Abstract
Class IB phosphoinositide 3-kinase γ (PI3Kγ) elicits various immunologic and cardiovascular responses; however, the molecular basis for this signal heterogeneity is unclear. PI3Kγ consists of a catalytic p110γ and a regulatory p87PIKAP (p87, also p84) or p101 subunit. Hitherto p87 and p101 are generally assumed to exhibit redundant functions in receptor-induced and G protein βγ (Gβγ)-mediated PI3Kγ regulation. Here we investigated the molecular mechanism for receptor-dependent p87/p110γ activation. By analyzing GFP-tagged proteins expressed in HEK293 cells, PI3Kγ-complemented bone marrow–derived mast cells (BMMCs) from p110γ-/- mice, and purified recombinant proteins reconstituted to lipid vesicles, we elucidated a novel pathway of p87-dependent, G protein–coupled receptor (GPCR)-induced PI3Kγ activation. Although p101 strongly interacted with Gβγ, thereby mediating PI3Kγ membrane recruitment and stimulation, p87 exhibited only a weak interaction, resulting in modest kinase activation and lack of membrane recruitment. Surprisingly, Ras-GTP substituted the missing Gβγ-dependent membrane recruitment of p87/p110γ by direct interaction with p110γ, suggesting the indispensability of Ras for activation of p87/p110γ. Consequently, interference with Ras signaling indeed selectively blocked p87/p110γ, but not p101/p110γ, kinase activity in HEK293 and BMMC cells, revealing an important crosstalk between monomeric and trimeric G proteins for p87/p110γ activation. Our data display distinct signaling requirements of p87 and p101, conferring signaling specificity to PI3Kγ that could open up new possibilities for therapeutic intervention.
Keywords: confocal life cell imaging, G protein, receptor signaling, mast cells
Chemotaxis, the release of antimicrobial molecules from granules or production of reactive oxygen species (ROS), displays some of the most potent defense mechanisms of the immune system. These immune responses are under spatial and temporal control of diverse stimuli including chemokines, chemotactic peptides, or nucleosides, which typically signal through the G protein–coupled receptor (GPCR)-dependent phosphoinositide 3-kinase γ (PI3Kγ) (1). Thus, gene-targeted mice lacking the catalytic subunit of PI3Kγ show severe defects in immune response and are completely protected against systemic anaphylaxis (2–5). Moreover, in models of rheumatoid arthritis, systemic lupus erythematosus, and atherosclerosis, loss of PI3Kγ activity resulted in partial or even complete protection against disease progression (6–8). Thus, PI3Kγ is considered a promising drug target for the treatment of chronic inflammation and allergy (9, 10). The vital role of PI3Kγ is not restricted to the immune system, however; it exhibits additional functions in the cardiovascular system (11–14). Although this reflects the ability of PI3Kγ to regulate multiple effects, elucidating how signal-specificity is achieved may facilitate the design of strategies for selective pharmacologic interventions.
To some degree, specificity within PI3K-dependent signaling is accomplished by the existence of 4 class I PI3K isoforms, all of which are regulated by transmembrane receptors and/or active Ras. Class I PI3Ks are heterodimers composed of a catalytic p110 and a regulatory subunit and are further subdivided into class IA and class IB (15, 16). Within the class IA PI3Ks, 3 distinct catalytic p110 isoforms contribute to heterogeneity and signal-specificity of PI3Ks: α, β, and δ (17–21). Although class IA PI3Kβ is under the control of GPCRs and receptor tyrosine kinases, PI3Kγ is considered to be exclusively GPCR-regulated and thus the only member of class IB (11, 19–28). PI3Kγ is composed of p110γ and the regulatory p101 or the novel p87PIKAP (p87, also called p84) subunit (29–32). The discovery of p87 opened the possibility of conferring PI3Kγ signal-specificity on the 2 regulatory subunits, but p87 is considered functionally identical to p101. Very recently, we demonstrated that p87/p110γ and p101/p110γ differ in their capability to regulate distinct adenosine-induced cellular functions (33). Although we correlated distinct cell responses elicited by the 2 dimers with the formation of PtdIns (3,4,5)P3 pools at different membrane compartments, the regulatory mechanism conferring this signal-specificity remains unknown (31–34).
In the present study, we demonstrate that p87 and p101 do not equivalently function as Gβγ adapters. Furthermore, we report that Ras is critical for distinct regulation of p87/p110γ and p101/p110γ, arguing for a specific role of Ras in GPCR-controlled class IB PI3K signaling.
Results
fMLP Selectively Activates PI3Kγ Heterodimers.
As a first step toward deciphering a molecular mechanism for PI3Kγ signal-specificity downstream of GPCRs, we examined the contribution of p87 and p101 in this process. PI3Kγ activity was induced by stimulating a prototypical GPCR expressed in HEK cells, formyl-methionyl-leucyl-phenylalanine (fMLP) receptor, and monitored by confocal laser scanning microscopy using a GFP-fused PH domain. PH-domain containing proteins bind to the major product of PI3K-activity, PtdIns (3,4,5)P3, resulting in their plasma membrane translocation, thereby providing a direct readout for PI3Kγ activity in cells (30, 31, 35, 36). As expected, solitarily expressed p110γ showed no GPCR-induced activation [supporting information (SI) Fig. S1A, Upper], whereas p101/p110γ was robustly activated (Fig. S1A, Lower) (35). In contrast to p101/p110γ, GPCR-induced stimulation of p87/p110γ led only to a half-maximal activation of the kinase (Fig. S1 A–C) (31). Similar results were obtained on direct stimulation of both heterodimers with Gβγ (Fig. 1A). We excluded differences in expression levels of p87 and p101 and improper dimerization of p87 and p110γ (Fig. S2 A and B); thus, the results indicate that the fMLP receptor discriminates between the 2 PI3Kγ dimers by means of distinct efficiencies of Gβγ to stimulate the dimers.
Fig. 1.
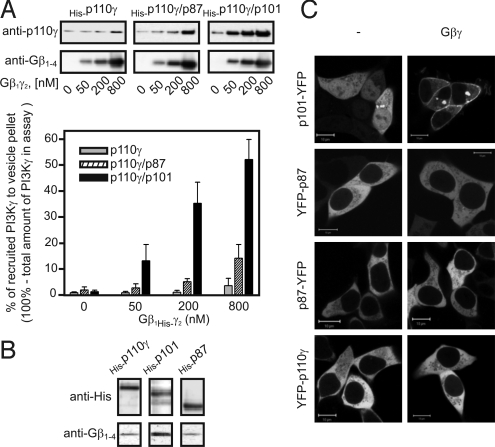
Activation of p87/p110γ and p101/p110γ by Gβγ. (A) Membrane recruitment of GFP-Grp1PH in living cells. HEK cells were transfected with plasmids encoding Gβγ, PI3Kγ (p110γ, p87/p110γ, or p101/p110γ), and GFP-Grp1PH as indicated above. The localization of GFP-Grp1PH is shown in representative, starved (18 h) HEK cells out of 3 independent experiments (cLSM slices of 0.8 μm). p101/p110γ activity led to a more pronounced redistribution of the PH domain compared with p87/p110γ activity. (Scale bar: 10 μm.) The histogram represents the statistic evaluation of the membrane translocation of GFP-Grp1PH in the corresponding experiments. The data represent the mean ± SD of 3 independent experiments analyzing 18 cells. (B) Purified PI3Kγ (i.e., p110γ, p87/p110γ, and p101/p110γ), stimulated with increasing concentrations of Gβγ using PtdIns (4,5)P2 as a substrate. 32P-labeled PtdIns (3,4,5)P3 was separated by thin-layer chromatography and quantified with a Fujifilm FLA-5000 imaging system. p101, but not p87, affects the potency with which Gβγ activates p110γ. Data points and EC50 values represent the mean of 3 independent experiments.
To determine the specific effect of Gβγ on p87/p110γ activity, we compared Gβγ-induced stimulation of PI3Kγ heterodimers using purified proteins reconstituted to phospholipid vesicles (Figs. 1 B and S2C). Gβγ moderately stimulated monomeric p110γ (EC50 of 214 ± 35.7 nM), whereas dimerization of p110γ with p101 resulted in a dramatic increase in the sensitivity of p110γ toward Gβγ (EC50 of 12 ± 2.2 nM and 60-fold improved efficiency; Fig. 1B). Surprisingly, p87 failed to mediate an appropriate sensitization of p110γ toward Gβγ. The potency of Gβγ to induce p110γ activation was not affected (EC50: 223 ± 49.7 nM), although it increased the efficiency slightly. These results certainly call into question the current view of equal Gβγ-mediated regulatory mechanisms for p87/p110γ and p101/p110γ. In addition, comparing the in vitro data with corresponding results obtained in cells revealed a clear discrepancy in p87/p110γ activity. Although the degree of activation of p87/p110γ and monomeric p110γ by Gβγ did not differ in vitro (Fig. 1B), both GPCR-induced and Gβγ-induced stimulation clearly resulted in significant activity of p87/p110γ, but not of p110γ, in cells (Figs. 1 A and S1A). Thus, we postulated that an additional regulator is involved in GPCR-induced p87/p110γ activation in vivo.
Endogenous GTP-Bound Ras is Indispensable for Activation of p87/p110γ.
The second known PI3Kγ activator, the small GTPase Ras, stimulates PI3Kγ through direct interaction with p110γ (Fig. S3 A–C) (37, 38). Thus, Ras represents an excellent candidate for coregulating p87/p110γ activity. To assess the impact of Ras on GPCR-induced p87/p110γ activation, we used a p110γ variant (referred to as p110γK251A) known to exhibit reduced H-Ras binding (37). Coimmunoprecipitation assays revealed a negligible binding of p110γK251A to constitutive active H-RasG12V compared with wild-type p110γ (Fig. S3D), and thus H-RasG12V could stimulate p110γK251A only marginally (Fig. S3E). Using this variant, fMLP-stimulated receptors almost completely failed to activate p87/p110γK251A, whereas, conversely, the degree of fMLP-induced p101/p110γK251A stimulation remained unchanged compared with the wild-type heterodimer (Fig. 2 A and B). In a second independent and complementary approach, we overexpressed the GTPase-activating protein (GAP) domain of neurofibromin 1 (NF1) to neutralize endogenous Ras activity. NF1 stimulates the GTPase activity of all p110γ-interacting Ras isoforms, thereby retaining Ras in its GDP-bound inactive form (Figs. S3F and S4C) (38–40). Although the fMLP-induced activation of p101/p110γ was unaffected by NF1, the p87/p110γ heterodimer failed to activate in the presence of NF1, resembling the situation seen with p110γK251A (Fig. 2 A and C). Equivalent results were obtained using overexpressed Gβγ to stimulate PI3Kγ (Fig. S4 A and B), while NF1 activity was confirmed in parallel aliquots of the transfected HEK cells (Fig. S3F). Because blockage of the Ras–p110γ interaction led to a loss of fMLP-induced p87/p110γ activity, but not of p101/p110γ activity, our data implicate that fMLP receptor–induced activation of p87/p110γ is mediated by Ras and Gβγ. In contrast, p101/p110γ is sufficiently activated by Gβγ, with an insignificant impact of Ras for activation. Thus, Ras is an indispensable coactivator of the p87/p110γ heterodimer.
Fig. 2.
Interaction of PI3Kγ with Ras is indispensable for fMLP-induced activation of p87/p110γ. (A) Membrane recruitment of GFP-Grp1PH in living cells. HEK cells were transfected with plasmids encoding fMLP-R, PI3Kγ (p110γ, p87/p110γ, p101/p110γ, p87/p110γK251A, or p101/p110γK251A), NF1, and GFP-Grp1PH, as indicated above. Shown are representative, starved (18 h) HEK cells (cLSM slices of 0.8 μm) out of 3 independent experiments before and after the addition of fMLP (1 μM; 3 min). Comparison of the PI3Kγ activities shows equal p101/p110γ and p101/p110γK251A activities, whereas p87/p110γK251A activity is significantly reduced compared with its wild-type p110γ counterpart. Consistently, in the presence of NF1, p101/p110γ activity is unaltered, whereas p87/p110γ activity is lost. (Scale bar: 10 μm.) (B) Kinetics of GFP-Grp1PH redistribution in the cells shown in A. (C) Histogram showing the statistical evaluation of 6 cells out of 6 independent experiments as shown in (A), and bar graphs showing mean ± SD values of the redistributed fluorescence on fMLP treatment (1 μM; 3 min).
GPCR-Induced p87/p110γ Activity Is Enhanced by Ras Overexpression.
To extend our findings, we coexpressed wild-type H-Ras in HEK cells (Fig. S5). fMLP-induced p87/p110γ activity was significantly enhanced (Fig. S5, Middle), although the basal activity was unaffected. Furthermore, the overexpression of wild-type Ras led to a maximal fMLP-induced activation of p87/p110γ, equal to that of p101/p110γ (Fig. S5C). This indicates specific roles for Gβγ and Ras during receptor-induced activation of p87/p110γ. This approach does not allow clarification of how Ras and Gβγ mechanistically contribute to the activation of p87/p110γ, however.
p87 Fails To Serve as a Gβγ Recruitment Adapter for PI3Kγ.
To tackle this issue, we used an established lipid vesicle pull-down assay (24) to gain deeper insight into Gβγ interaction with monomeric or heterodimeric p110γ. Gβγ recruited p87/p110γ to phospholipid vesicles only marginally compared with p101/p110γ (Fig. 3A). To estimate the affinity of p87 toward Gβγ, we copurified both entities from baculovirus-infected Sf9 cells. Only very little Gβγ was associated with p87, resembling the situation for p110γ, whereas Gβγ was strongly bound to p101 (Fig. 3B). The weak interaction between p87 and Gβγ appears to be insufficient for membrane recruitment of PI3Kγ in cells (Fig. 3C). These observations demonstrate that the PI3K adapter protein p87PIKAP unlikely serves as an adapter for Gβγ-dependent recruitment.
Fig. 3.
p87 does not function as a Gβγ adapter for PI3Kγ membrane recruitment. (A) Gβγ was tested for its ability to recruit p110γ, p87/p110γ, or p101/p110γ (each with 200 ng of p110γ) to phospholipid vesicles. Aliquots of pelleted phospholipid vesicles and supernatants were subjected to SDS/PAGE, followed by immunoblotting. Chemiluminescence signals were documented with a CCD camera. Data are given as mean ± SD of duplicate determinations in 3 independent experiments (Lower). Immunoblots of 1 representative experiment are shown (Upper). Only p101/p110γ was relevantly translocated to phospholipids by Gβγ. (B) Gβγ was coexpressed with His-p110γ, His-p101, or His-p87 in Sf9 cells and purified. Following purification on Ni2+-NTA Superflow resin, bound proteins were separated by SDS/PAGE and analyzed by immunoblotting. The amount of Gβγ copurified with p110γ and p87 is comparable, whereas that copurified with p101 was higher. (C) Subcellular distribution of fluorescently labeled PI3Kγ subunits. HEK cells were transfected with plasmids encoding Gβγ and N-terminally or C-terminally YFP-tagged PI3Kγ subunits (p110γ, p101, or p87), as indicated above. The localization of YFP-labeled PI3Kγ subunits in representative, starved (18 h) HEK cells (cLSM slices of 0.8 μm) out of 3 independent experiments is shown. Only p101 is sufficiently translocated to the plasma membrane by Gβγ. (Scale bar: 10 μm.)
H-Ras Recruits p110γ to Membranes.
The recruitment of the cytosolic PI3Kγ to the plasma membrane, which contains its regulators as well as its substrate, is mandatory for the activation of this enzyme. Thus, we hypothesized that active Ras may represent the missing link required for p87/p110γ membrane translocation. Indeed, constitutively active H-RasG12V, but not dominant negative H-RasS17N, induced a cellular redistribution of YFP-labeled p110γ to membranes, indicating that Ras-GTP is indeed capable of recruiting the lipid kinase in living cells (Fig. 4A). Furthermore, analysis of fixed HEK cells revealed a colocalization of H-RasG12V and YFP-p110γ at cellular membranes (Fig. 4B). In control cells, we demonstrated H-Ras–dependent recruitment of p87/p110γ and p101/p110γ, but not of solitarily expressed p87 or p101, to cell membranes, suggesting that recruitment of p87/p110γ is facilitated through a direct interaction of Ras with the catalytic p110γ subunit (Figs. S3A and S6). To further support the conclusion that membrane recruitment of p87/p110γ by Ras represents its prominent function, we used a previously generated p110γCAAX mutant known to constitutively localize p110γ to membranes (Fig. S2B) (35). Consistently coexpressed NF1 did not blunt Gβγ-induced stimulation of p87/p110γCAAX activity (Fig. 4C).
Fig. 4.
Active H-Ras recruits p110γ to cell membranes. (A) Subcellular distribution of fluorescently labeled p110γ in living cells. HEK cells were transfected with plasmids encoding H-Ras (H-RasG12V or H-RasS17N) and YFP-tagged p110γ as indicated above. The localization of YFP-labeled p110γ subunits in representative, starved (18 h) HEK cells (cLSM slices of 0.8 μm) out of 3 independent experiments is shown. p110γ translocated to the plasma membrane in the presence of H-RasG12V. (Scale bar: 10 μm.) (B) Colocalization of fluorescently labeled p110γ and immunostained H-Ras. HEK cells were transfected with plasmids encoding H-Ras (H-RasG12V or H-RasS17N) and YFP-tagged p110γ as indicated above. The localization of YFP-labeled p110γ (yellow) and immunostained H-Ras (red) is shown in representative fixed HEK cells from 3 independent experiments (cLSM slices of 0.8 μm). p110γ colocalized at the plasma membrane with H-RasG12V. (Scale bar: 10 μm.) (C) Membrane recruitment of GFP-Grp1PH in living cells. HEK cells were transfected with plasmids encoding Gβγ, PI3Kγ (p87/p110γ or p87/p110γCAAX), GFP-Grp1PH, and NF1 as indicated above. Shown are representative, starved (18 h) HEK cells (cLSM slices of 0.8 μm) from 4 independent experiments. Unlike p87/p110γ activity, p87/p110γCAAX activity was unchanged in the presence of NF1. (Scale bar: 10 μm.) The right panels present the quantification of the membrane translocation of GFP-Grp1PH in the corresponding experiments. The data represent the mean ± SD of 4 independent experiments analyzing 24 cells.
NF1 Selectively Inhibits Adenosine-Induced Activation of p87/p110γ in Murine Bone Marrow–Derived Mast Cells (BMMCs).
To validate our findings in more relevant cells, we used BMMCs from p110γ null mice. p110γ-/- BMMCs are devoid of both PI3Kγ catalytic and regulatory subunits, enabling specific analysis of either p87/p110γ or p101/p110γ on their complementation (33, 34). Attenuation of Ras activity by the coexpression of NF1 in PI3Kγ-reconstituted BMMCs led to a loss of adenosine-dependent phosphorylation of PKB/Akt in cells expressing p87/p110γ, but not in cells expressing p101/p110γ (Fig. 5).
Fig. 5.
Overexpressed NF1 blunts activation of p87/p110γ, but not p101/p110γ, in murine BMMCs. (A) PKB/Akt phosphorylation (pThr308 and pSer473) in BMMCs. p110γ and HA-p87 or HA-p101 were coexpressed with or without Flag-NF1 in p110γ null BMMCs. Transfected BMMCs were starved for 3 h and then stimulated with 2 μM adenosine (Ade) or 10 ng/mL of murine stem cell factor (SCF) for 2 min. Expression of proteins was verified. NF1-induced attenuation of Ras activity was validated using anti-phosphoMAPK (pMAPK) antibodies. Adenosine-induced p101/p110γ PKB phosphorylation was unchanged, whereas p87/p110γ PKB phosphorylation was depleted in the presence of NF1. At the same time, the phosphorylation of MAPK triggered by adenosine or SCF was affected by NF1. (B) Quantification of adenosine-triggered phosphorylation of PKB (on pSer473 and pThr308) and pMAPK from experiments as shown in (A). Values from NF1 cotransfected cells are expressed as percent of control (p110γ null BMMCs reconstituted with the indicated PI3Kγ complexes). Data are given as mean ± SEM (pSer473, n = 5; pThr308, n = 3; pMAPK, n = 4). *, P < .02.
Discussion
The signaling specificity of class IA PI3Ks is supported by the existence of 3 distinct catalytic subunits, whereas a comparable source for signaling specificity within the class IB PI3Ks, which includes only 1 isoform (i.e., PI3Kγ), is missing (1, 17–21, 41). Here we have demonstrated that Ras, in concert with Gβγ and the regulatory PI3Kγ subunits, provides a basis for PI3Kγ signal-specificity. This conclusion stems from 3 fundamental and unexpected findings. In contradiction to current thinking, (i) interaction of p87 with Gβγ differs significantly from that of p101, (ii) Ras is able to recruit p110γ to membranes, and (iii) Ras acts as an indispensable coregulator of p87/p110γ.
p87 Fails to Substitute for p101.
Most surprising was our observation that p87 failed to sensitize p110γ for Gβγ-dependent activation and to function as a recruitment adapter. These results were particularly amazing, because we discovered p87 as a homolog of p101 with significant sequence similarities not only within the predicted N-terminal p110γ, but also within the putative C-terminal Gβγ-binding region of p101 (30). In fact, based on the significant degree of homology within the Gβγ-binding region of p87 and p101, in combination with the initial results of a similar GPCR- or Gβγ-induced activation of p87/p110γ and p101/p110γ, we and others initially assumed that p87 functioned as a second Gβγ-recognizing recruitment adapter (31, 32, 34). However, a sequence similarity of only 24%, a lower degree of similarity within the putative Gβγ-binding region compared with the predicted p110γ-binding region of p101 and p87, and an attenuated activation of p87/p110γ by GPCRs or Gβγ (ref. 31 and this study) prompted us to reconsider our initial assumption. We compared the function of p87/p110γ not only with that of p101/p110γ, but also with that of monomeric p110γ. We found only a faint interaction of p87 with Gβγ in vitro, accounting for the inability of p87 to mediate Gβγ-induced membrane recruitment in cells, as well as for the missing sensitization of p110γ.
Ras Has a Recruiting Effect on PI3Kγ.
Our study demonstrates that in living cells, interaction of Ras with the catalytic p110γ subunit provokes membrane recruitment of p110γ, p87/p110γ, or p101/p110γ. This finding is striking, because the Ras-dependent recruitment of p110γ has been a subject of intense debate. Attempts to prove Ras-dependent p110γ membrane recruitment have failed, although membrane localization of Ras-GTP has been shown to be necessary for its activating impact on p110γ (38). Contrarily, genetic approaches suggest that p110γ-dependent cell mutagenesis is essentially regulated by Ras-driven membrane recruitment of p110γ (42). We cannot exclude the possibility that previous attempts to directly show Ras-dependent p110γ membrane recruitment might have failed because of methodologic limitations. To avoid this, we analyzed intact, living cells and indeed observed a Ras-dependent membrane accumulation of p110γ due to direct interaction. Certainly, our result is supported by the concept that a membrane-bound stimulator recruits its cytosolic effector to bring it in close contact with its substrate. Furthermore, our result is consistent with studies demonstrating membrane translocation as a major function of Ras on other effectors (43, 44). Thus, our data extend the current knowledge of Ras action on PI3Kγ, showing that Ras has both activating and membrane-recruiting effects on p110γ.
PI3Kγ Heterodimers Are Specifically Regulated by GPCRs.
Finally, we have shown that active Ras acts as an indispensable costimulator of p87/p110γ, whereas Gβγ is sufficient for activation of p101/p110γ. This conclusion is supported by 2 independent and complementary approaches, first by interfering with Ras binding by modification of p110γ and second by modulating Ras activity. Both diminished the regulatory impact of Ras while leaving Gβγ-dependent regulation untouched. Furthermore, these results are consistent with the observation that p87 failed to be recruited by Gβγ but instead was translocated by Ras via interaction with p110γ. In addition, using a constitutively membrane-bound p110γ mutant, we could strengthen our hypothesis that recruitment represents the predominant function of Ras in the activation of p87/p110γ.
Based on our data, we have proposed a model for distinct regulatory mechanisms of GPCR-induced activation of the 2 PI3Kγ heterodimers (Fig. 6). In this scenario, p87/p110γ is specifically regulated by Ras and Gβγ in an orchestrated fashion, where Ras is indispensable for membrane recruitment and thus activation of the lipid kinase. In contrast, p101/p110γ is essentially and sufficiently recruited and stimulated by Gβγ. Thus, distinct interactions of the 2 PI3Kγ heterodimers p101/p110γ and p87/p110γ with their upstream regulators define specific regulation of these heterodimers.
Fig. 6.
Hypothetical model of regulatory mechanisms of PI3Kγ heterodimers. The scheme illustrates the activation processes of p87/p110γ (Left) and p101/p110γ (Right). Interaction of p101 with Gβγ is sufficient to translocate p101/p110γ to the plasma membrane as a prerequisite for its activation. In contrast, p87/p110γ is not translocated to the plasma membrane by Gβγ; instead, translocation of p87/p110γ is accomplished by the interaction of p110γ with Ras-GTP. Based on the requirement of Ras-dependent recruitment of p87/p110γ, Ras constitutes an indispensable activator in the activation process of this specific isoform. Following recruitment of PI3Kγ, both p87/p110γ and p101/p110γ are allosterically activated. Consistent with the present knowledge, these distinct mechanisms might be the basis for the specific regulation of different PI3Kγ-dependent cellular effects (see the text for details). The data presented here provide evidence of differential regulatory mechanisms of the 2 PI3Kγ heterodimers.
This instantly raises questions about this model's physiological relevance. Interestingly, it has been recently proposed that Ras and Gβγ can simultaneously, but differentially, regulate distinct PI3Kγ effectors on activation by the same GPCR in neutrophils (45). Essentially, the data from that study imply that most of the GPCR-induced and PI3Kγ-regulated cellular effects in neutrophils are mediated via the Gβγ adapter p101 or via Ras binding, whereas the GPCR-induced production of ROS is mediated exclusively via Ras binding to p110γ. In addition, we have recently reported that adenosine-induced activation of the 2 PI3Kγ heterodimers addresses distinct cellular effects in BMMCs (33). The underlying regulatory mechanisms for this remain unclear, however (34). Here we provide evidence that Ras may contribute to the differential coupling of PI3Kγ heterodimers to downstream cell responses. It may be speculated that this is accomplished by directing the 2 PI3Kγ heterodimers to different membrane compartments by Ras (46, 47).
Taken together, our findings point to a unique and indispensable role of Ras for the GPCR-induced PI3Kγ-dependent production of ROS, as well as degranulation of mast cells. Thus, it is reasonable to speculate that ROS production and degranulation, in contrast to other PI3Kγ-dependent cellular effects, may be specifically regulated by Ras and p87/p110γ heterodimer (Fig. 6).
Conclusion
In conclusion, we have identified distinct regulatory mechanisms upstream of the 2 PI3Kγ dimers p87/p110γ and p101/p110γ, conferring isoform specificity within class IB PI3Ks. A promising future aim is to extend the assignment of these 2 PI3Kγ heterodimers to distinct cellular functions. This could lead to the development of pharmacologic strategies for more specific intervention in pathophysiologically relevant signaling pathways.
Experimental Procedures
Construction of Expression Plasmids.
The p87 expression plasmids were generated using mouse full-length cDNA (German Science Centre for Genome Research). Table S1 lists the primers, restriction enzymes, and vectors used for the cloning of p87, p110γ, YFP-H-Ras, H-RasG12V, and the Ras-GAP domain of NF1 constructs. H-Ras-wt and H-RasS17N in pcDNA3, as well as templates for the construction of YFP-H-Ras and H-RasG12V, were generous gifts from M. Schmidt and K. Giehl, respectively. The construction of all of the other plasmids has been described elsewhere (25, 31, 35).
Cell Culture, Transfection, and Infection.
HEK cells (German Resource Centre for Biological Material) were grown and transfected as described previously (30, 35, 36). Fall armyworm ovary cells (Sf9 cells; Invitrogen) were cultured and infected as described previously (25, 28, 48).
Immunofluorescence and Fluorescence Imaging.
Immunofluorescence staining was carried out as detailed previously (36) with minor modifications (see SI Text). Cell imaging was performed as described previously using a Zeiss LSM 510-META confocal laser scanning microscope (30, 35, 36).
Analysis of GFP-Grp1PH Translocation.
The subcellular distribution of GFP-Grp1PH was evaluated as detailed elsewhere (36). Significance was assessed using the paired Student t-test, with *, P < .05; **, P < .01; and ***, P < .005.
Purification and Copurification of Recombinant Gβ1γ2 and PI3Kγ From Sf9 Cells.
The purification of recombinant Gβ1γ2 and PI3Kγ has been detailed elsewhere (25, 28, 48). Copurification was performed as described previously (28) with modifications (see SI Text).
Phospholipid Vesicle Pull-Down Assay.
Determination of Gβ1γ2 and PI3Kγ association on phospholipid vesicles was performed as described previously (24) with modifications (see SI Text).
BMMC Cell Culture, Isolation, Differentiation, Nucleofection, and Stimulation.
BMMC cell culture, isolation, differentiation, nucleofection, and stimulation from PI3Kγ null mice (2) was performed as described previously (33) with modifications (see SI Text).
Supplementary Material
Acknowledgments.
We thank Wibke Ballhorn, Julie Mason, and Ilse Meyer for technical assistance. B.K. thanks Oleg Fedorchenko for assistance with the cloning of p87. We are grateful to Dr. Martina Schmidt and Dr. Peter Gierschik's laboratory for providing experimental tools. Valuable discussions with Drs. Phil Hawkins, Klaus Schulze-Osthoff, Len Stephens, Roger Williams, and colleagues from our department are greatly appreciated. This work was supported by the Deutsche Forschungsgemeinschaft, the Swiss National Science Foundation (Grants 31EM30-126143 and 310030_127574), and a Keystone Symposia Scholarship (from National Institutes of Health Grant 1R13CA139718-01, to B.K.).
Footnotes
The authors declare no conflicts of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0905506106/DCSupplemental.
References
- 1.Rommel C, Camps M, Ji H. PI3Kδ and PI3Kγ: Partners in crime in inflammation in rheumatoid arthritis and beyond? Nat Rev Immunol. 2007;7:191–201. doi: 10.1038/nri2036. [DOI] [PubMed] [Google Scholar]
- 2.Hirsch E, et al. Central role for G protein–coupled phosphoinositide 3-kinase γ in inflammation. Science. 2000;287:1049–1053. doi: 10.1126/science.287.5455.1049. [DOI] [PubMed] [Google Scholar]
- 3.Sasaki T, et al. Function of PI3Kγ in thymocyte development, T cell activation, and neutrophil migration. Science. 2000;287:1040–1046. doi: 10.1126/science.287.5455.1040. [DOI] [PubMed] [Google Scholar]
- 4.Laffargue M, et al. Phosphoinositide 3-kinase γ is an essential amplifier of mast cell function. Immunity. 2002;16:441–451. doi: 10.1016/s1074-7613(02)00282-0. [DOI] [PubMed] [Google Scholar]
- 5.Wymann MP, et al. Phosphoinositide 3-kinase γ: A key modulator in inflammation and allergy. Biochem Soc Trans. 2003;31:275–280. doi: 10.1042/bst0310275. [DOI] [PubMed] [Google Scholar]
- 6.Camps M, et al. Blockade of PI3Kγ suppresses joint inflammation and damage in mouse models of rheumatoid arthritis. Nat Med. 2005;11:936–943. doi: 10.1038/nm1284. [DOI] [PubMed] [Google Scholar]
- 7.Barber DF, et al. PI3Kγ inhibition blocks glomerulonephritis and extends lifespan in a mouse model of systemic lupus. Nat Med. 2005;11:933–935. doi: 10.1038/nm1291. [DOI] [PubMed] [Google Scholar]
- 8.Chang JD, et al. Deletion of the phosphoinositide 3-kinase p110γ gene attenuates murine atherosclerosis. Proc Natl Acad Sci USA. 2007;104:8077–8082. doi: 10.1073/pnas.0702663104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rückle T, Schwarz MK, Rommel C. PI3Kγ inhibition: Towards an “aspirin of the 21st century”? Nat Rev Drug Discov. 2006;5:903–918. doi: 10.1038/nrd2145. [DOI] [PubMed] [Google Scholar]
- 10.Marone R, Cmiljanovic V, Giese B, Wymann MP. Targeting phosphoinositide 3-kinase: Moving towards therapy. Biochim Biophys Acta. 2008;1784:159–185. doi: 10.1016/j.bbapap.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 11.Hirsch E, et al. Signaling through PI3Kγ: A common platform for leukocyte, platelet and cardiovascular stress sensing. Thromb Haemost. 2006;95:29–35. [PubMed] [Google Scholar]
- 12.Patrucco E, et al. PI3Kγ modulates the cardiac response to chronic pressure overload by distinct kinase-dependent and -independent effects. Cell. 2004;118:375–387. doi: 10.1016/j.cell.2004.07.017. [DOI] [PubMed] [Google Scholar]
- 13.Hirsch E, et al. Resistance to thromboembolism in PI3Kγ-deficient mice. FASEB J. 2001;15:2019–2021. doi: 10.1096/fj.00-0810fje. [DOI] [PubMed] [Google Scholar]
- 14.Macrez N, et al. Phosphoinositide 3-kinase isoforms selectively couple receptors to vascular L-type Ca2+ channels. Circ Res. 2001;89:692–699. doi: 10.1161/hh2001.097864. [DOI] [PubMed] [Google Scholar]
- 15.Hawkins PT, Anderson KE, Davidson K, Stephens LR. Signaling through Class I PI3Ks in mammalian cells. Biochem Soc Trans. 2006;34:647–662. doi: 10.1042/BST0340647. [DOI] [PubMed] [Google Scholar]
- 16.Fruman DA, Bismuth G. Fine-tuning the immune response with PI3K. Immunol Rev. 2009;228:253–272. doi: 10.1111/j.1600-065X.2008.00750.x. [DOI] [PubMed] [Google Scholar]
- 17.Okkenhaug K, Ali K, Vanhaesebroeck B . Antigen receptor signaling: A distinctive role for the p110δ isoform of PI3K. Trends Immunol. 2007;28:80–87. doi: 10.1016/j.it.2006.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graupera M, et al. Angiogenesis selectively requires the p110α isoform of PI3K to control endothelial cell migration. Nature. 2008;453:662–666. doi: 10.1038/nature06892. [DOI] [PubMed] [Google Scholar]
- 19.Guillermet-Guibert J, et al . The p110β isoform of phosphoinositide 3-kinase signals downstream of G protein–coupled receptors and is functionally redundant with p110γ. Proc Natl Acad Sci USA. 2008;105:8292–8297. doi: 10.1073/pnas.0707761105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ciraolo E, et al . Phosphoinositide 3-kinase p110β activity: Key role in metabolism and mammary gland cancer but not development. Sci Signal. 2008;1:ra3. doi: 10.1126/scisignal.1161577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jia S, et al. Essential roles of PI3K-p110β in cell growth, metabolism and tumorigenesis. Nature. 2008;454:776–779. doi: 10.1038/nature07091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stoyanov B, et al. Cloning and characterization of a G protein–activated human phosphoinositide-3 kinase. Science. 1995;269:690–693. doi: 10.1126/science.7624799. [DOI] [PubMed] [Google Scholar]
- 23.Kurosu H, et al. Heterodimeric phosphoinositide 3-kinase consisting of p85 and p110β is synergistically activated by the βγ subunits of G proteins and phosphotyrosyl peptide. J Biol Chem. 1997;272:24252–24256. doi: 10.1074/jbc.272.39.24252. [DOI] [PubMed] [Google Scholar]
- 24.Maier U, et al. Gβγ is a highly selective activator of phospholipid-dependent enzymes. J Biol Chem. 2000;275:13746–13754. doi: 10.1074/jbc.275.18.13746. [DOI] [PubMed] [Google Scholar]
- 25.Czupalla C, et al. Identification and characterization of the autophosphorylation sites of phosphoinositide 3-kinase isoforms β and γ. J Biol Chem. 2003;278:11536–11545. doi: 10.1074/jbc.M210351200. [DOI] [PubMed] [Google Scholar]
- 26.Tang X, Downes CP. Purification and characterization of Gβγ-responsive phosphoinositide 3-kinases from pig platelet cytosol. J Biol Chem. 1997;272:14193–14199. doi: 10.1074/jbc.272.22.14193. [DOI] [PubMed] [Google Scholar]
- 27.Hawkins PT, Stephens LR. PI3Kγ is a key regulator of inflammatory responses and cardiovascular homeostasis. Science. 2007;318:64–66. doi: 10.1126/science.1145420. [DOI] [PubMed] [Google Scholar]
- 28.Maier U, Babich A, Nürnberg B. Roles of non-catalytic subunits in Gβγ-induced activation of class I phosphoinositide 3-kinase isoforms β and γ. J Biol Chem. 1999;274:29311–29317. doi: 10.1074/jbc.274.41.29311. [DOI] [PubMed] [Google Scholar]
- 29.Stephens LR, et al. The Gβγ sensitivity of a PI3K is dependent upon a tightly associated adaptor, p101. Cell. 1997;89:105–114. doi: 10.1016/s0092-8674(00)80187-7. [DOI] [PubMed] [Google Scholar]
- 30.Voigt P, Brock C, Nürnberg B, Schaefer M. Assigning functional domains within the p101 regulatory subunit of phosphoinositide 3-kinase γ. J Biol Chem. 2005;280:5121–5127. doi: 10.1074/jbc.M413104200. [DOI] [PubMed] [Google Scholar]
- 31.Voigt P, Dorner MB, Schaefer M. Characterization of p87PIKAP, a novel regulatory subunit of phosphoinositide 3-kinase gamma that is highly expressed in heart and interacts with PDE3B. J Biol Chem. 2006;281:9977–9986. doi: 10.1074/jbc.M512502200. [DOI] [PubMed] [Google Scholar]
- 32.Suire S, et al. p84, a new Gβγ-activated regulatory subunit of the type IB phosphoinositide 3-kinase p110γ. Curr Biol. 2005;15:566–570. doi: 10.1016/j.cub.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 33.Bohnacker T, et al. PI3Kγ adaptor subunits define coupling to degranulation and cell motility by distinct PtdIns(3,4,5)P3 pools in mast cells. Sci Signal. 2009;2:ra27. doi: 10.1126/scisignal.2000259. [DOI] [PubMed] [Google Scholar]
- 34.Balla T. Finding partners for PI3Kγ: When 84 is better than 101. Sci Signal. 2009;2:e35. doi: 10.1126/scisignal.274pe35. [DOI] [PubMed] [Google Scholar]
- 35.Brock C, et al. Roles of Gβγ in membrane recruitment and activation of p110γ/p101 phosphoinositide 3-kinase γ. J Cell Biol. 2003;160:89–99. doi: 10.1083/jcb.200210115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Preuss I, Kurig B, Nürnberg B, Orth JH, Aktories K. Pasteurella multocida toxin activates Gβγ dimers of heterotrimeric G proteins. Cell Signal. 2009;21:551–558. doi: 10.1016/j.cellsig.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 37.Pacold ME, et al. Crystal structure and functional analysis of Ras binding to its effector phosphoinositide 3-kinase γ. Cell. 2000;103:931–943. doi: 10.1016/s0092-8674(00)00196-3. [DOI] [PubMed] [Google Scholar]
- 38.Suire S, Hawkins P, Stephens L. Activation of phosphoinositide 3-kinase γ by Ras. Curr Biol. 2002;12:1068–1075. doi: 10.1016/s0960-9822(02)00933-8. [DOI] [PubMed] [Google Scholar]
- 39.Ahmadian MR, Hoffmann U, Goody RS, Wittinghofer A. Individual rate constants for the interaction of Ras proteins with GTPase-activating proteins determined by fluorescence spectroscopy. Biochemistry. 1997;36:4535–4541. doi: 10.1021/bi962556y. [DOI] [PubMed] [Google Scholar]
- 40.Rodriguez-Viciana P, Sabatier C, McCormick F. Signaling specificity by Ras family GTPases is determined by the full spectrum of effectors they regulate. Mol Cell Biol. 2004;24:4943–4954. doi: 10.1128/MCB.24.11.4943-4954.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hirsch E, Costa C, Ciraolo E. Phosphoinositide 3-kinases as a common platform for multi-hormone signaling. J Endocrinol. 2007;194:243–256. doi: 10.1677/JOE-07-0097. [DOI] [PubMed] [Google Scholar]
- 42.Zhao L, Vogt PK. Class I PI3K in oncogenic cellular transformation. Oncogene. 2008;27:5486–5496. doi: 10.1038/onc.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song C, et al. Regulation of a novel human phospholipase C, PLCε, through membrane targeting by Ras. J Biol Chem. 2001;276:2752–2757. doi: 10.1074/jbc.M008324200. [DOI] [PubMed] [Google Scholar]
- 44.Stokoe D, Macdonald SG, Cadwallader K, Symons M, Hancock JF. Activation of Raf as a result of recruitment to the plasma membrane. Science. 1994;264:1463–1467. doi: 10.1126/science.7811320. [DOI] [PubMed] [Google Scholar]
- 45.Suire S, et al. Gβγs and the Ras binding domain of p110γ are both important regulators of PI3Kγ signalling in neutrophils. Nat Cell Biol. 2006;8:1303–1309. doi: 10.1038/ncb1494. [DOI] [PubMed] [Google Scholar]
- 46.Kranenburg O, Verlaan I, Moolenaar WH. Regulating c-Ras function: Cholesterol depletion affects caveolin association, GTP loading, and signaling. Curr Biol. 2001;11:1880–1884. doi: 10.1016/s0960-9822(01)00582-6. [DOI] [PubMed] [Google Scholar]
- 47.Furuchi T, Anderson RG. Cholesterol depletion of caveolae causes hyperactivation of extracellular signal–related kinase (ERK) J Biol Chem. 1998;273:21099–21104. doi: 10.1074/jbc.273.33.21099. [DOI] [PubMed] [Google Scholar]
- 48.Shymanets A, Ahmadian MR, Nürnberg B. Gβγ-copurified lipid kinase impurity from Sf9 cells. Protein Pept Lett. 2009;16:1053–1056. doi: 10.2174/092986609789055340. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.