Abstract
Genetic isolates have been successfully used in the study of complex traits, mainly because due to their features, they allow a reduction in the complexity of the genetic models underlying the trait. The aim of the present study is to describe the population of Campora, a village in the South of Italy, highlighting its properties of a genetic isolate. Both historical evidence and multi-locus genetic data (genomic and mitochondrial DNA polymorphisms) have been taken into account in the analyses. The extension of linkage disequilibrium (LD) regions has been evaluated on autosomes and on a region of the X chromosome. We defined a study sample population on the basis of the genealogy and exogamy data. We found in this population a few different mitochondrial and Y chromosome haplotypes and we ascertained that, similarly to other isolated populations, in Campora LD extends over wider region compared to large and genetically heterogeneous populations. These findings indicate a conspicuous genetic homogeneity in the genome. Finally, we found evidence for a recent population bottleneck that we propose to interpret as a demographic crisis determined by the plague of the 17th century. Overall our findings demonstrate that Campora displays the genetic characteristics of a young isolate.
Key Words: Genetic isolates, Genealogy, Haplotype analysis, Bottleneck
Introduction
Recently, there has been wide-spread discussion about the use of isolated human populations for the identification of genes responsible for complex traits [1, 2]. The usefulness of isolated populations in these studies has been widely shown [3,4,5,6,7,8,9]. Isolated populations originate from a restricted number of founders due either to a past migration event or to a past reduction in the population size (e.g. a bottleneck). Because of this founder effect and limited gene flow, it is possible that fewer risk alleles might underlie complex disorders in those populations. Moreover, the effect of genetic drift can be considerable [10, 11], causing an increase in the frequency and the attributable risk of particular alleles. The presence of inbreeding and extended genomic regions in linkage disequilibrium (LD), together with previous mentioned factors, contribute to make the genetic background extremely homogeneous. Such genetic homogeneity is a great advantage in the initial approach to gene mapping, even though it could be a disadvantage in the subsequent step of refining the identified genomic regions.
Members of isolated populations share a common environment and a very similar life-style, thus the environmental diversity is greatly reduced. The availability of extensive genealogical records can provide large genealogies, potentially highly informative for linkage analysis. Therefore, in these genetically and culturally homogeneous populations, a large proportion of individuals presenting a given trait are likely to share the same trait-predisposing gene inherited from a common ancestor. Finally, additional features such as the presence of extensive genealogical records, and possibility of standardized phenotypes [12] enhance the value of these populations for the studies of complex traits.
The main features of isolated populations have been extensively reviewed [13]. It is clear that every isolated population carries the signs of its own demographic history. Knowledge of the underlying population structure is essential to design studies for gene identification and the choice of statistical methods critically depends on features of the population [14, 15] such as the degree of isolation (ranging from ‘extreme’ to ‘mild’), the length of time that the population has remained isolated and the size of the founding nucleus. In the current literature about isolated populations, such demographic characteristics have been primarily evaluated for those representing extreme cases of isolation, such as the Amish or the Hutterites. On the other hand, the structure of only a handful of ‘mild’ isolated populations has been characterized, although a number of such isolates have been identified [16,17,18].
Here we describe the population in the village of Campora in South Italy, which suffered a bottleneck in the 17th century and has remained geographically isolated until the last century. In this population we have recently identified a locus associated with hypertension [9]. Moreover, our preliminary data indicate that linkage studies in the Campora population will also be a powerful tool to detect QTLs. In this paper, we trace the genetic history of Campora and establish its degree of isolation, applying both genealogy-based and genetic-based strategies. Our analysis provides a description of the Campora population as a model of a mild genetic isolate.
Historical Background
The area that today corresponds to the National Park of ‘Cilento and Vallo di Diano’, within which Campora is located (see map in fig. 1), was originally occupied by Greeks during the 8th century BC. In the middle of the 5th century BC, the Lucanians conquered the internal area without reaching the coast. Subsequently Lucanians were chased from this territory by the Greeks. The community of Campora was already present at the time of Lucanians but no further historical information is available until the 11th century [19].
Fig. 1.
Geographical location of Campora. The village is part of the National Park of ‘Cilento e Vallo di Diano’ of the region Campania, in the South of Italy.
In the 8th century, groups of monks coming from the Byzantine Empire reached the coast of the area that today corresponds to the park. In the 10th century, the monks were forced to move to the internal hilly region to elude the coastal invasions of Saraceni coming from the middle East. Once there, the monks organized groups of local people into villages. Among those was Campora, for which the arrival of monks has been dated at the beginning of the 11th century.
Presumably, the first nucleus of inhabitants was made of individuals of Greek and Lucanian origin employed in the agricultural activities of the monastery [19]. Subsequently, despite different dominations affecting the village after its foundation, none of them contributed to the population in terms of individuals. In the second half of the 16th century, there was a general scarcity of food in the area surrounding Campora. The famine lasted about one century and was followed by a severe epidemic of bubonic plague. The first registered case of plague in the area of Campora dates to the year 1656 in the nearby village of Novi Velia.
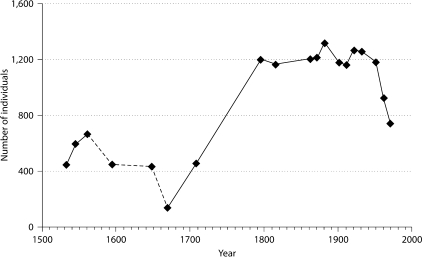
According to historical sources and owing to its geographical position, Campora experienced isolation from its foundation until the end of the World War II. A first wave of emigrants went to America at the end of 19th century while a second one, mainly directed to big cities in Italy, moved after World War II and is still occurring (fig. 2). The first wave was compensated by a high birth rate. However since the second half of the 20th century, births have decreased (data not shown) while emigration has been constant and has gradually reduced the number of individuals currently living in the village from 1,300 in 1880 to only 500 at present.
Fig. 2.
Population trend of Campora according to historical and civil census data from the 16th century to recent time. A strong reduction in population size is evident across one century (dashed line) since the middle of 16th century (due to famine and the 1656 plague epidemic) followed by a period of population expansion. It is also possible to notice a minor reduction at the end of the 19th century most likely corresponding to the first migration wave. Finally, since the beginning of 20th century migration diminished the number of individuals currently living in the village to 500 (not shown).
Subjects and Methods
Subjects and Genealogy
Extensive genealogical data from the 16th century up to the present day have been collected by consulting the Registry Office and the Parish archives. Additional information about emigrated people was obtained by directly querying inhabitants about their relatives. Comparison and integration of this information led to the accumulation of 10,737 individual records of which 1,719 are of living individuals.
Demographic data on Campora from the 16th to 18th century result from ‘stati delle anime’, a type of religious census present at that time. For later centuries, data derive from civil census. Exogamic marriages were counted from the registries of the Parish.
Matrilinear and patrilinear genealogical lines (GLs) were built by scanning the pedigree with a Perl algorithm to connect each individual with the parent of the same sex, proceeding until no further connections were possible.
The mean generation time (MGT) was calculated as the average age of individuals at the birth of his/her children. Individuals in the whole genealogical dataset were considered. We found a value of MGT = 32.5 ± 8.0 years (females MGT = 30.9 ± 5.5 years; males MGT = 34.1 ± 5.8 years).
All individuals participating in the study, recruited among both resident and immigrants, signed an informed consent in accordance with the Declaration of Helsinki (World Medical Association). The study was approved by the Ethics Committee of Azienda Sanitaria Locale Napoli 1.
DNA Preparation and Genotyping of Microsatellite Markers
Genomic DNA was extracted from 10 ml of peripheral blood using a Flexigene kit (Qiagen) following the manufacturer's instructions. Genotyping at 1,122 autosomal microsatellites (average marker spacing of 3.6 cM and mean marker heterozygosity of 0.70) was performed by deCODE genotyping service on 584 individuals. Mendelian inheritance inconsistencies were identified using the Pedcheck program [20].
On the Y chromosome, a set of seven microsatellites (DYS19, DYS385, DYS389, DYS390, DYS391, DYS392, DYS393) was analyzed. Primer sequences were obtained from the Genome Database (http://www.gdb.org). Polymerase chain reaction (PCR) cycling conditions were 95°C for 10 min, then thirty cycles of 95°C for 30″, annealing at 55°C (for DYS19, DYS390, DYS392, DYS393) or 57°C (for DYS391) or 62°C (for DYS385, DYS389) for 30′, then synthesis at 72°C for 30′, and finally 72°C for 7′. PCR products were loaded on a MegaBACE1000Flexi (Amersham) and genotype data were analyzed using Fragment Profiler software.
Six microsatellites in the Xq13 region (DXS983, DXS8092, DXS8037, DXS1225, DXS8082, DXS986) were considered for Linkage Disequilibrium (LD) analysis testing. Primer sequences were obtained from the Genome Database. PCR cycling conditions were 94°C for 2′, then 94°C for 30′, 60°C to 65°C (−0,5°C/cycle) for 30′, then 72°C for 30′ over 15 cycles; 96°C for 15′, 65°C for 30′, 72 °C for 30′ over 20 cycles and finally 72°C for 4′. PCR products were loaded on a MegaBACE1000Flexi (Amersham) and genotype data were analyzed using Fragment Profiler software.
mtDNA Analysis
Seven fragments were amplified from mtDNA. For each fragment, the position of the first base of the primer on the light (L) strand and on the heavy (H) strand, according to the reference sequence [21, 22] is: L15996-H16401; L1643-H1874; L6909-H7115; L8845-H9163; L10290-H10557; L9932-H10088; L15428-H15682. Haplogroups of mtDNA [23, 24] were determined through the analysis of restriction polymorphisms (in brackets) as follow: H (−10394 DdeI; −7025 AluI); T (−10394 DdeI; −15925 MspI; +15606 AluI; +13366BamHI); U (−10394 DdeI; +12308 Hin); V (−10394 DdeI; −4577 NlaIII); W (−10394 DdeI; −8994 HaeIII; +8249 AvaII); X (−10394 DdeI; −1715DdeI); I (−1715DdeI; −4529 HaeII; +8249 AvaII; +10028 AluI; +16389 BamHI); K (+12308 HinfI; −9052 HaeII); J (−16065 HinfI; −13704 BstNI); M (+10397 AluI); L (+3592 HpaI); preH (−10394 DdeI; +7025 AluI; +16517 HaeIII). Enzymatic reactions were carried out at 37°C for 90′ in a reaction volume of 20 μl using 6–8 μl purified PCR product/reaction.
Polymorphisms in the Hypervariable Region I (HVR-I) were determined by sequencing from nucleotide 15940 to 16383. Sequencing was done using BigDye Terminator Cycle Sequencing Ready Reaction (Applied Biosystems, Warrington, UK) and loaded on an ABI PRISM 377 DNA analyzer (PE Biosystems). Sequences were analysed using AutoAssembler software (Applied Biosystems, Warrington, UK).
Statistical Analyses
Coefficients of inbreeding (f) were evaluated from the genealogy using two different algorithms: the one proposed by Karigl [25] implemented in the KinInbCoef [26] (http://galton.uchicago.edu/∼mcpeek/software/CCtests) and the Stevens-Boyce algorithm [27] implemented in the KINSHIP module of the PEDSYS software (http://www.sfbr.org/pedsys/pedsys.html).
The Fisher test associated p value for the evaluation of LD on the X chromosome was determined using the Haploxt module in the GOLD package [28] (http://www.sph.umich.edu/csg/abecasis/GOLD/) in a sample of 63 unrelated males.
To assess disequilibrium between alleles from autosomal markers, we inferred haplotypes using Merlin (http://www.sph.umich.edu/csg/abecasis/Merlin/). We manage to infer the haplotype of 635 individuals, belonging to a 2,947-member pedigree. Among those 635 individuals, we chose 73 whose coefficient of kinship was <0.0625 (first cousin) using KinSamp, an algorithm that we developed that take into account the kinship matrix obtained by KinInbCoef to choose a sample of individuals allowing a degree of kinship defined by the user. We analyzed pairwise disequilibrium on haplotype data using the software miLD-2.1 (http://www.geneticepi.com/Research/software/software.html), which implements the calculation of a corrected D′ value (Dadj) [29]. Dadj is based on the traditional Lewontin's multiallelic measure of LD, the multiallelic D′, but it is corrected in order to minimize the effect of sample size and allele frequencies, allowing the comparison between samples with different sizes. The miLD-2.1 software also allows the estimation of the significance of LD through the MLD programme [30].
Intermarker distances were established on the basis of the DeCode sex-averaged maps using the Haldane map function.
Temporary excess of heterozygosity compared to the expected one in relation to the number of alleles at each locus was tested using BOTTLENECK [31] (http://www.montpellier.inra.fr/URLB/bottleneck). The 1,072 autosomal microsatellites available were tested for Hardy-Weinberg (HW) equilibrium using a test analogous to Fisher's exact test implemented in the Arlequin package (http://cmpg.unibe.ch/software/arlequin3/). HW equilibrium was tested in a sample of 80 individuals (assembled with KinSamp) whose coefficient of kinship was <0.0625 to avoid inference of relatedness in the calculation [32]. HW equilibrium was ascertained for 1,012 microsatellites that were grouped in 5 datasets and used in the successive calculations. Average marker spacing in each dataset is 17.5 cM. This marker spacing avoids overrepresentation in genomic regions that are in linkage disequilibrium, as recommended by the authors of BOTTLENECK. Allele frequencies at microsatellite loci were estimated using the BLUE estimator [33] and used as input for BOTTLENECK. The analysis was carried out under the Two-phased Model of Mutation (TPM) as model of microsatellites evolution. In the software the TPM model combines the Stepwise Mutation Model (SMM) and the Infinite Allele Model (IAM) in a percentage defined by the user. For the former model, the heterozygosity excess after a bottleneck has been demonstrated to be present for a consistent period of time [34], while under the SMM model the decline of heterozygosity is more rapid and thus not detectable by the software. Although the SMM model is considered to more faithfully represent the true process of evolution of microsatellites compared to the IAM model [35], most microsatellite data sets fit the TPM more better than the SMM or IAM model [36]. In our case, the SMM component in the TPM model was set to 90%.
Results
Identification of the Study Sample in the Population of Campora
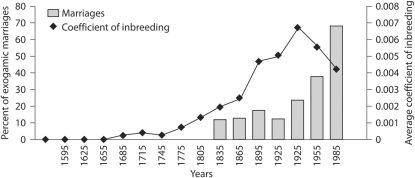
Using data of the parish archive from the 19th century to the present, we estimated the percentage of exogamic marriages (marriages with individuals from different villages) per generation (fig. 3), considering a mean generation time of 32 years. According to the data, exogamy in Campora through the 19th century remained below 20% but has increased since the beginning of the 20th century (last three generations). Exogamy values are consistent with those observed in another Italian isolate Talana [17]. We want to take into account exogamy in assembling a study sample and thus we considered the analysis of exogamic marriages as a rough estimate of the gene flow. We assembled the study sample among all living individuals for whom genealogical records are available (n = 1,719) including in it all those individuals deriving from ancestors that entered the pedigree before the last three generations, that is before exogamy started to break the isolation.
Fig. 3.
Percentage of exogamic marriages and inbreeding trend over time. Classes of 30 years have been considered and the midpoint of each class is represented on the x axis. Marriages were counted from marriage register in the church archive, while inbreeding was determined from the genealogy.
Matrilinear and patrilinear genealogical lineages (GLs) were determined on the whole genealogy, which includes 10,737 members distributed over four centuries. Each line begins with an ancestor and includes related descendents of the same sex. There is no overlapping among individuals of different lines. Each GL has been dated with the birth year of the ancestor. Table 1 shows the number of GLs starting in each of the four centuries included in the pedigree. Notably there has been a greater turnover of females compared to males due to patrilocal behaviour (tendency of females to move to the native village of the male after marriage) common in this area.
Table 1.
Distribution of genealogical lines through centuries. In bold are indicated the lineages from which derives the study sample population.
| Century | Matrilineages |
Patrilineages |
||
|---|---|---|---|---|
| total | with living descendant | total | with living descendant | |
| 17th | 39 | 13 | 22 | 20 |
| 18th | 111 | 18 | 23 | 21 |
| 19th | 59 | 18 | 37 | 29 |
| 17–19th | 209 | 49 | 82 | 70 |
| 20th | 101 | 97 | 165 | 147 |
In concordance with exogamy data, most of the matrilinear and patrilinear lineages with still living descendents began in the last century and only 46 female GLs and 70 male GLs were present before 1890 (table 1). These lines are represented today by 576 living females and 608 living males. These 1,184 living individuals, with at least one ancestor that entered the pedigree before 1890, constitute our study sample. Those lineages that do not have living descendants could have terminated because of emigration or because only descendants of the opposite sex were generated or because there were no descendants at all.
Individuals in the study sample (n = 1,184) represent 69% of the living individuals (n = 1,719) included in the genealogical data. The remaining portion of living individuals (n = 535) includes: subjects for whom a gender-specific parent-sib lineage was missed (15%) and immigrants that recently joined the village (16%).
The study sample was assembled considering matrilinear and patrilinear lineages with the aim of investigating about the founding nucleus of the village through the analyses of mtDNA and Y chromosome. However, although only these two special lines were used, we found out that they catch almost all the information of all the possible ascending genealogical lines (data not shown) and thus we used the same study sample also for the examination of the demographic structure.
A Few Founding Lineages Gave Rise to the Current Population
The genealogical information is limited to four centuries and therefore we cannot investigate the coalescence of GLs before the 17th century. We thus analyzed mitochondrial and Y chromosome DNA to verify the coalescence of GLs by grouping those who present the same haplotype in what we refer to as a ‘founding lineage’ (FL). It is important to notice that each lineage can include more than one individual, and that is why we talk about lineages and not in terms of individuals. Samples for the analysis were chosen according to GLs in order to sample almost all possible different haplotypes. In fact due to the high degree of relatedness among individuals, a ‘random’ sampling could easily lead to underestimation of the number of different haplotypes. For each GL, at least two individuals were chosen (if available) to assure the concordance of results within it. The number of female FLs was determined from the number of different mitochondrial DNA (mtDNA) haplotypes present in the sample. By the analysis of 92 sequences of individuals belonging to 46 GLs (2 for each female GL) of the HVR-I region, we described 27 different HVR-I types (table 2). In a similar study [37], Babalini and colleagues observed a similar paucity of haplotypes in a group of Croatian-Italians constituting a linguistic minority, compared to samples coming from open populations as indicated in table 3, where we have added Campora for comparison. It is worth mentioning that Campora is located within the open population of Campania reported in table 3.
Table 2.
Distribution of the HVR-I types into the examined haplogroups and polymorphisms of the HVR-I sequence
| Haplogroup (% a) | HVR-type | %a,b | 16017 | 16037 | 16069 | 16086 | 16093 | 16104 | 16111 | 16126 | 16129 | 16136 | 16145 | 16146 | 16148 | 16153 | 16163 | 16172 | 16183 | 16184 | 16186 | 16187 | 16189 | 16192 | 16193 | 16194 | 16195 | 16215 | 16218 | 16224 | 16225 | 16244 | 16249 | 16250 | 16257 | 16261 | 16262 | 16279 | 16285 | 16287 | 16294 | 16295 | 16299 | 16301 | 16312 | 16320 | 16321 | 16344 | 16353 | 16357 | 16363 | 16369 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 29.9 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | T | A | - | - | - | T | - | - | d | - | - | - | C | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | C | - | - | - | - | - | |
| 2 | 9.4 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | T | A | - | - | - | T | - | - | d | - | - | - | C | - | - | - | - | T | - | - | - | - | - | - | - | - | - | - | - | C | - | - | - | C | - | |
| 3 | 8.0 | - | - | - | - | - | - | - | - | - | - | A | - | - | - | - | T | A | - | - | - | T | - | - | d | - | - | - | C | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | C | -- | -- | - | - | - | |
| 4 | 7.6 | - | - | - | - | - | - | - | - | - | - | - | - | T | - | - | T | A | - | - | - | T | - | - | d | - | - | - | C | - | - - | - | T | - | - | - | - | - | - | - | - | - | C | A | C | - | - | - | - | - | ||
| H | 5 | 4.2 | - | G | - | - | - | - | - | - | - | - | - | - | - | - | - | T | A | - | - | - | T | - | - | d | - | - | - | C | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | C | -- | - | - | - | - |
| (61.6) | 6 | 0.7 | - | - | - | - | C | - | - | - | - | - | - | - | - | - | - | T | A | - | - | - | T | - | - | d | - | - | - | C | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | C | - | - | - | - | - |
| 7 | 0.5 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | T | A | - | - | - | - | - | d | - | - | - | C | - | - | - | - | - | - | - | - | - | - | - | - | G | - | - | - | C | - | - | - | C | - | ||
| 8 | 0.5 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | T | - | - | - | - | - | - | - | - | - | - | - | C | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | C | - | - | C | - | - | |
| 9 | 0.5 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | -- | - | - | - | - | - | C | - | - | - | - | T | - | - | - | - | - | G | - | - | - | - | - | C | - | C | - | - | - | |
| 10 | 0.3 | - | - | - | C | - | - | - | - | - | - | - | - | - | - | - | - | A | - | - | T | - | - | - | d | - | - | C | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | C | - | - | - | - | - | |
| T | 11 | 1.7 | - | - | - | - | - | - | - | C | - | - | - | - | - | - | G | T | A | - | - | - | - | - | - | d | - | - | - | C | - | C | - | - | - | - | - | - | - | - | - | T | - | - | C | - | C | - | - | - | - | - |
| (2.6) | 12 | 0.7 | - | - | - | - | - | - | - | C | - | - | - | - | - | - | G | T | A | - | - | - | - | - | - | d | - | - | - | C | - | C | - | - | T | - | - | - | - | - | - | T | - | - | C | - | C | - | - | - | - | - |
| 13 | 0.2 | - | - | - | - | - | - | - | C | - | - | - | - | - | - | G | T | A | - | T | - | - | - | - | - | - | - | - | C | - | - | - | - | - | - | - | - | - | - | - | - | T | - | - | - | C | - | - | C | - | - | |
| U | 14 | 1.4 | - | - | - | - | - | T | - | - | - | - | - | - | - | - | - | T | A | - | - | - | - | - | - | d | - | - | - | C | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | C | G | - | - | - | - |
| (1.4) | ||||||||||||||||||||||||||||||||||||||||||||||||||||
| preH | 15 | 12.8 | G | - | - | - | C | - | - | - | - | - | - | - | - | - | - | T | A | - | - | - | T | - | - | d | - | - | - | C | - | - | - | - | - | - | - | - | - | - | - | C | - | - | - | - | C | - | - | - | - | - |
| (17.7) | 16 | 4.9 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | T | A | - | - | - | T | - | - | d | - | - | - | C | - | - | - | - | - | - | - | - | - | - | - | C | - | - | - | - | C | - | - | - | - | - |
| X | 17 | 2.1 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | T | A | A | - | - | - | - | - | - | - | - | - | - | - | - | T | - | - | - | - | T | - | - | - | - | - | - | - | C | - | - | - | - | - | |
| (4.2) | 18 | 2.1 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | T | - | - | - | - | - | - | - | - | C | - | - | - | - | - | - | T | - | - | - | - | T | - | - | - | - | - | - | - | C | - | - | - | - | - |
| J | 19 | 1.4 | - | - | T | - | - | - | - | C | - | - | - | - | - | - | - | T | A | - | - | - | T | - | T | d | - | - | - | C | - | - | - | - | - | - | - | - | - | - | - | - | - | G | C | - | C | - | - | - | - | - |
| (2.9) | 20 | 1.2 | - | - | T | - | - | - | - | C | - | - | - | - | - | - | - | T | A | - | - | - | T | - | - | d | - | - | - | C | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | C | - | - | - | - | - |
| 21 | 0.3 | - | - | T | - | - | - | - | C | - | - | A | - | - | - | - | T | A | - | - | - | T | - | - | d | - | - | - | C | - | - | - | - | - | - | T | - | - | - | - | - | - | - | - | - | C | - | - | - | - | C | |
| K | 22 | 1.9 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | T | A | - | - | - | T | - | - | d | - | - | - | C | C | - | - | - | - | - | - | - | - | - | - | - | - | - | C | - | C | - | - | - | - | - |
| (4.2) | 24 | 1.4 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | T | A | - | - | - | T | T | - | d | - | - | - | - | C | - | - | - | - | - | - | - | G | G | - | - | - | - | C | - | C | - | - | - | - | - |
| 23 | 0.9 | - | - | - | - | C | - | - | - | - | - | - | - | - | - | - | T | A | - | - | - | T | - | - | d | - | - | - | C | C | - | - | - | - | - | - | - | - | - | - | - | - | - | C | - | C | - | - | - | - | - | |
| M | 25 | 2.1 | - | - | - | - | - | - | - | - | A | - | - | - | - | - | - | T | - | - | - | - | - | - | - | - | T | - | - | - | - | - | C | - | - | - | - | - | - | - | - | - | - | C | - | C | - | - | - | - | - | |
| (2.1) | ||||||||||||||||||||||||||||||||||||||||||||||||||||
| n.d. | 26 | 1.0 | - | - | - | - | - | - | - | - | A | - | - | - | - | - | - | T | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | C | - | - | - | - | - | - | - | - | - | - | C | - | C | - | - | - | - | - |
| (2.3) | 27 | 0.3 | - | - | - | - | - | - | T | - | - | - | - | - | - | A | - | T | A | - | - | - | T | - | - | d | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | A | C | - | - | - | - | - |
The position of polymorphisms are relative to the reference sequence. n.d. = not determined; d = deletion.
a Referred to the female population in the study sample (n = 567).
Relative to HVR-I type.
Table 3.
Number of different haplotypes in the HVR-I region in different populations
| Population | Features | Number of individuals | HVR-I type number |
|---|---|---|---|
| Campora | genetic isolate | 46a | 27 |
| Croatian-Italiansb | linguistic minority | 41 | 29 |
| Abruzzo-Moliseb | open population | 73 | 51 |
| Campaniab | open population | 48 | 41 |
| Laziob | open population | 52 | 37 |
| Pugliab | open population | 26 | 24 |
Here we report the number of different GLs because the sampling has been done according to GLs. The actual number of individuals is 92.
From Babalini et al. 2005.
Polymorphisms characterizing each HVR-I type are reported in table 2 where their position according to the reference sequence [21, 22] is indicated together with the percentage of living females that each type comprises. We interpret the presence of 27 different HVR-I types as evidence of 27 different FLs, with the most common including 29.9% of living females.
We also performed a study to ascertain which haplogroups, according to the main classification [23, 24] contain the different HVR-I types. We characterized a set of polymorphisms describing the haplogroups typical of European, Asian and African populations and we found nine different groups in the study sample (table 2). There was no African contribution, and only a minor Asian contribution (haplogroup M). As expected, the majority of females (61.4%) belongs to the H haplogroup, the most frequent in Europe. A small percentage indicated as ‘n.d.’ was of uncertain classification.
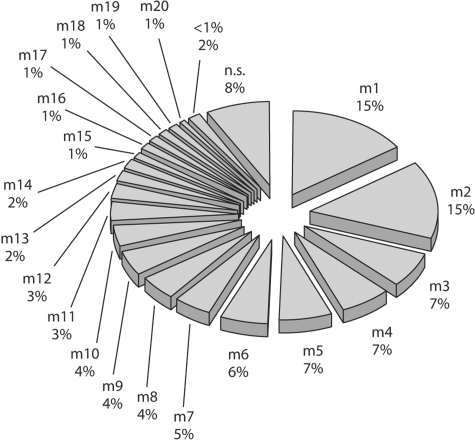
As was done for the female FLs, the male FLs were determined by counting the number of different haplotypes on the non-recombinant Y chromosome region in the male sample. We used an informative Y-STR core set (DYS19, DYS389I, DYS390, DYS391, DYS392, DYS393, DYS385) to define the haplotypes. We found 24 different haplotypes that suggest the presence of 24 FLs (fig. 4).
Fig. 4.
Male founding lineages (FLs). The percentage of the population in the study sample that each FL represents is indicated. The category ‘< 1%’ includes all those lineages whose descendents represent less than 1% of the male population. n.s. = not sampled.
Linkage Disequilibrium in Campora
It has been well demonstrated that isolated populations show extended regions of LD [38, 39]. Within the Campora population, we analysed a low recombination rate and non-coding DNA segment located on the Xq13 chromosome region [40] that has been previously characterized in both large and isolated populations [41,42,43,44]. In this region, tests for disequilibrium among six STRs spanning about 10 Mb were carried out in a sample of 63 unrelated males. Although in the Campora sample, the average number of alleles at each locus is not significantly reduced (data not shown), only 44 different haplotypes in 63 individuals were found. The resulting LD-associated p-values relative to the 15 possible pairs among the six STRs grouped according to the distance between markers are shown in table 4. The p-values from similar studies on other isolated (Saami, Gavoi) and large populations are also reported [41, 42]. It is evident that in Campora, like in the other isolated populations, a consistent number of marker pairs (12 out of 15) are in significant LD.
Table 4.
LD on the X chromosome
| Marker pairs | Markers distance |
SAAMIa (n = 54) | GAVOIb (n = 73) | Campora (n = 53) | Swedena (n = 41) | Sardiniab (n = 73) | UKb (n = 73) | Finland a (n = 80) | Estoniaa (n = 45) | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mb | cMa | ||||||||||
| DXS8092 | DXS8037 | 0.00 | 0.40 | 0.000 | 0.000 | 0.045 | 0.028 | 0.280 | 0.620 | 0.180 | 0.072 |
| DXS8092 | DXS986 | 1.01 | 0.20 | 0.000 | 0.000 | 0.001 | 0.618 | 0.322 | 0.884 | 0.092 | 0.143 |
| DXS1225 | DXS986 | 1.17 | 0.50 | 0.000 | 0.000 | 0.104 | 0.448 | 0.166 | 0.703 | 0.393 | 0.688 |
| DXS1225 | DXS8092 | 1.62 | 0.30 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| DXS8037 | DXS1225 | 3.98 | 0.00 | 0.091 | 0.008 | 0.009 | 0.242 | 0.710 | 0.647 | 0.836 | 0.488 |
| DXS8092 | DXS1225 | 3.98 | 0.40 | 0.000 | 0.000 | 0.008 | 0.676 | 0.921 | 0.320 | 0.283 | 0.120 |
| DXS8037 | DXS8092 | 4.14 | 0.30 | 0.012 | 0.004 | 0.002 | 0.033 | 0.630 | 0.002 | 0.238 | 0.625 |
| DXS8092 | DXS8092 | 4.14 | 0.10 | 0.000 | 0.000 | 0.001 | 0.102 | 0.319 | 0.492 | 0.044 | 0.065 |
| DXS983 | DXS8092 | 4.64 | 1.60 | 0.000 | 0.000 | 0.000 | 0.746 | 0.876 | 0.974 | 0.314 | 0.153 |
| DXS983 | DXS8037 | 4.68 | 2.00 | 0.300 | 0.000 | 0.407 | 0.924 | 0.036 | 0.149 | 0.683 | 0.104 |
| DXS8037 | DXS986 | 5.15 | 0.50 | 0.000 | 0.003 | 0.119 | 0.256 | 0.302 | 0.975 | 0.620 | 0.739 |
| DXS8092 | DXS986 | 5.15 | 0.10 | 0.000 | 0.000 | 0.006 | 0.332 | 0.125 | 0.940 | 0.331 | 0.100 |
| DXS983 | DXS1225 | 8.65 | 2.00 | 0.000 | 0.170 | 0.001 | 0.480 | 0.169 | 0.338 | 0.630 | 0.520 |
| DXS983 | DXS8092 | 8.82 | 1.70 | 0.000 | 0.245 | 0.050 | 0.082 | 0.142 | 0.243 | 0.565 | 0.730 |
| DXS983 | DXS986 | 9.82 | 1.50 | 0.000 | 0.003 | 0.042 | 0.400 | 0.825 | 0.253 | 0.829 | 0.468 |
Marker pairs are ordered according to the distance between markers. Significant LD associated p-values are in bold.
From Laan and Paabo, 1997.
From Zavattari et al., 2000.
Results obtained on the X chromosome have been confirmed also in the analysis relative to the autosomal part of the genome. LD-associated p-value and Dadj were evaluated among all possible pairs of syntenic markers and then marker pairs were grouped according to their recombination interval in classes as shown in table 5. For each class the average of the LD-associated measures is reported and compared with two other isolated populations: the GRIP from the South West Netherland [18] and the one of Palau from Oceania [45]. As shown in the table, when recombination intervals are <0.1, both the percentage of pairs in significant disequilibrium and the Dadj in Campora are doubled compared to the GRIP population and more than doubled compared to Palau. In classes of greater recombination interval, the trend changes and values become comparable among populations.
Table 5.
Genomewide LD in Campora compared to the isolated populations of Palau and GRIP
| Recombination interval | Number of marker pairs |
% Significant p-values |
Average Dadj ±SD |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Campora | GRIP | Palau | Campora | GRIP | Palau | Campora | GRIP | Palau | |
| (DOT)<0.02 | 325 | 65 | – | 64.6 | 35.4 | (DOT)– | 0.113±0.077 | 0.050±0.008 | – |
| (DOT)0.02–0.05 | 940 | 393 | – | 50.9 | 24.7 | (DOT)– | 0.076±0.069 | 0.037±0.003 | – |
| (DOT)0.05–0.1 | 1,827 | 775 | – | 27.1 | 17.7 | (DOT)– | 0.043±0.063 | 0.024±0.002 | – |
| (DOT)<0.1 | 3,092 | 1,233 | – | 38.3 | 20.8 | (DOT)16.2 | 0.060±0.07 | 0.030±0.001 | 0.031 |
| (DOT)0.1–0.2 | 4,206 | 1,705 | – | 12.0 | 9.0 | (DOT)11.6 | 0.015±0.055 | 0.010±0.001 | 0.019 |
| (DOT)0.2–0.3 | 5,142 | 2,124 | – | 6.9 | 6.4 | (DOT)11.6 | 0.004±0.053 | 0.003±0.001 | 0.017 |
| (DOT)0.3–0.4 | 6,827 | 2,720 | – | 6.8 | 4.3 | (DOT)7.1 | 0.003±0.053 | 0.000±0.001 | 0.012 |
| (DOT)>0.4 | 9,139 | 3,520 | – | 0.6 | 5.1 | (DOT)4.4 | 0.003±0.054 | 0.001±0.001 | 0.009 |
Data on Palau from Devlin B et al. (2001) and on GRIP from Aulchenko YS et al. (2004).
Overall, these results indicate that in the genome of the population of Campora, extended regions show significant LD as in other isolated and sub-isolated populations.
The Population of Campora Experienced a Bottleneck in the Past
The famine of the 16th century almost halved the population and the plague of 1656 halved it again so that in 1669 the population of Campora consisted of only 140 plague survivors (fig. 2) [19].
A temporary excess of heterozygosity, relative to that expected on the basis of the number of alleles, takes place when a bottleneck occurs. Such an excess is caused by the more rapid decline of the number of alleles compared to the decline of gene diversity (heterozygosity), as rare alleles are lost more quickly. The period of time during which it is possible to estimate the heterozygosity excess depends on the effective population size and on the extent of the population reduction at the bottleneck [34]. We assessed heterozygosity excess in the genome of the Campora population using 1,012 microsatellites in a sample of 584 individuals. The 1,012 loci were ascertained to be in Hardy-Weinberg equilibrium in a sample of 80 individuals taking into account for their relatedness. In table 6, we show the results of the bottleneck analysis under the TPM model in which the SMM component has been set to 90% and under the IAM. This last option is not representative of the model of microsatellites evolution but it has been considered only for the purpose of comparing datasets of Campora with the only other data available about human. These data belong to an expanding population, far removed from a bottleneck [31]. In Campora, an excess of heterozygosity is detected under both the TPM and the IAM models, suggesting that a bottleneck has occurred.
Table 6.
Detection of heterozygosity excess caused by the bottleneck
| Dataset | Campora |
Sardiniaa | |||||
|---|---|---|---|---|---|---|---|
| a | b | c | d | e | |||
| Sample size (2n) | 584 | 584 | 584 | 584 | 584 | 23 | |
| Number of loci | 204 | 202 | 203 | 201 | 203 | 10 | |
| Average observed heterozygosity ± SD | 0.727±0.117 | 0.725±0.110 | 0.727±0.109 | 0.723±0.121 | 0.719±0.121 | –b | |
| Average number of alleles ± SD | 9±2 | 8±2 | 9±2 | 8±2 | 9±2 | –b | |
| TPM | Sign test | excess | excess | excess | excess | excess | |
| SSM = 90% | exp | 120.29 | 119.19 | 119.46 | 118.13 | 118.86 | –b |
| obs | 137 | 147 | 145 | 142 | 135 | ||
| p value | 0.00984 | 0.00003 | 0.00013 | 0.00033 | 0.01195 | ||
| S.D.T. | excess | excess | excess | excess | excess | ||
| IT2I | 3.062 | 3.805 | 4.381 | 4.290 | 3.226 | –b | |
| p value | 0.00110 | 0.00007 | 0.00001 | 0.00001 | 0.00063 | ||
| Wilcoxon | excess | excess | excess | excess | excess | ||
| p value | 0.00001 | <10e-5 | <10e-5 | <10e-5 | <10e-5 | –b | |
| IAM | Sign test | excess | excess | excess | excess | excess | deficiency |
| exp | 117.99 | 116.95 | 111.28 | 115.77 | 116 | –b | |
| obs | 199 | 198 | 199 | 198 | 197 | ||
| p value | <10e-5 | <10e-5 | <10e-5 | <10e-5 | <10e-5 | 0.001 | |
| S.D.T. | excess | excess | excess | excess | excess | deficiency | |
| T2 | 17.0 | 17.3 | 17.6 | 17.4 | 17.0 | 15.1 | |
| p value | <10e-4 | <10e-5 | <10e-5 | <10e-5 | <10e-5 | <10e-5 | |
| Wilcoxon | excess | excess | excess | excess | excess | deficiency | |
| p value | <10e-4 | <10e-5 | <10e-5 | <10e-5 | <10e-5 | <10e-5 | |
exp/obs = Expected/observed number of loci with heterozygosity excess; excess/deficiency refers to the number of loci in hetero-zygosity.
Data from Cornuet and Luikart 1996.
Data not available.
The results of the genetic analysis were matched by the study of the genealogy. We estimated the percentage of the living population derived from FLs originating during the plague period. This period was defined as one generation after the first documented case of plague in the nearby village of Novi Velia (i.e. 32 years after the year 1656). FLs were dated according to the date of the most elderly GLs. We found that 82% of the 1,184 individuals in the study sample belong to ‘plague FLs’. In other words, it is legitimate to consider the Campora population as derived from the survivors of the bottleneck, some 13 generations ago.
Inbreeding in the Population of Campora
Average inbreeding (f) in the study sample (n = 1,184) was evaluated from the genealogy using a 3,906-member sub-pedigree that included all ancestors of living individuals and that was distributed over 17 generations and over four centuries. Two different computational methods were used and gave consistent results: 82% of the living population have a value of f different from zero; the average inbreeding is 0.00651 ± 0.00915. Furthermore, 0.93% of the population show a value of f > 0.0625 (first cousin), and 9.44% show f > 0.0156 (second cousin). The f value in the Campora population is compared with those of other populations in table 7 where the genealogy structure of the sample used in the analysis is also reported [46,47,48]. In Campora, the average value of the inbreeding coefficient is modest compared to that of populations that have experienced extreme isolation like the Hutterites [49]. The distribution of f in Campora is instead comparable to those of the two isolates from Sardinia. In addition, similar values of the inbreeding coefficients were estimated in other European genetic isolates, such as Wurtenburg and Val di Parma [50].
Table 7.
Inbreeding evaluated from the genealogy in different isolates
| Campora | Perdasdefogu | Talana | S-leut Hutteritesa | |
|---|---|---|---|---|
| Pedigree | ||||
| Founding lineages | (PLUS)53 | (PLUS)–b | (PLUS)44c | (PLUS)64 |
| Generations | (PLUS)17 | (PLUS)15d | (PLUS)16d | (PLUS)13 |
| Members | (PLUS)3,906 | (PLUS)2,506d | (PLUS)5,219d | (PLUS)1,623 |
| Sample population size | (PLUS)1,184 | (PLUS)821d | (PLUS)876e | (PLUS)806 |
| Inbreeding | ||||
| Mean ± SD | (PLUS)0.006±0.009 | (PLUS)0.010±0.021e | (PLUS)0.018±0.022e | (PLUS)0.034±0.015 |
| Median | (PLUS)0.004 | (PLUS)0.005 d | (PLUS)0.015e | (PLUS)–b |
| 1st quartile | (PLUS)0.001 | (PLUS)0.001 d | (PLUS)0.007e | (PLUS)–b |
| 3rd quartile | (PLUS)0.008 | (PLUS)0.010 d | (PLUS)0.021e | (PLUS)–b |
Features of the pedigree used in the calculations are reported.
From Weiss et al., 2005;
data not available;
from Angius et al., 2001;
from Falchi et al., 2004;
Angius A., personal communication.
We then calculated the average f per generation and plotted it together with the exogamic marriages as shown in figure 3. From the graph, it is evident that, since the bottleneck had occurred, f increased throughout the period of isolation but this trend changed when the recent gene flow occurred.
Discussion
In this study, we traced the genetic history of the population of Campora and we found evidence suggesting that Campora can be defined as a genetic isolate. The population features and the extent of isolation have been determined on the basis of consistent information coming from the analysis of historical, demographic and genetic data, as well as through comparisons with other populations.
According to historical data, the first considerable nucleus of the population appeared around the 11th century and was of Greek and Lucanian origin. Accurate demographic information is available for the last four centuries. On the basis of this information, we evaluated exogamy in the last three centuries. We observed that exogamy was present through the 18th and 19th centuries, although it has never been consistent (less than 20%) indicating that overall, the population remained subject to a constant but weak gene flow during its growth (mainly due to exogamic marriages from nearby villages). However, as exogamy had conspicuously increased in the last century, we decided to take it into account. Thus we planned a strategy to define a study sample, tracing back matrilineages and patrilineages in order to exclude those individuals whose ancestors entered the pedigree after 1890. We performed this study on the genealogy not only to define our study sample, but also to show that a ‘core’ population can be obtained from complex genealogies of populations in which isolation starts to decline. In fact, many populations, like Campora, have experienced periods of geographical isolation in the past and only recent exposure to migration. Such populations are probably going to lose their characteristic features in the near future, and therefore they deserve critical attention in the present day [51].
We estimated that in 96.7% of the study sample population there are only 17 and 20 female and male sex-specific haplotypes, respectively (10 out of 27 female FLs and 4 out of 24 male FLs have a number of descendents <1% of the total population) thus indicating that the population of Campora is genetically homogeneous. Moreover, there is a striking difference in the number of living descendents among FLs, most probably because of random sorting of alleles through genetic drift. The same difference has been found in a similar study in the village of Talana in Sardinia, where only eight Y chromosome haplotypes represent 70% of current males and ten mtDNA haplogroups represent 77% of current females [17]. Correspondingly, in Campora, 74% of the living males are represented by ten different haplotypes and seven mitochondrial haplotypes account for 76.6% of living females.
Evidence of genetic homogeneity also comes from the LD analysis. We have demonstrated that LD extends over wide regions in the genome of the population of Campora. Moreover the comparison with the GRIP population, whose founding nucleus is dated to the middle of the 18th century [18], suggests that the founding nucleus of Campora must be earlier to it. This is consistent with the historical hypothesis of the first settlement of Campora in the 11th century.
Using a dense map of microsatellite markers, we observed that the population recently experienced a bottleneck. To our knowledge, this is the first example of bottleneck evaluation in a human population based on such a large set of genetic markers. Most likely the bottleneck coincides with historical reports about the plague of the 17th century through which almost all the ancestors of the living individuals have passed. This is also suggested by the ‘dating’ of FLs, which shows that the living individuals derive from ancestors who were already present in the village before the plague and thus survived it. Thus due to the bottleneck, Campora can be considered a young isolate (< 20 generations) according to the Heutink and Oostra classification [51], despite the fact that its founding nucleus seems to be more ancient.
Inbreeding is present in the population (mean inbreeding coefficient is 0.006), but is not as high as in ‘extreme’ human isolates, like the S-leu Hutterites [49], which could be considered close to the upper limit for human populations. Campora instead can be considered a ‘mild’ isolate, like two other isolates from Sardinia [17, 47], where the average inbreeding in the population is also moderate. These kinds of isolates are certainly more common in humans and their usefulness in complex trait mapping has already been demonstrated.
A reduced number of founders and the presence of inbreeding provide evidence that isolation has occurred. Further, the progressive increase of inbreeding since the bottleneck suggests that mating was occurring mainly between individuals who were becoming more and more similar genetically; apparently, people from outside the village were only marginally participating in the mating. Hence, this inbreeding trend provides evidence that the population expanded under conditions of isolation, even though it is possible that, because of a partial availability of genealogical data in the 17th century, inbreeding in the first generations is slightly underestimated. We note that for the 19th and 20th centuries, inbreeding sharply decreases when exogamy rises, which is an argument in favour of the completeness of the genealogical data.
The determination of GLs from the genealogy was crucial to achieve many of the results in our present study. Due to the presence of the Catholic Church in Italy, written records of births, marriages, and deaths have been produced since the 17th century. Consequently, wide ranging genealogical information is available for many villages and provides a valuable resource for population genetics. We want to emphasize the role that genealogical information has played in our study, showing how it can integrate and support the genetic analyses. In fact, matrilinear and patrilinear GLs allowed us to assemble the study sample and played a key role in sampling for mtDNA and Y chromosome haplotype analyses. In addition, GLs tell us how successful was the DNA sampling. According to genealogical data, we managed to sample almost all the population. We found that the 9% of males for which no DNA is available are grouped in 25 different lineages, each lineage thus contributing to a very small number of males. In contrast, the female sampling was more successful: only about 2% of the females, corresponding only to three different lineages, could not be sampled.
With this work, we have demonstrated that Campora is a young and homogeneous isolate. We also proved the usefulness of the comparison of genealogical and genetic information to investigate the structure of human populations. These characteristics, together with the environmental uniformity and an accurate phenotype description, make this population a valuable resource for the study of complex traits.
Acknowledgements
We thank the population of the village of Campora for their kind cooperation, Don Guglielmo Manna for helping in the interaction with the population and the Institutions. We thank Lucio Luzzatto, Guido Barbujani, Claudia Angelini, Catherine Bourgain, Yurii S. Aulchenko, Mario Aversano, Francesco Cucca and Patrizia Zavattari for valuable comments and suggestions; Jim McGhee for reviewing the manuscript; Mrs. M. Terracciano for technical assistance. We also want to thank two anonymous Reviewer whose suggestions were helpful for the improvement of the manuscript. This work was supported by grants from Ente Parco Nazionale del Cilento e Vallo di Diano, the Associazione Italiana per la Ricerca sul Cancro (AIRC), the Assessorato Ricerca Regione Campania, the Fondazione Banco di Napoli to MGP.
References
- 1.Varilo T, Peltonen L. Isolates and their potential use in complex gene mapping efforts. Curr Opin Genet Dev. 2004;14:316–323. doi: 10.1016/j.gde.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 2.Sheffield VC, Stone EM, Carmi R. Use of isolated inbred human populations for identification of disease genes. Trends Genet. 1998;14:391–396. doi: 10.1016/s0168-9525(98)01556-x. [DOI] [PubMed] [Google Scholar]
- 3.Arcos-Burgos M, Palacio G, Sanchez JL, Londono AC, Uribe CS, Jimenez M, Villa A, Anaya JM, Bravo ML, Jaramillo N, Espinal C, Builes JJ, Moreno M, Jimenez I. Multiple sclerosis: Association to HLA DQalpha in a tropical population. Exp Clin Immunogenet. 1999;16:131–138. doi: 10.1159/000019105. [DOI] [PubMed] [Google Scholar]
- 4.Gianfrancesco F, Esposito T, Ombra MN, Forabosco P, Maninchedda G, Fattorini M, Casula S, Vaccargiu S, Casu G, Cardia F, Deiana I, Melis P, Falchi M, Pirastu M. Identification of a novel gene and a common variant associated with uric acid nephrolithiasis in a Sardinian genetic isolate. Am J Hum Genet. 2003;72:1479–1491. doi: 10.1086/375628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peltonen L, Jalanko A, Varilo T. Molecular genetics of the Finnish disease heritage. Hum Mol Genet. 1999;8:1913–1923. doi: 10.1093/hmg/8.10.1913. [DOI] [PubMed] [Google Scholar]
- 6.Sheffield VC. Use of isolated populations in the study of a human obesity syndrome, the Bardet-Biedl syndrome. Pediatr Res. 2004;55:908–911. doi: 10.1203/01.pdr.0000127013.14444.9c. [DOI] [PubMed] [Google Scholar]
- 7.Anaya JM, Correa PA, Mantilla RD, Arcos-Burgos M. Rheumatoid arthritis association in Colombian population is restricted to HLA-DRB1*04 QRRAA alleles. Genes Immun. 2002;3:56–58. doi: 10.1038/sj.gene.6363833. [DOI] [PubMed] [Google Scholar]
- 8.Anderson SL, Coli R, Daly IW, Kichula EA, Rork MJ, Volpi SA, Ekstein J, Rubin BY. Familial dysautonomia is caused by mutations of the IKAP gene. Am J Hum Genet. 2001;68:753–758. doi: 10.1086/318808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ciullo M, Bellenguez C, Colonna V, Nutile T, Calabria A, Pacente R, Iovino G, Trimarco B, Bourgain C, Persico MG. New susceptibility locus for hypertension on chromosome 8q by efficient pedigree-breaking in an Italian isolate. Hum Mol Genet. 2006;15:1735–1743. doi: 10.1093/hmg/ddl097. [DOI] [PubMed] [Google Scholar]
- 10.Pardo LM, MacKay I, Oostra B, van Duijn CM, Aulchenko YS. The effect of genetic drift in a young genetically isolated population. Ann Hum Genet. 2005;69(Pt 3):288–295. doi: 10.1046/j.1529-8817.2005.00162.x. [DOI] [PubMed] [Google Scholar]
- 11.Patton MA. Genetic studies in the Amish community. Ann Hum Biol. 2005;32:163–167. doi: 10.1080/03014460500075274. [DOI] [PubMed] [Google Scholar]
- 12.Peltonen L, Palotie A, Lange K. Use of population isolates for mapping complex traits. Nat Rev Genet. 2000;1:182–190. doi: 10.1038/35042049. [DOI] [PubMed] [Google Scholar]
- 13.Arcos-Burgos M, Muenke M. Genetics of population isolates. Clin Genet. 2002;61:233–247. doi: 10.1034/j.1399-0004.2002.610401.x. [DOI] [PubMed] [Google Scholar]
- 14.Bourgain C, Genin E. Complex trait mapping in isolated populations: Are specific statistical methods required? Eur J Hum Genet. 2005;13:698–706. doi: 10.1038/sj.ejhg.5201400. [DOI] [PubMed] [Google Scholar]
- 15.Wright AF, Carothers AD, Pirastu M. Population choice in mapping genes for complex diseases. Nat Genet. 1999;23:397–404. doi: 10.1038/70501. [DOI] [PubMed] [Google Scholar]
- 16.Vitart V, Biloglav Z, Hayward C, Janicijevic B, Smolej-Narancic N, Barac L, Pericic M, Klaric IM, Skaric-Juric T, Barbalic M, Polasek O, Kolcic I, Carothers A, Rudan P, Hastie N, Wright A, Campbell H, Rudan I. 3000 years of solitude: extreme differentiation in the island isolates of Dalmatia, Croatia. Eur J Hum Genet. 2006;14:478–487. doi: 10.1038/sj.ejhg.5201589. [DOI] [PubMed] [Google Scholar]
- 17.Angius A, Melis PM, Morelli L, Petretto E, Casu G, Maestrale GB, Fraumene C, Bebbere D, Forabosco P, Pirastu M. Archival, demographic and genetic studies define a Sardinian sub-isolate as a suitable model for mapping complex traits. Hum Genet. 2001;109:198–209. doi: 10.1007/s004390100557. [DOI] [PubMed] [Google Scholar]
- 18.Aulchenko YS, Heutink P, Mackay I, Bertoli-Avella AM, Pullen J, Vaessen N, Rademaker TA, Sandkuijl LA, Cardon L, Oostra B, van Duijn CM. Linkage disequilibrium in young genetically isolated Dutch population. Eur J Hum Genet. 2004;12:527–534. doi: 10.1038/sj.ejhg.5201188. [DOI] [PubMed] [Google Scholar]
- 19.Del Mercato P, Infante A. Cilento, uomini e vicende. Salerno: Reggiani Editore; 1980. [Google Scholar]
- 20.O'Connell JR, Weeks DE. PedCheck: A program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 22.Ingman M, Kaessmann H, Paabo S, Gyllensten U. Mitochondrial genome variation and the origin of modern humans. Nature. 2000;408:708–713. doi: 10.1038/35047064. [DOI] [PubMed] [Google Scholar]
- 23.Macaulay V, Richards M, Hickey E, Vega E, Cruciani F, Guida V, Scozzari R, Bonne-Tamir B, Sykes B, Torroni A. The emerging tree of West Eurasian mtDNAs: a synthesis of control-region sequences and RFLPs. Am J Hum Genet. 1999;64:232–249. doi: 10.1086/302204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Torroni A, Huoponen K, Francalacci P, Petrozzi M, Morelli L, Scozzari R, Obinu D, Savontaus ML, Wallace DC. Classification of European mtDNAs from an analysis of three European populations. Genetics. 1996;144:1835–1850. doi: 10.1093/genetics/144.4.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karigl G. A recursive algorithm for the calculation of identity coefficients. Ann Hum Genet. 1981;45(Pt 3):299–305. doi: 10.1111/j.1469-1809.1981.tb00341.x. [DOI] [PubMed] [Google Scholar]
- 26.Bourgain C, Hoffjan S, Nicolae R, Newman D, Steiner L, Walker K, Reynolds R, Ober C, McPeek MS. Novel case-control test in a founder population identifies P-selectin as an atopy-susceptibility locus. Am J Hum Genet. 2003;73:612–626. doi: 10.1086/378208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swedlund AC, Boyce AJ. Mating structure in historical populations: estimation by analysis of surnames. Hum Biol. 1983;55:251–262. [PubMed] [Google Scholar]
- 28.Abecasis GR, Cookson WO. GOLD – graphical overview of linkage disequilibrium. Bioinformatics. 2000;16:182–183. doi: 10.1093/bioinformatics/16.2.182. [DOI] [PubMed] [Google Scholar]
- 29.Aulchenko YS, Axenovich TI, Mackay I, van Duijn CM. miLD and booLD programs for calculation and analysis of corrected linkage disequilibrium. Ann Hum Genet. 2003;67(Pt 4):372–375. doi: 10.1046/j.1469-1809.2003.00041.x. [DOI] [PubMed] [Google Scholar]
- 30.Zaykin D, Zhivotovsky L, Weir BS. Exact tests for association between alleles at arbitrary numbers of loci. Genetica. 1995;96:169–178. doi: 10.1007/BF01441162. [DOI] [PubMed] [Google Scholar]
- 31.Cornuet JM, Luikart G. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics. 1996;144:2001–2014. doi: 10.1093/genetics/144.4.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bourgain C, Abney M, Schneider D, Ober C, McPeek MS. Testing for Hardy-Weinberg equilibrium in samples with related individuals. Genetics. 2004;168:2349–2361. doi: 10.1534/genetics.104.031617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McPeek MS, Wu X, Ober C. Best linear unbiased allele-frequency estimation in complex pedigrees. Biometrics. 2004;60:359–367. doi: 10.1111/j.0006-341X.2004.00180.x. [DOI] [PubMed] [Google Scholar]
- 34.Maruyama T, Fuerst PA. Population bottlenecks and nonequilibrium models in population genetics. II. Number of alleles in a small population that was formed by a recent bottleneck. Genetics. 1985;111:675–689. doi: 10.1093/genetics/111.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cornuet J, Luikart G. Empirical evaluation of a test for identifying recently bottlenecked populations from allele frequency data. Conservation Biol. 1998;12:228–237. [Google Scholar]
- 36.Di Rienzo A, Peterson AC, Garza JC, Valdes AM, Slatkin M, Freimer NB. Mutational processes of simple-sequence repeat loci in human populations. Proc Natl Acad Sci USA. 1994;91:3166–3170. doi: 10.1073/pnas.91.8.3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Babalini C, Martinez-Labarga C, Tolk HV, Kivisild T, Giampaolo R, Tarsi T, Contini I, Barac L, Janicijevic B, Martinovic Klaric I, Pericic M, Sujoldzic A, Villems R, Biondi G, Rudan P, Rickards O. The population history of the Croatian linguistic minority of Molise (Southern Italy): A maternal view. Eur J Hum Genet. 2005;13:902–912. doi: 10.1038/sj.ejhg.5201439. [DOI] [PubMed] [Google Scholar]
- 38.Ardlie KG, Kruglyak L, Seielstad M. Patterns of linkage disequilibrium in the human genome. Nat Rev Genet. 2002;3:299–309. doi: 10.1038/nrg777. [DOI] [PubMed] [Google Scholar]
- 39.Varilo T, Paunio T, Parker A, Perola M, Meyer J, Terwilliger JD, Peltonen L. The interval of linkage disequilibrium (LD) detected with microsatellite and SNP markers in chromosomes of Finnish populations with different histories. Hum Mol Genet. 2003;12:51–59. doi: 10.1093/hmg/ddg005. [DOI] [PubMed] [Google Scholar]
- 40.Kaessmann H, Heissig F, von Haeseler A, Paabo S. DNA sequence variation in a non-coding region of low recombination on the human X chromosome. Nat Genet. 1999;22:78–81. doi: 10.1038/8785. [DOI] [PubMed] [Google Scholar]
- 41.Laan M, Paabo S. Demographic history and linkage disequilibrium in human populations. Nat Genet. 1997;17:435–438. doi: 10.1038/ng1297-435. [DOI] [PubMed] [Google Scholar]
- 42.Zavattari P, Deidda E, Whalen M, Lampis R, Mulargia A, Loddo M, Eaves I, Mastio G, Todd JA, Cucca F. Major factors influencing linkage disequilibrium by analysis of different chromosome regions in distinct populations: demography, chromosome recombination frequency and selection. Hum Mol Genet. 2000;9:2947–2957. doi: 10.1093/hmg/9.20.2947. [DOI] [PubMed] [Google Scholar]
- 43.Latini V, Sole G, Doratiotto S, Poddie D, Memmi M, Varesi L, Vona G, Cao A, Ristaldi MS. Genetic isolates in Corsica (France): linkage disequilibrium extension analysis on the Xq13 region. Eur J Hum Genet. 2004;12:613–619. doi: 10.1038/sj.ejhg.5201205. [DOI] [PubMed] [Google Scholar]
- 44.Laan M, Wiebe V, Khusnutdinova E, Remm M, Paabo S. X-chromosome as a marker for population history: linkage disequilibrium and haplotype study in Eurasian populations. Eur J Hum Genet. 2005;13:452–462. doi: 10.1038/sj.ejhg.5201340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Devlin B, Roeder K, Otto C, Tiobech S, Byerley W. Genome-wide distribution of linkage disequilibrium in the population of Palau and its implications for gene flow in Remote Oceania. Hum Genet. 2001;108:521–528. doi: 10.1007/s004390100511. [DOI] [PubMed] [Google Scholar]
- 46.Fraumene C, Petretto E, Angius A, Pirastu M. Striking differentiation of sub-populations within a genetically homogeneous isolate (Ogliastra) in Sardinia as revealed by mtDNA analysis. Hum Genet. 2003;114:1–10. doi: 10.1007/s00439-003-1008-3. [DOI] [PubMed] [Google Scholar]
- 47.Falchi M, Forabosco P, Mocci E, Borlino CC, Picciau A, Virdis E, Persico I, Parracciani D, Angius A, Pirastu M. A genomewide search using an original pairwise sampling approach for large genealogies identifies a new locus for total and low-density lipoprotein cholesterol in two genetically differentiated isolates of Sardinia. Am J Hum Genet. 2004;75:1015–1031. doi: 10.1086/426155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weiss LA, Abney M, Cook EH, Jr, Ober C. Sex-specific genetic architecture of whole blood serotonin levels. Am J Hum Genet. 2005;76:33–41. doi: 10.1086/426697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weiss LA, Abney M, Parry R, Scanu AM, Cook EH, Jr, Ober C. Variation in ITGB3 has sex-specific associations with plasma lipoprotein(a) and whole blood serotonin levels in a population-based sample. Hum Genet. 2005;117:81–87. doi: 10.1007/s00439-004-1250-3. [DOI] [PubMed] [Google Scholar]
- 50.Crawford MH, Mielke JH, Morton NE (1982) Kinship and inbreeding in populations of Middle Eastern origin and controls pp 449–466, in Current Developments in Anthropological Genetics. Vol. II. Ecology and Population Structure., Press P, Editor. 1982: New York.
- 51.Heutink P, Oostra BA. Gene finding in genetically isolated populations. Hum Mol Genet. 2002;11:2507–2515. doi: 10.1093/hmg/11.20.2507. [DOI] [PubMed] [Google Scholar]