Abstract
Objective
SNP maps are becoming the gold standard for genetic markers, even for linkage analyses. However, because of the density of SNPs on most high throughput platforms, the resulting significant linkage disequilibrium (LD) can bias classical nonparametric multipoint linkage analyses. This problem may be even stronger in population isolates where LD can extend over larger distances and with a more stochastic pattern. We investigate the issue of linkage analysis with SNPs from the Affymetrix 500K GeneChip array in extended families from the isolated Hutterite population.
Methods
We minimized LD between SNPs by two methods based on a LD block pattern (Merlin and SNPLINK) and by MASEL, a new algorithm that we proposed to select SNP subsets with minimum LD and with no prior hypothesis about the LD pattern.
Results
Simulations, performed using the real LD pattern observed in the Hutterite population, show that sizeable inflation of linkage statistics persist when LD between SNPs is minimized by Merlin and SNPLINK. Inflation of linkage statistics is better controlled with MASEL.
Conclusion
In this population, it may be difficult to extract from standard GeneChip arrays a SNP map without LD-driven bias that is more informative than a dense microsatellite map.
Key Words: Linkage analysis, Dense SNP maps, Linkage disequilibrium, Isolated populations
Introduction
Dense SNP maps have become the gold standard for whole genome scans because they provide more accurate genotyping of denser and more informative maps at decreasing cost. Consequently, genome-wide association studies are quickly replacing genome-wide linkage scans. Still, there is a particular sampling scheme in which both association and linkage scans can be performed on the same samples: population-based sampling in population isolates with carefully reconstructed genealogy and in which virtually all affected individuals may be related to others. Because linkage and association provide non-redundant information, these two types of analyses are complementary.
However, the use of dense SNP maps for linkage analyses is never straightforward. Indeed, important linkage disequilibrium (LD) may be observed between SNPs and the presence of LD can increase type I error rates in classical nonparametric multipoint linkage analyses, especially when founder genotypes are missing [1, 2]. This issue may be even more pronounced in population isolates. In the large genealogies available and suitable for linkage analysis, some founder genotypes are inevitably missing. Further, the characteristics of population isolates that make them particularly useful for genetic mapping studies are the consequences of founder effects and the subsequent genetic drift. Both processes reduce the amount of variation, influence allele frequencies, and alter the pattern of LD among the variation that persists. In general, significant LD extends over longer distances and with a more stochastic pattern in these populations [3].
Many studies have assessed the effect of LD on type I error of linkage tests in outbred populations, considering nuclear families with simulated LD pattern [1, 2,4,5,6,7,8,9]. Recently, Kim et al. [10] investigated type I error rates in SNP maps of various density (and thus indirectly of different LD levels) with real and moderate LD patterns in multigenerational pedigrees with various missing genotype patterns. Li and Leal [11] further showed that inter-marker LD could increase false-positive evidence of linkage in the analysis of consanguineous pedigrees when genotype data were missing for any pedigree member within a consanguinity loop.
Different approaches have been suggested to perform linkage analysis with maps that include markers in LD. Some authors propose to model LD in the Lander-Green algorithm in clusters of SNPs [6] or between adjacent SNPs only [12]. Thomas [13] incorporates LD in Markov Chain Monte Carlo linkage analysis. An alternative approach has been to perform linkage analysis using subsets of SNPs with minimum LD. Hence, Allen-Brady et al. [14] remove SNPs in high LD through a principal component analysis approach. Bacanu [8] partitions the initial panel of SNPs in k subsets by selecting one marker every k markers, starting with the first SNP for the first subset, with the second SNP for the second subset, etc. Each subset is analysed separately and final linkage statistics are the standardized averages of linkage statistics associated with each subset. If this method allows to take into account all the SNPs of the initial panel, it severely increases the computational burden of the analyses as linkage computation has to be performed for several different maps. Finally, several authors retain only one SNP from each pair of SNPs in high LD or within cluster of SNPs in high LD, using different criteria to choose the SNP to be selected. For example, Schaid et al. [15] retain only the most informative SNP, Murray [16] removes the minimum number of SNPs, Goode et al. [4] select the SNPs which create the shortest gaps in the final map, while Webb et al. [17] select the middle SNP from each cluster of high-LD SNPs (SNPLINK).
Here, we investigate approaches to linkage analysis with a dense SNP map in extended families from the isolated Hutterite population. Our study is based on a sample of individuals phenotyped for asthma related traits and genotyped for both microsatellite markers and the 500K Affymetrix SNP chip. To evaluate the amount of linkage information that can be extracted from the SNP map when LD is minimized, we consider a 30 Mb region on chromosome 12 where we detected a linkage signal with microsatellite markers, using a novel multiple pedigree splitting approach [35] . We start by considering two methods based on a block description of the LD pattern, one modelling LD in the linkage analysis [6] and another selecting a subset of SNPs with minimum LD, SNPLINK [17]. Using simulations, we show that linkage statistics are still highly inflated using either of these two methods in the extended Hutterite families.
To address this issue, we propose a new method to minimize LD in linkage analysis that we name MASEL (MArker SElection for Linkage). MASEL belongs to the category of methods [4, 15, 16] in which SNPs are iteratively selected with minimum pair-wise LD, without assuming any prior hypothesis about the pattern of LD. The novelty of MASEL is to simultaneously control for three SNP selection criteria: number of SNPs in the map, informativity, and regularity of spacing. Simulations show that the inflation of linkage statistics is better controlled with MASEL than with the two previously considered approaches.
Material and Methods
Hutterite Data
Subjects and Phenotype
Many phenotypes have been evaluated in a sample of Hutterites from South Dakota, all related through a 3,028-member pedigree. The study subjects here are 667 individuals, linked through a multiple 13-generation pedigree that includes 1,840 individuals. The mean inbreeding coefficient in this sample is 0.034 (sd = 0.015), slightly greater than that of first-cousins once removed. The phenotype considered is based on the presence of self-reported asthma symptoms (at least two of the following three: cough, wheeze or shortness of breath). 132 individuals are affected in the sample and the disease prevalence is 0.22.
Microsatellite Data
Microsatellite genotyping was performed by the Mammalian Genotyping Service; 597 individuals were genotyped for both Marshfield sets 9 and 51 (658 autosomal microsatellites), 27 individuals for set 51 only (293 autosomal microsatellites) and 43 individuals for set 9 only (365 autosomal microsatellites). Six affected individuals are genotyped for set 51 only and four for set 9 only.
In a previous study [35], we used a multiple pedigree splitting approach to perform linkage analysis for asthma symptoms with the microsatellite markers. Several different sub-pedigree sets, resulting from breaking the 1,840-member Hutterite pedigree with GREFFA [18], were analysed. A linkage for asthma symptoms was detected on chromosome 12q21 using multipoint linkage analysis in several of these sub-pedigree sets.
In the present study, only individuals genotyped for both SNPs and microsatellites are of interest. Consequently, linkage with chromosome 12 using the microsatellite map was reanalyzed considering these individuals only (the results presented hereafter are thus slightly different from the results in Bellenguez et al. [35]). A maximum LOD score of 3.78 is detected at 95.17 cM (about 83 Mb) in a sub-pedigree set, which is comprised of 16 extended sub-pedigrees and includes 206 genotyped and 104 affected individuals. The mean number of affected relative pairs in these sub-pedigrees is 18.7; the mean information content [20] on chromosome 12 is 0.75. Evidence for linkage of asthma or asthma-related traits to chromosome 12q21 has been reported by many other studies [19,20,21,22,23,24,25,26,27,28].
SNP Data
The 667 subjects were genotyped using 500K Affymetrix SNP array in Chicago and for 1,126 nonsynonymous SNPs by the NHLBI-funded Resequencing and Genotyping Service at Johns Hopkins University. In what follows, we focus on the region between 70 and 100 Mb of chromosome 12 that includes the linkage signal detected with microsatellite markers. We only consider SNPs with minor allele frequency (MAF) greater than 5%, a Hardy Weinberg p-value test greater than 0.001, a call rate greater than 90% and genotyped for at least 85% of the individuals. The Hardy Weinberg test was corrected for inbreeding and population structure using gene-dropping simulations on the large Hutterite pedigree: the empirical distribution of the Hardy Weinberg test score in the replicates was used to estimate the p-value of the observed Hardy Weinberg test statistics. Mendelian inheritance inconsistencies were checked with the Pedcheck program [29] on the 1,840-member pedigree. Non mendelian inconsistencies were further checked with Merlin [30] on the 171 nuclear families (mean number of genotyped individuals in families is five) of the 1,840-member pedigree. The final SNP set was comprised of 2,228 SNPs across the 30 Mb region of interest on chromosome 12.
Computation of Linkage Disequilibrium
Linkage disequilibrium between SNPs was computed with Haploview [31] from 76 genotyped individuals iteratively selected from the 667 genotyped subjects so that the kinship between any pair of individuals did not exceed 0.1. In practice, we iteratively removed from the initial set of individuals the individual who had a kinship above 0.1 with the highest number of other individuals, until no individual pair with kinship above 0.1 remained.
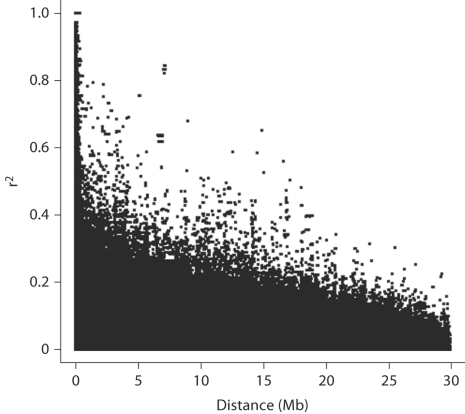
In what follows, we consider r2 as the measure of LD, shown to be better than D′ predicting inflation of linkage statistics due to LD [1, 7]. LD was computed between all SNP pairs of the 30 Mb region of chromosome 12. In this region, the mean r2 between all SNP pairs is 0.03 (sd = 0.05). The 25, 50 and 75% quantiles are 0.003, 0.014 and 0.037 respectively. The LD pattern in this region is shown in figure 1. Considering only adjacent SNPs, the mean r2 between SNP pairs is 0.41 (sd = 0.39) and the 25, 50 and 75% quantiles are 0.058, 0.235 and 0.874 respectively.
Fig. 1.
LD pattern of the SNPs between 70 and 100 Mb on chromosome 12. LD is measured by r2.
Linkage Analysis with LD
Block-Based Methods: Merlin and SNPLINK
We used the LD modelling in linkage analysis implemented in Merlin 1.1.2 [6]. Briefly, in this algorithm, clusters of SNPs are first defined and LD is then modelled by estimating haplotype frequencies within each cluster. Absence of LD is assumed among markers of different clusters and absence of recombination is assumed among markers of each cluster. We used an r2 criterion to define the clusters: markers for which pair-wise r2 exceeds a predefined threshold are grouped together with all intervening markers in a cluster.
SNPLINK [17] implements an algorithm that creates a SNP subset with minimum LD (that we call the linkage set) from a large initial SNP set (that we call the source set). It first defines blocks of adjacent markers in LD and then retains only the middle SNP from each block to build the linkage set. Two SNPs are considered in LD if their pair-wise LD is above a threshold defined by the user. We used the LD measures obtained from our 76-individual sample (see ‘Computation of linkage disequilibrium’ above) to select SNPs in the 30 Mb region of chromosome 12 with SNPLINK.
For both methods, an r2 threshold of 0.01 was used because it ensures the best LD control possible with the two approaches, while still allowing for good detection of the microsatellite linkage signal (see ‘Results’ below).
MASEL
We developed a SNP subset selection method that does not assume a LD block pattern. This method, MASEL, iteratively creates a SNP subset suitable for linkage analysis (referred to as the linkage set, as above) from a large initial SNP set (referred to as the source set, as above). Starting with two SNPs, the linkage set is created by selecting at each step a SNP of the source set that is not in LD with the SNPs already present in the linkage set. Two SNPs are considered in LD if their pair-wise LD is above a threshold T, specified by the user. A similar framework has been proposed by different authors that have suggested a variety of criteria to select the SNPs at each step. Schaid et al. [15] retain the most informative SNP, Goode et al. [4] select the SNP that minimizes inter-marker distance in the linkage set, and Murray [16] tries to maximize the number of SNPs selected. Since the quality of a linkage set depends on each of these criteria, MASEL considers all of them during the selection procedure.
Initialization
Initialization of the algorithm consists in selecting at least two SNPs for the linkage set. When possible, the initial linkage set is simply made up of all the SNPs of the source set that are not in LD with any other SNPs (pair-wise LD below the threshold T). If only one SNP meets this criterion, this marker is selected with the most polymorphic SNP of the source set that is not in LD with this initial SNP to make up the initial linkage set. If all the SNPs of the source set are in LD with at least one other SNP, the two most polymorphic SNPs that are not in LD one with the other are taken to initialize the algorithm.
After this initial step, the SNPs of the source set in LD with those in the linkage set are removed from the source set. During the initialization step, if many SNPs are equally informative, the one that removes the least number of SNPs from the source set is systematically preferred.
Algorithm
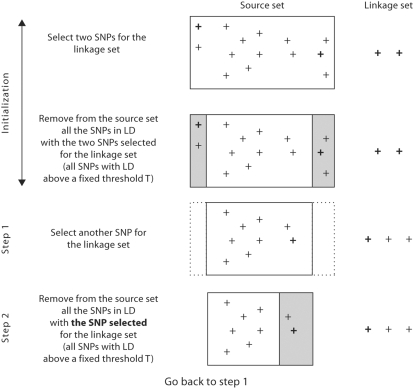
Each step of the algorithm consists in adding one SNP of the source set to the linkage set (step 1, fig. 2) and then removing from the source set the SNPs in LD with this selected SNP (step 2, fig. 2).
Fig. 2.
MASEL algorithm.
At each step 1 and for each SNP of the source set, the algorithm computes the variance of the distance between the SNPs of the linkage set that would be created if this SNP was selected and the number of SNPs that would be removed from the source set if this SNP was selected. Three ranks are then assigned to each SNP of the source set: rankdist corresponding to the inter-SNP distance variance; ranksize corresponding to the number of SNPs removed from the source set and rankhet corresponding to its minor allele frequency. rankdist and ranksize are built in increasing order while rankhetis built in decreasing order. rankdist = 1 for the SNP minimizing the inter-SNP distance variance, ranksize = 1 for the SNP minimizing the number of SNPs removed from the source set and rankhet = 1 for the SNP with the highest heterozygosity. Two SNPs can have an equal value for one or two criteria. In this case, SNPs are sorted according to the other criteria. For example, if two SNPs have the same MAF, the smallest rankhet is assigned to the SNP minimizing the inter-SNP distance variance.
Finally, the algorithm computes for each SNP of the source set:
M = whetrankhet + wdistrankdist + wsizeranksize
where whet, wdist and wsize are weights that are applied to each criterion. The SNP of the source set that minimizes M is finally added to the linkage set. Then, in step 2, SNPs with a LD above the threshold T with this selected SNP are removed from the source set. The algorithm is applied until no SNP remains in the source set.
MASEL thus tries to generate a linkage set made up of a high number of informative SNPs with pair-wise LD lower than the threshold T and uniformly distributed (the rationale for minimizing the inter-SNP distance variance). Four parameters have to be specified by the user: the LD threshold and the three criterion weights. This algorithm is implemented in a Perl program, available from the authors. Even if we present MASEL in the context of linkage analysis, the program can be used to select a SNP map for other analyses that require markers with minimum LD, as for example genome-based inbreeding estimation following the method proposed by Leutenegger et al. [33].
Linkage Analysis
Multipoint nonparametric linkage analysis was performed with the LOD score based on Spairs with the exponential model [34] in the same 16 Hutterite sub-pedigrees considered in the microsatellite analysis. All 2,228 SNPs of the region are used in Merlin 1.1.2 for the LD modelling option, whereas the linkage sets created with SNPLINK and MASEL are considered without the modelling LD option in Merlin. We make the assumptions that 1 Mb is approximately equivalent to 1 cM in the analysis.
Control of LD-Driven Bias
We designed two different simulation schemes under the null hypothesis of no linkage to assess whether false positive evidence of linkage still occurs when LD is minimized using Merlin, SNPLINK and MASEL. We underline that these two simulation schemes are not designed to assess the significance of the results obtained in our data. The first simulation scheme is based on nuclear families while the second uses extended families with a structure similar to that of the 16 Hutterite sub-pedigrees of our data. In both cases, a phenotype re-assignment is performed to simulate a trait unlinked to markers while keeping the observed genotypes. We thus consider the particular LD pattern of the Hutterite population and, since individual genotypes are not available for all markers, the missing genotype pattern of our data.
Simulations on Nuclear Families
With these simulations, we seek to assess the LD-driven bias through the comparison of linkage statistics computed under the null hypothesis of no linkage in nuclear families with missing parental genotypes to those computed when parental genotypes are available, the latter is used as the reference.
82 nuclear families with both parents genotyped are available in the 1,840-member Hutterite pedigree. Sibship sizes range from 2 to 10. Phenotype assignment is applied on the 359 children of these families by assigning to each child a value from a uniform distribution between 0 and 1. The child is considered affected if this value is below 0.22, the prevalence of the disease in our data.
5,000 replicates were performed. For each of them, two runs of linkage analysis were carried out on informative families (families with at least two affected children). The first considers parental genotypes while in the second, parental genotypes are set as unknown. In both cases, the statistic of interest (LOD) is computed with Merlin several times: on the linkage set created by SNPLINK, on the linkage sets created by MASEL, on the microsatellite map, and on all the SNPs (using in this latter case the LD modelling option with r2 = 0.01). To ensure comparable information content in the 30 Mb region of interest between the SNP and microsatellite analyses, all microsatellites of chromosome 12 were used in the multipoint computation.
For each analysis, the maximum LOD score is identified in each replicate. The empirical distributions of these maximum LOD values are used to estimate the empirical significance thresholds at 5% (ST) in each situation of family configuration and marker map.
We note that we compare results obtained on the same replicates considering parental genotypes available or not. Thus, the fact that the number of affected individuals varies between replicates should not have impact our conclusions.
Simulations on Extended Families
These simulations aim to assess LD-driven bias in family structures similar to those of the 16 Hutterite extended families of our data. In these simulations, the results obtained on the microsatellites are used for comparison.
Contrary to the 82 nuclear families we considered in the previous simulation scheme, the 16 extended families of our data were selected so as to include a great number of affected individuals. Because 104 of the 206 genotyped individuals of the 16 extended Hutterite sub-pedigrees are affected, the phenotype assignment could not be performed in an optimal way in those families (an important proportion of individuals would be considered as affected in agreement with their real observed phenotype. The region under study being linked to the phenotype, an important proportion of replicates would be closed to the alternative hypothesis). The phenotype assignment procedure described for the nuclear families is thus applied on the 667 genotyped individuals of the 1,840-member Hutterite pedigree. For each replicate, the 1,840-member pedigree is broken in extended families with GREFFA [18], setting GREFFA parameters to the values used to create the 16 Hutterite extended families of our data. Only replicates for which all sub-pedigrees have a bit size under 23 are analysed (considering bit size equals 2n – f, with n the number of non-founders and f the number of founders). For each of these replicates, linkage analysis is performed on the extended families for the linkage set created by SNPLINK, the linkage sets created by MASEL and the microsatellite map. Because the complexity of the extended families is intractable by the Merlin LD modelling option for many replicates, these simulations could not be conducted for the whole SNP map. Empirical significance thresholds at 5% associated to the maximum LOD scores are then estimated on 5,000 replicates for the maps considered.
In this procedure, affected individuals differ between replicates. Consequently, the number and the structures of the extended families also vary. However, similarly to the note we made on simulations performed on nuclear families, we compare the significance thresholds obtained with the microsatellite and the SNP maps on the same replicates. Consequently, the fact that the number of affected individuals and families differ between replicates will not matter.
Besides, to generate the extended families for the simulations, we used the same fixed parameter set as the one used to create the 16 extended Hutterite families of our data. This ensures a comparable complexity of families between those used in the simulations and those available in our data. We thus expect that the conclusions made according to the simulations will apply to our data.
Results
Block-Based Methods: Merlin and SNPLINK
The 2,228 SNPs in our region of interest are modelled in 113 SNP clusters by Merlin in the 16 extended Hutterite sub-pedigrees. A maximum LOD score of 4.36 is detected at 84.59 Mb with a region-wide mean information content of 0.91 (table 1).
Table 1.
Maximum LOD scores and mean information content (IC) for the microsatellite map, the SNPLINK linkage set, the MASEL linkage set created with the weighting scheme (1, 1, 1) and a r2 threshold of 0.04 and the SNP map with LD modelled by Merlin
| Merlin r2 = 0.01 | SNPLINK r2 = 0.01 | MASEL (1, 1, 1) r2 = 0.04 | Microsatellites | |
|---|---|---|---|---|
| Max LOD (position in Mb) | (DOT)4.36 (84.59) | (DOT)4.37 (86.22) | (DOT)3.67 (87.46) | (DOT)3.78 (83) |
| Mean information content | (DOT)0.91 | (DOT)0.90 | (DOT)0.80 | (DOT)0.73 |
The linkage set selected using SNPLINK is comprised of 168 SNPs (see table 2 for details). A maximum LOD score of 4.37 is detected at 86.22 Mb with this map, the mean information content being 0.9 (table 1).
Table 2.
Characteristics of the linkage set created by SNPLINK
| Number of SNPs | 168 |
| Median MAF | |
| [lower quartile – upper quartile] | 0.20 [0.10–0.38] |
| Mean MAF (sd) | 0.24 (0.14) |
| Mean inter-SNP distance in Mb (sd) | 0.18 (0.19) |
| Maximum inter-SNP distance in Mb | 0.96 |
| Median r2 | |
| [lower quartile – upper quartile] | 0.013 [0.003–0.035] |
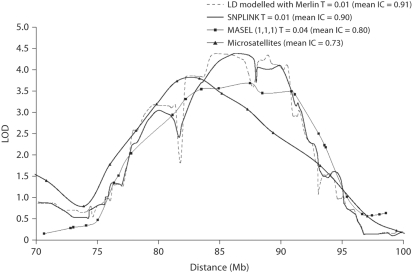
In both cases, the linkage signals are stronger and the information content greater than with the microsatellite map (see fig. 3 and table 1). This may result either from the greater information content using SNP maps or from residual LD between SNPs that may have falsely increased these statistics. These alternative hypotheses can be distinguished by the simulations performed under the null hypothesis (table 3 and 4).
Fig. 3.
LOD curves and mean information content (IC) for the microsatellite map, the SNPLINK linkage set, the MASEL linkage set created with the weighting scheme (1, 1, 1) and a r2 threshold of 0.04 and the SNP map with LD modelled by Merlin.
Table 3.
Significance thresholds at 5% using simulations on nuclear families with parental genotypes available or missing
| Significance threshold at 5% | ||
|---|---|---|
| Parental genotypes | ||
| available | missing | |
| Microsatellite markers | (DOT)1.30 | (DOT)1.41 |
| SNPLINK linkage set (r2 threshold 0.01) | (DOT)1.46 | (DOT)2.19 |
| LD modelling with Merlin (r2 threshold 0.01) | (DOT)1.50 | (DOT)2.27 |
| MASEL linkage set | ||
| (1, 1, 1), T = 0.03 | (DOT)1.20 | (DOT)1.42 |
| (1, 1, 1), T = 0.04 | (DOT)1.32 | (DOT)1.65 |
| (1, 1, 1), T = 0.05 | (DOT)1.38 | (DOT)1.88 |
| (1, 1, 1), T = 0.07 | (DOT)1.42 | (DOT)2.05 |
| (1, 1, 1), T = 0.10 | (DOT)1.44 | (DOT)2.11 |
| (1, 1, 1), T = 0.15 | (DOT)1.48 | (DOT)2.16 |
Results are given for the microsatellite markers, the SNPLINK linkage set, the SNP map with LD modelled by Merlin and the linkage sets created with MASEL using the weighting scheme (1, 1, 1) with r2 thresholds T of 0.03, 0.04, 0.05, 0.07, 0.1 and 0.15.
Table 4.
Significance thresholds at 5% using simulations on extended families
| Significance threshold at 5% | |
|---|---|
| Microsatellite markers | (DOT)1.59 |
| SNPLINK linkage set (r2 threshold 0.01) | (DOT)2.31 |
| MASEL linkage set | |
| (1, 1, 1), T = 0.03 | (DOT)1.60 |
| (1, 1, 1), T = 0.04 | (DOT)1.64 |
| (1, 1, 1), T = 0.05 | (DOT)1.89 |
| (1, 1, 1), T = 0.07 | (DOT)1.99 |
| (1, 1, 1), T = 0.10 | (DOT)2.21 |
| (1, 1, 1), T = 0.15 | (DOT)2.38 |
Results are given for the microsatellite markers, the SNPLINK linkage set and the linkage sets created with MASEL using the weighting scheme (1, 1, 1) with r2 thresholds T of 0.03, 0.04, 0.05, 0.07, 0.1 and 0.15.
When simulations under the null are performed on the nuclear families with available parental genotypes (table 3), significance thresholds for a type I error rate of 0.05 are slightly higher for the SNP maps than for the microsatellite map (ST of 1.30, 1.46 and 1.50 for the microsatellite map, the SNPLINK linkage set and the SNP map with modelled LD respectively). This may be due to the greater density of the SNP maps and, therefore, the higher information content of those maps. Further, as we consider the real observed genotypes, parental genotypes may not be available for all markers. Thus, a small linkage bias might be generated in the presence of LD because of these missing data.
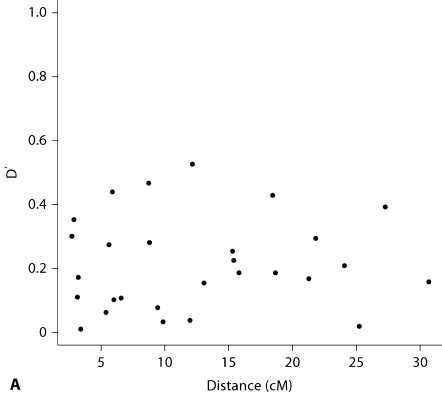
When parental genotypes are removed, we observe an increase of the significance thresholds for the SNPLINK linkage set and the SNP map with modelled LD, even if linkage information is reduced. This increase is similar for both SNP maps (+0.73 for the SNPLINK linkage set and +0.77 when LD is modelled) and suggests that linkage statistics are still falsely inflated when LD is minimized using Merlin or SNPLINK. Note that in these analyses the value of the significance threshold at 5% also slightly increases with the microsatellite map (+0.11). Indeed, although spaced 4.39 cM apart on average, residual LD is present among the microsatellites in this small and highly inbred population (see fig. 4A, B).
Fig. 4.
LD pattern for the microsatellites converted into biallelic markers (most frequent allele renumbered 1, and all others pooled). LD is measured by D′ (A) or by r2 (B).
Simulations in the extended families confirm the results in nuclear families for the SNPLINK linkage set (table 4). Indeed, we observe a +0.72 increase between the significance thresholds of the SNPLINK linkage set and the microsatellite map (ST of 1.59 for the microsatellite map and of 2.31 for the SNPLINK linkage set). This increase is notably more important than the one observed in the nuclear families with parental genotypes available and explained by the difference in information content between the two maps. It is also comparable to the +0.78 increase observed in the nuclear families and caused by missing parental genotypes (ST of 1.41 for the microsatellite map and of 2.19 for the SNPLINK linkage set). Thus, the increase in significance threshold between the microsatellite map and the SNPLINK linkage set observed in extended families is certainly due to residual LD in the SNPLINK linkage set.
A sizeable inflation of linkage statistics is thus expected when LD is minimized using Merlin or SNPLINK in our SNP map, even with a very stringent r2 threshold of 0.01. Our simulations suggest that the +0.59 (for SNPLINK) and +0.58 (for LD modelling) increase in maxLOD on the real data (fig. 3 and table 1) compared to the microsatellite map may be completely explained by the residual LD in the maps. The block structure of LD on which these two approaches rely does not seem to be an appropriate model for linkage analysis in the Hutterite population, even when weak constraints on LD (here, we used r2 = 0.01) are considered to define the blocks. The remaining LD among SNPs in the linkage set created by SNPLINK (median r2 between all selected SNPs of 0.013 as shown in table 2) is large enough to produce a bias in the linkage analysis.
MASEL
Linkage Set Characteristics and Linkage Results
Different linkage sets of the 2,228 SNPs available in the 30 Mb region were generated with MASEL, using six different r2 thresholds T of 0.03, 0.04, 0.05, 0.07, 0.1 and 0.15 and four different weighting schemes for each of the six LD thresholds. Only small variations of the criterion weights were considered to ensure that all criteria are taken into account. These four different weighting schemes are (1, 1, 1), (1, 1, 2), (1, 2, 1), (2, 1, 1), using the notation (whet, wdist, wsize).
Four linkage sets, all created with T = 0.04, but each with a different weighting scheme, are characterized in table 5, through the number of SNPs they include, the mean and median MAF of the SNPs in the set, the mean, standard deviation and maximum inter-SNP distance in the set and the median r2 between SNPs of the set. Mean information content of the linkage sets and maximum LOD scores detected on the 16 Hutterite sub-pedigrees with each set are also given. The same statistics are summarized for the linkage sets created with the other r2 thresholds T in table 6. For each LD threshold, we present the variation ranges of the statistics, corresponding to the four weighting schemes.
Table 5.
Characteristics of the linkage sets created with MASEL with a r2 threshold T of 0.04 and four different weighting schemes
| r2 threshold T = 0.04 | ||||
|---|---|---|---|---|
| Weight set (whet, wdist, wsize) | 111 | 211 | 121 | 112 |
| Number of SNPs | 24 | 24 | 25 | 28 |
| Median MAF [lower quartile – upper quartile] | 0.44 [0.22–0.47] | 0.44 [0.27–0.48] | 0.41 [0.21–0.47] | 0.31 [0.18–0.45] |
| Mean MAF (sd) | 0.36 (0.15) | 0.37 (0.14) | 0.34 (0.16) | 0.30 (0.16) |
| Mean inter-SNP distance in Mb (sd) | 1.21 (0.83) | 1.24 (0.84) | 1.15 (0.72) | 1.10 (0.78) |
| Maximum inter-SNP distance in Mb | 3.38 | 3.63 | 3.38 | 3.38 |
| Median r2 [lower quartile – upper quartile] | 0.008 [0.002–0.018] | 0.007 [0.002–0.020] | 0.008 [0.002–0.018] | 0.008 [0.002–0.019] |
| Mean information content | 0.80 | 0.80 | 0.80 | 0.80 |
| Maximum LOD score (location in Mb) | 3.67 (87.456) | 3.76 (90.339) | 3.78 (87.456) | 3.70 (87.310) |
Table 6.
Characteristics of the linkage sets created with MASEL with different r2 thresholds T
| r2 threshold T | 0.03 | 0.04 | 0.05 | 0.07 | 0.1 | 0.15 |
|---|---|---|---|---|---|---|
| Number of SNPs range | (NDASH)17–21 | (NDASH)24–28 | (NDASH)33–37 | (NDASH)50–55 | (NDASH)93–100 | (NDASH)183–191 |
| Mean MAF range | (NDASH)0.32–0.37 | (NDASH)0.30–0.37 | (NDASH)0.30–0.35 | (NDASH)0.29–0.33 | (NDASH)0.29–0.30 | (NDASH)0.29–0.29 |
| Median MAF range | (NDASH)0.35–0.45 | (NDASH)0.31–0.44 | (NDASH)0.28–0.40 | (NDASH)0.28–0.39 | (NDASH)0.30–0.36 | (NDASH)0.28–0.31 |
| Mean inter-SNP distance range | (NDASH)1.45–1.85 | (NDASH)1.10–1.24 | (NDASH)0.81–0.93 | (NDASH)0.55–0.61 | (NDASH)0.30–0.32 | (NDASH)0.16–0.16 |
| sd inter-SNP distance range | (NDASH)0.96–1.10 | (NDASH)0.72–0.84 | (NDASH)0.55–0.61 | (NDASH)0.36–0.51 | (NDASH)0.24–0.26 | (NDASH)0.13–0.16 |
| Maximum inter-SNP distance range | (NDASH)3.40–4.09 | (NDASH)3.38–3.63 | (NDASH)1.95–2.47 | (NDASH)1.62–2.18 | (NDASH)1.17–1.51 | (NDASH)0.76–0.94 |
| Median r2 range | (NDASH)0.006–0.007 | (NDASH)0.007–0.008 | (NDASH)0.008–0.010 | (NDASH)0.009–0.010 | (NDASH)0.01–0.011 | (NDASH)0.011–0.012 |
| Mean information content range | (NDASH)0.74–0.77 | (NDASH)0.80–0.80 | (NDASH)0.83–0.84 | (NDASH)0.86–0.87 | (NDASH)0.89–0.89 | (NDASH)0.90–0.90 |
| Maximum LOD score range | (NDASH)2.66–3.49 | (NDASH)3.67–3.78 | (NDASH)3.75–3.98 | (NDASH)3.78–4.31 | (NDASH)4.41–4.70 | (NDASH)4.62–4.70 |
| Location range of maximum LOD score (Mb) | (NDASH)84.91–90.45 | (NDASH)87.31–90.34 | (NDASH)82.93–87.45 | (NDASH)84.84–87.76 | (NDASH)87.46–87.76 | (NDASH)87.76–87.84 |
Characteristics of the linkage sets built using the same r2 threshold T but with different weighting schemes are very similar (table 5 and 6). Even if the median MAF varies slightly between the linkage sets generated with the same T, the information content is not modified.
The number of SNPs, the information content and linkage statistics increase with the r2 threshold T used (table 6). In all cases, only a small part of the SNPs available in the source set are considered in the linkage sets. Using the least stringent r2 threshold T of 0.15, the maximum number of SNPs included in the linkage sets is 191 (8.6 % of the number of SNPs in the source set). With the most stringent r2 threshold T of 0.03, only 17 to 21 SNPs are included in the linkage sets (less than 1% of the number of SNPs in the source set).
Compared to the other linkage sets, the linkage sets created with T = 0.03 show a low information content (minimum of 0.74 compared to 0.80 with the linkage sets created with T = 0.04) and the linkage signal is not always detected (maximum LOD scores between 2.66 and 3.49). Results are much better with the linkage sets generated with T = 0.04, yielding a maximum number of SNPs of 28 and maximum LOD scores between 3.67 and 3.78, similar to that obtained on the microsatellite map (maximum LOD score of 3.78 for the microsatellites).
Control of LD-Driven Bias
To assess whether false positive evidence of linkage still occurs when LD is minimized with MASEL, significance thresholds at 5% of the maximum LOD score that can be observed in nuclear and extended families were estimated from the replicates generated under the null hypothesis to evaluate SNPLINK. These replicates were analysed with the linkage sets created using the weighting scheme (1, 1, 1) for the six LD thresholds. Results of these simulations are given in table 3 and 4.
When parental genotypes are available, significance thresholds at 5% increase with the r2 threshold T used to generate the linkage sets, and therefore with the number of markers in the linkage sets (table 3), but are relatively close to the microsatellite threshold. When parental genotypes are missing in nuclear families, the increase in threshold is always larger than the one observed with the microsatellite map. It varies from +0.22 when T = 0.03 to +0.68 when T = 0.15. LD-driven bias may thus still occur, even when using a stringent T of 0.03, but this inflation is reduced as compared to the results obtained with Merlin (ST increase of 0.77, table 3) or SNPLINK (ST increase of 0.73, table 3).
Simulations on extended families (table 4) confirm this increase in significance threshold with the linkage sets created with T higher than 0.05. However, for the linkage sets generated with T = 0.03 and T = 0.04, significance thresholds (1.60 and 1.64 respectively) are very similar to the one observed for the microsatellite map (ST of 1.59). Thus, the increase seems to be well controlled in extended families for the SNP map created with T = 0.04, contrary to what was observed in nuclear families with parental genotypes missing: the difference between significance thresholds obtained on this linkage set and on the microsatellite map is greater in the simulations performed on nuclear families with no parental genotypes available (difference of 0.24, table 3) than on those performed on extended families (difference of 0.05, table 4). This may be due to a smaller impact of LD because more individuals are genotyped in extended families compared to nuclear families with all parental genotypes missing.
Discussion
In this study, we evaluate linkage bias due to LD in SNP maps in the special case of the highly complex Hutterite population. We considered a 30 Mb region on chromosome 12 where a linkage signal for asthma symptoms was detected using microsatellite markers. Simulations performed using the real LD pattern observed in the Hutterites showed that sizeable inflation of linkage statistics may still occur when LD is minimized by Merlin or SNPLINK. In our data, LD is not well controlled by these methods, which are both based on LD block patterns. Because of its history, the Hutterite population may indeed show a more stochastic pattern of LD that is not well captured by blocks.
To allow for better control of LD among the SNPs, we developed MASEL, which does not assume a block-like pattern of LD. MASEL iteratively generates a linkage set by selecting SNPs with pair-wise LD below a threshold T. Simulations show that inflation of linkage statistics is better controlled with MASEL than with Merlin or SNPLINK in the Hutterite pedigree, provided that a very stringent LD threshold is used. On the other hand, compared to Merlin or SNPLINK, fewer SNPs are considered in linkage analysis when LD is minimized with MASEL, which has an impact on the information content (table 1).
The discussion on the r2 threshold highlights the key question related to linkage analysis with dense SNP maps in populations with extended and stochastic patterns of LD: is it possible to extract a SNP map at least as informative as the microsatellite map but free of LD-driven bias? From the results we present on nuclear families with all parental genotypes missing, the answer is unfortunately ‘no’. A LD threshold as low as 0.03 still creates a LD-driven bias that is slightly higher than the one observed with the microsatellite map. Yet, the number of SNPs in the corresponding linkage set is so low that linkage detection is not as good as the one obtained with the microsatellites.
Fortunately, the situation is more favourable when considering extended families available in this type of population. In family structures similar to those of the 16 Hutterite extended families, LD-driven bias is well controlled with the linkage sets generated by MASEL at a r2 threshold of 0.04. A smaller impact of LD is indeed expected in extended families because they include non affected genotyped individuals who provide additional information about founder genotypes. Hence, Huang et al. [2] nearly eliminate false positive evidence of linkage by adding two unaffected siblings in nuclear families with one affected sib-pair and missing parental genotypes. Kim et al. [10] also found that the severity of the type I error inflation increases as the amount of missing genotype data increases.
Linkage signals detected by the linkage sets generated by MASEL with a r2 threshold of 0.04 are all located in the same region, between 87 and 90 Mb. The spatial variance of the linkage statistics is thus limited among the linkage sets. These linkage statistics are similar to that obtained with the microsatellite map. This, in turn, means that the correlations among SNPs on standard genotyping platforms are sufficiently high in this population that obtaining a linkage map that is more informative than a dense microsatellite map may be difficult, if not impossible, to extract. Indeed, it seems that LD level between markers must be very low to avoid bias in linkage statistics. Thus, even if only small differences in LD are observed between some isolated populations and large populations, this may be sufficient to lead to a substantial reduction of the number of SNPs that can be considered for linkage analysis. However, even if no additional linkage information can be gained from SNPs compared to microsatellites in these populations, a gain in power should arise from the possibility of conducting linkage and association studies using the same set of markers.
New SNP panels can include more than one million SNPs and consequently more SNPs in LD. We thus expect that MASEL will remove even more SNPs from those panels as compared to what we observe with the 500K SNP chip to ensure no bias due to LD when analysing the linkage map. However, MASEL will probably extract denser and more informative maps thanks to the greatest amount of information available in those panels.
Acknowledgement
We thank Ying Sun for helping with data preparation. This work was supported in part by NIH grants HL56399, HL66533, and HL85197 to C.O., and the NHLBI-funded Marshfield Genotyping Service (Marshfield, Wisconsin) and Resequencing and Genotyping Center at Johns Hopkins University (Baltimore, Maryland).
References
- 1.Boyles AL, Scott WK, Martin ER, Schmidt S, Li YJ, Ashley-Koch A, Bass MP, Schmidt M, Pericak-Vance MA, Speer MC, Hauser ER. Linkage disequilibrium inflates type I error rates in multipoint linkage analysis when parental genotypes are missing. Hum Hered. 2005;59:220–227. doi: 10.1159/000087122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang Q, Shete S, Amos CI. Ignoring linkage disequilibrium among tightly linked markers induces false-positive evidence of linkage for affected sib pair analysis. Am J Hum Genet. 2004;75:1106–1112. doi: 10.1086/426000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bourgain C, Genin E, Quesneville H, Clerget-Darpoux F. Search for multifactorial disease susceptibility genes in founder populations. Ann Hum Genet. 2000;64:255–265. doi: 10.1046/j.1469-1809.2000.6430255.x. [DOI] [PubMed] [Google Scholar]
- 4.Goode EL, Badzioch MD, Jarvik GP. Bias of allele-sharing linkage statistics in the presence of intermarker linkage disequilibrium. BMC Genet. 2005;6(suppl 1):S82. doi: 10.1186/1471-2156-6-S1-S82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levinson DF, Holmans P. The effect of linkage disequilibrium on linkage analysis of incomplete pedigrees. BMC Genet. 2005;6 (suppl 1):S6. doi: 10.1186/1471-2156-6-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abecasis GR, Wigginton JE. Handling marker-marker linkage disequilibrium: Pedigree analysis with clustered markers. Am J Hum Genet. 2005;77:754–767. doi: 10.1086/497345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xing C, Sinha R, Xing G, Lu Q, Elston RC. The affected-/discordant-sib-pair design can guarantee validity of multipoint model-free linkage analysis of incomplete pedigrees when there is marker-marker disequilibrium. Am J Hum Genet. 2006;79:396–401. doi: 10.1086/506331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bacanu SA. Multipoint linkage analysis for a very dense set of markers. Genet Epidemiol. 2005;29:195–203. doi: 10.1002/gepi.20089. [DOI] [PubMed] [Google Scholar]
- 9.Cho K, Yang Q, Dupuis J. Handling linkage disequilibrium in linkage analysis using dense single-nucleotide polymorphisms. BMC Proc. 2007;1:S161. doi: 10.1186/1753-6561-1-s1-s161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim Y, Duggal P, Gillanders EM, Kim H, Bailey-Wilson JE. Examining the effect of linkage disequilibrium between markers on the type I error rate and power of nonparametric multipoint linkage analysis of two-generation and multigenerational pedigrees in the presence of missing genotype data. Genet Epidemiol. 2008;32:41–51. doi: 10.1002/gepi.20260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li B, Leal SM. Ignoring intermarker linkage disequilibrium induces false-positive evidence of linkage for consanguineous pedigrees when genotype data is missing for any pedigree member. Hum Hered. 2008;65:199–208. doi: 10.1159/000112367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Albers C, Kappen H. Modeling linkage disequilibrium in exact linkage computations: A comparison of first-order markov approaches and the clustered-markers approach. BMC Proc. 2007;1:S159. doi: 10.1186/1753-6561-1-s1-s159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thomas A. Towards linkage analysis with markers in linkage disequilibrium by graphical modelling. Hum Hered. 2007;64:16–26. doi: 10.1159/000101419. [DOI] [PubMed] [Google Scholar]
- 14.Allen-Brady K, Horne B, Malhotra A, Teerlink C, Camp N, Thomas A. Analysis of high-density single-nucleotide polymorphism data: Three novel methods that control for linkage disequilibrium between markers in a linkage analysis. BMC Proceedings. 2007;1:S160. doi: 10.1186/1753-6561-1-s1-s160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schaid DJ, Guenther JC, Christensen GB, Hebbring S, Rosenow C, Hilker CA, McDonnell SK, Cunningham JM, Slager SL, Blute ML, Thibodeau SN. Comparison of microsatellites versus single-nucleotide polymorphisms in a genome linkage screen for prostate cancer-susceptibility loci. Am J Hum Genet. 2004;75:948–965. doi: 10.1086/425870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murray SS. Evaluation of linkage disequilibrium and its effect on non-parametric multipoint linkage analysis using two high density single-nucleotide polymorphism map- ping panels. BMC Genet. 2005;6(suppl 1):S85. doi: 10.1186/1471-2156-6-S1-S85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Webb EL, Sellick GS, Houlston RS. Snplink: Multipoint linkage analysis of densely distributed snp data incorporating automated linkage disequilibrium removal. Bioinformatics. 2005;21:3060–3061. doi: 10.1093/bioinformatics/bti449. [DOI] [PubMed] [Google Scholar]
- 18.Falchi M, Forabosco P, Mocci E, Borlino CC, Picciau A, Virdis E, Persico I, Parracciani D, Angius A, Pirastu M. A genomewide search using an original pairwise sampling approach for large genealogies identifies a new locus for total and low-density lipoprotein cholesterol in two genetically differentiated isolates of Sardinia. Am J Hum Genet. 2004;75:1015–1031. doi: 10.1086/426155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES. Parametric and nonparametric linkage analysis: A unified multipoint approach. Am J Hum Genet. 1996;58:1347–1363. [PMC free article] [PubMed] [Google Scholar]
- 20.Barnes KC, Freidhoff LR, Nickel R, Chiu YF, Juo SH, Hizawa N, Naidu RP, Ehrlich E, Duffy DL, Schou C, Levett PN, Marsh DG, Beaty TH. Dense mapping of chromosome 12q13.12-q23.3 and linkage to asthma and atopy. J Allergy Clin Immunol. 1999;104:485–491. doi: 10.1016/s0091-6749(99)70398-2. [DOI] [PubMed] [Google Scholar]
- 21.Barnes KC, Neely JD, Duffy DL, Freidhoff LR, Breazeale DR, Schou C, Naidu RP, Levett PN, Renault B, Kucherlapati R, Iozzino S, Ehrlich E, Beaty TH, Marsh DG. Linkage of asthma and total serum ige concentration to markers on chromosome 12q: Evidence from Afro-Caribbean and Caucasian populations. Genomics. 1996;37:41–50. doi: 10.1006/geno.1996.0518. [DOI] [PubMed] [Google Scholar]
- 22.Nickel R, Wahn U, Hizawa N, Maestri N, Duffy DL, Barnes KC, Beyer K, Forster J, Bergmann R, Zepp F, Wahn V, Marsh DG. Evidence for linkage of chromosome 12q15-q24.1 markers to high total serum ige concentrations in children of the German multicenter allergy study. Genomics. 1997;46:159–162. doi: 10.1006/geno.1997.5013. [DOI] [PubMed] [Google Scholar]
- 23.Celedon JC, Soto-Quiros ME, Avila L, Lake SL, Liang C, Fournier E, Spesny M, Hersh CP, Sylvia JS, Hudson TJ, Verner A, Klanderman BJ, Freimer NB, Silverman EK, Weiss ST. Significant linkage to airway responsiveness on chromosome 12q24 in families of children with asthma in Costa Rica. Hum Genet. 2007;120:691–699. doi: 10.1007/s00439-006-0255-5. [DOI] [PubMed] [Google Scholar]
- 24.CSGA. A genome-wide search for asthma susceptibility loci in ethnically diverse populations. The collaborative study on the genetics of asthma (CSGA) Nat Genet. 1997;15:389–392. doi: 10.1038/ng0497-389. [DOI] [PubMed] [Google Scholar]
- 25.Dizier MH, Besse-Schmittler C, Guilloud-Bataille M, Annesi-Maesano I, Boussaha M, Bousquet J, Charpin D, Degioanni A, Gormand F, Grimfeld A, Hochez J, Hyne G, Lockhart A, Luillier-Lacombe M, Matran R, Meunier F, Neukirch F, Pacheco Y, Parent V, Paty E, Pin I, Pison C, Scheinmann P, Thobie N, Vervloet D, Kauffmann F, Feingold J, Lathrop M, Demenais F. Genome screen for asthma and related phenotypes in the French egea study. Am J Respir Crit Care Med. 2000;162:1812–1818. doi: 10.1164/ajrccm.162.5.2002113. [DOI] [PubMed] [Google Scholar]
- 26.Wjst M, Fischer G, Immervoll T, Jung M, Saar K, Rueschendorf F, Reis A, Ulbrecht M, Gomolka M, Weiss EH, Jaeger L, Nickel R, Richter K, Kjellman NI, Griese M, von Berg A, Gappa M, Riedel F, Boehle M, van Koningsbruggen S, Schoberth P, Szczepanski R, Dorsch W, Silbermann M, Wichmann HE, et al. A genome-wide search for linkage to asthma. German asthma genetics group. Genomics. 1999;58:1–8. doi: 10.1006/geno.1999.5806. [DOI] [PubMed] [Google Scholar]
- 27.Yokouchi Y, Nukaga Y, Shibasaki M, Noguchi E, Kimura K, Ito S, Nishihara M, Yamakawa-Kobayashi K, Takeda K, Imoto N, Ichikawa K, Matsui A, Hamaguchi H, Arinami T. Significant evidence for linkage of mite-sensitive childhood asthma to chromosome 5q31-q33 near the interleukin 12 b locus by a genome-wide search in Japanese families. Genomics. 2000;66:152–160. doi: 10.1006/geno.2000.6201. [DOI] [PubMed] [Google Scholar]
- 28.Blumenthal MN, Rich SS, King R, Weber J. Approaches and issues in defining asthma and associated phenotypes map to chromosome susceptibility areas in large Minnesota families. The collaborative study for the genetics of asthma (CSGA) Clin Exp Allergy. 1998;28(suppl 1):51–55. doi: 10.1046/j.1365-2222.1998.0280s1051.x. discussion 65–56. [DOI] [PubMed] [Google Scholar]
- 29.O’Connell JR, Weeks DE. Pedcheck: A program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin – rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 31.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 32.Ober C. Perspectives on the past decade of asthma genetics. J Allergy Clin Immunol. 2005;116:274–278. doi: 10.1016/j.jaci.2005.04.039. [DOI] [PubMed] [Google Scholar]
- 33.Leutenegger AL, Prum B, Genin E, Verny C, Lemainque A, Clerget-Darpoux F, Thompson EA. Estimation of the inbreeding coefficient through use of genomic data. Am J Hum Genet. 2003;73:516–523. doi: 10.1086/378207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kong A, Cox NJ. Allele-sharing models: Lod scores and accurate linkage tests. Am J Hum Genet. 1997;61:1179–1188. doi: 10.1086/301592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bellenguez C, Ober C, Bourgain C. Genet Epidemiol 2008 Oct 6. A multiple splitting approach to linkage analysis in large pedigrees identifies a linkage to asthma on chromosome 12. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]