Abstract
The question of how far pollen can move between plants has implications for topics as diverse as habitat fragmentation, conservation management, and the containment of genetically modified crops. The monoecious African fig tree Ficus sycomorus L. relies on the small, short-lived, night-flying, host-specific fig wasp Ceratosolen arabicus Mayr for pollination. We used microsatellite markers to characterize a geographically isolated riparian population of F. sycomorus growing along the Ugab River in the Namib Desert, Namibia, together with paternity analysis of seedlings from known mothers, to map pollen movement within this population. In this way we tracked insect movements between individually recognizable trees by means of their pollen cargo and documented the movement of C. arabicus between known trees separated by more than 160 km, with a mean distance for confirmed successful pollination events of 88.6 km. The predominant observed movement of pollinators was in a westerly direction, toward the sea, reflecting seasonal nighttime wind direction and the wind-borne dispersal of fig wasps. Our results suggest the existence of an extensive panmictic population of trees that are well suited to overcome the effects of geographical isolation.
Keywords: Ficus sycomorus, fig wasp, gene flow, spatial structure, pollen dispersal
Gene flow is central in shaping population demography and determining the spatial and temporal scale at which populations respond to selection. The mode of dispersal is an important factor, determining where species persist and how they extend their ranges (1). For most plant species, this is a passive process. However, many plant species depend on insects to transport their pollen, and so the behavior of their particular pollinators determines how far pollen is transported. Pollen flow is commonly skewed toward short distances of just a few meters, reflecting insect behavior and the spacing of the plants (2, 3), but some obligate out-breeding plant species form widely scattered populations, with individuals growing far apart. Long-distance dispersal can be mediated by wind, water, or animal vectors and is normally associated with specific but rare events (4) rather than through frequent long-distance movements. Pollen dispersal distances for some species have been found to range from hundreds of meters (5–11) to kilometers (12, 13). Long-distance pollen flow between fig trees (Ficus spp., Moraceae) is particularly well documented (14, 15). Each of the 800 or so species of fig tree is pollinated exclusively by one or a few host-specific fig wasps (Agaonidae) (16). Fig wasps are small, delicate, weak-flying, and short-lived insects (17) that use the wind to carry them between fig trees in open woodland (18) or in rainforests, where many species are transported by the fast-flowing air found above the general canopy (19, 20). Fig trees and their wasps are also effective colonizers of islands, and low levels of genetic differentiation at microsatellite loci between mainland and island populations suggest gene flow can occur over distances of tens of kilometers (15, 21).
Nason et al. (14) were the first to use molecular approaches to detect long-distance dispersal of fig wasps. Their findings, obtained by means of parental reconstruction of single foundress progeny arrays, revealed that incoming pollen at focal trees must have come from many different individuals. Given the low densities of each species, this could only be possible if the wasps were routinely covering distances of more than 10 km. Our article extends the work of Nason et al. by determining the distances traveled by fig wasps between known individual trees. Their study took place in a Central American rainforest, where the tracking of pollen flow between individuals across many square kilometers of tropical forest would present a formidable logistical challenge. We therefore sought an alternative habitat, where all of the trees over a large area could be sampled more easily.
The African fig tree Ficus sycomorus L. is widespread throughout Southern Africa, but it is restricted to riverine habitats in the more arid parts of its range. It is a large, monoecious tree that produces crops ranging from a few tens to many thousands of figs that are pollinated by the nocturnal fig wasp Ceratosolen arabicus Mayr. Flowering in F. sycomorus is largely asynchronous between trees, and synchronous within trees, forcing the fig wasps to fly between trees to find a receptive host. As adult fig wasps survive 48 h or less, this must occur quickly. On entering a fig, the wasp's wings become detached, preventing subsequent dispersal to other trees.
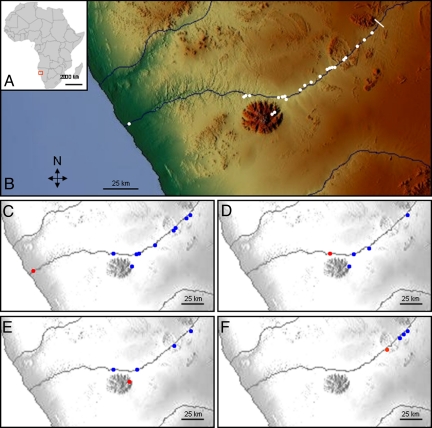
Our study site followed the course of the Ugab River and its White Lady tributary in Namibia (Fig. 1 A and B), which contains a linearly-distributed and geographically isolated population of F. sycomorus. The river flows for ≈450 km, crossing the gravel plains of the North Namib Desert and ending at the Skeleton Coast. A rainfall gradient from west to east results in a gradual transformation from extreme desert by the coast to dry savannah habitat inland. With the help of volunteer assistants and guides, we walked three times along the last 253 km of the lower Ugab River, mapping all of the F. sycomorus individuals that we found, starting from the river mouth (21°07.5′ S, 13°38.7′ E) and ending 240-km inland (20°30.5′ S, 15°16.0′ E). The surveys were carried out during the winter, when prevailing nighttime winds are in an easterly direction (22). Because F. sycomorus is restricted to a narrow band on the banks and river bed, we could expect to find and map every mature tree. F. sycomorus was absent from the last 13 km of our transect, but more trees are known to be present further inland. Specifically, across the desert, the Huab is the nearest river to the Ugab, but they are separated by 50 km at their closest point. We used direct microsatellite-based paternity analysis within the Ugab's apparently isolated population of F. sycomorus to identify the pollen parents of seedlings from known mothers. Tracking the movements of the fig wasps via their pollen cargo revealed both the distance and direction of travel.
Fig. 1.
Distribution of trees within the study site and pollen movements between them. (A) Position of the field site in Namibia, Southern Africa. (B) Distribution of adult trees mapped along the Ugab River. Individuals or clusters of trees are represented by white dots (for map coordinates see Table S1). Paternity assignments relating to individual female parents (red) and assigned pollen donors (blue) as follows: (C) Tree-1 mother, (D) Tree-3 mother, (E) Tree-17 mother, (F) Tree-41 mother. The base map was obtained from www.maps-for-free.com.
Results
Within our study site (see Fig. 1 A and B), 81 mature trees were identified and all except two inaccessible individuals were mapped by global positioning system (Table S1). Leaf material was collected and preserved for DNA extraction. The straight-line distance between neighboring mapped trees ranged from a few meters to 84 km (see Fig. 1B and Table S1). Crop sizes ranged from less than a hundred to several thousand figs. There was no evidence that large numbers of figs had aborted because of absence of pollination. Despite its isolation, Tree-1, which was 12-km inland from the coast and 81.6 km from its nearest neighbor, had a large crop of mature pollinated figs, from which we collected seeds for paternity analysis. The following year, the entire population was resurveyed twice and seeds from trees upstream of Tree-1 were collected. Of these samples, seed from nine trees were successfully germinated to yield 90 seedlings, 39 from Tree-1 and 51 from other trees. These seedlings were genotyped using five microsatellite markers developed for F. sycomorus (23) plus one from a congener (24). The genotypes of the 79 mature trees and their offspring are given in Tables S2 and S3.
Analysis of data from the 79 genotyped adult trees identified 61 distinct alleles from the six loci. The mean expected heterozygosity and mean observed heterozygosity were 0.708 and 0.663, respectively (Table 1). Four of the loci showed significant departures from Hardy-Weinberg proportions after sequential Bonferroni correction for multiple comparisons (25), but the presence of null alleles was identified for only one locus and there was no evidence of allelic drop-out or preferential amplification of alleles. The absence of mother-offspring mismatches further argues against high null-allele frequencies. We detected no structure in the population (see below), making Wahlund effects or biparental inbreeding unlikely. Therefore, the observed inbreeding coefficient value (FIS = 0.026, excluding the locus with null alleles) is most likely to be a result of selfing (implying a selfing rate of ≈5%, although none were detected in our seedling samples). The confirmed paternity exclusions were not based upon single genotypes of the problematic locus and the confidence of our assignments using this marker combination remained high. The combined exclusion probability of the markers, where one parent is known, was >99%. None of the loci were in linkage disequilibrium.
Table 1.
Pollination distances
| Mother tree ID | Nearest neighbor (m) | Number of offspring genotyped | Number assigned paternity | Number of fathers contributing | Minimum observed distance (km) | Maximum observed distance (km) | Mean distance (km) | Standard deviation |
|---|---|---|---|---|---|---|---|---|
| 1 | 81,600 | 37 | 19 | 12 | 85.3 | 164.7 | 140.1 | 6.6 |
| 3 | 2,000 | 13 | 5 | 4 | 24.2 | 80.8 | 43.7 | 18.6 |
| 4 | 1,000 | 5 | 2 | 2 | 24.6 | 78.5 | 51.6 | 19.0 |
| 6 | 1,000 | 3 | 2 | 2 | 71.4 | 75.7 | 73.6 | 1.5 |
| 8 | 2,300 | 2 | 1 | 1 | 51.0 | 51.0 | 51.0 | 0.0 |
| 13 | 1 | 1 | 0 | — | — | — | — | — |
| 17 | 1 | 5 | 4 | 4 | 14.2 | 71.5 | 38.3 | 2.7 |
| 28 | 3 | 11 | 2 | 2 | 20.6 | 57.4 | 39.0 | 13.0 |
| 40 | 5 | 2 | 1 | 1 | 27.9 | 27.9 | 27.9 | 0.0 |
| 41 | 5 | 11 | 4 | 3 | 22.3 | 29.1 | 25.9 | 0.1 |
Data are presented by maternal trees, numbered from the coast moving inland, with nearest-neighbor distances for comparison.
Rousset's genetic distance measure (âr) (26) was calculated from the genotypic data obtained for the 79 trees using the software SPAGeDi v1.2 (27), but no significant correlation with spatial distance was found. Spatial clustering of individuals was also investigated using a Bayesian approach implemented in BAPS 4.14 software (28). The number of populations inferred by the optimal partition was always one when a maximum of between one and five populations was considered. Analysis using Structure v2.2 (29) also suggested a single population.
Paternity analysis of progeny collected over the two seasons of field work was then used to define the patterns of gene flow within this spatially delimited population. We used a categorical paternity analysis method, CERVUS v3 (30), to assign paternity to seedlings grown from the seed collected from individual maternal trees. Paternity analysis of all 90 genotyped seedlings from known mothers resolved the pollen-parent for 40 (Table S4). No occurrence of self-fertilization was detected. All assignments were at high logarithm of odds (LOD) scores (range 3.49–8.14; critical LOD score 3.37) with no mismatches. Five of the offspring left unassigned had multiple matching-paternal genotypes, the remaining 45 had no paternal match for trees in the sample. A range of analysis parameters in CERVUS (see Methods) yielded the same outcome. We estimated the number of different pollen donors contributing to offspring for which we could not assign a father by reconstruction of paternal genotypes from the unassigned progeny arrays, and counted the number of different alleles at each locus to reveal the minimum number of donors. Based on locus MFC2, which had 21 alleles in the adult sampled population, at least 10 unidentified fathers were needed to account for the Tree-1 offspring and 14 for the whole set of offspring. We also estimated the number of unsampled fathers using locus Fsyc 6, which has 10 alleles in the adult sampled population, and this suggested at least four different unsampled fathers for Tree-1 offspring and six for all of the unassigned offspring. Given that only two mature trees within the focal population were not genotyped, the cases of unassigned paternity must reflect the contribution of pollen from trees located outside the study area.
The closest distance between assigned parents was 14.2 km; the furthest pair was separated by 164.7 km (see Table 1) and the mean distance for all confirmed assignments was 88.6 km (SD 54.9, median 79.6 km). These long-distance pollen movements are on an unprecedented scale. The most extreme pollen-movement distances observed came from analysis of seedlings from Tree-1 (Fig. 1C and see Table 1). Thirty-seven genotyped seedlings came from this tree and we assigned paternity to 19 of them. Distances between Tree-1 and the assigned fathers ranged from 85.3 km to 164.7 km (mean 140.1 km, SD 26.2, median 150.8) (see Table 1). A consequence of the geographical location of this tree was that all potential fathers were located toward the east of our study area. Other trees in the population have neighbors both to their east and west, but paternity analysis of progeny from these trees revealed that all but one of the male parents (see Fig. 1 D–F for examples) were located to the east of the maternal trees, resulting in predominantly unidirectional gene flow; nearest neighbors were rarely the fathers of the seedlings.
Fifty-three seedlings were obtained from fruit collected from trees upstream of Tree-1 in the second survey year; of these, 21 were assigned paternity. The distances between mothers and assigned fathers for these seedlings (see Table 1) ranged from 14.2 to 80.8 km, with a mean distance of 42.0 km (SD 22.0, median 29.1 km). The standard deviation of parent-offspring distances σ, a key parameter which, together with effective population density, determines the expected scale of genetic structure in a continuous population, was 97.6 km for all assignments across the two seasons and 12.7 km for the second season alone.
To counter possible effects of false-paternity exclusion because of null alleles in locus Fsyc13, we performed the paternity analysis including only five loci. In this way, we were able to resolve the pollen parent of 29 seedlings out of 90, with predominantly unidirectional pollen flow and mean dispersal of 84.3 km including Tree-1 or 43.4 km excluding Tree-1. We also used a simple exclusion approach to estimate the number of offspring for which all putative fathers could be excluded by heterozygous loci alone, therefore avoiding the potential influence of null alleles at any locus. Using this model-free approach, we calculated that there are 22 offspring for which all sampled adult trees can be excluded as fathers.
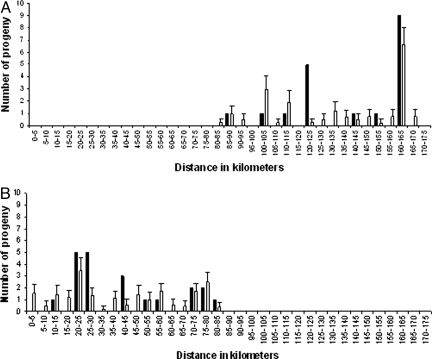
There was a striking pattern of declining mean pollination distances between trees toward the east of the survey area (see Table 1), reflecting the higher tree densities further inland and the overwhelmingly wind-borne westerly movement of pollinators. Our data may underestimate mean pollination distances between the more inland trees because unidentified distant pollen donors from outside the study area may also be contributing to paternity. We therefore estimated the underlying distribution of these movements using a maximum-likelihood method (31) that allows for a proportion of paternity from outside the study area and compares a variety of forms of the dispersal distribution. However, no model of finite dispersal provided a better fit than the null-model of panmixia, either with or without the outlying Tree-1 included in the dataset. We simulated the expected distribution of paternities under panmixia for each dataset, where we defined the mothers to match the observed data and allowed pollen movement up and down the river with no limit on distance (Fig. 2). The observed distributions of pollination distances are consistent with these expectations because the mean simulated displacement of fathers toward the east of the relevant mothers was 26.6 km, with an observed mean distance of 35.7 km. Moreover, the data obtained in 2001 reveal that 95% of the identified fathers were east of the mothers, a value that was never exceeded in 1,000 simulations, indicating that the null hypothesis of nondirectional pollination can be rejected with P < 0.001.
Fig. 2.
Distribution of paternity assignments. The observed numbers of progeny assigned (solid bars) and simulated numbers of assignments (± SD) (open bars) under the assumption of panmixia, with mothers and numbers of offspring defined by the observed data and fathers chosen at random from sampled trees up- or down-stream of the mother, with no limitation on distance, in 5-km distance increments for (A) year 2000 data (maternal Tree-1 only) and (B) year 2001 data.
Discussion
Our data demonstrate the movement of individual insects over unprecedented distances and define pollination events between known parent plants over distances >160 km, with a mean dispersal distance of 88.6 km. Within-crop synchrony and between-tree asynchrony in F. sycomorus, as is typical of many fig trees, ensured cross-pollination and meant that pollination events were often not between nearest neighbors. We chose this population because of its apparent isolation, but the diversity of genotypes among the seedlings with unassigned paternity cannot be accounted for by the two inaccessible trees within our study. This conclusion suggests that there must be a source of pollen from additional trees located outside our study area, which is also implied by the estimate of unsampled fathers in the paternal reconstruction. It is therefore likely that pollination events of at least the distance to the nearest neighboring water courses of 50 km, but possibly even greater than the observed distance of 164 km, were occurring.
The positive FIS values in the adult population suggest some inbreeding. Although we see no evidence for selfing or biparental inbreeding in our study of contemporary gene flow, it may be that the sampled adult population is a remnant from one of much greater density, where related individuals had greater opportunity to interbreed. Higher densities facilitate the possibility of near-neighbor matings because of the close proximity of trees at the correct stage of the fig reproductive cycle. There is anecdotal evidence for a recent decline in the abundance of fig trees in the study area. Alternatively, selfing may be more common in the summer when fig crops are larger and less synchronized than in the winter, when our sample was collected. Easterly winds in the summer may also reduce pollen flow from inland populations.
Pollination over such long distances has resulted in a single unstructured population that extends for at least 250 km along the Ugab and perhaps much further. F. sycomorus shares a monoecious breeding system with the strangler figs studied by Nason et al. (14, 32). Fig wasps associated with monoecious fig species are thought to transport pollen over greater distances than those associated with dioecious fig trees, which are often smaller in size, producing small, asynchronous fig crops (33, 34). Pollen dispersal ranges of 1 to 2 km have been inferred for the dioecious F. pumila in China (35), but genetic studies of two pollinators of dioecious fig trees from Indonesia nonetheless found low levels of population subdivision between mainland and island populations over 40 km apart (15), suggesting that the nature of the breeding system is a weak predictor of gene-flow distances in fig trees.
How does C. arabicus manage to pollinate F. sycomorus trees over 100 km apart? The slow flight speed of fig wasps (36) precludes active, directed flight in the time available during a 48-h adult lifespan. Wind-borne dispersal is therefore required, which offers no control of their necessarily downwind direction of movement. Once fig wasps are close to a tree that is releasing species-specific volatile attractants, they can then seek sheltered areas with low wind speeds that permit directed flight (18, 36). The varying flight heights of fig wasps (34) show that this mode of insect dispersal is not truly passive (37). Predominantly easterly winds occur at night over the central Namib from May to October (22). The winds average ≈10 km per hour, but can reach 60 km per hour, making possible dispersal events of more than 100 km within a single night. The observed east-to-west direction of pollination flow reflects these wind conditions. Intriguingly, at other times of the year the predominant nighttime winds in the area are westerly. This meteorological phenomenon leads us to predict a seasonal reversal in the direction of pollen flow between the trees.
Along with other tropical trees, many fig tree populations are being rapidly fragmented (38), but their pollination system and wind-mediated dispersal capabilities (14) may provide them with exceptional resistance to habitat loss, population fragmentation, and large-scale perturbations (14, 39). The recent return of elephants to the Ugab, after being absent for many years, has resulted in the deaths of numerous F. sycomorus, resulting in the increasing isolation of the remaining trees, exacerbated by low recruitment. Although the dispersal ability of C. arabicus means that isolated down-river individuals will still be pollinated, the likelihood of pollinators successfully finding a fig must scale with down-wind distances between trees, and their population dynamics may limit long-term persistence in the Ugab and other increasingly fragmented landscapes.
Routine dispersal on the scale seen here has implications for population differentiation, local adaptation, and speciation among small insects in general (17). How typical is the dispersal ability of fig wasps? Any slow-flying insects that fly up into an air column will be carried downwind for as long as they remain airborne, but such behavior is not accidental and among rain forest parasitoids (Chalcidoidea), there are clear differences in the preferred flight heights of different families (19), implying differences in behavior that will affect their capacity to be carried by the wind. “Slatkin's paradox” describes the contrast between direct and indirect measures of gene flow seen in many small insects (6, 40). For example, the predominantly flightless plant-hopper Tumidagena minuta shows levels of population differentiation that are comparable with mobile insects, even though most individuals are incapable of flight and so dispersal distances are typically very low (6, 41). The paradox may be resolved if differentiation is low because of recent range expansions or if levels of dispersal are often seriously underestimated. Our extraordinarily high estimates of fig wasp movement, based on genes in the pollen they carry, could not have been achieved by methods such as mark-recapture. This suggests that a resolution of the paradox lies in underestimation of the dispersal abilities of small insects. If this is correct, the lack of genetic differentiation in other insects implies that fig wasp dispersal may not be exceptional.
Methods
Sampling.
F. sycomorus growing in the study area were numbered and global positioning system coordinates recorded. Young, fresh leaf tissue (5 g) was collected from each tree, the midrib was removed with scissors, and then placed in plastic vials containing 30-ml saturated NaCl-CTAB. Fruit was collected from each tree where possible. Ripe fruits were pressed flat, air-dried in the sun, and placed in paper bags containing silica gel. Seeds were subsequently germinated in a heated glasshouse at 22 °C, 65 to 80% relative humidity. After 3 weeks, seedlings were transferred to pots on capillary matting containing a 2:1:1 mix of compost (John Innes No. 3), perlite, and grit and maintained at 20 °C.
DNA Extraction and PCR Analysis.
DNA extraction and PCR analysis were performed as described in ref. 23.
Microsatellite Markers.
Microsatellite genotyping was performed using six loci, five of which were developed for this species (23) and one for a congener (24). Scoring of the genotype data were performed without knowing which offspring belonged to which mother to avoid type-1 errors and controls were included in all runs to allow repetition and ensure reproducibility and reliability of results. For summary statistics relating to these loci see the SI Text and Table S5.
Population Differentiation and Structure.
The data were tested for null alleles and other problems with scoring using Microchecker (42). Estimations of F statistics and departures from Hardy-Weinberg Equilibrium were implemented in Genepop v3.4 (43). Population structure was investigated using the programs BAPs 4.3 (28) and Structure v2.2 (29). In Structure, data were analyzed with no population priors under the “no admixture model” and repeated for the “admixture model.”
Rousset's genetic distance measure (a), which is analogous to the FST/(1-FST) ratio but using pairs of individuals rather than populations, was calculated in the software SPAGeDi v1.2 (27), such that the slope of the regression with distance provides an estimate of gene-dispersal distances.
Paternity Analysis.
Paternity analysis was performed using the program CERVUS v3 (30). We repeated simulations in CERVUS five times and determined the mean critical LOD score. Paternity analysis of the 90 genotyped seedlings with known mothers resolved the pollen-parent for 40 at the 80% confidence level estimated in CERVUS, 5 of which were at the higher confidence level of 95%. For detailed description of the analysis parameters applied see the SI Text.
Simulations.
We simulated the expected distribution of paternities under panmixia by drawing the observed number of mothers at random from the total set of 79 sampled trees and assigning paternities at random (excluding the possibility of self-fertilization) to give the observed numbers of offspring per mother; the analysis was repeated 1,000 times. We also simulated pollination distances (1,000 replicates) by restricting mothers to those observed and, to test the null hypothesis of nondirectional pollen flow, selected fathers from either east or west of the mother with unlimited distance (see Fig. 2).
Supplementary Material
Acknowledgments.
We thank Raleigh International volunteers for help with fieldwork, Martin Lappage for plant maintenance in Leeds, Terry Burke and Deborah Dawson (NERC Biomolecular Analysis Facility) for assistance in microsatellite marker development and genotyping, and the Natural Environment Research Council for supporting this aspect of the work. We also thank Hirohisa Kishino and Mark Beaumont for assistance with data analysis, Deborah Charlesworth, Terry Burke, and Mike Shaw for comments on early drafts, and three anonymous reviewers and the journal editor for their input. The work was funded by a Biotechnology and Biological Sciences Research Council studentship (to S.A.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. E.A.H. is a guest editor invited by the Editorial Board
This article contains supporting information online at www.pnas.org/cgi/content/full/0902213106/DCSupplemental.
References
- 1.Butlin RK, Bridle JR, Schluter D, editors. Speciation and Patterns of Diversity. Cambridge: Cambridge Univ Press; 2009. [Google Scholar]
- 2.Levin DA, Kerster HW. Gene flow in seed plants. Evol Biol. 1974;7:139–159. [Google Scholar]
- 3.Garcia C, Jordano P, Godoy JA. Contemporary pollen and seed dispersal in a Prunus mahaleb population: Patterns in distance and direction. Mol Ecol. 2007;16:1947–1955. doi: 10.1111/j.1365-294X.2006.03126.x. [DOI] [PubMed] [Google Scholar]
- 4.Nathan R. Long-distance dispersal of plants. Science. 2006;313:786–788. doi: 10.1126/science.1124975. [DOI] [PubMed] [Google Scholar]
- 5.Rousset F. Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics. 1997;145:1219–1228. doi: 10.1093/genetics/145.4.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Slatkin M. Gene flow and the geographic structure of natural populations. Science. 1987;236:787–792. doi: 10.1126/science.3576198. [DOI] [PubMed] [Google Scholar]
- 7.Wright S. The general structure of populations. Ann Eugenic. 1951;15:323–354. doi: 10.1111/j.1469-1809.1949.tb02451.x. [DOI] [PubMed] [Google Scholar]
- 8.Smouse PE, Dyer RJ, Westfall RD, Sork VL. Two-generation analysis of pollen flow across a landscape. I. Male gamete heterogeneity among females. Evolution. 2001;55:260–271. doi: 10.1111/j.0014-3820.2001.tb01291.x. [DOI] [PubMed] [Google Scholar]
- 9.Austerlitz F, et al. Using genetic markers to estimate the pollen dispersal curve. Mol Ecol. 2004;13:937–954. doi: 10.1111/j.1365-294x.2004.02100.x. [DOI] [PubMed] [Google Scholar]
- 10.Chase MR, Moller C, Kesseli R, Bawa KS. Distant gene flow in tropical trees. Nature. 1996;383:398–399. [Google Scholar]
- 11.Robledo-Arnuncio JJ, Gil L. Patterns of pollen dispersal in a small population of Pinus sylvestris L. revealed by total exclusion paternity analysis. Heredity. 2005;94:13–22. doi: 10.1038/sj.hdy.6800542. [DOI] [PubMed] [Google Scholar]
- 12.White GM, Boshier DH, Powell W. Increased pollen flow counteracts fragmentation in a tropical dry forest: An example from Swietenia humilis Zuccarini. Proc Natl Acad Sci USA. 2002;99:2038–2042. doi: 10.1073/pnas.042649999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dick CW, Etchelecu G, Austerlitz F. Pollen dispersal of tropical trees (Dinizia excelsa: Fabaceae) by native insects and African honeybees in pristine and fragmented Amazonian rainforest. Mol Ecol. 2003;12:753–764. doi: 10.1046/j.1365-294x.2003.01760.x. [DOI] [PubMed] [Google Scholar]
- 14.Nason JD, Herre EA, Hamrick JL. The breeding structure of a tropical keystone plant resource. Nature. 1998;391:685–687. [Google Scholar]
- 15.Zavodna M, et al. Pollinating fig wasps: Genetic consequences of island recolonization. J Evolution Biol. 2005;18:1234–1243. doi: 10.1111/j.1420-9101.2005.00937.x. [DOI] [PubMed] [Google Scholar]
- 16.Herre EA, Jander KC, Machado CA. Evolutionary ecology of figs and their associates: Recent progress and outstanding puzzles. Annu Rev Ecol Evol S. 2008;39:439–458. [Google Scholar]
- 17.Compton SG. Sailing with the wind: dispersal by small flying insects. In: Bullock D, editor. Dispersal Ecology. British Ecological Society, Blackwell; 2002. [Google Scholar]
- 18.Ware AB, Compton SG. Dispersal of adult female fig wasp 2. Movements between trees. Entomol Exp Appl. 1994;73:231–238. [Google Scholar]
- 19.Compton SG, Ellwood MDF, Davis AJ, Welch K. The flight heights of chalcid wasps (Hymenoptera, Chalcidoidea) in a lowland Bornean rain forest: fig wasps are the high fliers. Biotropica. 2000;32:515–522. [Google Scholar]
- 20.Harrison RD, Rasplus JY. Dispersal of fig pollinators in Asian tropical rain forests. J Trop Ecol. 2006;22:631–639. [Google Scholar]
- 21.Compton SG, Ross SJ, Thornton IWB. Pollinator limitation of fig tree reproduction on the island of Anak-Krakatau (Indonesia) Biotropica. 1994;26:180–186. [Google Scholar]
- 22.Tyson PD, Seely MK. Local winds over the central Namib. S Afr Geogr J. 1980;62:135–150. [Google Scholar]
- 23.Ahmed S, Dawson DA, Compton SG, Gilmartin PM. Characterization of microsatellite loci in the African fig Ficus sycomorus L. (Moraceae) Mol Ecol Notes. 2007;7:1175–1177. [Google Scholar]
- 24.Khadari B, Hochu I, Santoni S, Kjellberg F. Identification and characterization of microsatellite loci in the common fig (Ficus carica L.) and representative species of the genus Ficus. Mol Ecol Notes. 2001;1:191–193. [Google Scholar]
- 25.Rice WR. Analyzing tables of statistical tests. Evolution. 1989;43:223–225. doi: 10.1111/j.1558-5646.1989.tb04220.x. [DOI] [PubMed] [Google Scholar]
- 26.Rousset F. Genetic differentiation between individuals. J Evolution Biol. 2000;13:58–62. [Google Scholar]
- 27.Hardy OJ, Vekemans X. SPAGEDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes. 2002;2:618–620. [Google Scholar]
- 28.Corander J, Waldmann P, Sillanpaa MJ. Bayesian analysis of genetic differentiation between populations. Genetics. 2003;163:367–374. doi: 10.1093/genetics/163.1.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marshall TC, Slate J, Kruuk EB, Pemberton JM. Statistical confidence for liklihood-based paternity inference in natural populations. Mol Ecol. 1998;7:637–655. doi: 10.1046/j.1365-294x.1998.00374.x. [DOI] [PubMed] [Google Scholar]
- 31.Oddou-Muratorio S, Klein EK, Austerlitz F. Pollen flow in the wild service tree, Sorbus torminalis (L) Crantz II Pollen dispersal and heterogeneity in mating success inferred from parent-offspring analysis. Mol Ecol. 2005;14:4441–4452. doi: 10.1111/j.1365-294X.2005.02720.x. [DOI] [PubMed] [Google Scholar]
- 32.Nason JD, Herre EA, Hamrick JL. Paternity analysis of the breeding structure of strangler fig populations: Evidence for substantial long-distance wasp dispersal. J Biogeogr. 1996;23:501–512. [Google Scholar]
- 33.Harrison RD. Repercussions of El Nino: Drought causes extinction and the breakdown of mutualism in Borneo. P Roy Soc Lond B Bio. 2000;267:911–915. doi: 10.1098/rspb.2000.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harrison RD. Fig wasp dispersal and the stability of a keystone plant resource in Borneo. P Roy Soc Lond B Bio. 2003;270:S76–S79. doi: 10.1098/rsbl.2003.0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang R, et al. Spatial genetic structure and restricted gene flow in a functionally dioecious fig, Ficus pumila L var pumila (Moraceae) Popul Ecol. 2009;51:307–315. [Google Scholar]
- 36.Ware AB, Compton SG. Dispersal of adult female fig wasps 1. Arrivals and departures. Entomol Exp Appl. 1994;73:221–229. [Google Scholar]
- 37.Reynolds AM, Reynolds DR. Aphid aerial density profiles are consistent with turbulent advection amplifying flight behaviours: abandoning the epithet ‘passive’. P Roy Soc B Bio. 2009;276:137–143. doi: 10.1098/rspb.2008.0880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mawdsley NA, Compton SG, Whittaker RJ. Population persistence, pollination mutualisms, and figs in fragmented tropical landscapes. Conserv Biol. 1998;12:1416–1420. [Google Scholar]
- 39.Bronstein JL, Hossaert-McKey M. Hurricane Andrew and a Florida fig pollination mutualism—resilience of an obligate interaction. Biotropica. 1995;27:373–381. [Google Scholar]
- 40.Mallet J. Gene flow. In: Woiwod IP, Reynolds DR, Thomas CD, editors. Insect Movement: Mechanisms and Consequences. CAB International; 2001. pp. 337–360. [Google Scholar]
- 41.Peterson MA, Denno RF, Robinson L. Apparent widespread gene flow in the predominantly flightless planthopper Tumidagena minuta. Ecol Entomol. 2001;26:629–637. [Google Scholar]
- 42.Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. MICRO-CHECKER: Software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes. 2004;4:535–538. [Google Scholar]
- 43.Raymond M, Rousset F. Genepop (Version-1.2)—Population genetics software for exact tests and ecumenicism. J Hered. 1995;86:248–249. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.