Abstract
The synaptic vesicle protein synaptobrevin engages with syntaxin and SNAP-25 to form the SNARE complex, which drives membrane fusion in neuronal exocytosis. In the SNARE complex, the SNARE motif of synaptobrevin forms a 55-residue helix, but it has been assumed to be mostly unstructured in its prefusion form. NMR data for full-length synaptobrevin in dodecylphosphocholine micelles reveals two transient helical segments flanked by natively disordered regions and a third more stable helix. Transient helix I comprises the most N-terminal part of the SNARE motif, transient helix II extends the SNARE motif into the juxtamembrane region, and the more stable helix III is the transmembrane domain. These helices may have important consequences for SNARE complex folding and fusion: helix I likely forms a nucleation site, the C-terminal disordered SNARE motif may act as a folding arrest signal, and helix II likely couples SNARE complex folding and fusion.
Keywords: structure, dynamics, membrane fusion, SNARE proteins, NMR
Synaptic release of neurotransmitter requires the docking and fusion of synaptic vesicles at the presynaptic membrane. The fusion reaction is thought to be mediated by the formation of the SNARE complex from its components syntaxin-1a and SNAP-25 in the target membrane and synaptobrevin-2 (Syb) in the vesicle membrane (1). These three proteins together form a complex that consists of a thermally very stable coiled-coil four-helix bundle, as revealed by the crystal structure of the soluble SNARE core complex (2). SNAP-25 contributes two helices to this complex and syntaxin and synaptobrevin each contribute one. More recently, the structure of the postfusion cis-SNARE complex with its C-terminal transmembrane (TM) domain extensions was solved at 3.4 Å resolution (3). Most interestingly, the helices continue through the juxtamembrane linker region and into the membrane, suggesting that force could be transmitted through this region into the membrane leading to membrane bending and eventually membrane fusion. This interesting finding immediately raises the question: how exactly is the assembly and folding of the four-helix bundle coupled into the membrane and how does the energy derived from this reaction ultimately fuse two different membranes into one? A common notion is that an initial trans-SNARE complex forms by pairing vesicle and target membrane SNAREs from their N-terminal ends and progressively folds in a zipper-like fashion toward the C-terminal TM domains. This reaction is thought to pull the two membranes into closer contact until, at some stage they merge into a single membrane. However, structural data on a trans-SNARE complex do not yet exist. It is also not yet known whether zipper-folding of trans-SNARE complexes progresses smoothly and continuously into the membrane or whether this reaction is discontinuous and segmented in some fashion.
To understand how the SNARE complex is formed and to find possible reaction intermediates, several structural studies of SNARE proteins in isolation or in binary complexes have been performed. The soluble domains of neuronal Syb (residues 1–96) (4, 5) and its yeast homolog Snc1 (6) have been found by CD and NMR spectroscopy to be mostly unstructured in aqueous solution. The TM domain of Syb was polymorphic and showed different degrees of α-helical and β-sheet secondary structures depending on environmental conditions when modeled as a short peptide comprising Syb residues 97–112 and analyzed by FTIR and CD spectroscopy (7). A more recent FTIR and CD study of the full-length Syb(1–116) concluded that the TM domain is an α-helix that is inclined 30–40° from the membrane normal and that the entire cytoplasmic domain is natively disordered (8). The cytoplasmic domain of Syb(1–116) was also found to be disordered when analyzed by site-directed spin label EPR spectroscopy (9). NMR, CD, and EPR studies showed that Syb assumes more structure when complexed with soluble syntaxin fragments (4, 5, 9).
To elucidate structural features of full-length synaptobrevin in the presence of a membrane-mimetic environment, we have used solution NMR spectroscopy to study the structure and dynamics of Syb(1–116) in lipid micelles. The results indicate helical propensities not only in the TM domain but also in regions at the beginning and end of the SNARE motif, which likely serve, respectively, as a nucleation site for SNARE complex formation and as an element to couple SNARE complex folding with fusion.
Results
Secondary Structure of Lipid-Bound Synaptobrevin.
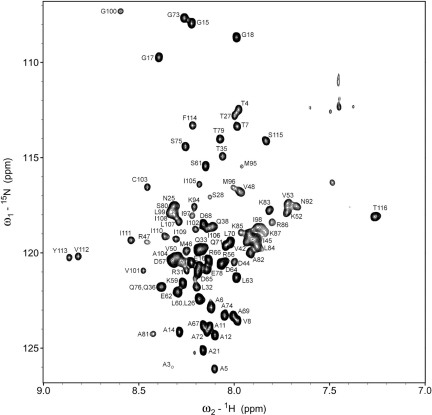
A typical HSQC spectrum of 15N,13C-labeled Syb(1–116) in dodecylphosphocholine (DPC) micelles is shown in Fig. 1. Sample conditions were optimized as described in SI Experimental Procedures and yielded the highest quality spectra at a protein:lipid ratio of 1:200 and a recording temperature of 45 °C. Variable peak intensities in this spectrum indicate that different residues may experience different rotational correlation times, possibly because of different degrees of association with the lipid micelles. Of the 108 non-proline residues, 106 were assigned from the Cα, Cβ, and CO backbone connectivities obtained from HNCA, HNCACB, HNCO, HN(CA)CO, and CBCA(CO)NH experiments and a 15N-edited NOESY experiment. Met-1 and Asp-40 were the only residues that remained only partially assigned (Cα and Cβ assignments complete). We also studied the shorter membrane-bound fragment Syb(60–116), which was independently completely assigned from triple-resonance experiments (Fig. S1). The chemical shifts of Syb(60–116) were very similar to those of the corresponding residues in Syb(1–116), except for residues 60–63, which are internal in full-length Syb, but form the N terminus of the fragment.
Fig. 1.
15N-1H HSQC spectrum of Syb(1–116) in DPC micelles at pH 6.0 and 45 °C, measured at 600 MHz. Assignments of backbone amides are denoted by one letter amino acid abbreviations followed by their sequence numbers.
The Cα and Cβ chemical shifts were used to estimate elements of secondary structure of Syb(1–116). Fig. 2A shows three-residue averaged (ΔCα–ΔCβ) secondary chemical shifts plotted as a function of amino acid sequence number. The pattern in Fig. 2A clearly indicates that there are three regions that likely form α-helices: residues 36–54 (helix I), 77–88 (helix II), and 93–115 (helix III). Helix I covers a good portion of the N-terminal half of the SNARE motif, which encompasses residues 28–84. Helix II covers the C-terminal end of the SNARE motif, but extends about one turn longer into the juxtamembrane domain. Helix III corresponds to the TM domain.
Fig. 2.
Three-bond averaged secondary chemical shifts, where (ΔCα − ΔCβ)i = 1/3(ΔCi-1α + ΔCiα + ΔCi+1α − ΔCi−1β − ΔCiβ − ΔCi+1β), versus residue sequence numbers i. Each individual ΔC value on the right hand side of the equation is the chemical shift deviation from its respective random coil value. (A) Syb(1–116) in DPC micelles, (B) Syb(1–96) in DPC micelles, and (C) Syb(1–96) in aqueous solution. The SNARE motif of synaptobrevin as defined by the crystal structure of the SNARE complex (2) is shown as a box and helices I, II, and III are indicated by blue, green, and red colors in the sequence on the Top.
Comparison with the Soluble Fragment Syb(1–96).
An earlier NMR study of human Syb(1–96) indicated that this soluble fragment is unstructured, except for perhaps a hint of some helical structure near the C terminus (5). We prepared the same fragment of rat synaptobrevin and essentially reproduced the earlier data (Fig. 2C). The HSQC spectrum of Syb(1–96) in buffer revealed almost uniform peak intensities and linewidths (Fig. S2). All resonances were assigned from triple-resonance experiments. Except for a few residues around the single amino acid substitution from the human to the rat sequence (Ala-8 becomes a Val), the chemical shifts of the human and rat datasets are in excellent agreement.
When the same fragment was measured in the presence of DPC micelles, the NMR spectra changed dramatically. Many resonances disappeared in the HSQC spectrum at 18 °C, but gradually reappeared at higher temperatures. This process was fully reversible. A representative fully assigned (from triple-resonance backbone experiments) HSQC spectrum collected at 45 °C is shown in Fig. S3. As with Syb(1–116), peak intensities and linewidths varied, indicative of different rotational correlation times for different residues. The secondary chemical shift plot (Fig. 2B) indicates a significant induction of secondary structure by the presence of DPC micelles. The remarkable resemblance of secondary chemical shifts between Syb(1–116) and Syb(1–96) in DPC shows that the structure of the soluble domain of Syb is very similar in DPC micelles, irrespective of whether it is anchored in micelles with a TM domain or not.
Backbone Dynamics of Free and Lipid-Bound Synaptobrevin.
To probe the backbone dynamics, heteronuclear {1H}-15N NOEs of Syb(1–116) in DPC micelles were recorded. Since the theoretical rigid limit for {1H}-15N NOEs at 800 MHz is 0.86 (10), values greater than approximately 0.5 indicate restricted backbone motions, and smaller positive and negative values indicate extended backbone motions on the ps-ns time-scale. All three proposed α-helices showed relatively high {1H}-15N NOE values (Fig. 3B). The average values were 0.54, 0.51, and 0.78 for residues comprising helices I, II, and III, respectively. According to these data, the TM region is the most rigid of these segments. Helices I and II are more flexible, although they are clearly not as dynamic as the rest of protein. Based on the negative heteronuclear NOEs, the proline-rich N terminus is particularly dynamic.
Fig. 3.
Backbone dynamics of Syb(1–116) measured at 800 MHz. (A) Top: helices, as defined in this manuscript; Bottom: predicted domain arrangement of synaptobrevin, (B) Heteronuclear {1H}-15N NOEs, (C) 15N-R1, (D) 15N-R2, and (E) water-amide proton exchange. In (E), the residues undergoing fast (<10 ms) exchange are given a value of +1, and the residues undergoing slow (>80 ms) or no exchange are given a value of −1.
These conclusions are further supported by NMR relaxation experiments. Backbone 15N spin-lattice (Fig. 3C) and spin-spin (Fig. 3D) relaxation rates were measured. Helices I and III exhibited decreased R1 rates averaging 1.24 s−1 and 0.77 s−1, respectively, compared to helix II and nonhelical residues (1.77 s−1 and 1.67 s−1, respectively). By contrast, helices I, II, and III had R2 rates of 14.2, 12.1, and 23.5 s−1, respectively, compared to 5.71 s−1 for disordered regions. The relatively high R2 and low R1 values of helices I and III indicate that these helices experience long rotational correlation times meaning that they likely rotate together with the micelle, whereas helix II has a shorter correlation time, and therefore moves more independently. The relatively high R2 rate coupled with a lack of reduction of the R1 rate of helix II also indicates that this helix may undergo conformational exchange on the μs-ms time-scale (11).
To further probe the conformational stability of the three helical segments of Syb(1–116), we measured amide hydrogen-exchange with water by performing a CLEANEX-PM experiment (12). Fig. 3E shows residues that exhibit hydrogen-exchange within 10 ms with an assigned value of +1 and residues that do not exchange after 80 ms with an assigned value of −1. Evidently, helices I and III are quite stable on this time-scale, whereas helix II and the juxtamembrane domain are solvent accessible, as is the rest of the protein.
Hydropathy and Helical Wheel Analysis.
A hydropathy and hydrophobic moment analysis (13) of the synaptobrevin sequence reveals that, as expected, the TM domain shows up as the most hydrophobic part and that a highly amphipathic helical region is centered on residue 44 (14) (Fig. S4a). If this region is modeled on a helical wheel, it is clear that one face of the helix is quite hydrophobic and the other face is hydrophilic and highly charged (Fig. S4b). Consistent with a micelle-induced formation of a stable α-helix in the N-terminal, but not in the C-terminal half of the SNARE motif, the hydrophobic moment and hydropathy profiles for the SNARE domain (residues 28–85) indicate that the C-terminal half of the domain is less amphipathic and more hydrophilic than the N-terminal half.
Structure of Synaptobrevin in DPC Micelle.
One hundred structures of Syb(1–116) in DPC micelles were calculated from TALOS-derived dihedral angle, NOE-derived distance, and hydrogen-bond restraints as described in SI Experimental Procedures. The 20 lowest energy structures were analyzed and the structural statistics are summarized in Table S1. As expected from the described chemical shift and dynamical data, the structure of synaptobrevin is characterized by three helices connected by two flexible natively disordered domains and a highly flexible proline-rich N-terminal tail (Fig. 4). The TM helix (helix III) comprising residues 93–115 is a couple of residues longer at the N terminus than what might be expected from simple hydropathy analysis. It is slightly bent with about six helical turns that align very well in all structures (Fig. 4D) (backbone rmsd of 0.62 ± 0.18 Å). Not surprisingly, the interfacial residues Leu-93, Lys-94, Ser-115, and Thr-116 are a little less well ordered compared to the rest of the helix.
Fig. 4.
Twenty lowest energy structures of Syb(1–116) in DPC micelles. Residues 36–54 are blue, 77–88 are green, 93–115 are red, and the rest are yellow. (A) Residues 36–116 aligned along TM domain (residues 93–115). (B) Helix I (residues 36–54) aligned (rmsd 1.03 ± 0.29 Å) (C) Helix II (residues 77–88) aligned (rmsd 0.81 ± 0.29 Å). (D) Transmembrane helix III (residues 93–115) aligned (rmsd 0.62 ± 0.18 Å). N-terminals are at the Top in B, C, and D.
The next most prominent helix is the amphipathic helix I that spans residues 36–54, with a backbone rmsd of 1.03 ± 0.29 Å (Fig. 4B). Helix II, which comprises the end of the SNARE motif and leads into the juxtamembrane domain, i.e., residues 77–88, is well defined (Fig. 4C) (rmsd 0.81 ± 0.29 Å), although it undergoes quite fast amide hydrogen-exchange on the 10-ms time scale (Fig. 3E).
It is interesting to note that helices II and III are connected by only four residues, namely Trp-89, Trp-90, Lys-91, and Asn-92, which apparently form a flexible hinge at the micelle surface, and by inference, at the membrane-water interface. This hinge allows helix II to emanate from the micelle or membrane surface in many different directions as is evident from Fig. 4A.
Most of the C-terminal half of the SNARE motif (i.e., residues 55–76) is natively disordered. Even though the N-terminal half of the SNARE motif (helix I) interacts with the micelle and presumably the membrane surface and, therefore, has to bend back toward an interface, this helix samples many different orientations relative to the TM domain owing to the flexible nature of the C-terminal portion of the SNARE motif.
Fig. 5 shows a ribbon diagram of the lowest energy structure of Syb(1–116) in DPC micelles. One can position this structure so that the amphipathic helix I lies at a membrane interface, helix III is on the hydrophobic side of the interface and rest of the molecule is almost completely on the hydrophilic side of the interface, as shown in Fig. 5. Not all structures in the structural ensemble can be positioned in this way. Due to the flexible nature of the molecule and the lack of long-range structural restraints many structures have their amphipathic helices projected into different directions relative to a hypothetical membrane plane. Since our data were collected in DPC micelles at a protein:lipid ratio of 1:200 and since a typical DPC micelle contains ≈40–80 lipid molecules (15), our sample contains an approximate 3- to 5-fold molar excess of micelles over protein, potentially allowing one protein to interact with more than one micelle that may not assume fixed spatial orientations relative to one another. We have attempted to measure residual dipolar couplings in several different aligning media, but were unsuccessful to prepare appropriate samples. However, even if such experiments had been successful, we suspect that the distribution of relative orientations of the helices of synatpobrevin would still be very broad due to the dynamic nature of the intervening unstructured regions.
Fig. 5.
Ribbon representation of lowest energy structure of Syb(1–116) in DPC micelles. This structure conforms well to the plane of a membrane, the interface of which is shown as a line. Helices I, II, and III are colored in blue, green, and red as in the sequence and secondary chemical shift plots of Fig. 2. For simplicity, the natively disordered N-terminal tail (residues 1–35) is not shown. A version of Fig. 5 with residues 1–35 included is shown in Fig. S6.
Discussion
The structure of full-length synaptobrevin-2 in DPC micelles is segmented, dynamical, and characterized by varied structural and motional properties along the sequence. Previously, structures of postfusion cis-SNARE complexes comprising all three synaptic SNARE proteins or structural studies on only the soluble fragments of individual SNARE proteins have been completed (2, 3, 5, 6, 16, 17). Although based on earlier NMR studies, it was expected that the cytoplasmic domain of membrane-bound synaptobrevin would be natively disordered, we found that lipid-bound synaptobrevin contains substantial structure not only in its TM domain, but also in its SNARE motif and juxtamembrane domains even before it engages with other SNAREs to form the fusion complex. We have used the chemical shift-based secondary structure propensity method (18) to estimate fractional helical populations and find average values of 0.73 ± 0.18, 0.65 ± 0.18, and 1.2 ± 0.2 for helices I, II, and III, respectively (a value of 1 indicates a fully formed stable α-helix and a value of 0.5 means that 50% of the ensemble is α-helical). The high value for helix III is likely due to the fact that the chemical shift database used in this method is almost completely from soluble proteins and, therefore may not be appropriate for helix III. The lower values for helices I and II indicate that their conformational ensembles are predominantly helical, but that they also contain substantial unstructured fractions.
Although a previous CD and FTIR study of membrane-bound synaptobrevin emphasized the unstructured nature of its soluble domain, experiments with a shorter fragment comprising residues 74–116 is consistent with our results (8). The authors found approximately 27 residues of this fragment to be helical, which is clearly more than required for the transmembrane domain and within experimental error similar to the sum of our helices II and III, especially taking into account that helix II may not be formed all of the time. The previous EPR study on full-length synaptobrevin did not indicate helical residues in the SNARE motif (9), but the data could also be interpreted as forming a transient helix in this region. In fact, their spectra are very similar to EPR spectra of transient helical regions in the GCN4 coiled coil transcriptional activator (19) and in linker segments of rhodopsin (20). Similarly, residues 84–88 clearly show a helical period, but residues 89–92 do not, although the authors interpreted the data as a continuous interfacial helix in this region. In summary, we think that the CD data for synaptobrevin in micelles (8) and the raw EPR data for synaptobrevin in bilayers (9) are completely consistent with the current NMR data for synaptobrevin in DPC micelles and the interpretation in terms of the transient helices I and II as indicated in Figs. 4 and 5.
Interestingly, the N-terminal half of the SNARE motif of synaptobrevin is structured as an amphipathic helix when associated with membrane-mimicking micellar lipids, but the C-terminal half of the SNARE motif remains natively disordered even in the presence of lipids. The N-terminal SNARE motif helix is induced by the presence of the lipid micelle because it is unstructured in the absence of lipids, confirming previous results (5, 6). Since synaptobrevin in synaptic vesicles is always near a membrane interface, the N-terminal SNARE motif helix observed here may also be present in its natural physiological environment. The hydrophobic face of helix I consists of the same residues which form the coiled coil heptad repeat and face the interior of the SNARE complex (2, 3). The existence of helix I (and II) is not an artifact of the high DPC concentration used in these experiments because experiments at a 40-fold lower concentration essentially produced the same result (Fig. S5). Similarly, adding 12% of the negatively charged detergent SDS to mimic negatively charged lipids in the synaptic membranes did not alter our results either (Fig. S5). A second important structural feature is a nascent α-helix at the transition from the SNARE motif to the 10-residue juxtamembrane domain (residues 85–94) that links the SNARE motif (residues 28–84) with the TM domain (residues 95–116). Even before forming a complex with cognate SNAREs, this segment adopts a helical structure in its DPC-bound prefusion form. Residues 77–88 form a helix that extends four more residues from layer +8 of the SNARE motif into the juxtamembrane domain. Similarly, the TM domain is N-terminally two residues longer than what might be expected from hydropathy analysis and includes Lys-94 whose charged side chain likely snorkels up to the micelle and membrane surface. This leaves only residues Trp-89–Asn-92 as an unstructured linker between the extended SNARE motif and TM domains.
Although a DPC micelle is not a lipid bilayer, DPC still has the same zwitterionic phosphocholine headgroup as the major membrane phospholipid phosphatidylcholine. Therefore, it is generally considered a mild detergent that largely preserves native structures of membrane proteins. Indeed, DPC is a preferred lipid for determining structures of membrane proteins by solution NMR (21–25). However, and despite the earlier NMR successes with DPC, caution is still advised when extrapolating some aspects of our synaptobrevin structure to situations in lipid bilayers. Although we believe that the general conclusions of the current work also carry over to lipid bilayers, there may be details especially in the interfacial juxtmembrane region of the protein that may not be exactly the same in the two environments. Future studies using additional methods will be needed to substantiate or refute some of these details.
The dynamics experiments shed further light on the conformational flexibility of different regions of the synaptobrevin structure. The TM helix is clearly the most rigid and stable of all three helical segments. Based on its inability to undergo amide hydrogen-exchange, the association of this helix with lipid micelles is strong. Helices I and II exhibit different dynamical behaviors. Motions on the ps-ns time-scale, as well as amide hydrogen-exchange on the 10 ms time-scale provide strong evidence that helix I, but not helix II interacts with the micelle surface. Based on the R1 and R2 relaxation behavior, helix II may also undergo conformational exchange on the μs-ms time-scale.
Helix I as a Nucleation Site for trans-SNARE Complex Formation?
The existence of nascent helix I before synaptobrevin forms a complex with t-SNAREs may have significant biological consequences. It is generally accepted that SNAREs zipper from the N- toward the C-terminal ends and that this folding reaction drives membrane fusion (26–29). In accordance with this notion, the N-terminal SNARE motif helix I of synaptobrevin observed here may serve as a nucleation site for trans-SNARE complex formation. A preformed helix in this region would provide a mechanism to (a) speed up the process of forming the initial trans-SNARE complex and (b) increase the fidelity of starting SNARE complex folding at the N-terminal rather than the C-terminal end. In general, folding can be induced by binding or assembly might occur by conformational selection or some combination of the two (30). An example of conformational selection has been described in detail for the folding of the GCN4 leucine zipper coiled coil (31). If SNARE complex folding follows the model of GCN4 coiled coil folding, a preformed N-terminal helix would be kinetically advantageous.
Since the C-terminal portion of the synaptobrevin SNARE motif has a lower propensity to form an α-helix and, by inference, to pair with t-SNARE helices, zipper-folding of the trans-SNARE complex may transiently stop or be slowed down when it reaches this region of the protein. Thus, the C-terminal portion of the SNARE motif may serve as a “stop-folding” signal. This folding-arrested trans-SNARE complex may be present at the docked, but not yet fused stage of the overall reaction. An additional trigger, perhaps provided by synaptotagmin (un)binding and/or complexin dissociation, may push complex formation over the final energy barrier to proceed to completion and ultimately to membrane fusion. This model is in fact very similar to, and supports a model recently proposed for the function of complexin as a mediator of two-stage SNARE complex folding (32, 33).
Further support for this model comes from folding studies, in which the assembly and disassembly of SNARE complexes with synaptobrevin fragments of different lengths was investigated (28). In these studies, SNARE complexes with Syb25–96, Syb35–96, Syb42–96, Syb49–96, or Syb60–96 were formed and then challenged with Syb1–96. Interestingly, Syb1–96 could not replace peptides covering the N-terminal portion of helix I (Syb25–96 and Syb35–96) in the complex, but was able to exchange peptides that covered the more C-terminal portions of the SNARE motif (Syb42–96, Syb49–96, and Syb60–96). These studies and a subsequent detailed thermodynamic analysis of the interaction of various SNARE fragments (34) strongly indicate that the N-terminal portion of the sequence (helix I) contributes more to the stability of the complex than the C-terminal portions of the SNARE complex, again supporting the nucleation-propagation model of SNARE complex folding. Electrophysiological experiments using progressive SNARE layer mutants of SNAP-25 also favor a two-stage model implying separate nucleation and propagation phases of the folding reaction (29).
A structure-function study of a SNARE complex with a single site mutation near the C-terminal end of the syntaxin SNARE motif is also consistent with a lower stability and folding rate of the C-terminal compared to the N-terminal end of SNARE complex formation (35). The layer +7 T254I mutation enhances constitutive and evoked fusion and also changes the structure to one that is more similar to SNARE complexes associated with constitutive fusion.
Helix II as a Coupler of trans- to cis-SNARE Complex Conversion and Membrane Merger?
Nascent helix II extends from the last SNARE motif layer +8 to the juxtamembrane domain of uncomplexed synaptobrevin. Down-stream from the above mentioned “stop-folding” signal, this helix may well transmit force from SNARE complex folding into the membrane and thus coerce the two membranes to fuse once the folding signal arrives at this stage. The subsequent 4-residue linker comprises two Trps, one Lys, and one Asn. Tryptophans are known to have a strong affinity (approximately 2 kcal/mol/residue) for membrane interfaces, which is further amplified when they occur in pairs or in tandem with other aromatic residues (36). Lysines also have a relatively long aliphatic chain before the terminal amino group and therefore can be buried quite deep in a membrane interface. Thus, this very short flexible linker region appears to be ideally designed to convert a trans-SNARE into a cis-SNARE complex, which must be coupled with merging the two membranes because the TM domains of synaptobrevin and syntaxin reside in the same membrane in the postfusion cis-SNARE complex. Flexibility is needed to tilt and accommodate the folding of the trans-SNARE complex between the two approaching membranes and shortness is needed for efficient coupling between folding and membrane merger, possibly by injecting the linker Trp residues into the adjacent membrane. Trp-89 and Trp-90 have been claimed by some investigators to regulate SNARE assembly in membranes (9), but this notion is controversial (37).
Deletion or insertion of a small number of residues including insertion of two helix-breaking prolines in the juxtamembrane region has little or no effect on SNARE-mediated fusion, but insertion of longer flexible linkers has progressively inhibitory effects on SNARE-mediated liposome fusion in vitro (37, 38). However, quite mild and short (39), as well as more dramatic long insertions (40) have significant effects on Ca2+-evoked exocytosis in cellular settings. The study by Kesavan et al. (39) clearly shows that Ca2+ sensitivity is not affected by the specifics of the juxtamembrane linker region, but that down-stream fusion pore initiation and expansion on the millisecond timescale critically depends on it. This suggests that membrane proximity of the SNARE motif and rapid propagation of SNARE folding through the juxtamembrane region is more critical for fast millisecond time scale fusion than it is for slow fusion that is typical in in vitro liposome fusion assays. Fast in vitro fusion assays, such as one recently developed in our laboratory (41) should shed more light on this possibility and should help reconcile the mentioned discrepancies on the mechanistic role of the linker region.
Very recently, the structure of the full-length SNARE complex including the TM domains of syntaxin and synaptobrevin was solved (3). Most interestingly, the SNARE motif helical bundle of syntaxin and synaptobrevin continues uninterrupted into the helical TM regions of these two proteins. Apparently, the transition from the pre- to the postfusion structure involves helical folding of the last four residues between the SNARE motif and TM domain of synaptobrevin. Therefore, this final folding step appears to be the major link between SNARE zippering and membrane fusion. It will be interesting to see if syntaxin in its uncomplexed form or complexed with SNAP-25 only also has a flexible linker that is as short as that of synaptobrevin. If so, this would provide an efficient mechanical device to couple SNARE complex folding with the unification of two initially separate membranes.
Materials and Methods
Expression and purification of Syb(1–116) and Syb(1–96) from Rattus norvegicus were performed as described in refs. 42 and 43. The final optimized NMR samples of Syb(1–116) were prepared from 13C and 15N-labeled proteins containing 200 mM dodecylphosphocholine (DPC), 150 mM NaCl, 5 mM DTT, 1 mM EDTA, 5% D2O, and 20 mM Mes, pH 6.0. For the backbone and side-chain resonance assignments, a total of 10 different types of 3-D experiments were collected at 45 °C on 600 or 800 MHz spectrometers. 15N-edited NOESY spectra with a mixing time of 120 ms, heteronuclear {1H}-15N NOE, 15N-T1, and T2 measurements, and water-amide proton exchange experiments were collected at 800 MHz. All spectra were processed and analyzed with NMRPipe (44) and Sparky (45). Indirect dimensions in the 3-D experiments were processed with forward-backward linear prediction. For structural calculations, dihedral angle restraints predicted from the TALOS program (46), NOE distances from NOESY experiments, and alpha-helical backbone hydrogen bonds were used as inputs in the program Xplor-NIH (47). Details are provided in SI Experimental Procedures and Fig. S7.
Supplementary Material
Acknowledgments.
This work was supported by National Institutes of Health grant P01 GM72694.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2KOG).
This article contains supporting information online at www.pnas.org/cgi/content/full/0908317106/DCSupplemental.
References
- 1.Jahn R, Scheller RH. SNAREs–engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- 2.Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 3.Stein A, Weber G, Wahl MC, Jahn R. Helical extension of the neuronal SNARE complex into the membrane. Nature. 2009;460:525–528. doi: 10.1038/nature08156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fasshauer D, et al. Structural changes are associated with soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor complex formation. J Biol Chem. 1997;272:28036–28041. doi: 10.1074/jbc.272.44.28036. [DOI] [PubMed] [Google Scholar]
- 5.Hazzard J, Sudhof TC, Rizo J. NMR analysis of the structure of synaptobrevin and of its interaction with syntaxin. J Biomol NMR. 1999;14:203–207. doi: 10.1023/a:1008382027065. [DOI] [PubMed] [Google Scholar]
- 6.Fiebig KM, Rice LM, Pollock E, Brunger AT. Folding intermediates of SNARE complex assembly. Nat Struct Biol. 1999;6:117–123. doi: 10.1038/5803. [DOI] [PubMed] [Google Scholar]
- 7.Langosch D, et al. Peptide mimics of SNARE transmembrane segments drive membrane fusion depending on their conformational plasticity. J Mol Biol. 2001;311:709–721. doi: 10.1006/jmbi.2001.4889. [DOI] [PubMed] [Google Scholar]
- 8.Bowen M, Brunger AT. Conformation of the synaptobrevin transmembrane domain. Proc Natl Acad Sci USA. 2006;103:8378–8383. doi: 10.1073/pnas.0602644103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kweon DH, Kim CS, Shin YK. Regulation of neuronal SNARE assembly by the membrane. Nat Struct Biol. 2003;10:440–447. doi: 10.1038/nsb928. [DOI] [PubMed] [Google Scholar]
- 10.Kay LE, Torchia DA, Bax A. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: Application to staphylococcal nuclease. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 11.Jarymowycz VA, Stone MJ. Fast time scale dynamics of protein backbones: NMR relaxation methods, applications, and functional consequences. Chem Rev. 2006;106:1624–1671. doi: 10.1021/cr040421p. [DOI] [PubMed] [Google Scholar]
- 12.Hwang TL, van Zijl PC, Mori S. Accurate quantitation of water-amide proton exchange rates using the phase-modulated CLEAN chemical EXchange (CLEANEX-PM) approach with a Fast-HSQC (FHSQC) detection scheme. J Biomol NMR. 1998;11:221–226. doi: 10.1023/a:1008276004875. [DOI] [PubMed] [Google Scholar]
- 13.Eisenberg D, Weiss RM, Terwilliger TC. The helical hydrophobic moment: A measure of the amphiphilicity of a helix. Nature. 1982;299:371–374. doi: 10.1038/299371a0. [DOI] [PubMed] [Google Scholar]
- 14.Jahn R, Sudhof TC. Synaptic vesicles and exocytosis. Annu Rev Neurosci. 1994;17:219–246. doi: 10.1146/annurev.ne.17.030194.001251. [DOI] [PubMed] [Google Scholar]
- 15.Kallick DA, Tessmer MR, Watts CR, Li CY. The use of dodecylphosphocholine micelles in solution NMR. J Magn Reson B. 1995;109:60–65. doi: 10.1006/jmrb.1995.1146. [DOI] [PubMed] [Google Scholar]
- 16.Antonin W, et al. Crystal structure of the endosomal SNARE complex reveals common structural principles of all SNAREs. Nat Struct Biol. 2002;9:107–111. doi: 10.1038/nsb746. [DOI] [PubMed] [Google Scholar]
- 17.Chen X, et al. Three-dimensional structure of the complexin/SNARE complex. Neuron. 2002;33:397–409. doi: 10.1016/s0896-6273(02)00583-4. [DOI] [PubMed] [Google Scholar]
- 18.Marsh JA, Singh VK, Jia Z, Forman-Kay JD. Sensitivity of secondary structure propensities to sequence differences between alpha- and gamma-synuclein: Implications for fibrillation. Protein Sci. 2006;15:2795–2804. doi: 10.1110/ps.062465306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Columbus L, Hubbell WL. Mapping backbone dynamics in solution with site-directed spin labeling: GCN4–58 bZip free and bound to DNA. Biochemistry. 2004;43:7273–7287. doi: 10.1021/bi0497906. [DOI] [PubMed] [Google Scholar]
- 20.Altenbach C, et al. Structural features and light-dependent changes in the cytoplasmic interhelical E-F loop region of rhodopsin: A site-directed spin-labeling study. Biochemistry. 1996;35:12470–12478. doi: 10.1021/bi960849l. [DOI] [PubMed] [Google Scholar]
- 21.Arora A, Abildgaard F, Bushweller JH, Tamm LK. Structure of outer membrane protein A transmembrane domain by NMR spectroscopy. Nat Struct Biol. 2001;8:334–338. doi: 10.1038/86214. [DOI] [PubMed] [Google Scholar]
- 22.Oxenoid K, Chou JJ. The structure of phospholamban pentamer reveals a channel-like architecture in membranes. Proc Natl Acad Sci USA. 2005;102:10870–10875. doi: 10.1073/pnas.0504920102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang B, Tamm LK. Structure of outer membrane protein G by solution NMR spectroscopy. Proc Natl Acad Sci USA. 2007;104:16140–16145. doi: 10.1073/pnas.0705466104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou Y, et al. NMR solution structure of the integral membrane enzyme DsbB: Functional insights into DsbB-catalyzed disulfide bond formation. Mol Cell. 2008;31:896–908. doi: 10.1016/j.molcel.2008.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Horn WD, et al. Solution nuclear magnetic resonance structure of membrane-integral diacylglycerol kinase. Science. 2009;324:1726–1729. doi: 10.1126/science.1171716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Melia TJ, et al. Regulation of membrane fusion by the membrane-proximal coil of the t-SNARE during zippering of SNAREpins. J Cell Biol. 2002;158:929–940. doi: 10.1083/jcb.200112081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fasshauer D, Margittai M. A transient N-terminal interaction of SNAP-25 and syntaxin nucleates SNARE assembly. J Biol Chem. 2004;279:7613–7621. doi: 10.1074/jbc.M312064200. [DOI] [PubMed] [Google Scholar]
- 28.Pobbati AV, Stein A, Fasshauer D. N- to C-terminal SNARE complex assembly promotes rapid membrane fusion. Science. 2006;313:673–676. doi: 10.1126/science.1129486. [DOI] [PubMed] [Google Scholar]
- 29.Sorensen JB, et al. Sequential N- to C-terminal SNARE complex assembly drives priming and fusion of secretory vesicles. EMBO J. 2006;25:955–966. doi: 10.1038/sj.emboj.7601003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wright PE, Dyson HJ. Linking folding and binding. Curr Opin Struct Biol. 2009;19:31–38. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steinmetz MO, et al. Molecular basis of coiled-coil formation. Proc Natl Acad Sci USA. 2007;104:7062–7067. doi: 10.1073/pnas.0700321104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xue M, et al. Distinct domains of complexin I differentially regulate neurotransmitter release. Nat Struct Mol Biol. 2007;14:949–958. doi: 10.1038/nsmb1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giraudo CG, et al. Alternative zippering as an on-off switch for SNARE-mediated fusion. Science. 2009;323:512–516. doi: 10.1126/science.1166500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wiederhold K, Fasshauer D. Is assembly of the SNARE complex enough to fuel membrane fusion? J Biol Chem. 2009;284:13143–13152. doi: 10.1074/jbc.M900703200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lagow RD, et al. Modification of a hydrophobic layer by a point mutation in syntaxin 1A regulates the rate of synaptic vesicle fusion. PLoS Biol. 2007;5:e72. doi: 10.1371/journal.pbio.0050072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hong H, et al. Role of aromatic side chains in the folding and thermodynamic stability of integral membrane proteins. J Am Chem Soc. 2007;129:8320–8327. doi: 10.1021/ja068849o. [DOI] [PubMed] [Google Scholar]
- 37.Siddiqui TJ, et al. Determinants of synaptobrevin regulation in membranes. Mol Biol Cell. 2007;18:2037–2046. doi: 10.1091/mbc.E07-01-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McNew JA, et al. The length of the flexible SNAREpin juxtamembrane region is a critical determinant of SNARE-dependent fusion. Mol Cell. 1999;4:415–421. doi: 10.1016/s1097-2765(00)80343-3. [DOI] [PubMed] [Google Scholar]
- 39.Kesavan J, Borisovska M, Bruns D. v-SNARE actions during Ca(2+)-triggered exocytosis. Cell. 2007;131:351–363. doi: 10.1016/j.cell.2007.09.025. [DOI] [PubMed] [Google Scholar]
- 40.Deak F, Shin OH, Kavalali ET, Sudhof TC. Structural determinants of synaptobrevin 2 function in synaptic vesicle fusion. J Neurosci. 2006;26:6668–6676. doi: 10.1523/JNEUROSCI.5272-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Domanska MK, et al. Single vesicle millisecond fusion kinetics reveals number of SNARE complexes optimal for fast SNARE-mediated membrane fusion. J Biol Chem. 2009;284:32158–32166. doi: 10.1074/jbc.M109.047381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fasshauer D, Eliason WK, Brunger AT, Jahn R. Identification of a minimal core of the synaptic SNARE complex sufficient for reversible assembly and disassembly. Biochemistry. 1998;37:10354–10362. doi: 10.1021/bi980542h. [DOI] [PubMed] [Google Scholar]
- 43.Schuette CG, et al. Determinants of liposome fusion mediated by synaptic SNARE proteins. Proc Natl Acad Sci USA. 2004;101:2858–2863. doi: 10.1073/pnas.0400044101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 45.Goddard TD, Kneller DG. San Francisco: Univ of California; 2008. SPARKY, NMR Assignment and Integration Software. Version 3.114. [Google Scholar]
- 46.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 47.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J Magn Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.