Phosphopantetheine is a fundamental feature in many biological acyl transfer reactions. The molecule is found imbedded within coenzyme A (CoA), as well as on a post-translationally modified, conserved serine of acyl carrier protein (ACP). Both CoA and ACP play essential roles as acyl group donor substrates in several reactions associated with intermediary metabolism and cell membrane assembly in living organisms.1 Comparative genomics, using phylogenetic profiling, has concluded that the synthesis of phosphopantetheine-containing molecules from common metabolic precursors is fundamentally conserved across all domains of life.2 Genome-wide transposon mutagenesis studies carried out in E. coli have also revealed the essential nature of genes involved in vitamin B5 metabolism (coaABCDE).3, 4 The critical nature of phosphopantetheine-containing molecules to the integrity and viability of cells makes the biosynthetic pathway leading to the production of these compounds an intriguing target for antimicrobial development.
Phosphopantothenolycysteine synthetase (PPCS) plays a vital role in the conversion of pantothenate (vitamin B5) to CoA and is responsible for installing the biologically reactive cystamine moiety contained within the phosphopantetheine prosthetic group.5, 6 PPCS activity is essential to every kingdom of life and can be broadly classified by three types: Type I PPCS are found in a majority of bacteria and archaea, are CTP specific, and are expressed as the C-terminal domain of a bifunctional protein fusion in conjunction with phosphopantothenolycysteine decarboxylase (PPCDC).6, 7 Type II PPCS are found mainly in eukaryotes, can use either ATP or CTP to support catalysis, and are expressed as a monofunctional enzyme.3, 8 Type III PPCS, found in a smaller subset of bacteria, are also expressed monofunctionally and have recently been shown to be CTP specific.9
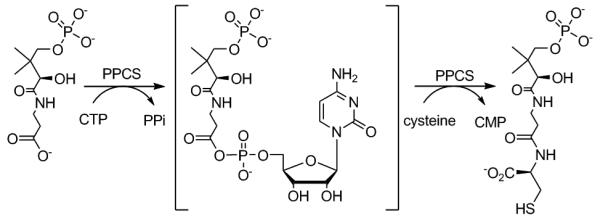
Chemically, PPCS catalysis proceeds via an acyl nucleotide activated intermediate, analogous to aminoacyl-tRNA synthetases (Figure 1).9, 10 The topical antibiotic mupirocin inhibits bacterial isoleucyl tRNA synthetase by mimicking binding contacts of the isoleucyl adenylate intermediate.11 Kinetic and structural studies have shown that the nucleobase binding sites of bacterial PPCS (Types I & III) differ greatly from that of the human Type II enzyme.8, 10, 12 We have therefore designed and synthesized several inhibitors which mimic the acyl cytidylate formed during bacterial PPCS catalysis, which utilize exclusively CTP to support catalysis. In the design of our intermediate mimics, we removed the electrophilic carbonyl of the phosphopantothenoyl cytidylate to create stable phosphodiesters 3 and 4. Alternatively, the carbonyl carbon was left intact in analogues 7 and 8, and a nonhydrolysable sulfamate isostere used in place of the bridging phosphate of the phosphopantothenoyl cytidylate mixed anhydride linkage.
Figure 1.
Reaction catalyzed by phosphopantothenoylcysteine synthetase.
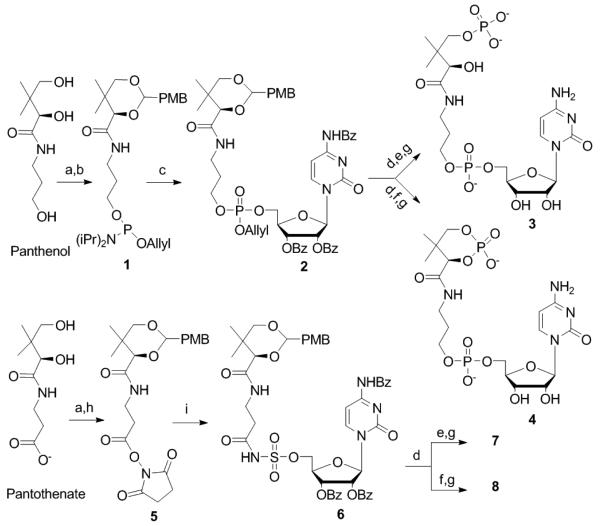
The synthesis of the phosphodiester mimics begins with the protection of D-panthenol as a p-methoxybenzylidene (PMB) acetal, followed by a tetrazole-mediated phosphitylation at the open primary alcohol to give phosphite 1 (Figure 2).13, 14 Phosphite 1 is then coupled in a similar manner to the 5′-hydroxy group of a tribenzoyl protected cytidine, followed by an in situ oxidation using CSO to yield the phosphodiester 2.13, 14 After removal of the p-methoxybenylidene, the open primary alcohol is phosphitylated using pyridinium HCl as the activator at −20°C and oxidized in situ to give the protected terminal phosphate.15 However, if tetrazole-mediated phosphitylation of the opened diol was employed at room temperature, subsequent oxidation gave the 1,3-cyclic phosphate. The global deprotection of both the terminal and cyclic phosphate analogues is accomplished in two sequential steps to give the desired products 3 and 4.16, 17
Figure 2.
Synthesis of inhibitors: a) (MeO)2CHC6H4OMe, CSA, DMF b) Allyl-O-P[N(i-Pr2)]2, 5-(Ethylthio)-1H-tetrazole, DCM c) 1) 5-(Ethylthio)-1H-tetrazole, HO-2′,3′,N4-tribenzoyl cytidine, CH3CN 2) CSO, 0°C d) 80% AcOH e) 1) pyridinium HCl, β-cyanoethyl-O-P[N(iPr2)]2, CH3CN, −20°C 2) CSO, 0°C f) 1) 5-(Ethylthio)-1H-tetrazole, β-cyanoethyl-O-P[N(i-Pr2)]2, CH3CN 2) CSO, 0°C g) 1) TMSCl, DBU, CH3CN 2) NH4OH, 55°C h) NHS, DCC, THF i) Cs2CO3, NH2SO2-2′,3′,N4-tribenzoyl cytidine, DMF.
A similar strategy was employed in the synthesis of the sulfamate analogues. D-pantothenic acid was protected as a PMB acetal and converted to NHS ester 5. Sulfamoyl tribenzoyl cytidine, obtained by sulfamoyl chloride treatment of tribenzoyl cytidine, was then linked to the activated NHS ester in the presence of Cs2CO3.18-20 Compound 6 was subjected to the aforementioned sequence of PMB deprotection, phosphitylation and oxidation, and global deprotection to generate the sulfamate analogues 7 and 8.
Phosphodiester 3 proved to be the most potent PPCS inhibitor, showing nanomolar IC50 towards both Types I and III bacterial enzymes and 145-1000 fold selectivity for bacteria PPCS over the human enzyme (Table 1). Similar selectivity is seen with compound 4, which differs from 3 by the cyclization of the terminal phosphate moiety, albeit with a large decrease in potency. Both compounds 7 and 8, containing the internal sulfonamide linkage, display micromolar IC50 towards bacterial PPCS with 20-740 fold selectivity for the bacterial enzymes.
Table 1.
IC50 of compounds against Types I, II, & III PPCSs.
| hsPPCS (II) | efPPCS (III) | spPPCS (III) | ecPPCS (I) | |
|---|---|---|---|---|
| 3 | 10 μM (1) | 65 nM (9) | 10 nM (2) | 68 nM (9) |
| 4 | 2.7 mM (0.2) | 18 μM (4) | 13 μM (4) | 3.0 μM (0.3) |
| 7 | 200 μM (11) | 2.7 μM (0.2) | 3.9 μM (0.2) | 270 nM (3) |
| 8 | 5.9 mM (0.6) | 181 μM (7) | 279 μM (27) | 16 μM (5) |
hs=human, ef=E. faecalis, sp=S. pneumoniae, ec=E. coli. Assays were performed in triplicate. Standard error shown in ( ).
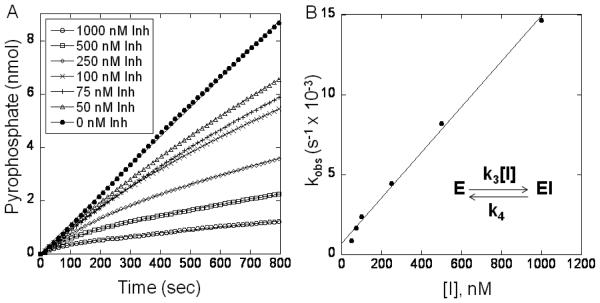
Since a full steady state kinetic characterization of efPPCS has recently been performed, this enzyme was used to determine the kinetic mechanism and inhibition constant for the most potent inhibitor, 3.9 Upon addition of various concentrations of 3 to the PPCS assay a pattern of slow-onset inhibition was observed (Figure 3). Subsequent plotting of the kobs against inhibitor concentration resulted in data points with a linear relationship. This is indicative of a single-step enzyme inhibition mechanism characterized by slow association and slow dissociation of compound 3 (Figure 3). The slope of the line in Figure 3B gives a k3app = 1.42 × 104 M−1s−1, and the y-intercept gives a k4 = 7.02 × 10−4s−1, giving a Kiapp = 49 nM. Taking into account the noncompetitive mode of inhibition of our inhibitors, Kiapp can be converted to Ki using the Cheng-Prusoff equation (see Supporting Information). Therefore, 3 exhibits a Ki = 24 nM for efPPCS.
Figure 3.
Slow-onset, tight-binding inhibition of E. faecalis PPCS by compound 3. A) Enzyme reactions (run in triplicate) were initiated by the addition of efPPCS. Concentrations of compound 3 are displayed in the legend. B) kobs obtained from the fit of the inhibition progress curves is plotted against the concentration of compound 3.
The compounds reported herein represent the first reported inhibitors of PPCS. While very effective against the isolated enzymes, these compounds exhibit no inhibitory effects against bacterial growth, most likely due to lack of cellular penetration as a result of their physiochemical properties. However, in vitro these compounds show a marked selectivity towards both types of bacterial PPCS, providing a foundation for the possible development of broad spectrum antimicrobial agents. Efforts to cocrystallize these inhibitors with all three types of PPCS are currently being investigated. With these studies we hope to gain insight into the binding determinants of selectivity and potency which could be capitalize upon to design the next generation of inhibitors. Also, previous attempts at obtaining crystal structures of PPCS with substrate L-cysteine bound at the active site have not been successful.10 Because our compounds mimic the phosphopantothenoyl cytidylate intermediate but are catalytically incompetent, it is possible that we could capture a ternary crystal complex with PPCS, inhibitor, and L-cysteine, which would provide a clear depiction as to the mechanism of PPCS's selectivity for L-cysteine.21
Supplementary Material
Acknowledgment
We thank Prof. Bruce Palfey for helpful discussions. This work was supported by the University of Michigan, College of Pharmacy (UM-COP). J.D.P. was supported in part by a National Institutes of Health Chemistry and Biology Interface Training Grant and in part by the Fred Lyons, Jr. Fellowship administer by UM-COP. J.Y. was supported in part by a U.S. Department of Homeland Security Fellowship administered by the Oak Ridge Institute for Science & Education.
Footnotes
Supporting Information Available. Complete Ref. 4, Synthetic and biochemical experimental procedures, compound spectroscopic characterization, and equations for inhibition constant determination. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Magnuson K, Jackowski S, Rock CO, Cronan JE., Jr. Microbiol Rev. 1993;57(3):522–542. doi: 10.1128/mr.57.3.522-542.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Genschel U. Mol Biol Evol. 2004;21(7):1242–1251. doi: 10.1093/molbev/msh119. [DOI] [PubMed] [Google Scholar]
- 3.Daugherty M, Polanuyer B, Farrell M, Scholle M, Lykidis A, de Crecy-Lagard V, Osterman A. J Biol Chem. 2002;277(24):21431–21439. doi: 10.1074/jbc.M201708200. [DOI] [PubMed] [Google Scholar]
- 4.Gerdes SY, et al. J Bacteriol. 2002;184(16):4555–4572. doi: 10.1128/JB.184.16.4555-4572.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown GM. J Biol Chem. 1959;234(2):370–378. [PubMed] [Google Scholar]
- 6.Strauss E, Kinsland C, Ge Y, McLafferty FW, Begley TP. J Biol Chem. 2001;276(17):13513–13516. doi: 10.1074/jbc.C100033200. [DOI] [PubMed] [Google Scholar]
- 7.Kupke T, Schwarz W. J Biol Chem. 2006;281(9):5435–5444. doi: 10.1074/jbc.M510056200. [DOI] [PubMed] [Google Scholar]
- 8.Manoj N, Strauss E, Begley TP, Ealick SE. Structure. 2003;11(8):927–936. doi: 10.1016/s0969-2126(03)00146-1. [DOI] [PubMed] [Google Scholar]
- 9.Yao J, Patrone JD, Dotson GD. Biochemistry. 2009;48(12):2799–2806. doi: 10.1021/bi802240w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stanitzek S, Augustin MA, Huber R, Kupke T, Steinbacher S. Structure. 2004;12(11):1977–1988. doi: 10.1016/j.str.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 11.Nakama T, Nureki O, Yokoyama S. J Biol Chem. 2001;276(50):47387–47393. doi: 10.1074/jbc.M109089200. [DOI] [PubMed] [Google Scholar]
- 12.Yao J, Dotson G. Biochimica et Biophysica Acta - Proteins and Proteomics. 2009 Accepted for publication. [Google Scholar]
- 13.Cohen SB, Halcomb RL. J Org Chem. 2000;65(19):6145–6152. doi: 10.1021/jo000646+. [DOI] [PubMed] [Google Scholar]
- 14.Michalski J, Dabkowski W. New Aspects in Phosphorus Chemistry IV. 2004:43–47. [Google Scholar]
- 15.Imoto S, Patro JN, Jiang YL, Oka N, Greenberg MM. J Am Chem Soc. 2006;128(45):14606–14611. doi: 10.1021/ja065525r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evans DA, Gage JR, Leighton JL. J Org Chem. 1992;57(7):1964–1966. [Google Scholar]
- 17.Manoharan M, Lu Y, Casper MD, Just G. Org Lett. 2000;2(3):243–246. doi: 10.1021/ol9910518. [DOI] [PubMed] [Google Scholar]
- 18.Somu RV, Boshoff H, Qiao C, Bennett EM, Barry CE, 3rd, Aldrich CC. J Med Chem. 2006;49(1):31–34. doi: 10.1021/jm051060o. [DOI] [PubMed] [Google Scholar]
- 19.Rolf A, Günter B. Chemische Berichte. 1958;91(6):1339–1341. [Google Scholar]
- 20.Okada M, Iwashita S, Koizumi N. Tetrahedron Letters. 2000;41(36):7047–7051. [Google Scholar]
- 21.Strauss E, Begley TP. Chembiochem. 2005;6(2):284–286. doi: 10.1002/cbic.200400340. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.