromeric (SMN2) copies that contain 8 exons. SMN2 differs from SMN1 by 5 nucleotide substitutions that do not alter the amino acid sequence of the SMN protein but result in aberrant splicing of exon 7, which encodes 16 amino acids at the carboxy terminal end. Spinal muscular atrophy is characterized by reduced levels of full-length SMN protein due to deletions or mutations in SMN1. SMN2 cannot compensate for the deficiency of the SMN protein due to deletion of SMN1 because the SMN2 gene product is about 25% native SMN protein and more predominantly (about 75%) an aberrantly spliced form of truncated and unstable SMN protein that lacks the exon 7 region.4 However, a functional role for the exon 7 splice variant of SMN (SMNΔ7) is unclear. It has been reported that the SMN oligomerization defect that results from increased expression of SMNΔ7 correlates with severity of spinal muscular atrophy.5 Furthermore, an exon 7 C to T transition at codon 280, a translationally silent variance, is sufficient to dictate exon 7 alternative splicing.6 Therefore, a substantial reduction in SMN native protein levels results in degeneration of lower motor neurons in patients with spinal muscular atrophy. However, which complications are associated with spinal muscular atrophy and how SMN might function in a cell-specific manner are poorly understood. In this review, we focus on skeletal complications and evidence implicating a functional role for SMN in bone remodeling in patients with spinal muscular atrophy.
Skeletal Complications in Patients With Spinal Muscular Atrophy
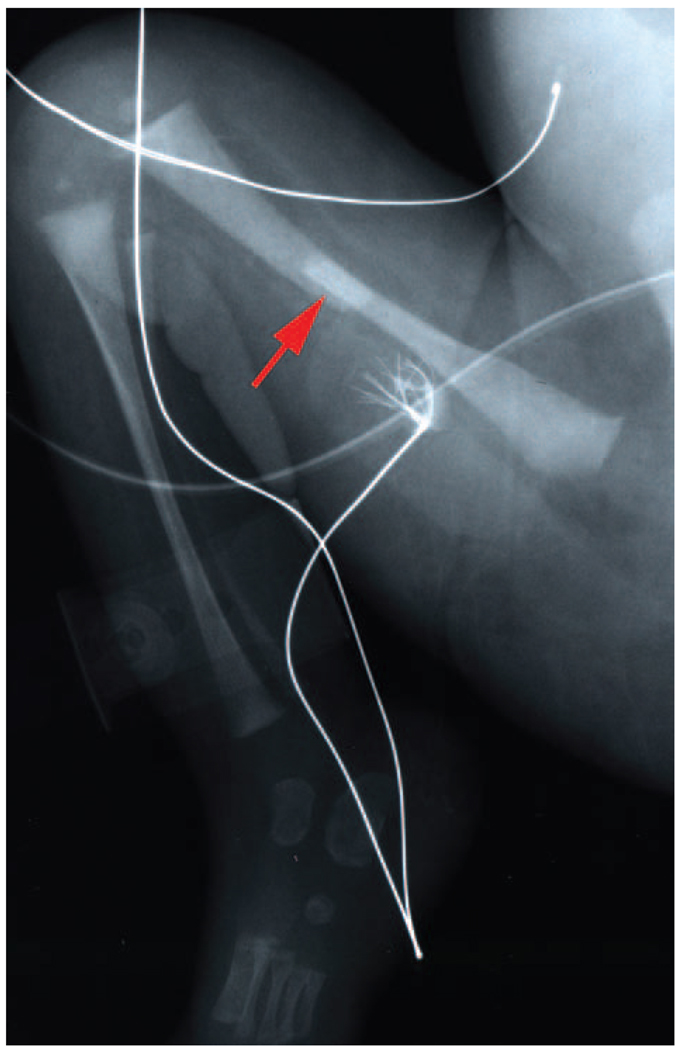
Although spinal muscular atrophy is characterized by degeneration of lower motor neurons manifesting predominantly with hypotonia and muscular weakness, patients with this disorder often demonstrate severe osteopenia and sometimes experience recurrent fractures associated with minimal trauma. In addition, a subset of severely affected infants demonstrates congenital bone fractures (Figure 1). Individual case reports have noted hypercalcemia and hypercalciuria in these patients, presumably due to altered bone turnover secondary to reduced muscular activity.7–9 A severe spinal muscular atrophy variant condition has been described, characterized by congenital bone fractures and extremely thin ribs but without contractures.7 Recently, it has also been reported that bone mineral density is normal in most young children with spinal muscular atrophy; however, bone mineral density seems to decrease notably with increasing age, without a definite direct relation to a substantial change in motor function.10 Clearly, this is an issue that warrants further study in this population.
Figure 1.
The arrow indicates a bone fracture in a newborn with spinal muscular atrophy. The radiograph is courtesy of the University of Miami and the University of Maryland Brain and Tissue Bank for Developmental Disorders.
It is well known that osteoporosis is a complication of spinal cord injury; however, the mechanism of pathogenesis is considered to be due to disuse or unloading of skeleton. 11 Excess bone loss leads to fracture risk in these patients. Therefore, it may be important to examine the status of bone resorption markers such as excretion of N-telopeptides in the urine and bone formation markers such as bone-specific alkaline phosphatase activity levels in the serum to assess bone turnover in children with spinal muscular atrophy. Enhanced levels of urine N-telopeptides necessitate the use of antiresorptive agents such as bisphosphonates and denosumab antibody12 to prevent excess bone loss and fracture risk, enhancing the quality of life in children with spinal muscular atrophy.
Skeletal Abnormalities in a Mouse Model for Spinal Muscular Atrophy Disease
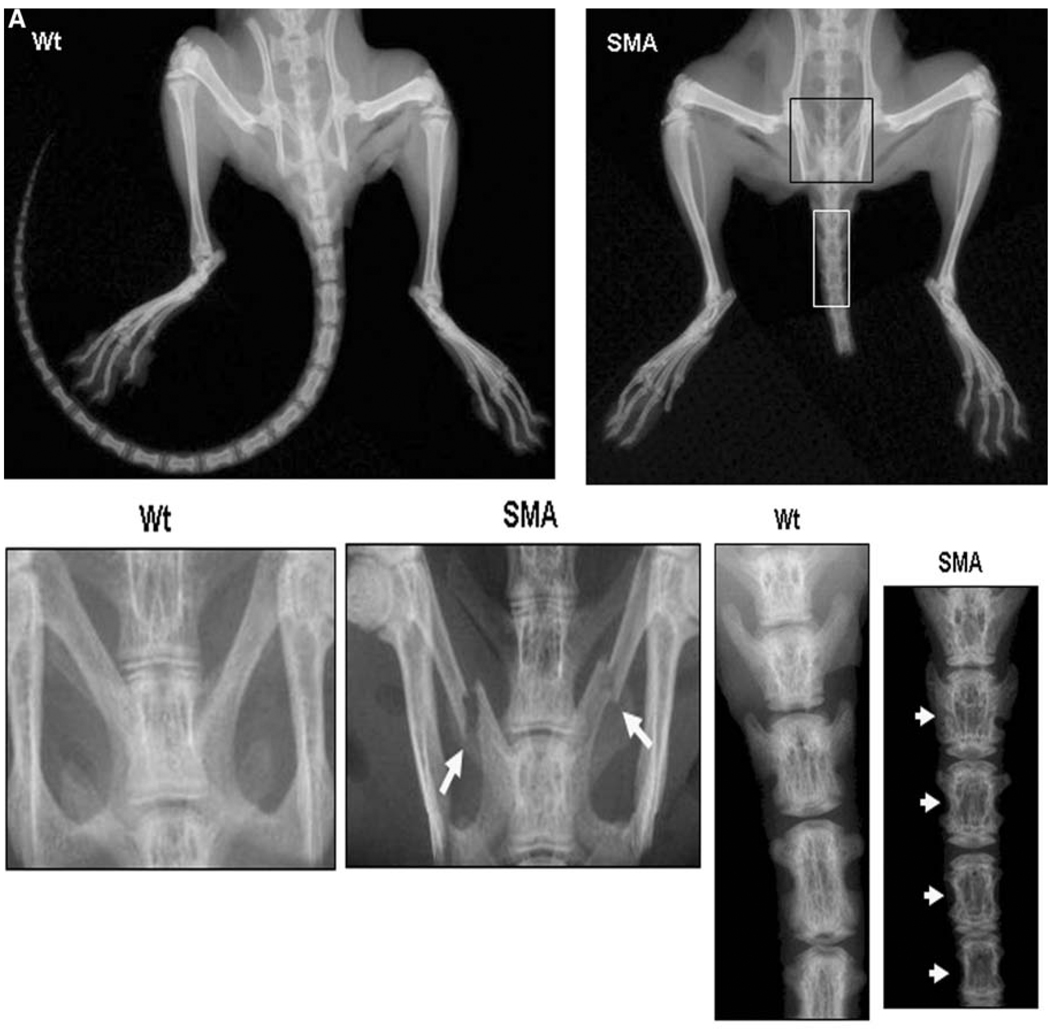
The mouse genome contains a single copy of the gene encoding survival motor neuron protein (Smn) (equivalent to human SMN1). Mouse Smn is 82% identical to human SMN, and aberrant splicing of SMNΔ7 does not occur in mouse tissues because they lack the SMN2 copy present in the human genome.13 Mice with homozygous Smn disruption display massive cell death during early embryonic development, indicating that Smn is necessary for cellular survival and function.14 Furthermore, transgenic mice harboring human SMN2 in the Smn−/− background (Smn−/−SMN2) showed pathologic changes in the spinal cord and skeletal muscles similar to those of patients with spinal muscular atrophy,15,16 demonstrating that SMN2 expression in Smn−/− mice rescued embryonic lethality and that the rescued mice had spinal muscular atrophy Smn−/−SMN2 mice demonstrated degeneration of tails, which were notably shorter than those of control mice. We further examined skeletal phenotype in these mice by radio-logic and dual energy x-ray absorptiometry analysis. As shown in Figure 2, Smn−/−SMN2 mice demonstrated skeletal abnormalities in the lower body, with caudal vertebra notably reduced in number, size, and bone quality compared with those of wild-type mice. Furthermore, these mice demonstrated pelvic bone fractures (Figure 2A). Dual energy x-ray absorptiometry analysis identified a substantial decrease in total bone area, bone mineral content, and bone mineral density in these mice (Figure 2B and C). These results are consistent with the changes observed on dual energy x-ray absorptiometry imaging in many subjects with spinal muscular atrophy and with the observation of bone fractures in patients with the most severe congenital form of the disease. Expression of SMN and SMNΔ7 in bone-resorbing human osteoclasts was recently reported.17 Collectively, these studies indicate a functional role for SMN in skeletal development and bone remodeling.
Figure 2.
Skeletal phenotype in Smn−/−SMN2 mice.15 (A) Radiologic assessment of 4-week-old Smn−/−SMN2 (Jackson Laboratories, Bar Harbor, ME) mice shows short tails compared with wild-type mice. A high-resolution (3x) radiogram indicates pelvic bone fractures and poorly developed caudal vertebra in spinal muscular atrophy mice.
(B) Dual energy x-ray absorptiometry analyses of total animal bone area and total spine for bone area, bone mineral content, and bone mineral density in spinal muscular atrophy mice. (C) Dual energy x-ray absorptiometry analysis of caudal vertebra region for bone area, bone mineral content, and bone mineral density (n = 3, P < .05).
SMN Function and Protein Interaction
The osteoclast is hematopoietic in origin and is the primary bone-resorbing cell derived from the monocyte and macrophage lineage. The receptor activator of nuclear factor kappa B ligand, a member of the tumor necrosis factor family, is expressed on marrow stromal and osteoblast cells in response to several osteotropic factors and is crucial for osteoclast precursor differentiation to form multinucleated osteoclasts, which resorb bone. Macrophage colony-stimulating factor is required for proliferation, survival, and expression of receptor activator of nuclear factor kappa B in osteoclast precursors. The interaction between receptor activator of nuclear factor kappa B ligand and receptor activator of nuclear factor kappa B results in activation of various signaling cascades during osteoclast development and bone resorption.18 The osteoclast is an autocrine, paracrine, and intracrine regulatory cell that produces factors such as interleukin 6, annexin II, transforming growth factor β, and osteoclast inhibitory peptide 1/human stem cell antigen, which affect its formation and activity.19
Using an expression-cloning approach, a novel intracellular protein, osteoclast stimulatory factor, that is produced by the osteoclasts to enhance osteoclast formation and bone resorption activity was previously isolated.20 It is a 28-kDa peptide that contains c-Src homology 3 domain and ankyrin repeats that implicated osteoclast stimulatory factor participation in cellular signaling; furthermore, recombinant osteoclast stimulatory factor demonstrated high affinity binding to c-Src tyrosine kinase, an important regulator of osteoclast activity. Osteoclast stimulatory factor interaction with c-Src results in secretion of soluble stimulators of osteoclast formation, which suggests a possible role for osteoclast stimulatory factor in the bone resorption process through protein interactions with cellular signal transduction machinery.
Because osteoclast stimulatory factor expression is ubiquitous, yeast 2-hybrid screening using a HeLa cell complementary DNA library to identify proteins that interact with osteoclast stimulatory factor was previously performed to further identify participants in the osteoclast stimulatory factor signaling cascade.17 SMN interaction with the c-Src homology 3 domain in osteoclast stimulatory factor was identified, and in vitro affinity binding investigations further mapped osteoclast stimulatory factor binding to the SMN exon 6 region. Coimmunoprecipitation of osteoclast stimulatory factor with an anti-SMN antibody further confirmed the specificity of the interaction between osteoclast stimulatory factor and SMN. Osteoclast stimulatory factor interaction with the c-Src homology 3/c-Src homology 2 domain was also identified.20 However, c-Src does not immunoprecipitate with an anti-SMN antibody. These data suggest that osteoclast stimulatory factor and SMN may play a central role in the cellular signal transduction cascade, leading to the release of soluble factors that stimulate osteoclast formation and activity (Figure 3). It is possible that the deletions or point mutations that include exons 6 and 7 of the SMN protein may affect the osteoclast stimulatory factor and SMN signaling mechanism in bone-resorbing osteoclasts, as well as in other cell types in children with spinal muscular atrophy. Therefore, our data further suggest SMN protein participation in osteoclast stimulatory factor-induced osteoclast formation and bone resorption activity. By using antisense constructs to osteoclast stimulatory factor and SMN, respectively, SMN-conditioned media stimulate osteoclast formation analogous to osteoclast stimulatory factor, and osteoclast stimulatory activity can be blocked. These data confirm the participation of SMN in the expression of an osteoclast stimulator through an interaction with the osteoclast stimulatory factor.
Figure 3.
Osteoclast stimulatory factor and SMN protein interactions in osteoclasts. Osteoclast stimulatory factor contains a proline-rich sequence at the amino terminus, followed by an SH3 domain and ankyrin repeats. Osteoclast stimulatory factor interacts with c-Src through a proline-rich sequence. Osteoclast stimulatory factor SH3 domain binds to SMN at the exon 6 encoding region. Osteoclast stimulatory factor and SMN signaling release soluble factors that stimulate osteoclast formation and survival.17
The identity of downstream targets and soluble factors released in response to osteoclast stimulatory factor and SMN signaling is unknown. Neutralizing antibodies to known osteotropic factors such as interleukin 1, interleukin 6, and tumor necrosis factor α did not block osteoclast stimulatory factor–induced osteoclastogenesis.20 However, it is possible that osteoclast stimulatory factor and SMN signaling may result in production of osteotropic and cell survival factors such as parathyroid hormone–related peptide, enhancing the survival of cell types. It is also unclear whether SMN may directly affect the basic transcription machinery through interaction with specific transcription factors that would induce gene expression of such factors. Expression of the SMN2 gene product SMNΔ7 in osteoclasts was reported17; however, the interaction of SMNΔ7 with osteoclast stimulatory factor and its functional role in associated signaling are unknown. More recently, it has been shown that osteoclast stimulatory factor is a novel component of adhesion sites and is associated with Casitas B-lineage lymphoma and Src signaling molecules.21 Casitas B-lineage lymphoma is a major substrate for the Src family of tyrosine kinases and plays an important role in Src-mediated migration and function of osteoclasts.22 Therefore, SMN2-rescued Smn−/− mice would be a potential model to study the role of osteoclast stimulatory factor and SMN in osteoclast development and bone resorption.15
SMN is ubiquitous in expression and has been shown to interact with several other proteins. It has been shown to interact with B-cell lymphoma 2, and coexpression of SMN with B-cell lymphoma 2 prevented B-cell lymphoma 2-associated X protein-induced or Fas type I membrane protein-mediated apoptosis, although SMN had only weak antiapoptotic activity.23 In addition, SMN has been shown to interact with the SMN interacting protein 1 protein complex.24 Although the exact mechanisms for SMN-SMN interacting protein 1 nuclear translocation are unclear, it is evident that the SMN-SMN interacting protein 1 complex is directly involved in the biogenesis of spliceosomal small nuclear ribonucleoproteins. Recently, it was reported that SMN might regulate gene transcription through interaction with corepressor mSin3A.25 It is also unclear if SMNΔ7 could translocate to the nucleus and whether SMN or SMNΔ7 directly regulates gene transcription at the cellular level. Liu et al26 previously identified that the P2 peptide region of the SMN protein carboxyl terminus (amino acids 240–267) was involved in the interaction with the small nuclear ribonuclear core protein. Previously, it was shown that the profilins bind to and colocalize with SMN in nuclear gems and that this interaction may be involved in the neuron-specific effects associated with spinal muscular atrophy disease.27 Other proteins reported to interact with SMN include transcription factor E2 (nuclear transcription activator of papilloma virus) and far-upstream binding protein.28,29 However, it is unclear whether these protein interactions and functions are affected by the low SMN levels that exist in patients with spinal muscular atrophy or in the spinal muscular atrophy mouse model. SMN protein has been shown to interact with the zinc finger protein 1 and colocalize in small subnuclear structures called gems and Cajal bodies. Decreased zinc finger protein 1 expression prevents SMN localization to nuclear bodies, indicating that zinc finger protein 1 is required for nuclear localization of SMN.30 Furthermore, SMN has been shown to interact with the tumor suppressor protein p53 and colocalize in nuclear Cajal bodies, implicating a potential role in cellular apoptosis.31 Taken together, these data suggest that SMN deficiency could affect bone cell function and that signaling cascades associated with SMN may play an important role in skeletal complications of spinal muscular atrophy disease.
Conclusions
Skeletal complications of spinal muscular atrophy involve congenital bone fractures. SMN is ubiquitously expressed and interacts with cellular signaling molecules such as osteoclast stimulatory factor that are expressed in bone-resorbing osteoclasts. SMN interaction with osteoclast stimulatory factor may play an important role in a novel signaling cascade that induces stimulators of osteoclast formation. Studies pertaining to SMN-osteoclast stimulatory factor signaling may provide additional insights into the often severe osteopenia and increased fracture risk associated with spinal muscular atrophy and may identify novel targets for therapeutic interventions against skeletal complications to enhance quality of life in these patients.
Acknowledgements
This study was supported by grants DE 12603 and C06 RR015455 from the Extramural Research Facilities Program of the National Center for Research Resources, National Institutes of Health. Dr Reddy is a recipient of the Department of Defense Medical Research Award.
Footnotes
This study was presented at the Neurobiology of Disease in Children: Symposium on Spinal Muscular Atrophy, in conjunction with the 35th Annual Meeting of the Child Neurology Society; October 18, 2006; Pittsburgh, Pennsylvania.
References
- 1.Swoboda KJ, Prior TW, Scott CB, et al. Natural history of denervation in SMA: relation to age, SMN2 copy number, and function. Ann Neurol. 2005;57(5):704–712. doi: 10.1002/ana.20473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hausmanowa-Petrusewicz I, Vrbová G. Spinal muscular atrophy: a delayed development hypothesis. Neuroreport. 2005;16(7):657–661. doi: 10.1097/00001756-200505120-00001. [DOI] [PubMed] [Google Scholar]
- 3.Lefebvre S, Burglen L, Reboullet S, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80(1):155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 4.Le TT, Pham LT, Butchbach ME, et al. SMNΔ7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum Mol Genet. 2005;14(6):845–857. doi: 10.1093/hmg/ddi078. [DOI] [PubMed] [Google Scholar]
- 5.Lorson CL, Strasswimmer J, Yao JM, et al. SMN oligomerization defect correlates with spinal muscular atrophy severity. Nat Genet. 1998;19(1):63–66. doi: 10.1038/ng0598-63. [DOI] [PubMed] [Google Scholar]
- 6.Lorson CL, Hahnen E, Androphy EJ, Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc Natl Acad Sci U S A. 1999;96(11):6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Felderhoff-Mueser U, Grohmann K, Harder A, et al. Severe spinal muscular atrophy variant associated with congenital bone fractures. J Child Neurol. 2002;17(9):718–721. doi: 10.1177/088307380201700915. [DOI] [PubMed] [Google Scholar]
- 8.Kelly TE, Amoroso K, Ferre M, Blanco J, Allinson P, Prior TW. Spinal muscular atrophy variant with congenital fractures. Am J Med Genet. 1999;87(1):65–68. doi: 10.1002/(sici)1096-8628(19991105)87:1<65::aid-ajmg13>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 9.Khawaja K, Houlsby WT, Watson S, Bushby K, Cheetham T. Hypercalcaemia in infancy: a presenting feature of spinal muscular atrophy. Arch Dis Child. 2004;89(4):384–385. doi: 10.1136/adc.2003.028225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kinali M, Banks LM, Mercuri E, Manzur AY, Muntoni F. Bone mineral density in a paediatric spinal muscular atrophy population. Neuropediatrics. 2004;35(6):325–328. doi: 10.1055/s-2004-830366. [DOI] [PubMed] [Google Scholar]
- 11.Jiang SD, Jiang LS, Dai LY. Mechanisms of osteoporosis in spinal cord injury. Clin Endocrinol (Oxf) 2006;65(5):555–565. doi: 10.1111/j.1365-2265.2006.02683.x. [DOI] [PubMed] [Google Scholar]
- 12.Geusens P, Reid D. Newer drug treatments: their effects on fracture prevention. Best Pract Res Clin Rheumatol. 2005;19(6):983–989. doi: 10.1016/j.berh.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 13.Bergin A, Kim G, Price DL, Sisodia SS, Lee MK, Rabin BA. Identification and characterization of a mouse homologue of the spinal muscular atrophy-determining gene, survival motor neuron. Gene. 1997;204(1–2):47–53. doi: 10.1016/s0378-1119(97)00510-6. [published correction appears in Gene. 1998;213(1–2):227] [DOI] [PubMed] [Google Scholar]
- 14.Schrank B, Götz R, Gunnersen JM, et al. Inactivation of the survival motor neuron gene, a candidate gene for human spinal muscular atrophy, leads to massive cell death in early mouse embryos. Proc Natl Acad Sci U S A. 1997;94(18):9920–9925. doi: 10.1073/pnas.94.18.9920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsieh-Li HM, Chang JG, Jong YJ, et al. A mouse model for spinal muscular atrophy. Nat Genet. 2000;24(1):66–70. doi: 10.1038/71709. [DOI] [PubMed] [Google Scholar]
- 16.Monani UR, Sendtner M, Coovert DD, et al. The human centromeric survival motor neuron gene (SMN2) rescues embryonic lethality in Smn(−/−) mice and results in a mouse with spinal muscular atrophy. Hum Mol Genet. 2000;9(3):333–339. doi: 10.1093/hmg/9.3.333. [DOI] [PubMed] [Google Scholar]
- 17.Kurihara N, Menaa C, Maeda H, Haile DJ, Reddy SV. Osteoclast-stimulating factor interacts with the spinal muscular atrophy gene product to stimulate osteoclast formation. J Biol Chem. 2001;276(44):41035–41039. doi: 10.1074/jbc.M100233200. [DOI] [PubMed] [Google Scholar]
- 18.Ross FP, Teitelbaum SL. αvβ3 and macrophage colony-stimulating factor: partners in osteoclast biology. Immunol Rev. 2005;208(1):88–105. doi: 10.1111/j.0105-2896.2005.00331.x. [DOI] [PubMed] [Google Scholar]
- 19.Reddy SV. Regulatory mechanisms operative in osteoclasts. Crit Rev Eukaryot Gene Expr. 2004;14(4):255–270. doi: 10.1615/critreveukaryotgeneexpr.v14.i4.20. [DOI] [PubMed] [Google Scholar]
- 20.Reddy S, Devlin R, Menaa C, et al. Isolation and characterization of a cDNA clone encoding a novel peptide (OSF) that enhances osteoclast formation and bone resorption. J Cell Physiol. 1998;177(4):636–645. doi: 10.1002/(SICI)1097-4652(199812)177:4<636::AID-JCP14>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 21.Szymkiewicz I, Destaing O, Jurdic P, Dikic I. SH3P2 in complex with Cbl and Src. FEBS Lett. 2004;565(1–3):33–38. doi: 10.1016/j.febslet.2004.03.100. [DOI] [PubMed] [Google Scholar]
- 22.Sanjay A, Houghton A, Neff L, et al. Cbl associates with Pyk2 and Src to regulate Src kinase activity, αvβ3 integrin-mediated signaling, cell adhesion, and osteoclast motility. J Cell Biol. 2001;152(1):181–195. doi: 10.1083/jcb.152.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwahashi H, Eguchi Y, Yasuhara N, Hanafusa T, Matsuzawa Y, Tsujimoto Y. Synergistic anti-apoptotic activity between Bcl-2 and SMN implicated in spinal muscular atrophy. Nature. 1997;390(6658):413–417. doi: 10.1038/37144. [DOI] [PubMed] [Google Scholar]
- 24.Lefebvre S, Burlet P, Liu Q, et al. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat Genet. 1997;16(3):265–269. doi: 10.1038/ng0797-265. [DOI] [PubMed] [Google Scholar]
- 25.Zou J, Barahmand-pour F, Blackburn ML, Matsui Y, Chansky HA, Yang L. Survival motor neuron (SMN) protein interacts with transcription corepressor mSin3A. J Biol Chem. 2004;279(15):14922–14928. doi: 10.1074/jbc.M309218200. [DOI] [PubMed] [Google Scholar]
- 26.Liu Q, Fischer U, Wang F, Dreyfuss G. The spinal muscular atrophy disease gene product, SMN, and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell. 1997;90(6):1013–1021. doi: 10.1016/s0092-8674(00)80367-0. [DOI] [PubMed] [Google Scholar]
- 27.Giesemann T, Rathke-Hartlieb S, Rothkegel M, et al. A role for polyproline motifs in the spinal muscular atrophy protein SMN: profilins bind to and colocalize with SMN in nuclear gems. J Biol Chem. 1999;274(53):37908–37914. doi: 10.1074/jbc.274.53.37908. [DOI] [PubMed] [Google Scholar]
- 28.Strasswimmer J, Lorson CL, Breiding DE, et al. Identification of survival motor neuron as a transcriptional activator–binding protein. Hum Mol Genet. 1999;8(7):1219–1226. doi: 10.1093/hmg/8.7.1219. [DOI] [PubMed] [Google Scholar]
- 29.Williams BY, Hamilton SL, Sarkar HK. The survival motor neuron protein interacts with the transactivator FUSE binding protein from human fetal brain. FEBS Lett. 2000;470(2):207–210. doi: 10.1016/s0014-5793(00)01320-x. [DOI] [PubMed] [Google Scholar]
- 30.Gangwani L, Mikrut M, Theroux S, Sharma M, Davis RJ. Spinal muscular atrophy disrupts the interaction of ZPR1 with the SMN protein. Nat Cell Biol. 2001;3(4):376–383. doi: 10.1038/35070059. [DOI] [PubMed] [Google Scholar]
- 31.Young PJ, Day PM, Zhou J, Androphy EJ, Morris GE, Lorson CL. A direct interaction between the survival motor neuron protein and p53 and its relationship to spinal muscular atrophy. J Biol Chem. 2002;277(4):2852–2859. doi: 10.1074/jbc.M108769200. [DOI] [PubMed] [Google Scholar]