Abstract
Although DNA replication is often imagined as a regular and continuous process, the DNA polymerase enzyme is a complicated machine and can pause upon encountering physical and chemical barriers. We used single molecule measurements to make a detailed characterization of this behavior as a function of the template's secondary structure and the sequence context. Strand displacement replication through a DNA hairpin by single DNA polymerase molecules was measured in real time with near single base resolution and physiological concentrations of nucleotides. These data enabled the measurement of the intrinsic “speed limit” of DNA polymerase by separating the burst synthesis rate from pausing events. The strand displacement burst synthesis rate for Escherichia coli DNA Polymerase I (KF) was found to be an order of magnitude faster than the reported bulk strand displacement rate, a discrepancy that can be accounted for by to sequence specific pausing. The ability to follow trajectories of single molecules revealed that the burst synthesis rate is also highly stochastic and varies up to 50-fold from molecule to molecule. Surprisingly, our results allow a unified explanation of strand displacement and single strand primer extension synthesis rates.
Keywords: FRET, pausing, DNA replication, DNA hairpin
Fast and accurate DNA replication is required to ensure the faithful transfer of genetic information to daughter cells. Replication forks can be slowed or paused by encountering DNA-binding ligands (1), genomic tRNA gene sites (2), stalled ternary complexes of RNA polymerase (3), and DNA-bound protein (4–5). DNA polymerase can also pause due to specific DNA sequences, including palindromic DNA capable of forming hairpin secondary structure (6–8), slow zones (9), and trinucleotide repeats of (CGG)n/(CCG)n or (CTG)n/(CAG)n (10–11). While these factors are thought to prevent fork movement along the template by steric hindrance and may be regulated by the replisome itself, other factors, including temperature, contaminants, nucleotide analogs (12), template tension (13–14), and nonclassic pause sites such as Pyr-G-C (15), may also interfere with steps in the DNA polymerase reaction pathway.
To measure the intrinsic “speed limit” of DNA polymerase itself, there must be sufficient spatial and temporal resolution to separate pausing events from burst synthesis. Although the kinetics of the Klenow fragment of Escherichia coli DNA Polymerase I (Pol I(KF)) have been studied using a variety of approaches (13, 15–20), there are conflicting reports of the synthesis rate and it has not yet been possible to dissect the interplay between pausing and burst synthesis. Rapid quenching and stopped-flow bulk techniques have determined that during primer extension synthesis, the first nucleotide is incorporated by Pol I(KF) with a rate ranging from 40–87 nt sec−1 (16–17, 19) while subsequent nucleotides are incorporated at a slower rate of 13–15 nt sec−1 (17, 20). This bulk processive rate is twice as fast as that found by single molecule primer extension synthesis rate measurements (7 nt sec−1) (13). Primer extension synthesis is generally thought to be faster than strand-displacement synthesis, and this idea is supported by a bulk rapid quenching study which found that the strand displacement synthesis rate of Pol I(KF) was even slower at 1.2 nt sec−1 (20).
We developed a FRET-based approach (21) to study single polymerase molecules undergoing strand displacement replication through a DNA hairpin to further investigate this conundrum between the various reported rates. This assay allowed us to measure polymerase activity in real time at saturating nucleotide concentrations with near single base resolution, thereby enabling separation of burst synthesis rates from pausing effects. Our results show that the only literature strand displacement synthesis rate for Pol I(KF) (20) is not the true burst synthesis rate because it likely includes sequence specific pausing effects due to the template used in the experiments. These pausing events are not unique to Pol I(KF) and were also observed with ϕ29 DNA polymerase. The existence of the pausing effect is known from bulk studies (15), but without the characterization of its heterogeneity and kinetics enabled by the single molecule experiments described here, its central role in determining the speed limit of DNA polymerase was not previously appreciated. The burst synthesis strand displacement rates obtained from our single molecule measurements were also highly heterogeneous from molecule to molecule, with rates varying from 1 nt sec−1 to over 50 nt sec−1. This variation in rates was previously hidden due to the ensemble averaging of bulk measurements and could only be accessed with a single molecule approach. Surprisingly, our characterization of the pausing heterogeneity in strand displacement also allows us to explain the discrepancies in rates found in primer extension experiments and shows that strand displacement rates are not slower than primer extension rates.
Results and Discussion
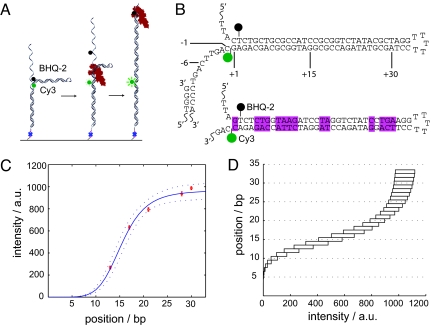
A 259-nt single-stranded DNA molecule was designed and synthesized containing an internal double-stranded 33-bp hairpin flanked by 94-nt single-stranded tails, one of which had a 3′-biotin group (Fig. 1A). The 3′ base of the hairpin contained an internal Cy3 FRET donor and the 5′ base contained an internal nonfluorescent FRET acceptor (Black Hole Quencher-2, BHQ-2) so that when the hairpin is fully folded, quenching of Cy3 by BHQ-2 prevents any fluorescence emission (Fig. 1B). The length of the hairpin was chosen to make full use of the dynamic range for this FRET pair: when the last base pair of the hairpin is broken due to strand displacement replication, the distance between fluorophores is just over twice the Förster radius (R0 = 5.02 nm, 33 bp of dsDNA = 11.2 nm). An 80-nt primer was annealed to the 3′ end of the template and the DNA constructs were attached to the surface of a glass coverslip via a 3′ biotin-streptavidin linker. The coverslip was placed on an inverted Nikon TE-2000S TIRF microscope and images were collected with a 60 × 1.45 NA objective at 5 Hz with a Photometrics Cascade II EMCCD. Each field of view contained, on average, over 1,000 individually resolvable single molecules (for full experimental details, see Materials and Methods).
Fig. 1.
FRET can be used to observe the real-time position of single DNA polymerase molecules. (A) Cartoon of the DNA template used to measure real-time polymerase kinetics. Primed 259 bp DNA molecules containing internal 33-bp hairpins and flanking 94 bp tails were immobilized on a glass surface through a biotin-streptavidin linker. DNA replication through the hairpin resulted in a reduction in FRET efficiency between Cy3 and BHQ-2, giving rise to an increase in Cy3 fluorescence. (B) Sequence and structure of the two DNA templates used in these experiments. The pink boxes highlight the differences in sequence between the sample (Top) and the control (Bottom) hairpins. Sequence numbering refers to the template position awaiting nucleotide addition. (C) Cy3 fluorescence recovery data for primer extension calibration experiments. The position along the x axis refers to the polymerase position along the stem of the hairpin and the error bars are the SEM. Dashed lines are 95% confidence intervals. (D) Intensity-to-position curve based on (C) to convert an arbitrary intensity to a template position. Each rectangle represents the 95% confidence intervals for the expected intensity at each polymerase position along the template.
To correlate the FRET signal with template position, a series of primer extension experiments were performed by adding DNA polymerase and only 3 of the 4 deoxynucleotide triphosphates (dNTPs). After the first extension and imaging step, the flow cell was thoroughly washed to remove any unincorporated nucleotides, and the process was repeated multiple times with a different set of 3 dNTPs. This allowed us to extend the primer to known positions on the template and observe the corresponding reduction in FRET signal as the donor and acceptor were forced apart. Before extension begins, the distance R between the 2 dyes is small and the FRET efficiency is EFRET = 1/[1+(R/R0)6] ≈ 1. As the primer extends and the hairpin unwinds, R increases and EFRET decreases, leading to recovery of Cy3 signal. The observed Cy3 intensity is thus given by ICY3 = IMAX (1-EFRET) = IMAX [1−(1/[1+(R/R0)6])], where IMAX is the maximum Cy3 intensity achievable when the dyes are as far apart as possible (assuming 0.34 nm per base pair, the maximum dsDNA separation is 2 × 33bp + 5bp loop = 71 bp ≈ 24 nm). A weighted least-squares fit of the data to this equation gave IMAX = 987 a.u. and R0 = 15.3 bp = 5.2 nm, which is consistent with the vendor's reported Förster radius for Cy3 and BHQ-2. There is some width to the distributions of Cy3 intensities at each position due to signal to noise variation and Cy3-BHQ-2 orientation effects (supporting information (SI) Fig. S1 and Table S1). However, by performing this alignment procedure on an ensemble of records at each position, a calibration curve relating the observed fluorescent intensity and the polymerase position along the template was created (Fig. 1C). Using this calibration curve and its 95% confidence limits, any arbitrary fluorescent intensity could be converted into a corresponding polymerase location along the template (Fig. 1D).
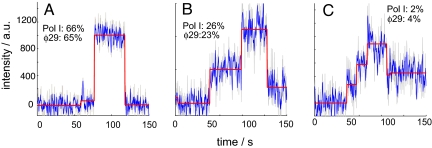
The real-time kinetics of DNA replication were examined in the presence of all four dNTPs with two different DNA polymerases: the Klenow fragment of DNA Polymerase I (exo−) from E. coli (Pol I(KF)) and the replicative polymerase from the Bacillus subtilis bacteriophage ϕ29. The trajectories for both polymerases exhibited heterogeneous behaviors that could be classified into 4 categories: fast replication without pausing (Fig. 2A), fast replication with a single pause (Fig. 2B), fast replication with multiple pauses (Fig. 2C), and slow replication. Slow replication occurred in less than 6% of the traces, and while its origin is unclear, it could stem from the template sticking to the surface or unfavorable polymerase-surface interactions. Approximately 66% of the DNA polymerases extended the entire template without a pause while about 28% of the polymerases paused at least once. The pauses occurred at highly stereotyped positions and we took advantage of the single molecule approach to measure both the distribution between pausing and nonpausing polymerase molecules and the relationship between the pauses and the burst synthesis speeds of the polymerase, data that would be impossible to observe with conventional bulk techniques.
Fig. 2.
Single molecule trajectories of extension through the hairpin exhibited four distinct patterns. We observed rapid extension without pausing (A), rapid extension with a single pause (B), and rapid extension with multiple pauses (C) before photobleaching. The gray curves are the raw data, the blue curves are the smoothed data over 5 raw data time points, and the red steps are generated by the step-fitting algorithm (see Materials and Methods). A total of 221 trajectories for Pol I(KF) and 207 trajectories for ϕ29 were analyzed. The percentages show the fraction of trajectories for each polymerase that demonstrated that particular pattern at 23 °C. Approximately 6% of the trajectories exhibited gradual intensity increases that could not be classified into any of these three categories.
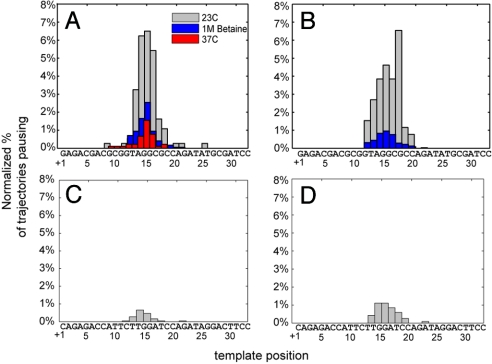
The positional accuracy of individual trajectories depended on both template location and signal-to-noise with a maximum attainable resolution of approximately 2 bp. For example, a well-behaved trajectory that paused with a mean intensity of 562 ± 47 a.u. (SD) was called at +15 or +16 bp; a noisier trajectory that paused with a mean intensity of 447 ± 168 a.u. (SD) was called at +14 ± 2 bp. By aligning an ensemble of trajectories for Pol I(KF) and ϕ29, polymerase pause positions were localized with near single base accuracy over the template from +8 bp to +18 bp (Fig. 3 A and B). Outside of this window the overlapping confidence limits of the calibration curve prevented pause localization accuracy better than ±3 bp. For Pol I(KF), over 85% of the pauses were located between +13 and +17 bp, a region that is GC-rich and includes a Pyr-G-C motif at +16 bp. Similarly, over 82% of the ϕ29 pauses were located between +14 and +18 bp. Pyr-G-C motifs at +8 bp and +26 bp showed weak pausing for Pol I(KF) and no pausing for ϕ29, but the identification accuracy of pause events is lower at those locations.
Fig. 3.
Pause frequency is sequence dependent and can be suppressed with betaine or elevated temperature. The mean intensity of each pause was normalized based on the final extension intensity and the frequency of pausing for the given conditions, and then mapped to the template sequence. (A) and (C) show Pol I(KF) data for sample and control sequences; (B) and (D) show the data for ϕ29.
A significant reduction in the frequency of pausing for both enzymes was observed with the addition of 1 M betaine to the DNAP/dNTP mix (Fig. 3 A and B). Betaine is a zwitteronic osmoprotectant that is thought to alter DNA stability so that GC-rich regions melt at AT-rich temperatures (22), a finding that has led to its inclusion in PCR formulations for improved amplification of difficult templates (23–24). Betaine has also been used in bulk studies to suppress replication pausing at Pyr-G-C sequences (15). Increasing the temperature of the Pol I(KF) reaction from 23 °C to 37 °C also reduced the frequency of pausing significantly, which is in agreement with previous bulk studies by Mytelka et al. (15). These authors suggested that pauses at Pyr-G-C sequences might be caused by difficulties in the polymerase fingers-closing conformational change, as at the time this transition was thought to be rate-limiting and the most sensitive to changes in temperature. However, a recent report (25) shows that the slow prechemistry step is likely not the fingers-closing transition, raising the possibility that these pauses are associated with an earlier DNA template rearrangement step that might be sequence dependent.
To verify that the observed pausing was sequence dependent and not due to the hairpin secondary structure, a control DNA molecule was constructed with the same overall structure and length as the original (Fig. 1B Top) but with a different stem sequence (Fig. 1B Bottom). The control sequence removed all three occurrences of the Py-G-C motif while maintaining as much similarity to the original sequence as possible. Importantly, only 2 bases were changed in the region where most pausing was found to occur: 5′-TAGGCGCCA-3′ was changed to 5′-TAGGATCCA-3′. Both polymerases showed over a 5-fold reduction in pause frequency with the control sequence compared to the original sequence, with less than 3% of the trajectories showing residual pausing (Fig. 3 C and D). The source of the residual control sequence pausing could be the high GC content (66%) from +15 to +20 where the majority of the pauses occurred. In addition, Mytelka et al. (15) found that Pol I(KF) paused at two nonconsensus sequences that were within 1 nt of the Pyr-G-C consensus (TCC and TGT). The control sequence did contain a TCC motif at +18 which may have contributed to some weak pausing. The control also had a TGG motif at +14, which is within 1 nt of the consensus and was not previously identified to cause pausing, but may have also contributed. Taken together, the control experiments suggest that the secondary structure of the hairpin was not playing a role in the observed pausing. It also suggests that the central 5′-CG-3′ motif, either by itself or in the context of the surrounding sequence, was responsible for causing the majority of the pauses.
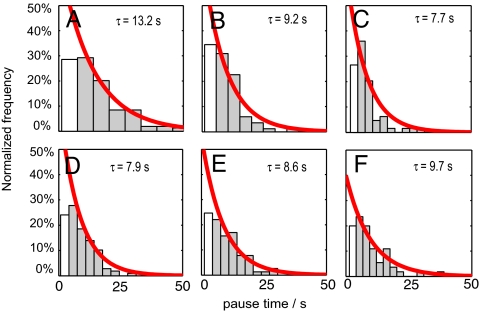
The distribution of pause lifetimes is consistent with single-step Poisson statistics. At 23 °C Pol I(KF) exhibited a mean pause lifetime of 13.2 sec for the sample template (Fig. 4A). In the reduced population of trajectories that showed pausing even with the addition of betaine or heat, the mean pause lifetime decreased 30% (Fig. 4B and Fig. S2). A similar reduction in pause lifetime was observed for Pol I(KF) acting on the control template (Fig. 4C). The mean pause lifetimes for ϕ29 with the sample template were less than for Pol I(KF) (Fig. 4E), and a reduction in lifetimes was not observed with betaine addition (Fig. 4F) or on the control sequence (Fig. 4G). Short time period pauses were likely undersampled due to limitations of the sampling bandwidth and signal to noise, which resulted in some of the pause lifetime distributions to have a distinct rise and decay, and so they were excluded from the fit. The pause intensities had a mean signal to noise ratio of 2.6 while the full extension intensities had a mean signal to noise ratio of 5.6 (Fig. S3). Pause frequencies and lifetimes are impossible to obtain through ensemble studies and this knowledge may help constrain the biophysical models of the polymerase kinetic pathway.
Fig. 4.
Pause lifetimes for Pol I(KF) and ϕ29 were measured under different conditions for the sample and control sequences. The red curves are normalized single exponential fits given by f = τ−1 exp(−τ/τ), where τ is the mean pause lifetime. Bins excluded from the fit due to undersampling are shown in white. (A) Pol I(KF) at 23 °C with the sample template; (B) Pol I(KF) at 23 °C with 1 M betaine with the sample template; (C) Pol I(KF) at 23 °C with the control template, (D) ϕ29 at 23 °C with the sample template, (E) ϕ29 at 23 °C with 1 M betaine with the sample template, and (F) ϕ29 at 23 °C with the control template.
The pauses seen here are clearly distinct from those observed in previous single molecule DNA polymerase experiments. T7 and Pol I(KF) polymerases have been shown to exhibit long pauses of heterogeneous length under template tension (13–14). These pauses were thought to be due to fluctuating hairpins, exogenous DNA hybridization, or template damage. The entire T7 replisome was also shown to halt leading-strand synthesis due to primase activity on the lagging strand (26). Similar experiments with the E. coli replisome identified a relationship between pause times and core polymerase concentration, suggesting that pauses can be caused by enzyme dissociation events (27). A recent study on real-time DNA sequencing with single ϕ29 polymerase molecules identified pause sites that corresponded to regions with predicted template secondary structure (28). However, these experiments were not performed at saturating dNTP concentrations, which complicates the interpretation. We show here that ϕ29 and Pol I(KF) are susceptible to sequence-dependent pausing at saturating dNTP concentrations and that these pauses are not due to template secondary structure.
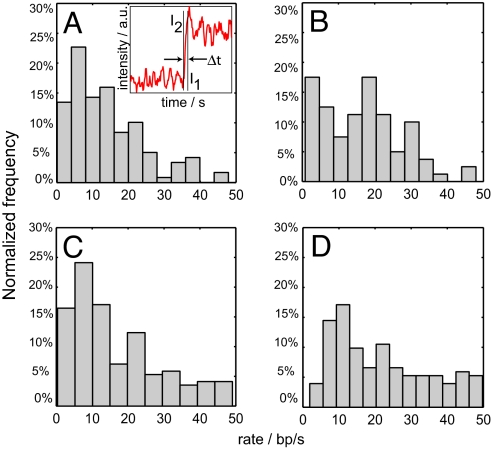
DNA synthesis is therefore a combination of high activity rapid synthesis that reflects the intrinsic “speed limit” of DNA polymerase interspersed with low activity pauses; our single molecule measurements enable us to separate these contributions and measure their kinetics. DNA synthesis burst rates were obtained by calculating the slopes of the trajectories between pauses (Fig. 5A Inset; see Materials and Methods). The time-averaged strand displacement rate for Pol I(KF) at 23 °C without betaine was 14 ± 3 nt sec−1 (SEM) (Fig. 5A). The mean rate increased with either the addition of betaine [17 ± 2 nt sec−1 (SEM), Fig. 5B] or with increasing temperature; at 37 °C, Pol I(KF) had a mean replication rate of 17 ± 3 nt sec−1 (SEM) (Fig. 5C). For ϕ29 at 23 °C, a wide distribution of synthesis burst rates was found ranging from 1 nt sec−1 to over 50 nt sec−1 with a mean rate of 28 ± 5 nt sec−1 (SEM) (Fig. 5D). Our window of sensitivity and data acquisition rate placed an upper limit of approximately 50 nt sec−1 on our rate measurements, with faster rates being discarded. This is likely why our ϕ29 rate is slightly less than the reported ϕ29 bulk rates of 38 nt sec−1 (29).
Fig. 5.
DNA replication rates were measured for single DNA polymerase molecules under different conditions. For trajectories that paused, only data in the intervals between pauses was analyzed; for trajectories that did not pause, data over the entire interval was analyzed. Rates were determined by calculating the average intensities at the start and finish of active replication through the hairpin, converting the intensities to polymerase positions, and then calculating the slope (A, Inset). The normalized distribution of rates are shown for (A) Pol I(KF) at 23 °C (mean 14 ± 3 nt sec−1), (B) Pol I(KF) with 1 M betaine at 23 °C (mean 17 ± 2 nt sec−1), (C) Pol I(KF) at 37 °C (mean 17 ± 3 nt sec−1), and (D) ϕ29 at 23 °C (mean 28 ± 5 nt sec−1).
A recent bulk study of Pol I(KF) reported a strand displacement synthesis rate over an 18-bp template of 1.2 nt sec−1 (20), which is an order of magnitude slower than our time-averaged burst synthesis rate. However, the template used in the bulk study contained three Pyr-G-C pause motifs. Based on the results of our single molecule experiments, we would predict Pol I(KF) to have 1.3 sec of synthesis time and 10.3 sec of pause time over this template, giving an estimated ensemble rate of 1.6 nt sec−1, which in good agreement with their observations.
Although strand displacement synthesis and single stranded primer extension synthesis are expected to have substantial differences in their energetics, we compared our results to experiments that measured single stranded primer extension synthesis and found surprising agreement. Our observed rate of 14 nt sec−1 is consistent with bulk primer extension experiments (17, 20). The rates measured here are 2- to 3-fold faster than a Pol I(KF) single molecule measurement (13), which was a low tension force spectroscopy study on a long template that did not have sufficient time resolution to separate pauses from bursts. Assuming that Pyr-G-C pause sites occur once every 32 bp in a random template and that the pause motif is 25% efficient, our data predict that the expected net synthesis rate over long templates is approximately 6 nt sec−1, which is in agreement with the previous single molecule measurement (7 nt sec−1).
For Pol I(KF) molecules that paused, the mean replication rate over the template sequence up until the pause site was 11 ± 1 nt sec−1 (SEM); after the pause site, the mean rate increased to 18 ± 2 nt sec−1 (SEM). Similar to Pol I(KF), ϕ29 molecules that paused underwent faster replication after arrest [41 ± 3 nt sec−1 (SEM)] than before [24 ± 4 nt sec−1 (SEM)]. Accelerating synthesis rates were also observed in trajectories that did not pause (Fig. S4). The small difference in rates before and after pausing may be due to the transition near the end of the hairpin from strand displacement replication to primer extension replication, or it could be an artifact related to the presence of the fluorophore in the template. Given the predictive accuracy of our results described above, we believe this change is more likely the replication transition than a dye-enzyme effect.
There was a surprising amount of heterogeneity in the strand displacement synthesis rates for both Pol I(KF) and ϕ29 from molecule to molecule over identical sequences (Fig. 5), with values ranging from 1 nt sec−1 to over 50 nt sec−1. The stochasticity of the synthesis rates is hidden in time-averaged or bulk rates and can only be revealed with a single molecule approach. The source of this heterogeneity could be of great interest for future polymerase studies and has not received significant attention in the literature to date.
One potential drawback of a quencher-based approach is that blinking fluorophores might incorrectly be interpreted as real extension events. If a Cy3-Cy5 FRET pair were used, this would certainly be a concern as Cy5 is known to exhibit frequent blinking (30–31). In this FRET system, a false positive extension could be caused by either BHQ-2's quenching ability switching off before extension begins or a dark Cy3 fluorophore switching on after extension has finished. BHQ-2 blinking was not significantly observed while imaging folded hairpins in the absence of polymerase or dNTPs and has not been reported in the literature. Cy3 fluorophores have a dark-state lifetime of approximately 2 sec and the on-blink and off-blink rates have been reported to be 0.36 and 0.26 times per molecule over 120 sec, respectively (32). Given the infrequent blinking and short off times of Cy3, the probability of it being off during any real extension is very low. It is therefore not surprising that only approximately 1% of the trajectories exhibited identifiable blinking after full extension (signal going from on to off and then back to on). Importantly, for the kinetics experiments we observed intermediate intensity pauses in approximately 50% of the trajectories, a phenomenon that cannot be produced by fluorophore blinking. The calibration experiments (Fig. 1C) with polymerases stalled on the template did not show any significant deviation from the expected FRET behavior for a single acceptor system, so protein-induced quenching was not a significant source of error.
The real-time single molecule observation of Pol I(KF) revealed that the polymerase's intrinsic strand displacement “speed limit” is an order of magnitude faster than reported by previous bulk processive synthesis measurements. Replication was found to be interrupted by heterogeneous sequence-dependent pauses that were independent of template secondary structure, and knowledge of these pause frequencies and lifetimes would otherwise be impossible to obtain from ensemble studies. Importantly, the pause kinetics can account for the discrepancy between our time-averaged Pol I(KF) single molecule strand displacement rate and the reported bulk strand displacement rate. The pauses were highly localized to a short Pyr-G-C sequence motif and were relieved with the addition of heat or betaine, suggesting that they may be associated with a structural template rearrangement step during the polymerase reaction pathway. Pausing at Pyr-G-C motifs is thought to be fairly ubiquitous but the pause strength can be dependent on the surrounding sequence and other environmental factors. The assay described here could be used to explore how pause strength varies with surrounding sequence or to explore the pausing of other sequence motifs. Our single molecule assay also revealed up to 50-fold variation in the synthesis rates between polymerase molecules replicating identical templates. Future single molecule experiments should help fully elucidate the source of this rate stochasticity, the mechanism of DNA polymerase sequence-dependent pausing, and provide insight into how other endogenous and exogenous factors act to slow or stall replication forks.
Materials and Methods
DNA Hairpin Construction.
DNA hairpins were constructed through the stepwise enzymatic ligation of 4 separate synthetic oligonucleotides (Integrated DNA Technologies and Operon). In the first step, oligo A (5′-Phos-TCA TAG CCA GAT GCC CAG AGA TTA GAG CGC ATG ACA AGT AAA GGA CGG TT-Biotin-3′) and oligo B (5′-Phos-CGG ATG GCG CAG CAG AG[iCy3]C AGT TCA GTC CCA CCG ACG TTT GGT CAG TT-3′) were annealed to the complementary oligo Z (5′-AAC CGT CCT TTA CTT GTC ATG CGC TCT AAT CTC TGG GCA TCT GGC TAT GAT GTT GAT GGA ACT GAC CAA ACG TCG GTG GG-3′) in 20 mM Tris, 100 mM NaCl, pH 8.0 by heating to 95 °C for 5 min and slowly cooling at 0.1 °C/s to 4 °C. 20 units of E. coli DNA ligase and 1X E. coli DNA ligase reaction buffer (New England Biolabs) were then added and the reaction was held at 16 °C for 3 h, after which the ligase was inactivated by holding the reaction at 65 °C for 20 min. Oligo C (5′-Phos-AGT GAC GCC AAC GCA ATT AC[BHQ2-dT] CTG CTG CGC CAT CCG CGG TCT ATA CGC TAG GTT TTT CCT AGC GTA TAG ACC G-3′), oligo D (5′-GCC CTG AGA GAG TTG CAG CAA GCG GTC CAC GCT GGT TTG CCC CAG CAG GCG AAA ATC CTG TTT GAT GGT GGT TCC GAA AT-3′), and oligo Y (5′-TGC GTT GGC GTC ACT ATT TCG GAA CCA CCA TCA AAC AGG ATT TTC GCC TGC TGG GGC AAA CCA GCG TGG ACC GCT TGC TGC AAC TCT CTC AGG GC-3′) were added to the solution along with 100 units of Ampligase DNA ligase in 1X Ampligase Reaction Buffer (Epicentre Biotechnologies). The solution was then heated to 95 °C for 5 min to melt all of the strands, rapidly cooled to 89 °C, slowly cooled at 0.1 °C/s to 65 °C, held at 65 °C for 2 min, and then reheated to 95 °C for 1 min. This cooling and reheating process was repeated 20 times to ensure high stringency ligation. The DNA ligation mixture was then loaded on a 15% TBE-urea denaturing polyacrylamide gel (Invitrogen) and run at 175V for 1 h in a 65 °C water bath. The gel was stained with SYBR Green I (Invitrogen) for 20 min, the band corresponding to the 259 bp DNA product was excised from the gel, and the DNA was purified as described in Invitrogen's GeneTrapper manual. The control sequence hairpin was synthesized as above but with oligo B replaced with (5′-Phos-AGG ATC TTA CCA GAG AC/ iCy3/C AGT TCA GTC CCA CCG ACG TTT GGT CAG TTC CAT CAA CA-3′) and oligo C replaced with (5′-AGT GAC GCC AAC GCA ATT AC[BHQ2-T] CTC TGG TAA GAT CCT AGG TCT ATC CTG AAG GTT TTT CCT TCA GGA TAG ACC T-3′).
Single Molecule Kinetics Measurement.
Glass surfaces (Precision Glass & Optics, D-263T cut glass, 0.145 mm, 2“x1” 40/20 surface quality) were RCA cleaned and a plastic hybriwell (Grace Biolabs) was placed on the surface to create a fluidic chamber. The surface was incubated with 0.2 mg/ml biotinylated-BSA in 1X PBS for 20 min, washed extensively with 1X PBS, and then incubated with 0.5 mg/ml neutravidin in 1X PBS for 20 min. Purified DNA hairpin was annealed with a 5′ biotinylated primer (oligo Z) as described above, diluted to 10 pM in 1X PBS, and incubated on the surface for 20 min. Surfaces were imaged on an inverted Nikon TE2000S microscope with a 60 × 1.45 NA PlanApo TIR objective, a low-fluorescence immersion oil (n = 1.515), and an automated precision stepper motor stage (Mad City Labs). A Nikon TIRF attachment in conjunction with a 532 nm diode-pumped sold-state laser (MeshTel) and a custom laser-launch system (ThorLabs) was used for illumination. A Cy3 filter set (HQ535/50, Q565LP, HQ610/75, Chroma) and a Photometrics Cascade II EMCCD were used for filtering and image acquisition. Immediately before enzyme addition, surfaces were imaged to ensure the hairpin was properly folded as evidenced by the absence of Cy3 emission. For the premature chain termination reactions, an enzyme mixture consisting of 1X polymerase reaction buffer (New England Biolabs), 100 μM of dATP, dTTP, dCTP, 10 units of DNA polymerase (Klenow exo−, New England Biolabs), 0.1 mg/ml glucose oxidase, 0.2 mg/ml catalase, 10% wt/wt glucose, and 1 mM Trolox was flowed into the chamber. After 5 min of incubation at room temperature, the surface was washed extensively with 1X PBS and 20 different fields of view were imaged. Fluorescent intensities at this stage represented a polymerase position at −6 bp with respect to the hairpin (i.e., the polymerase active site is 6 bp away from the first bp of the hairpin stem). This process was repeated 6 times with the dNTP mixes shown in Table S1.
For the real-time kinetics experiments, surfaces were prepared as described above. An enzyme mixture consisting of 1X polymerase reaction buffer (New England Biolabs), 100 μM dNTPs, 10 units of DNA polymerase (Klenow exo− or ϕ29, New England Biolabs), 0.1 mg/ml glucose oxidase, 0.2 mg/ml catalase, 10% wt/wt glucose, and 200 μM Trolox was flowed into the chamber. Images were immediately acquired every 200 ms for approximately 20 min. Freshly prepared 1 M betaine was added to the above mixture for the betaine experiments. For the real-time temperature experiments, a commercial flow cell with internal heating elements (CFCS2, Bioptechs) was used in conjunction with an external temperature controller to hold the sample temperature at 37 °C.
Image Analysis.
Custom software was written in MATLAB to automatically identify and track the XY coordinates of single molecules using a previously published particle tracking algorithm (33). A 3 × 3 pixel grid surrounding each molecule was integrated and the 5 × 5 pixel perimeter was used as to estimate the background. Raw and smoothed net intensity counts for each trajectory were then plotted and manually analyzed. Steps were identified and distinguished from gradual nonstepped growth using a custom MATLAB step-fitting algorithm (34). This algorithm generates “S-values” which are a measure for the quality of a step-fit for a given number of steps over an entire trajectory, and the maximum value of S should correspond to the best fit. For noisy trajectories, step trains containing a slight excess of steps were chosen to ensure that all true steps were included. Some trajectories like 1C were scored as having multiple distinct pauses using this algorithm. Dwell times were calculated as the time between steps. To improve the accuracy of the rate calculations, trajectories were oversampled 20-fold and fit to a cubic spline in MATLAB. Synthesis rates were then calculated by measuring the trajectory slope between the mean intensities of 2 consecutive pauses or a pause and full extension. Due to the approximately 10 bp window of sensitivity and the 5 Hz acquisition rates, synthesis rates in excess of 50 nt/sec were discarded.
Supplementary Material
Acknowledgments.
The authors would like to thank Stavros Stavrakis for helpful discussions. This work was supported in part by the Department of Defense Advanced Research Projects Agency Optofluidics Center, the Howard Hughes Medical Institute, and the National Human Genome Research Institute (Grant 5R01HG003594–04).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0907404106/DCSupplemental.
References
- 1.Smolina IV, Demidov VV, Frank-Kamenetskii MD. Pausing of DNA Polymerases on duplex DNA templates due to ligand binding in vitro. J Mol Biol. 2003;326:1113–1125. doi: 10.1016/s0022-2836(03)00044-5. [DOI] [PubMed] [Google Scholar]
- 2.Deshpande AM, Newlon CS. DNA replication fork pause sites dependent on transcription. Science. 1996;272:1030–1033. doi: 10.1126/science.272.5264.1030. [DOI] [PubMed] [Google Scholar]
- 3.Krasilnikova MM, Samadashwily GM, Krasilnikov AS, Mirkin SM. Transcription through a simple DNA repeat blocks replication elongation. EMBO J. 1998;17:5095–5102. doi: 10.1093/emboj/17.17.5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dalgaard JZ, Klar AJS. swi1 and swi3 perform imprinting, pausing, and termination of DNA replication in S. pombe. Cell. 2000;102:745–751. doi: 10.1016/s0092-8674(00)00063-5. [DOI] [PubMed] [Google Scholar]
- 5.Takeuchi Y, Horiuchi T, Kobayashi T. Transcription-dependent recombination and the role of fork collision in yeast rDNA. Genes Dev. 2003;17:1497–1506. doi: 10.1101/gad.1085403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bedinger P, Munn M, Alberts BM. Sequence-specific pausing during in vitro DNA replication on double- stranded DNA templates. J Biol Chem. 1989;264:16880–16886. [PubMed] [Google Scholar]
- 7.LaDuca RJ, Fay PJ, Chuang C, McHenry CS, Bambara RA. Site-specific pausing of deoxyribonucleic acid synthesis catalyzed by 4 forms of Escherichia coli DNA polymerase III. Biochemistry. 1983;22:5177–5188. doi: 10.1021/bi00291a018. [DOI] [PubMed] [Google Scholar]
- 8.Lemoine FJ, Degtyareva NP, Lobachev K, Petes TD. Chromosomal translocations in yeast induced by low levels of DNA polymerase: A model for chromosome fragile sites. Cell. 2005;120:587–598. doi: 10.1016/j.cell.2004.12.039. [DOI] [PubMed] [Google Scholar]
- 9.Cha RS, Kleckner N. ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science. 2002;297:602–606. doi: 10.1126/science.1071398. [DOI] [PubMed] [Google Scholar]
- 10.Kang S, Ohshima K, Shimizu M, Amirhaeri S, Wells RD. Pausing of DNA synthesis in vitro at specific loci in CTG and CGG triplet repeats from human hereditary disease genes. J Biol Chem. 1995;270:27014–27021. doi: 10.1074/jbc.270.45.27014. [DOI] [PubMed] [Google Scholar]
- 11.Samadashwily GM, Raca G, Mirkin SM. Trinucleotide repeats affect DNA replication in vivo. Nat Genet. 1997;17:298–304. doi: 10.1038/ng1197-298. [DOI] [PubMed] [Google Scholar]
- 12.Tasara T, et al. Incorporation of reporter molecule-labeled nucleotides by DNA polymerases. II. High-density labeling of natural DNA. Nucl Acids Res. 2003;31:2636–2646. doi: 10.1093/nar/gkg371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maier B, Bensimon D, Croquette V. Replication by a single DNA polymerase of a stretched single-stranded DNA. Proc Natl Acad Sci USA. 2000;97:12002–12007. doi: 10.1073/pnas.97.22.12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wuite GJL, Smith SB, Young M, Keller D, Bustamante C. Single-molecule studies of the effect of template tension on T7 DNA polymerase activity. Nature. 2000;404:103–106. doi: 10.1038/35003614. [DOI] [PubMed] [Google Scholar]
- 15.Mytelka DS, Chamberlin MJ. Analysis and suppression of DNA polymerase pauses associated with a trinucleotide consensus. Nucl Acids Res. 1996;24:2774–2781. doi: 10.1093/nar/24.14.2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Astatke M, Grindley NDF, Joyce CM. How E. coli DNA polymerase I (klenow fragment) distinguishes between deoxy- and dideoxynucleotides. J Mol Biol. 1998;278:147–165. doi: 10.1006/jmbi.1998.1672. [DOI] [PubMed] [Google Scholar]
- 17.Dahlberg ME, Benkovic SJ. Kinetic mechanism of DNA polymerase I(Klenow fragment): Identification of a second conformational change and evaluation of the internal equilibrium constant. Biochemistry. 1991;30:4835–4843. doi: 10.1021/bi00234a002. [DOI] [PubMed] [Google Scholar]
- 18.Kuchta RD, Mizrahi V, Benkovic PA, Johnson KA, Benkovic SJ. Kinetic mechanism of DNA polymerase I (Klenow) Biochemistry. 1987;26:8410–8417. doi: 10.1021/bi00399a057. [DOI] [PubMed] [Google Scholar]
- 19.Purohit V, Grindley NDF, Joyce CM. Use of 2-Aminopurine Fluorescence To Examine Conformational Changes during Nucleotide Incorporation by DNA Polymerase I (Klenow Fragment)†. Biochemistry. 2003;42:10200–10211. doi: 10.1021/bi0341206. [DOI] [PubMed] [Google Scholar]
- 20.Singh K, Srivastava A, Patel SS, Modak MJ. Participation of the fingers subdomain of Escherichia coli DNA polymerase I in the strand displacement synthesis of DNA. J Biol Chem. 2007;282:10594–10604. doi: 10.1074/jbc.M611242200. [DOI] [PubMed] [Google Scholar]
- 21.Weiss S. Measuring conformational dynamics of biomolecules by single molecule fluorescence spectroscopy. Nat Struct Mol Biol. 2000;7:724–729. doi: 10.1038/78941. [DOI] [PubMed] [Google Scholar]
- 22.Rees WA, Yager TD, Korte J, Von Hippel PH. Betaine can eliminate the base pair composition dependence of DNA melting. Biochemistry. 1993;32:137–144. doi: 10.1021/bi00052a019. [DOI] [PubMed] [Google Scholar]
- 23.Henke W, Herdel K, Jung K, Schnorr D, Loening SA. Betaine improves the PCR amplification of GC-rich DNA sequences. Nucl Acids Res. 1997;25:3957–3958. doi: 10.1093/nar/25.19.3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baskaran N, et al. Uniform amplification of a mixture of deoxyribonucleic acids with varying GC content. Genome Res. 1996;6:633–638. doi: 10.1101/gr.6.7.633. [DOI] [PubMed] [Google Scholar]
- 25.Joyce CM, et al. Fingers-closing and other rapid conformational changes in DNA polymerase I (Klenow fragment) and their role in nucleotide selectivity. Biochemistry. 2008;47:6103–6116. doi: 10.1021/bi7021848. [DOI] [PubMed] [Google Scholar]
- 26.Lee J-B, et al. DNA primase acts as a molecular brake in DNA replication. Nature. 2006;439:621–624. doi: 10.1038/nature04317. [DOI] [PubMed] [Google Scholar]
- 27.Tanner NA, et al. Single-molecule studies of fork dynamics in Escherichia coli DNA replication. Nat Struct Mol Biol. 2008;15:170–176. doi: 10.1038/nsmb.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eid J, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2008;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 29.Soengas MS, Gutiérrez C, Salas M. Helix-destabilizing activity of φ29 single-stranded DNA binding protein: Effect on the elongation rate during strand displacement DNA replication. J Mol Biol. 1995;253:517–529. doi: 10.1006/jmbi.1995.0570. [DOI] [PubMed] [Google Scholar]
- 30.Rasnik I, McKinney SA, Ha T. Nonblinking and long-lasting single-molecule fluorescence imaging. Nat Meth. 2006;3:891–893. doi: 10.1038/nmeth934. [DOI] [PubMed] [Google Scholar]
- 31.Chandran RS, John SE, Amit M. Long time scale blinking kinetics of cyanine fluorophores conjugated to DNA and its effect on F[o-umlaut]rster resonance energy transfer. J Chem Phys. 2005;123:224708. doi: 10.1063/1.2136157. [DOI] [PubMed] [Google Scholar]
- 32.Aitken CE, Marshall RA, Puglisi JD. An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophys J. 2008;94(5):1826–1835. doi: 10.1529/biophysj.107.117689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crocker JC, Grier DG. Methods of digital video microscopy for colloidal studies. J Colloid Interface Sci. 1996;179:298–310. [Google Scholar]
- 34.Kerssemakers JWJ, et al. Assembly dynamics of microtubules at molecular resolution. Nature. 2006;442:709–712. doi: 10.1038/nature04928. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.