Abstract
The relative importance of local ecological and larger-scale historical processes in causing differences in species richness across the globe remains keenly debated. To gain insight into these questions, we investigated the assembly of plant diversity in the Cerrado in South America, the world's most species-rich tropical savanna. Time-calibrated phylogenies suggest that Cerrado lineages started to diversify less than 10 Mya, with most lineages diversifying at 4 Mya or less, coinciding with the rise to dominance of flammable C4 grasses and expansion of the savanna biome worldwide. These plant phylogenies show that Cerrado lineages are strongly associated with adaptations to fire and have sister groups in largely fire-free nearby wet forest, seasonally dry forest, subtropical grassland, or wetland vegetation. These findings imply that the Cerrado formed in situ via recent and frequent adaptive shifts to resist fire, rather than via dispersal of lineages already adapted to fire. The location of the Cerrado surrounded by a diverse array of species-rich biomes, and the apparently modest adaptive barrier posed by fire, are likely to have contributed to its striking species richness. These findings add to growing evidence that the origins and historical assembly of species-rich biomes have been idiosyncratic, driven in large part by unique features of regional- and continental-scale geohistory and that different historical processes can lead to similar levels of modern species richness.
The uneven distribution of species diversity across the globe and the occurrence of biodiversity hotspots with high concentrations of species are well established (1–3). However, the underlying causes of differences in species diversity and composition among biomes and the processes that have prompted accumulation of high species diversity in some areas remain poorly understood (4–6). Correlations between species richness and annual energy input, water supply, and physiographic complexity suggest that climatic and other environmental factors are major determinants of species diversity (7). Indeed, it has been shown that these factors can accurately predict the locations of most global diversity hotspots and account for the latitudinal gradient of species richness (7). However, such insights contribute little to our understanding of the historical assembly of species-rich biomes and the larger-scale evolutionary processes that have generated global patterns of diversity (4, 8, 9). Little is known about the historical and geographical assembly of species-rich biomes (5), in terms of when, how quickly, and from where the species and lineages that make up different biomes have been recruited and how they subsequently evolved in situ. This lack of data on geographical and temporal patterns of species diversification, especially in the tropics where most diversity resides, makes it difficult to assess why there are so many species in these areas, to what extent variation in diversity can be attributed to regional and continental-scale historical contingencies (4, 10, 11), and to compare patterns among different species-rich biomes (9, 12).
Recent discussion has highlighted the potential role of phylogenetic niche conservatism in shaping regional species pools (8, 13) and explaining diversity gradients (11). Prominent examples of large-scale niche or biome conservatism have been documented for the tropical–temperate biotas (11), mangroves (14), southern hemisphere extratropical biomes (13), Andean grasslands (8, 15), and seasonally dry tropical forests (16–18). However, the extent to which this tendency to retain ancestral ecology across lineages has influenced species composition in the most species-rich tropical biomes is unknown.
Recent insights into the historical assembly of species diversity and biomes have come from time-calibrated phylogenies for biome-specific lineages (5, 9, 19–23). Phylogenies have potential for reconstructing transitions from precursor to modern biotas and identifying the underlying factors that drive these processes (4, 11, 24). However, the sparse sampling of lineages and species in such studies (5, 24) has limited these insights to a few well-studied areas such as the Cape Floristic Region (12, 20).
The Cerrado.
To address these questions, we have investigated plant diversification in the Cerrado of South America by using a comparative phylogenetic approach. The Cerrado is a floristically diverse savanna that covers more than two million km2 of Central Brazil and parts of Bolivia and Paraguay (Fig. 1). This region has been recognized as a global biodiversity hotspot with more than 10,000 plant species, of which 44% are endemic (1, 25, 26). Despite the floristic and global conservation importance of the Cerrado, little is known about the origins and diversification of its flora. Indeed, hypotheses about Cerrado origins range from the early Cretaceous (27), wherein Cerrado lineages were suggested to be possible precursors of the adjacent Amazonian and Atlantic rain forests, to the Holocene (28). Here we evaluate these alternatives, but we particularly focus on the hypothesis that the origin of the biome coincided with the rise to dominance of flammable C4 grasses within the past ten million years (6, 29).
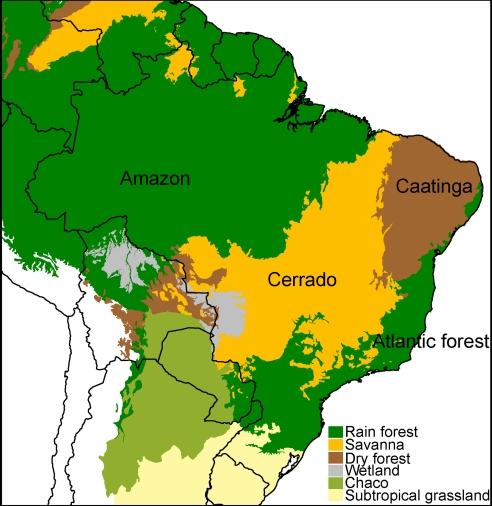
Fig. 1.
Map of major vegetation types in South America showing the location of the Cerrado surrounded by a diverse array of other biomes.
In common with other tropical savannas, the Cerrado is dominated by C4 grasses which take advantage of high light and warm wet summers to rapidly accumulate biomass, which becomes flammable in the long dry winters, prompting fires, generally several times in a decade (30, 31). The synergy between fast (re)growth and flammability allows grasses to outcompete trees and shrubs, maintaining the open canopy typical of savannas in areas where, without fire, forests would dominate (31). Diverse fire adaptations are the hallmark of the endemic Cerrado flora (32). These adaptations include functionally herbaceous or woody geoxylic suffrutices with enlarged underground xylopodia or lignotubers, thick corky bark, pachycaul rosulate shrubs and trees with sparse branching, thick shoots and leaves concentrated at shoot tips, perennial herbs, and specialized flowering and fruiting phenologies (27, 32, 33). These adaptations are largely lacking in the flora of adjacent biomes where fire has been less important over evolutionary time scales (29). C4 grasses originated and started to diversify 32–25 Mya in several independent lineages (34). However, evidence from isotopic signatures of fossil mammal teeth (paleodiets) and paleosols, charcoal particles, and pollen from sites on different continents suggests that these lineages only became ecologically dominant 4–8 Mya (35–37). This near-synchronous expansion of grass-dominated vegetation in different parts of the world is regarded as marking the onset of the modern savanna biome (38).
To test the hypothesis that the evolution of the Cerrado flora followed the ecological expansion of flammable C4 grasses and to investigate the significance of fire adaptations as a selective advantage for Cerrado diversification, we assembled time-calibrated phylogenies for plant groups that include Cerrado elements. Our sample includes four datasets representing lineages rich in Cerrado endemics (Mimosa and Microlicieae, Melastomataceae) and lineages sparsely represented in the Cerrado (Lupinus and Andira), as well as diverse life forms including trees, shrubs, and perennial herbs. Two of the phylogenies (Mimosa and Andira) are published for the first time. Central to the study is a new phylogeny for the species-rich (>500 species) legume genus Mimosa, the second-largest plant genus in the Cerrado (26, 39). In addition, new analyses are presented of published data for the legume genus Lupinus (15) and the tribe Microlicieae (40). These phylogenies are used to infer divergence times and ancestral biomes for Cerrado lineages, investigate the evolution of fire adaptations, and ultimately to build an evolutionary picture of the historical assembly of species in the Cerrado.
Results and Discussion
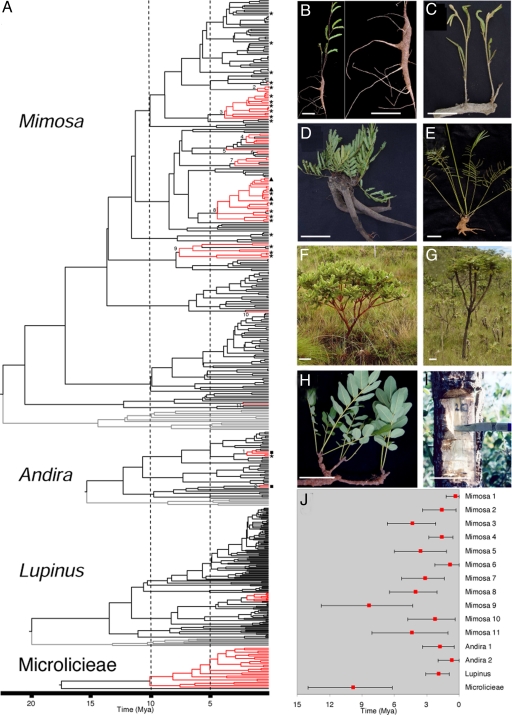
Our analyses identified 15 Cerrado lineages, 11 in Mimosa, two in Andira and one each in Lupinus and Microlicieae (Fig. 2A). Most of these clades are robustly supported and represent groups where fire-adapted endemics make up the majority of species (see Table 1 and SI Appendix for descriptions of each lineage). In three of the study groups, Cerrado lineages form clades that are deeply nested within genera. In two, there are multiple independent Cerrado lineages. These findings are in line with floristic data showing the preponderance of species-level endemism and lack of endemic genera in the Cerrado (32, 33). Within Mimosa, Cerrado lineages originated independently at least 11 times (Fig. 2A). Two of these clades (Mimosa 3 and 8) are notably rich in fire-adapted species resulting from in situ Cerrado diversification and together represent ≈75% of all Cerrado species of Mimosa. Notably, both these species-rich Mimosa radiations are estimated to have arisen approximately 4 Mya. The diversity of life forms in these two clades (Fig. 2 B–G), including functionally herbaceous subshrubs, prostrate herbs, wand-like subshrubs and pachycaul rosette trees, reflects the diverse ways that species have evolved to tolerate fire disturbance. Similar multiple independent origins of Cerrado species are apparent in Andira, with two recent clades both having two species. The largest Cerrado clade sampled here, representing ≈200 Cerrado endemics (40), is nested within the tribe Microlicieae. However, it is clear that this was not the only incursion of Melastomataceae into the Cerrado, with several other fire-adapted Cerrado lineages in this family (e.g., in Miconia, Cambessedesia, and Tibouchina).
Fig. 2.
Evolution of the fire-adapted Cerrado flora. (A) Chronograms for Mimosa, Andira, Lupinus, and Microlicieae showing 15 Cerrado lineages (red). Outgroups are depicted in gray. Symbols for fire adaptations: * = subshrub growing from xylopodium; ▴ = pachycaul treelet; ■ = thick corky bark. Numbered nodes correspond to Cerrado lineages. The Microlicieae phylogeny has been pruned to show just the Cerrado lineage and to fit within the time scale under investigation. Expanded phylogenies, including terminal names and support values, are presented in the SI Appendix. (B–I) Photographs illustrate the diversity of life forms and fire adaptations found in Mimosa clades 3 (B and C) and 8 (D–G), and Andira (H and I). (B) Wand-like subshrub with a xylopodium, Mimosa pseudoradula. (C) Functionally herbaceous subshrub with a horizontal underground xylopodium, Mimosa venatorum. (D) Functionally herbaceous subshrub growing from a massive xylopodium, Mimosa speciosissima. (E) Functionally herbaceous wand-like subshrub growing from xylopodium, Mimosa ulei. (F) Rosulate shrub, Mimosa oligosperma. (G) Pachycaul treelet with few thick branches, Mimosa splendida. (H) A branch of the geoxylic suffrutex or “underground tree”, A. humilis. (I) Thick corky bark, Andira cordata (scale bars = 10 cm). (J) Divergence-time estimates for 15 Cerrado lineages (crown nodes). Mean and 95% credibility intervals derived from 2.7 × 104 samples of a Bayesian analysis.
Table 1.
Diversity and origin of 15 Cerrado lineages sampled in this study
| Lineage | Total*/sampled species | Proportion of lineage in the Cerrado (%) | Age (Mya) (95%CI) | Life form/fire adaptation | Ancestral biome |
|---|---|---|---|---|---|
| Mimosa 1 | 1/1 | 100 | 0.4 (0.0–1.2) | subshrub with xylopodium | equivocal |
| Mimosa 2 | 3/3 | 100 | 1.6 (0.3–3.4) | subshrub with xylopodium | equivocal |
| Mimosa 3 | 34/16 | 92 | 4.4 (2.2–6.7) | subshrub with xylopodium | dry forest |
| Mimosa 4 | 11/6 | 73 | 1.6 (0.6–2.8) | shrub | equivocal |
| Mimosa 5 | 2/1 | 50 | 3.6 (1.2–6.0) | shrub | equivocal |
| Mimosa 6 | 4/2 | 100 | 0.9 (0.0–2.3) | shrub, tree | equivocal |
| Mimosa 7 | 8/6 | 87 | 3.2 (1.4–5.3) | subshrub with xylopodium | equivocal |
| Mimosa 8 | 50/26 | 100 | 4.1 (2.0–6.5) | xylopodium, pachycaul tree | equivocal |
| Mimosa 9 | 27/8 | 81 | 8.4 (4.3–12.8) | subshrub with xylopodium | dry forest |
| Mimosa 10 | 1/1 | 100 | 2.3 (0.4–4.8) | tree | dry forest (Caatinga) |
| Mimosa 11 | 1/1 | 100 | 4.4 (1.0–8.1) | tree | dry forest (Caatinga) |
| Andira 1 | 3/3 | 67 | 1.8 (0.5–3.4) | ″underground tree″ | rain forest (Amazon) |
| Andira 2 | 2/2 | 100 | 0.7 (0.0–2.0) | tree with thick corky bark | rain forest (Amazon) |
| Lupinus | 11/5 | 100 | 1.9 (0.9–3.1) | perennial herb, ericoid shrub | subtropical grassland |
| Microlicieae | 200/25 | 90 | 9.8 (6.2–14.0) | ericoid shrub, xylopodium | wetland |
*In some cases, the total number of species in a lineage was estimated by assigning unsampled taxa to groups based on taxonomic affinities.
The estimated ages of Cerrado lineages span the late Miocene to Pliocene from 9.8 to 0.4 Mya, with the majority less than 4 Mya (Fig. 2J), broadly coinciding with the hypothesized expansion of C4 grass-dominated savanna biomes (38). Some Cerrado lineages apparently slightly predate hypothesized C4 grass dominance, but the majority are younger (Fig. 2J), and this is compatible with fire as a common underlying trigger prompting the evolution of endemic Cerrado species and lineages, although the precise dynamics of this are clearly complex (30, 31, 38). One factor influencing very recent (Quaternary) evolution of Cerrado lineages (e.g., in Andira) may have been dry glacial periods, when the Cerrado was likely to have been more extensive. The derived nature of Cerrado lineages found here is in line with analyses of the legume family as a whole that showed that savanna elements worldwide are in general recently derived from dry or wet forest ancestors (18).
The idea that the Cerrado lineages evolved in response to increased C4 grass dominance and fire frequency is reinforced by the occurrence of fire adaptations in these groups (Fig. 2, Table 1). Fire adaptations in Andira include thick corky bark and a geoxylic suffruticose growth form in Andira humilis, a subshrub with an extensive network of underground woody shoots (“underground tree”), a life form unique within this otherwise arborescent genus (41) (Fig. 2 H and I and SI Appendix). Within Mimosa, fire adaptations include: functionally herbaceous perennials with xylopodia (Fig. 2 B–E) that evolved multiple times independently from an ancestral shrubby habit; unusual pachycaul treelets with few branches, thick shoots and leaves clustered at shoot apices (Fig. 2 F and G and SI Appendix); and crowded, persistent stipules that protect shoot apices from fire. The evolution of fire-adapted life forms is strongly correlated with Cerrado occurrence in Mimosa (likelihood ratio statistic = 30.30, P < 0.01) but much rarer or absent in sister lineages growing in adjacent wet and seasonally dry tropical forest. Indeed, seasonally dry tropical forest (the “succulent biome” sensu in ref. 18), which is the ancestral biome for several Cerrado lineages (see below), although sharing a long dry season with the Cerrado, is characterized by sparse occurrence of grasses, abundant Cactaceae and other succulents, and lack of natural fires (18, 29, 37). The large, fire-adapted Microlicieae clade is also nested within a family that predominates in fire-free (rain forest and swamp) habitats (discussed in ref. 40). It is notable that similar fire adaptations are common across other plant families in the Cerrado. For example, of 301 plant species in a one-hectare plot in the southern Cerrado, 94 species across 64 genera and 37 families have xylopodia (32).
Adaptive shifts between ecological zones can play an important role in the generation of species diversity (4, 11). Our results alongside wider floristic data (32, 42) show that fire adaptation and Cerrado occurrence are phylogenetically labile with multiple independent lineages at shallow depths within species-level phylogenies and across genera in disparate plant families. These frequent adaptive shifts suggest that fire does not pose a significant adaptive barrier to shifts between biomes. In comparison, shifts from tropical into temperate biomes and mangrove vegetation have been infrequent (4, 8), suggesting that adaptations to high salt and substrate anoxia or frost are more complex to evolve. In the case of the Cerrado, the boundaries appear to have been porous to the ingress and recruitment of lineages from a range of fire-free wet and dry forest vegetation types. Of significance is the intermingling of open, fire-prone savanna with patches of closed, less fire-prone riverine gallery forest, which share few species in common but which can harbor both fire-adapted and fire-sensitive congeners and even sister species (43). Phenotypic plasticity in fire traits (e.g., the normally stunted A. humilis can sometimes form a shrub or treelet in the absence of fire; ref. 41) is a further indication of the potential ease of the evolutionary transitions associated with fire adaptation. We suggest that the ease of fire adaptation has been significant in generating high species diversity in the Cerrado, although the ingress of a lineage into this fire-prone habitat has not necessarily led to significant or rapid in situ diversification. The 10 largest genera (1,474 spp.) make up only 13% of the Cerrado flora (26), and many of the dominant Cerrado tree genera (e.g., Qualea and Caryocar) are represented by only few species.
Several authors have pointed to affinities between the woody flora of the Cerrado and the Atlantic and Amazon rain forests (32, 42–44). For example, of 121 woody species dominant in the Brazilian Cerrado, 99 belong to predominantly rain forest genera (6). These affinities are borne out by wet forest–Cerrado transitions implied by the phylogeny of Andira (SI Appendix) and undoubtedly many other groups not sampled here. However, the sister groups and putative ancestral areas and ecologies of Cerrado lineages identified in our phylogenies are diverse and include geographically adjacent seasonally dry tropical forests, subtropical grasslands, and wetlands, as well as wet forests (Table 1 and SI Appendix). In seven of 15 lineages, ancestral areas were recorded as equivocal, but even for these, the options with the highest probabilities frequently included dry and wet forest. These results agree with floristic data that suggest that neotropical savannas comprise a mixture of elements of various provenance and floristic affinities (42). The Cerrado is bordered by a diverse array of biomes (Fig. 1) including the Amazon and Atlantic wet forests, tropical and subtropical dry thorn scrub (Caatinga and Chaco), subtropical grasslands, and wetlands. The proximity of these diverse vegetation types that have all contributed to the recruitment of Cerrado lineages suggests that this privileged position in the heart of South America may have played a role in the Cerrado's striking species richness.
The in situ assembly of the endemic rich Cerrado flora via frequent recent adaptive shifts to resist fire stands in contrast to the widespread support for ideas that lineages tend to maintain their ancestral ecologies (8, 11, 13) and that dispersal of preadapted lineages has played an important role in the assembly of regional species pools (8). For example, the species-rich páramo biome in the Andes includes many northern, temperate plant lineages that were able to disperse and to take advantage of similar ecological conditions to diversify rapidly in the Andes (8, 15, 45). Similarly, many seasonally dry tropical forest clades show evidence of phylogenetic niche conservatism among disjunct areas across the tropics (16, 18). In these cases, it has apparently been easier to switch geography than ecology (8). However, this does not appear to be the case for the Cerrado, where niche evolution from adjacent closed, fire-free habitats to open, fire-prone habitats, cross-cutting other abiotic niche dimensions, has apparently been prevalent in the evolution of the Cerrado flora.
This study is an initial attempt to assemble a representative set of time-calibrated phylogenies for Cerrado plants. The lineages discovered represent just 3–4% of the Cerrado flora. Despite this sparse sampling, there are indications that our results are representative of the flora as a whole. First, the study groups include species-rich and species-poor lineages as well as diverse life forms. Second, the preponderance of species, as opposed to generic endemism and prevalence of fire adaptations amongst these endemics, support the idea that Cerrado lineages are derived and recent. Finally, preliminary phylogenies for other plant groups including Styrax (46), Viguiera (47), Ruellia (48), and Manihot (49), also show evidence of recently derived Cerrado lineages, multiple independent Cerrado lineages (in Ruellia, Manihot, and Styrax) and associated fire adaptations (e.g., xylopodia in Ruellia and Viguiera) (SI Appendix).
The emerging picture of Cerrado origins is of recent diversification of endemic plant lineages that took place during the late Miocene and early Pliocene, driven by the common trigger of fire adaptation and facilitated by ease of fire adaptation across plant groups from the diverse biomes immediately surrounding the Cerrado. This picture is very different from what is known about the origins and diversification of plants in one of the best-studied model hotspots, the Cape Floristic Province in South Africa, where plant lineages are estimated to range from less than 5–46 Mya, with no evidence for simultaneous initiation driven by a single trigger (20, 50). Linder (9) classified these older Cape lineages as mature radiations, in which stability over time has produced numerous endemic genera. In contrast, the Cerrado seems to be the product of more recent diversification with very few endemic genera. These contrasting scenarios suggest that the origins of species-rich biomes and underlying causes of high species diversity can be highly idiosyncratic, driven as much by unique features of regional- and continental-scale history and physiography as by any more universal ecological processes at more local scales (4, 9, 10). If species-rich biomes do indeed have very different histories, then understanding the differences between biomes, and hence the mechanisms responsible for the origin and maintenance of the variation in species richness over the globe, will be crucial for the interpretation of global biodiversity gradients across space (e.g., refs. 8 and 45) and time (e.g., refs. 51 and 52).
Methods
Datasets and Sampling.
Our sample of plant lineages was constrained by the paucity of species-level phylogenies that include Cerrado elements. Of the published phylogenies for plant groups that include Cerrado species, Styrax (Styracaceae) (46), Viguiera (Asteraceae) (47), Microlicieae (Melastomataceae) (40), Ruellia (Acanthaceae) (48), Lupinus (Leguminosae) (15), and Manihot (Euphorbiaceae) (49), only two (Lupinus and Microlicieae) were judged to be well enough sampled, resolved and supported, and amenable to time calibration for more intensive analysis of Cerrado origins (SI Appendix). New DNA sequence data were generated for two legume genera, Mimosa and Andira. Our largest dataset is for the species-rich genus Mimosa (chloroplast region trnD-trnT), for which ≈50% of the >500 species were sampled, including 92 of 189 species listed for the Cerrado (39), most of these being narrow, fire-adapted endemics. For Andira, a nearly fully sampled dataset of ITS sequences was assembled, including multiple accessions for some species and all four species that grow in the Cerrado. The Lupinus dataset used here (15) comprises ITS and LEGCYC1A nuclear DNA sequences for 98 species, including five of the 11 Cerrado endemics. Data for Microlicieae include sequences for three plastid loci (rbcL, rpl16, and ndhF) and 59 species. The published Microlicieae data (40) were reanalyzed with 23 additional taxa, for which only one data partition (rpl16) was available, to increase taxon sampling and accuracy of divergence time estimates. Laboratory methods and taxa sampled are presented in the SI Appendix.
Phylogenetic Analyses and Dating.
Phylogenetic trees and divergence times for Cerrado nodes were estimated by using an uncorrelated relaxed molecular clock approach implemented in BEAST, version 1.4.8 (53). All analyses were conducted by using the uncorrelated lognormal relaxed clock, assuming a general time-reversible model, with invariable sites and among-site rate heterogeneity (GTR+I+Γ). Results were assessed to have reached stationarity and convergence by using Tracer, version 1.4 (54), and data from multiple runs were combined after exclusion of burn-in trees.
Divergence-time estimates for the legume genera, Mimosa, Andira, and Lupinus were based on a new family-wide analysis of matK data containing 839 terminals. This large dataset was assembled from previous alignments (55, 56) with the incorporation of 101 additional mimosoid legume sequences (see ref. 57 and additional refs. in the SI Appendix), thereby increasing taxon sampling well beyond previous legume studies. The dating procedure was built upon previous studies (55, 56) that used a set of well-documented fossils for calibration but with the addition of two new mimosoid fossil constraints. In total, 23 fossils were used as minimum age constraints (lognormal prior distribution, mean = 0, stdev = 1, in BEAST) (SI Appendix). The legume stem node was allowed to vary between 60 and 70 Mya (uniform prior distribution). Twenty individual chains of 107 generations were run, and after the exclusion of 2 × 106 generations (burn-in from each chain), the results were combined. Age estimates for five nodes (mean and 95% credibility intervals) from this analysis were used as calibration points for subsequent species-level analyses of Mimosa, Andira, and Lupinus by using a normal distribution prior, with mean and standard deviation set to the corresponding estimate.
For the Microlicieae dataset, three fossils provided minimum age constraints (40): (i) the oldest Melastomataceae fossil (53 Mya, Melastomataceae crown node), (ii) the oldest tribe Melastomeae fossil (23 Mya, Melastomeae crown node), and (iii) a 30-Mya fossil leaf assigned to the Merianieae crown node. These constraints were imposed as lognormal distributions as described above. The age of the core Microlicieae was previously estimated to be 3.7 Mya (40), which is considerably younger than the divergence time reported here (9.8 Mya). This discrepancy can be attributed to the denser taxon sampling used here, given the known bias toward younger age estimates caused by undersampling (58).
Analyses of the four study group datasets comprised three independent runs of 107 generations each, which all converged to the same posterior and were combined, resulting in final aggregates of 2.7 × 107 generations (after exclusion of burn-in trees). Fully annotated phylogenies, including terminal taxon names are presented for each study group (SI Appendix).
Ancestral Biome Reconstruction and the Evolution of Fire Adaptation.
Ancestral character reconstruction analysis was used to identify Cerrado lineages, infer their putative ancestral biomes, and investigate the evolution of fire adaptations. Ecologies or “biomes” of occurrence (seasonally dry tropical forest, rain forest, wetland, savanna, subtropical grassland, temperate, Mediterranean, tropical montane, widespread) for sampled species were assigned based on species distribution data from taxonomic accounts (SI Appendix) and field observations, coded as discrete character states and optimized under a maximum parsimony criterion (“Unordered”) onto the 50% majority rule consensus tree by using Mesquite (59). To account for topological uncertainty, the procedure “Trace over trees” was used to summarize reconstructions over a set of 540 Bayesian trees sampled at stationarity. This approach was used to define Cerrado lineages by identifying crown nodes assigned unequivocally to the “savanna” biome. The same procedure was used to identify ancestral biomes for Cerrado lineages as the most frequent state at the stem nodes subtending each clade. Estimating ages of Cerrado lineages by using stem, rather than crown, nodes generates estimates 2.5 Mya older on average and therefore does not alter the conclusions of the study. Optimizations were also performed by using a maximum-likelihood approach in Mesquite under the Mk1 model, as used in biome reconstructions in other studies (13), but this alternative approach did not alter the results.
The evolution of morphological traits related to fire was investigated in a similar way. The presence/absence of thick corky bark in Andira and the evolution of fire-adapted life forms in Andira and Mimosa were analyzed. For these two genera, species were coded for life form (herb, shrub, vine/liana, functionally herbaceous subshrub with xylopodium, woody shrub with xylopodium, pachycaul treelet, tree), and optimized as described above. We used a comparative analysis of discrete characters (60) to test the correlation between the evolution of fire-adapted life forms and Cerrado occurrence in Mimosa. This method (implemented in Mesquite) searches for evidence of correlated evolutionary change in two discrete characters by using a likelihood ratio test statistic to discriminate between two models fitted to the data—one for independent evolution of the two characters and the other allowing for correlated evolution. We ran 100 Monte Carlo simulations to test if the likelihoods of the two models are statistically different. Data matrices for biome of occurrence and morphology, along with character-state optimizations and detailed methods, are presented in SI Appendix.
Supplementary Material
Acknowledgments.
We thank Euan James, the Millenium Seed Bank Project, Sergio de Faria, Valquíria Dutra, Maria Schifino–Wittmann, Nair Dahmer, Ruth Eastwood, Joan Sutherland, Tiina Särkinen, Gwilym Lewis, Gerardo Vasconcelos, Carolyn Proença, Stephen Harris, Angélica Martínez, Sara Camargo, Janaína Nascimento, Terry Pennington, Melissa Luckow, John Wood, Carlos Reynel, Aniceto Daza, Rob Forrest, Margoth Atahuachi and Chris Fagg for help with field collections or provision of plant material, Anne Bruneau for part of the matK alignment, Tiina Särkinen and Greg Kenicer for unpublished matK sequences, David Spence and Neil Caithness, Oxford University E-Science Centre for help running BEAST on the National Computing Grid, Matt Lavin and two reviewers for comments on the manuscript, and the authorities in Peru, Bolivia, Mexico, and Brazil (IBAMA authorization project 02001.007621/2005) for permission to collect plant material. M.F.S. was supported by the Clarendon Fund, Wolfson College, and the Systematics Association during his PhD at the University of Oxford, C.S. was supported by the British Schools and Universities Foundation during her MSc at the Royal Botanic Garden Edinburgh, and C.E.H. was supported by the Royal Society.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. FJ542758-FJ542808 and FJ981975-FJ982245).
This article contains supporting information online at www.pnas.org/cgi/content/full/0903410106/DCSupplemental.
References
- 1.Myers N, Mittermeier RA, Mittermeier CG, Fonseca GAB, Kent J. Biodiversity hotspots for conservation priorities. Nature. 2000;403:853–858. doi: 10.1038/35002501. [DOI] [PubMed] [Google Scholar]
- 2.Lamoreux JF, et al. Global tests of biodiversity concordance and the importance of endemism. Nature. 2006;440:212–214. doi: 10.1038/nature04291. [DOI] [PubMed] [Google Scholar]
- 3.Gaston KJ. Global patterns in biodiversity. Nature. 2000;405:220–227. doi: 10.1038/35012228. [DOI] [PubMed] [Google Scholar]
- 4.Ricklefs RE. Evolutionary diversification and the origin of the diversity-environment relationship. Ecology. 2006;87:S3–S13. doi: 10.1890/0012-9658(2006)87[3:edatoo]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 5.Pennington RT, Cronk QCB, Richardson JA. Introduction and synthesis: Plant phylogeny and the origin of major biomes. Philos Trans R Soc London Ser B. 2004;359:1455–1464. doi: 10.1098/rstb.2004.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pennington RT, Richardson JE, Lavin M. Insights into the historical construction of species-rich biomes from dated plant phylogenies, neutral ecological theory and phylogenetic community structure. New Phytol. 2006;172:605–616. doi: 10.1111/j.1469-8137.2006.01902.x. [DOI] [PubMed] [Google Scholar]
- 7.Kreft H, Jetz W. Global patterns and determinants of vascular plant diversity. Proc Natl Acad Sci USA. 2007;104:5925–5930. doi: 10.1073/pnas.0608361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donoghue MJ. A phylogenetic perspective on the distribution of plant diversity. Proc Natl Acad Sci USA. 2008;105:11549–11555. doi: 10.1073/pnas.0801962105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Linder HP. Plant species radiations: where, when, why? Philos Trans R Soc London Ser B. 2008;363:3097–3105. doi: 10.1098/rstb.2008.0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qian H, Ricklefs RE. Large-scale processes and the Asian bias in species diversity of temperate plants. Nature. 2000;407:180–182. doi: 10.1038/35025052. [DOI] [PubMed] [Google Scholar]
- 11.Wiens JJ, Donoghue MJ. Historical biogeography, ecology and species richness. Trends Ecol Evol. 2004;19:639–644. doi: 10.1016/j.tree.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 12.Sauquet H, et al. Contrasted patterns of hyperdiversification in Mediterranean hotspots. Proc Natl Acad Sci USA. 2009;106:221–225. doi: 10.1073/pnas.0805607106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crisp MD, et al. Phylogenetic biome conservatism on a global scale. Nature. 2009;458:754–756. doi: 10.1038/nature07764. [DOI] [PubMed] [Google Scholar]
- 14.Ricklefs RE, Schwarzbach AE, Renner SS. Rate of lineage origin explains the diversity anomaly in the world's mangrove vegetation. Am Nat. 2006;168:805–810. doi: 10.1086/508711. [DOI] [PubMed] [Google Scholar]
- 15.Hughes C, Eastwood R. Island radiation on a continental scale: Exceptional rates of plant diversification after uplift of the Andes. Proc Natl Acad Sci USA. 2006;103:10334–10339. doi: 10.1073/pnas.0601928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lavin M, et al. Metacommunity process rather than continental tectonic history better explains geographically structured phylogenies in legumes. Philos Trans R Soc London Ser B. 2004;359:1509–1522. doi: 10.1098/rstb.2004.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pennington RT, Lavin M, Oliveira–Filho A. Woody plant diversity, evolution and ecology in the tropics: Perspectives from seasonally dry tropical forests. Ann Rev Ecol Evol Syst. 2009;40:437–457. [Google Scholar]
- 18.Schrire BD, Lavin M, Lewis GP. Global distribution patterns of the Leguminosae: Insights from recent phylogenies. Biol Skr. 2005;55:375–422. [Google Scholar]
- 19.Davis CC, Webb CO, Wurdack KJ, Jaramillo CA, Donoghue MJ. Explosive radiation of Malpighiales supports a mid-Cretaceous origin of modern tropical rain forests. Am Nat. 2005;165:E36–E65. doi: 10.1086/428296. [DOI] [PubMed] [Google Scholar]
- 20.Linder HP. Evolution of diversity: The Cape flora. Trends Plants Sci. 2005;10:536–541. doi: 10.1016/j.tplants.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 21.Becerra JX. Timing the origin and expansion of the Mexican tropical dry forest. Proc Natl Acad Sci USA. 2005;102:10919–10923. doi: 10.1073/pnas.0409127102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoder AD, Nowak MD. Has vicariance or dispersal been the predominant biogeographic force in Madagascar? Only time will tell. Ann Rev Ecol Evol Syst. 2006;37:405–431. [Google Scholar]
- 23.Pennington RT, et al. Historical climate change and speciation: Neotropical seasonally dry forest plants show patterns of both Tertiary and Quaternary diversification. Philos Trans R Soc London Ser B. 2004;359:515–537. doi: 10.1098/rstb.2003.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hawkins JA. Using phylogeny to investigate the origins of the Cape flora: The importance of taxonomic, gene and genome sampling strategies. Divers Distrib. 2006;12:27–33. [Google Scholar]
- 25.Kier G, et al. Global patterns of plant diversity and floristic knowledge. J Biogeog. 2005;32:1107–1116. [Google Scholar]
- 26.Mendonça RC, et al. Vascular flora of the Cerrado biome: Checklist with 12,356 species. In: Sano SM, Almeida SP, Ribeiro JF, editors. Cerrado: Ecology and Flora. Vol 2. Brasília, Brazil: Embrapa Cerrados/Embrapa Informação Tecnológica; 2008. pp. 421–1279. [Google Scholar]
- 27.Ratter JA, Ribeiro JF, Bridgewater S. The Brazilian Cerrado vegetation and threats to its biodiversity. Ann Bot. 1997;80:223–230. [Google Scholar]
- 28.Ledru MP. Late Quaternary history and evolution of the Cerrado as revealed by palynological records. In: Oliveira PS, Marquis RJ, editors. The Cerrados of Brazil. New York: Columbia Univ Press; 2002. pp. 33–50. [Google Scholar]
- 29.Pennington RT, Lewis GP, Ratter JA. An overview of the plant diversity, biogeography and conservation of neotropical savannas and seasonally dry forests. In: Pennington RT, Lewis GP, Ratter JA, editors. Neotropical Savannas and Seasonally Dry Forests: Plant Diversity, Biogeography, and Conservation. Boca Raton, FL: CRC Press; 2006. pp. 1–29. [Google Scholar]
- 30.Scholes RJ, Archer SR. Tree-grass interactions in savannas. Ann Rev Ecol Syst. 1997;28:517–544. [Google Scholar]
- 31.Bond WJ, Keeley JE. Fire as a global ‘herbivore’: The ecology and evolution of flammable ecosystems. Trends Ecol Evol. 2005;20:387–394. doi: 10.1016/j.tree.2005.04.025. [DOI] [PubMed] [Google Scholar]
- 32.Gottsberger G, Silberbauer–Gottsberger I. Life in the Cerrado, a South American Tropical Seasonal Ecosystem. Vol. 1. Origin, Structure, Dynamics and Plant Use. Ulm, Germany: Reta Verlag; 2006. [Google Scholar]
- 33.Eiten G. The Cerrado vegetation of Brazil. Bot Rev. 1972;38:201–341. [Google Scholar]
- 34.Christin PA, et al. Oligocene CO2 decline promoted C-4 photosynthesis in grasses. Curr Biol. 2008;18:37–43. doi: 10.1016/j.cub.2007.11.058. [DOI] [PubMed] [Google Scholar]
- 35.Cerling TE, et al. Global vegetation change through the Miocene/Pliocene boundary. Nature. 1997;389:153–158. [Google Scholar]
- 36.Latorre C, Quade J, McIntosh WC. The expansion of C-4 grasses and global change in the late Miocene: Stable isotope evidence from the Americas. Earth Planet Sci Lett. 1997;146:83–96. [Google Scholar]
- 37.Jacobs BF, Kingston JD, Jacobs LL. The origin of grass-dominated ecosystems. Ann Missouri Bot Gard. 1999;86:590–643. [Google Scholar]
- 38.Beerling DJ, Osborne CP. The origin of the savanna biome. Glob Change Biol. 2006;12:2023–2031. [Google Scholar]
- 39.Simon MF, Proença C. Phytogeographic patterns of Mimosa (Mimosoideae, Leguminosae) in the Cerrado biome of Brazil: An indicator genus of high-altitude centers of endemism? Biol Conserv. 2000;96:279–296. [Google Scholar]
- 40.Fritsch PW, Almeda F, Renner SS, Martins AB, Cruz BC. Phylogeny and circumscription of the near-endemic Brazilian tribe Microlicieae (Melastomataceae) Am J Bot. 2004;91:1105–1114. doi: 10.3732/ajb.91.7.1105. [DOI] [PubMed] [Google Scholar]
- 41.Pennington RT. Monograph of Andira (Leguminosae-Papilionoideae) Systematic Botany Monographs (Am Soc Plant Taxonomists, Ann Arbor) 2003;64:1–143. [Google Scholar]
- 42.Sarmiento G. The savannas of tropical America. In: Bourlière F, editor. Ecosystems of the World: Tropical Savannas. Amsterdam, Netherlands: Elsevier; 1983. pp. 245–288. [Google Scholar]
- 43.Prance GT. The phytogeography of savanna species of neotropical Chrysobalanaceae. In: Furley PA, Proctor J, Ratter JA, editors. Nature and dynamics of forest-savanna boundaries. New York, USA: Chapman & Hall; 1992. pp. 295–330. [Google Scholar]
- 44.Oliveira–Filho AT, Fontes MAL. Patterns of floristic differentiation among Atlantic forests in southeastern Brazil and the influence of climate. Biotropica. 2000;32:793–810. [Google Scholar]
- 45.Moore BR, Donoghue MJ. Correlates of diversification in the plant clade Dipsacales: Geographic movement and evolutionary innovations. Am Nat. 2007;170:S28–S55. doi: 10.1086/519460. [DOI] [PubMed] [Google Scholar]
- 46.Fritsch PW. Phylogeny and biogeography of the flowering plant genus Styrax (Styracaceae) based on chloroplast DNA restriction sites and DNA sequences of the internal transcribed spacer region. Mol Phylogenet Evol. 2001;19:387–408. doi: 10.1006/mpev.2001.0933. [DOI] [PubMed] [Google Scholar]
- 47.Schilling EE, Da Costa FB, Lopes NP, Heise PJ. Brazilian species of Viguiera (Asteraceae) exhibit low levels of ITS sequence variation. Edinburgh J Bot. 2000;57:323–332. [Google Scholar]
- 48.Tripp EA. Evolutionary relationships within the species-rich genus Ruellia (Acanthaceae) Syst Bot. 2007;32:628–649. [Google Scholar]
- 49.Chacón J, Madriñán S, Debouck D, Rodriguez F, Tohme J. Phylogenetic patterns in the genus Manihot (Euphorbiaceae) inferred from analyses of nuclear and chloroplast DNA regions. Mol Phylogenet Evol. 2008;49:260–267. doi: 10.1016/j.ympev.2008.07.015. [DOI] [PubMed] [Google Scholar]
- 50.Edwards D, Hawkins JA. Are Cape floral clades the same age? Contemporaneous origins of two lineages in the genistoids s. l. (Fabaceae) Mol Phylogenet Evol. 2007;45:952–970. doi: 10.1016/j.ympev.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 51.McKenna DD, Farrell BD. Tropical forests are both evolutionary cradles and museums of leaf beetle diversity. Proc Natl Acad Sci USA. 2006;103:10947–10951. doi: 10.1073/pnas.0602712103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jaramillo C, Rueda MJ, Mora G. Cenozoic plant diversity in the Neotropics. Science. 2006;311:1893–1896. doi: 10.1126/science.1121380. [DOI] [PubMed] [Google Scholar]
- 53.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rambaut A, Drummond AJ. Tracer v1.4. 2007. [Accessed June 15, 2008]. http://tree.bio.ed.ac.uk/software/tracer/
- 55.Lavin M, Herendeen PS, Wojciechowski MF. Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the Tertiary. Syst Biol. 2005;54:575–594. doi: 10.1080/10635150590947131. [DOI] [PubMed] [Google Scholar]
- 56.Bruneau A, Mercure M, Lewis GP, Herendeen PS. Phylogenetic patterns and diversification in the caesalpinioid legumes. Botany. 2008;86:697–718. [Google Scholar]
- 57.Luckow M, Miller JT, Murphy DJ, Livshultz T. A phylogenetic analysis of the Mimosoideae (Leguminosae) based on chloroplast DNA sequence data. In: Klitgaard BB, Bruneau A, editors. Advances in Legume Systematics, part 10: High Level Systematics. Kew, United Kingdom: Royal Botanic Gardens; 2003. pp. 197–220. [Google Scholar]
- 58.Linder HP, Hardy CR, Rutschmann F. Taxon sampling effects in molecular clock dating: An example from the African Restionaceae. Mol Phylogenet Evol. 2005;35:569–582. doi: 10.1016/j.ympev.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 59.Maddison WP, Maddison DR. Mesquite: A modular system for evolutionary analysis. 2007. [Accessed April 20, 2008]. Version 2.01, Available at: http://mesquiteproject.org.
- 60.Pagel M. Detecting correlated evolution on phylogenies: A general method for the comparative-analysis of discrete characters. Proc R Soc London Ser B. 1994;255:37–45. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.