Abstract
Introduction
Angiogenesis is an important process in the development of destructive synovial pannus in rheumatoid arthritis (RA). The ITGAV +gene encodes a cell cycle-associated antigen, integrin ανβ 3, which plays a role in RA angiogenesis. Previously, two independent studies identified an association between the major allele of the ITGAV single-nucleotide polymorphism (SNP) rs3738919 and RA. We therefore tested this association in an independent study using New Zealand (NZ) and Oxford (UK) RA case control samples.
Methods
We compared genotype frequencies in 740 NZ Caucasian RA patients and 553 controls genotyped for rs3738919, using a polymerase chain reaction-restriction fragment length polymorphism assay. A TaqMan genotyping SNP assay was used to type 713 Caucasian RA patients and 515 control samples from Oxford for the rs3738919 variant. Association of rs3738919 with RA was tested in these two sample sets using the chi-square goodness-of-fit test. The Mantel-Haenszel test was used to perform a meta-analysis, combining the genetic results from four independent Caucasian case control cohorts, consisting of 3,527 cases and 4,126 controls. Haplotype analysis was also performed using SNPs rs3911238, rs10174098 and rs3738919 in the Wellcome Trust Case Control Consortium, NZ and Oxford case control samples.
Results
We found no evidence for association between ITGAV and RA in either the NZ or Oxford sample set (odds ratio [OR] = 0.88, Pallelic = 0.11 and OR = 1.18, Pallelic = 0.07, respectively). Inclusion of these data in a meta-analysis (random effects) of four independent cohorts (3,527 cases and 4,126 controls) weakens support for the hypothesis that rs3738919 plays a role in the development of RA (ORcombined = 0.92, 95% confidence interval 0.80 to 1.07; P = 0.29). No consistent haplotype associations were evident.
Conclusions
Association of ITGAV SNP rs7378919 with RA was not replicated in NZ or Oxford case control sample sets. Meta-analysis of these and previously published data lends limited support for a role for the ITGAV in RA in Caucasians of European ancestry.
Introduction
Rheumatoid arthritis (RA) is a common systemic autoimmune disease characterised by chronic synovial inflammation leading to the formation of invasive, destructive pannus. The extensive formation of new blood vessels within the affected joint (hyperangiogenesis) is an important component of pannus formation [1].
A heritable component to RA is supported by twin studies [2] and the markedly increased sibling recurrence risk (λs = 5 to 7.2) compared with the general population [3]. Association with the HLA-DRB1 locus is well established, with many other loci also incriminated. These include the protein tyrosine phosphatase non-receptor 22 (PTPN22) gene [4,5], cytotoxic T-lymphocyte antigen 4 (CTLA4) [6], an intergenic region on human chromosome 6 [7,8], signal transducer and activator of transcription 4 (STAT4) [9], the tumour necrosis factor receptor-associated factor 1 region (TRAF/C5) [7,10,11] and CD40, CCL21 and IL2RB [12,13].
The integrin ανβ3 (ITGAV) locus contains 30 exons spanning more than 90 kb of genomic DNA on human chromosome 2q31. It encodes the αν subunit of the cell cycle-associated antigen, integrin ανβ3, which plays a major role in RA angiogenesis. Angiogenesis is stimulated in RA by the increased metabolic demand of the pathologically active synovial tissues [1,14-16]. Potentially, inhibition of this angiogenesis might suppress the destructive activities of pannus and even control disease activity [17]. This is supported by studies in animal models in which injection of ανβ3 antagonists has shown inhibition of neovascularisation and attenuation of joint inflammation [18].
A previous genome-wide linkage scan suggested 19 non-HLA regions contributing to RA in a French population [19]. One of these regions, on human chr2q31, contains the ITGAV gene (CD51). Subsequently, Jacq and colleagues [20] demonstrated an association between RA and the ITGAV rs3738919 C allele in a French Caucasian population (odds ratio for allele frequency difference [ORallelic] = 0.77, 95% confidence interval [CI] 0.63 to 0.94). Significant association with the rs3738919 C allele was also demonstrated from imputed data by the Wellcome Trust Case Control Consortium (WTCCC) (ORallelic = 0.91, 95% CI 0.83 to 1.00) [21]. Collectively, these studies suggest a role for ITGAV in RA and justify further investigation of this locus. We therefore tested this association in an independent study using New Zealand (NZ) and Oxford (UK) RA case control samples.
Materials and methods
Study subjects
The NZ population-based Caucasian sample consists of 740 RA patients fulfilling the American College of Rheumatology (ACR) criteria for RA [22]. Of the patients for whom data were available, 34.3% (234/683) were male, 82.9% (538/649) were rheumatoid factor (RF)-positive, 68.1% (275/404) were anti-cyclic citrullinated peptide (anti-CCP)-positive and 79.4% (576/725) carried the HLA-DRB1 shared epitope. Ethical approval for recruitment of cases was given by the New Zealand Multi-Region Ethics Committee, and recruitment of the controls was approved by the Lower South Ethics Committee. All patients provided written informed consent for the collection of samples and subsequent analysis. The control sample consisted of 553 NZ European Caucasians (226/552 male; 40.9%) with no history of autoimmune disease.
The 713 UK patients were recruited in Oxford, with informed consent from attendees at the rheumatology outpatient clinic at the Nuffield Orthopaedic Centre. All fulfilled the 1987 ACR criteria for RA; the average age of onset was 48 years, 28% were male, 77% were positive for RF and 77% carried the HLA-DRB1 shared epitope. Healthy ethnically matched controls (n = 515) were recruited from the same locale as that of blood donors. Approval for the study was given by the Oxford Research Ethics Committee (OxRec number C02.032).
DNA extraction and genotyping
DNA was extracted from peripheral blood samples of the RA patients and controls by means of guanidine isothiocyanate-chloroform extraction methods. NZ study participants were genotyped for the ITGAV single-nucleotide polymorphism (SNP) rs3738919 using a polymerase chain reaction-restriction fragment length polymorphic SNP genotyping assay as follows: forward primer CACTTTCTGTAAATTAGTGTTAGATCAAAAGG and reverse primer GCTTATAACTCACAATTCAGATTTTTGCC (primers from Sigma-Genosys, Sydney, Australia). The C allele (major allele) of the rs3738919 product (286 base pairs) was digested using the AluI restriction enzyme to form fragments of 223 and 63 base pairs. Oxford study participants were genotyped for rs3738919 using the TaqMan genotyping assay C___1278131_1, and both the NZ and Oxford samples were genotyped for the rs10174098 and rs3911238 ITGAV variants using the TaqMan genotyping assays C__30567648_10 and C___7617051_10, respectively, from Applied Biosystems (Scoresby, Australia).
Although the imputed rs3738919 data were available and were previously reported by Ahnert and Kirsten [21], we reimputed rs3738919 genotypes in order to provide information on imputation parameters. RA case and control genotypes were imputed from the WTCCC dataset using IMPUTE software [23]. Genotypes were imputed from 89% of cases (n = 1,659) and 90% of controls (n = 2,639) using a 7-Mb region and a calling threshold of 0.7.
Statistical analysis
An a priori power calculation was made based on the combined French [20] and WTCCC [21] OR (ORallelic = 1.16), using 800 cases and 600 controls with a major allele frequency of 65%. The power to detect association of rs3738919 to RA using either the NZ or Oxford sample set was estimated to be 47%, using α = 0.05. ITGAV SNPs were tested for deviation from Hardy-Weinberg equilibrium in both the control and RA samples using a chi-square goodness-of-fit test. The significance of differences in the minor allele frequency between RA patients and controls, and stratified patients, was assessed using the chi-square goodness-of-fit test.
Of the five ITGAV SNPs (rs3911238, rs2887827, rs10174098, rs13006571 and rs16828163) genotyped by the WTCCC and rs3738919, only three (rs3911238, rs10174098 and rs3738919) were required to tag all major haplotypes defined by the six variants. Haplotype association analysis was performed using the SHEsis software package [24], which uses a full-precise-iteration algorithm to construct haplotypes and estimate haplotype frequencies.
Meta-analysis combining the French [20], WTCCC [25], NZ and Oxford samples was performed using STATA version 8.0 (StataCorp LP, College Station, TX, USA). The Mantel-Haenszel test was used to estimate the average conditional common OR between the four independent sample sets and to test for any heterogeneity between the four groups using both fixed and random effects. R software [26] was used to perform the Cochran-Armitage trend test to determine recessive, additive and dominant trend values (Table 1).
Table 1.
Allele and genotype distribution of rs3738919
| Cohort | Case, number (frequency) | Control, number (frequency) | P value | OR (95% CI) |
|---|---|---|---|---|
| New Zealand | ||||
| Minor allele | 501 (0.339) | 408 (0.369) | 0.11 | 0.88 (0.75-1.03) |
| Genotype 1,1 | 326 (0.38) | 210 (0.38) | - | 1 |
| 1,2 | 327 (0.44) | 278 (0.50) | 0.021 | 0.76 (0.60-0.96) |
| 2,2 | 87 (0.12) | 65 (0.12) | 0.43 | 0.86 (0.60-1.24) |
| HWE | 0.72 | 0.061 | ||
| Dominant | 0.028 | |||
| Additive | 0.10 | |||
| Recessive | 0.99 | |||
| Oxford, UK | ||||
| Minor allele | 506 (0.355) | 329 (0.319) | 0.068 | 1.17 (0.99-1.39) |
| Genotype 1,1 | 300 (0.42) | 235 (0.46) | - | 1 |
| 1,2 | 319 (0.45) | 231 (0.45) | 0.52 | 1.08 (0.85-1.38) |
| 2,2 | 93 (0.13) | 49 (0.10) | 0.044 | 1.49 (1.01-2.19) |
| HWE | 0.60 | 0.47 | ||
| Dominant | 0.22 | |||
| Additive | 0.069 | |||
| Recessive | 0.055 | |||
| WTCCC | ||||
| Minor allele | 1,141 (0.344) | 1,928 (0.365) | 0.044 | 0.91 (0.83-1.00) |
| Genotype 1,1 | 708 (0.43) | 1,044 (0.40) | - | 1 |
| 1,2 | 761 (0.46) | 1,262 (0.48) | 0.079 | 0.89 (0.78-1.01) |
| 2,2 | 190 (0.11) | 333 (0.13) | 0.094 | 0.84 (0.69-1.03) |
| HWE | 0.50 | 0.11 | ||
| Dominant | 0.043 | |||
| Additive | 0.041 | |||
| Recessive | 0.26 | |||
| Jacq et al. [20] | ||||
| Minor allele | 292 (0.352) | 343 (0.413) | 0.01 | 0.77 (0.63-0.94) |
| Genotype 1,1 | 166 (0.40) | 148 (0.36) | - | 1 |
| 1,2 | 206 (0.50) | 191 (0.46) | 0.80 | 0.96 (0.71-1.29) |
| 2,2 | 43 (0.10) | 76 (0.18) | 0.002 | 0.50 (0.33-0.78) |
| HWE | 0.0718 | 0.30 | ||
| Dominant | 0.20 | |||
| Additive | 0.0096 | |||
| Recessive | 0.0011 | |||
| Combined | ||||
| Minor allele | 2,440 (0.346) | 3,008 (0.365) | 0.014 | 0.92 (0.86-0.98) |
| Genotype 1,1 | 1,500 (0.43) | 1,637 (0.40) | - | 1 |
| 1,2 | 1,614 (0.46) | 1,966 (0.48) | 0.028 | 0.90 (0.81-0.99) |
| 2,2 | 413 (0.12) | 523 (0.13) | 0.046 | 0.86 (0.74-1.00) |
| HWE | 0.51 | 0.083 | ||
| Dominant | 0.011 | |||
| Additive | 0.013 | |||
| Recessive | 0.20 |
CI, confidence interval; HWE, Hardy-Weinberg equilibrium P value; OR, odds ratio; WTCCC, Wellcome Trust Case Control Consortium.
Results
Analysis of rs3738919 in New Zealand and Oxford Caucasian rheumatoid arthritis samples
We genotyped rs3738919 across the NZ and Oxford RA case control cohorts and found no evidence for an association between ITGAV and RA (Pallelic = 0.11; ORallelic = 0.88 [95% CI 0.75 to 1.03] and Pallelic = 0.07; ORallelic = 1.17 [95% CI 0.99 to 1.39], respectively) (Table 1). The direction of the NZ allele distribution is consistent with the previous French [20] and WTCCC [21] data, with the minor allele under-represented in the case groups in all three cohorts (Table 1). However, the allele distribution in the Oxford RA cohort differs in that the minor allele is over-represented in the patient group compared with the control group (Table 1).
Haplotype analysis
Association analysis using our imputed genotypes for rs3738919 (P = 0.04) from the WTCCC data were consistent with the previous report of an association of this SNP with RA in the WTCCC [21]. In our analysis, we obtained imputed genotypes from 89% of the WTCCC case control subjects. Presumably, this reflects the influence of the low linkage disequilibrium that exists between rs3738919 and the genotyped SNPs (0.01 <r2 < 0.54 in CEU [Centre d'Etude du Polymorphisme Humain Utah] HapMap [27]) on confidence calls of imputed genotypes. Haplotype analysis (Table 2) was performed using the actual genotypes from ITGAV SNPs rs3911238 and rs10174098 (Additional data file 1) and the imputed genotypes for rs3738919 (Table 1). One susceptibility haplotype (1-1-1), containing the major allele at each of the three SNPs, was significantly over-represented in cases compared with controls (OR = 1.15, 95% CI 1.02 to 1.30; P = 0.021).
Table 2.
Three-marker haplotype analysis of ITGAV single-nucleotide polymorphisms (rs10174098, rs3911238, rs3738919) in the Wellcome Trust Case Control Consortium, New Zealand and Oxford sample sets
| rs10174098 | rs3911238 | rs3738919 | Case, number (frequency) | Control, number (frequency) | P value | OR (95% CI) |
|---|---|---|---|---|---|---|
| WTCCC | ||||||
| 1 | 1 | 2 | 866 (0.261) | 1,451 (0.275) | 0.19 | 0.94 (0.85-1.03) |
| 2 | 1 | 1 | 858 (0.259) | 1,382 (0.262) | 0.82 | 0.99 (0.90-1.09) |
| 1 | 2 | 1 | 792 (0.239) | 1,226 (0.232) | 0.43 | 1.04 (0.94-1.16) |
| 1 | 1 | 1 | 524 (0.158) | 741 (0.140) | 0.021 | 1.15 (1.02-1.30) |
| 2 | 1 | 2 | 219 (0.066) | 405 (0.077) | 0.070 | 0.85 (0.72-1.01) |
| New Zealand | ||||||
| 1 | 1 | 2 | 357 (0.244) | 260 (0.263) | 0.29 | 0.90 (0.75~1.09) |
| 2 | 1 | 1 | 364 (0.249) | 240 (0.243) | 0.73 | 1.03 (0.86~1.25) |
| 1 | 2 | 1 | 359 (0.246) | 223 (0.225) | 0.26 | 1.12 (0.92~1.35) |
| 1 | 1 | 1 | 241 (0.165) | 170 (0.172) | 0.60 | 0.95 (0.76~1.17) |
| 2 | 1 | 2 | 110 (0.075) | 73 (0.074) | 0.91 | 1.02 (0.75~1.39) |
| Oxford, UK | ||||||
| 1 | 1 | 2 | 310 (0.256) | 252 (0.253) | 0.80 | 1.03 (0.85-1.24) |
| 2 | 1 | 1 | 306 (0.253) | 262 (0.262) | 0.67 | 0.96 (0.79-1.16) |
| 1 | 2 | 1 | 335 (0.277) | 251 (0.252) | 0.16 | 1.15 (0.95-1.39) |
| 1 | 1 | 1 | 158 (0.130) | 162 (0.162) | 0.04 | 0.78 (0.62-0.99) |
| 2 | 1 | 2 | 78 (0.064) | 57 (0.057) | 0.47 | 1.14 (0.80-1.62) |
Only haplotypes with frequency of at least 0.05 in controls are shown. CI, confidence interval; HWE, Hardy-Weinberg equilibrium; OR, odds ratio; WTCCC, Wellcome Trust Case Control Consortium.
Variants rs3911238 and rs10174098 were typed over the NZ and Oxford sample sets, and association of three-marker haplotypes was examined (Additional data file 1 and Table 2). There was no support for a positive association of the 1-1-1 haplotype with RA in either the NZ or Oxford sample set. There was a significant protective effect for this haplotype in the Oxford samples (OR = 0.78, 95% CI 0.62 to 0.99; P = 0.042).
Meta-analysis of ITGAV in the four independent case control sample sets
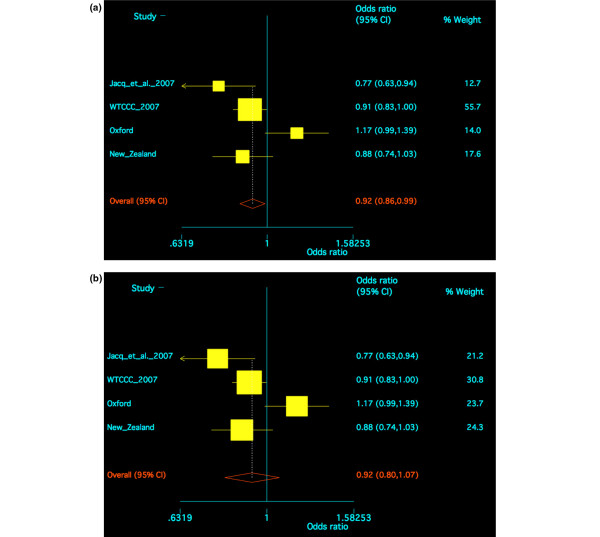
Meta-analysis of all four sample sets, using a fixed effects model, revealed some evidence for an association of rs3738919 with RA (OR = 0.92, 95% CI 0.86 to 0.99; P = 0.021) (Figure 1a). Because of evidence for heterogeneity between the sample sets (Breslow-Day P = 0.011), a random effects model was also used for meta-analysis; this did not provide evidence for an association of rs3738919 with RA (OR = 0.92, 95% CI 0.80 to 1.07; P = 0.29) (Figure 1b). The Oxford sample set is significantly different from the other three sample sets at rs3738919; however, the Oxford patient sample set did not have any large differences in relation to gender (Oxford 28% male, NZ 34%, WTCCC 25%, France 13%), RF status (Oxford 77% positive, NZ 83%, WTCCC 84%, France 75%) or inheritance of the shared epitope (Oxford 77% positive, NZ 79%, WTCCC 79%, France 79%).
Figure 1.
Meta-analysis of the ITGAV single-nucleotide polymorphism rs3738919 in four independent rheumatoid arthritis Caucasian cohorts: Jacq and colleagues [20], Wellcome Trust Case Control Consortium (WTCCC) [25], New Zealand and Oxford sample sets. (a) Fixed effects model. (b) Random effects model. CI, confidence interval.
Stratification according to subphenotype
The NZ and Oxford samples were stratified according to gender, RF and shared epitope status (Table 3). The stratification results did not reveal any significant differences in the rs3738919 genotype distribution for RA patients. There was a difference in the rs3738919 genotype distribution of male and female patients in the NZ cases (P = 0.026) (Table 3). However, a significant difference was not evident in either the Oxford or WTCCC case sample set (P = 0.55 and 0.80, respectively) (Table 3).
Table 3.
Subphenotype analysis of rs3738919 in rheumatoid arthritis patients
| CC, number (frequency) | CA, number (frequency) | AA, number (frequency) | P value | |
|---|---|---|---|---|
| New Zealand | ||||
| Gender | ||||
| Male | 117 (0.500) | 90 (0.385) | 27 (0.115) | 0.026 |
| Female | 177 (0.394) | 216 (0.481) | 56 (0.125) | |
| RF | ||||
| Yes | 232 (0.431) | 239 (0.444) | 67 (0.125) | 0.75 |
| No | 49 (0.441) | 51 (0.459) | 11 (0.100) | |
| SE | ||||
| Yes | 241 (0.418) | 269 (0.467) | 66 (0.115) | 0.05 |
| No | 75 (0.503) | 53 (0.356) | 21 (0.141) | |
| Oxford, UK | ||||
| Gender | ||||
| Male | 89 (0.434) | 86 (0.420) | 30 (0.146) | 0.55 |
| Female | 211 (0.416) | 233 (0.460) | 63 (0.124) | |
| RF | ||||
| Yes | 227 (0.411) | 247 (0.447) | 78 (0.141) | 0.34 |
| No | 69 (0.454) | 68 (0.447) | 15 (0.099) | |
| SE | ||||
| Yes | 226 (0.410) | 254 (0.461) | 71 (0.129) | 0.47 |
| No | 74 (0.457) | 66 (0.407) | 22 (0.136) | |
| WTCCC | ||||
| Gender | ||||
| Male | 180 (0.438) | 187 (0.455) | 44 (0.107) | 0.80 |
| Female | 528 (0.423) | 574 (0.460) | 146 (0.117) |
RF, rheumatoid factor; SE, shared epitope; WTCCC, Wellcome Trust Case Control Consortium.
Discussion
There was no evidence supporting a role for the ITGAV SNP rs3738919 in the etiology of RA when the NZ and the Oxford case control sample sets were analysed separately (P = 0.11 and 0.07, respectively) (Table 1). Trends for association were observed in both sets of samples, but in opposing directions (OR = 0.86 and 1.18, respectively). When all of the available sample sets were analysed together in a random effects model owing to the heterogeneity caused by the Oxford sample set, there was no longer evidence for an association of rs3738919 with RA (OR = 0.92; P = 0.29). To further investigate a potential association of RA with rs3738919, genotyping in a very large cohort will be required. The work previously undertaken by Thomson and colleagues [8] in confirming an association with RA of the Chr6q23 locus is an excellent example of how this can be achieved. A sample set of this size would have 69% power (α = 0.05; OR = 0.92) to detect association at rs3738919.
The rs3738919 ITGAV variant previously showed no association with RA in Japanese case-control samples [28]. These data were not included in our meta-analysis given that the major allele was present at a considerably different frequency in Japanese controls (0.92) than in Caucasian controls (France, 0.58; OXFORD, 0.64; and NZ, 0.63). It is already clear that there are genetic differences in susceptibility to RA between the Japanese and Caucasian populations. For example, the HLA-DRB1*0405 allele is most strongly associated with RA in Japanese patients [29] whereas the *0401 and *0404 alleles are more associated with RA in Caucasians [30]. Other genes showing population-specific effects in RA are the R620W variant of the protein tyrosine phosphatase, PTPN22, associated with Caucasian RA [4,5] but monomorphic in the Japanese population [31]; CTLA4, associated with RA in Caucasian [6] but not associated with RA in Japanese patients [32]; peptidylarginine deiminase type 4, PADI4, associated in Japanese patients [33,34] but very weakly in Caucasians [6,35,36]; and the Fc receptor-like 3 gene variant, FCRL3-169C, associated with RA in Japanese patients [37,38] and in one Caucasian study [39] but not in other Caucasian studies [40,41]. In conclusion, we have not been able to provide further support for the involvement of ITGAV in the etiology of RA in Caucasians. However, it is important that further genotyping be done in a large independent cohort to confirm whether ITGAV plays a role in RA.
Conclusions
In Caucasians, meta-analysis of 3,527 cases and 4,126 controls does not provide further evidence for a role of the ITGAV SNP rs3738919 in the development of RA.
Abbreviations
ACR: American College of Rheumatology; CI: confidence interval; CTLA4: cytotoxic T-lymphocyte antigen 4; ITGAV: integrin ανβ3; NZ: New Zealand; OR: odds ratio; ORallelic: odds ratio for allele frequency difference; PTPN22: protein tyrosine phosphatase non-receptor 22; RA: rheumatoid arthritis; RF: rheumatoid factor; SNP: single-nucleotide polymorphism; WTCCC: Wellcome Trust Case Control Consortium.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
JEH-M and TRM helped to design the study, oversee its execution, and prepare the manuscript. KAR, AJP-G and MEM provided technical support. PG, AAH, PBBJ, LKS and PH helped to provide clinical recruitment and analyse data. ND, JH and BPW helped to provide clinical recruitment, analyse data, and prepare the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Allele and genotype distribution of rs10174098 and rs3911238. OR, odds ratio; CI, confidence interval; HWE, Hardy-Weinberg equilibrium; UK, United Kingdom; WTCCC, Wellcome Trust Case Control Consortium.
Contributor Information
Jade E Hollis-Moffatt, Email: jade.hollis-moffatt@otago.ac.nz.
Kerry A Rowley, Email: kerry.rowley@otago.ac.nz.
Amanda J Phipps-Green, Email: mandy.phipps-green@otago.ac.nz.
Marilyn E Merriman, Email: marilyn.merriman@otago.ac.nz.
Nicola Dalbeth, Email: n.dalbeth@auckland.ac.nz.
Peter Gow, Email: PGow@middlemore.co.nz.
Andrew A Harrison, Email: andrew.harrison@otago.ac.nz.
John Highton, Email: john.highton@otago.ac.nz.
Peter BB Jones, Email: peter.jones@qehealth.co.nz.
Lisa K Stamp, Email: lisa.stamp@cdhb.govt.nz.
Pille Harrison, Email: Pille.Harrison@ndorms.ox.ac.uk.
B Paul Wordsworth, Email: Paul.Wordsworth@ndm.ox.ac.uk.
Tony R Merriman, Email: tony.merriman@stonebow.otago.ac.nz.
Acknowledgements
We thank all of the people who agreed to participate in our study and the Wellcome Trust Case Control Consortium for access to genotype data; Rachel Rodger, Helen Simkins, Wan Rohani Wan Taib and Cushla McKinney for assistance with laboratory work and data analysis; New Zealand research nurses Gael Hewett and Jill James for assistance in recruiting patients; and the Health Research Council of New Zealand and Arthritis New Zealand for their financial support of this work. This work was funded in part by a departmental grant from the Nuffield Department of Orthopaedics, Rheumatology and Musculoskeletal Sciences, University of Oxford, Oxford, UK.
References
- Paleolog EM. Angiogenesis in rheumatoid arthritis. Arthritis Res. 2002;4:S81–90. doi: 10.1186/ar575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silman AJ, MacGregor AJ, Thomson W, Holligan S, Carthy D, Farhan A, Ollier WE. Twin concordance rates for rheumatoid arthritis: results from a nationwide study. Br J Rheumatol. 1993;32:903–907. doi: 10.1093/rheumatology/32.10.903. [DOI] [PubMed] [Google Scholar]
- Wordsworth P, Bell J. Polygenic susceptibility in rheumatoid arthritis. Ann Rheum Dis. 1991;50:343–346. doi: 10.1136/ard.50.6.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begovich AB, Carlton VE, Honigberg LA, Schrodi SJ, Chokkalingam AP, Alexander HC, Ardlie KG, Huang Q, Smith AM, Spoerke JM, Conn MY, Chang M, Chang SY, Saiki RK, Catanese JJ, Leong DU, Garcia VE, McAllister LB, Jeffery DA, Lee AT, Batliwalla F, Remmers E, Criswell LA, Seldin MF, Kastner DL, Amos CI, Sninsky JJ, Gregersen PK. A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am J Hum Genet. 2004;75:330–337. doi: 10.1086/422827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simkins HM, Merriman ME, Highton J, Chapman PT, O'Donnell JL, Jones PB, Gow PJ, McLean L, Pokorny V, Harrison AA, Merriman TR. Association of the PTPN22 locus with rheumatoid arthritis in a New Zealand Caucasian cohort. Arthritis Rheum. 2005;52:2222–2225. doi: 10.1002/art.21126. [DOI] [PubMed] [Google Scholar]
- Plenge RM, Padyukov L, Remmers EF, Purcell S, Lee AT, Karlson EW, Wolfe F, Kastner DL, Alfredsson L, Altshuler D, Gregersen PK, Klareskog L, Rioux JD. Replication of putative candidate-gene associations with rheumatoid arthritis in >4,000 samples from North America and Sweden: association of susceptibility with PTPN22, CTLA4, and PADI4. Am J Hum Genet. 2005;77:1044–1060. doi: 10.1086/498651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plenge RM, Seielstad M, Padyukov L, Lee AT, Remmers EF, Ding B, Liew A, Khalili H, Chandrasekaran A, Davies LR, Li W, Tan AK, Bonnard C, Ong RT, Thalamuthu A, Pettersson S, Liu C, Tian C, Chen WV, Carulli JP, Beckman EM, Altshuler D, Alfredsson L, Criswell LA, Amos CI, Seldin MF, Kastner DL, Klareskog L, Gregersen PK. TRAF1-C5 as a risk locus for rheumatoid arthritis--a genomewide study. N Engl J Med. 2007;357:1199–1209. doi: 10.1056/NEJMoa073491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson W, Barton A, Ke X, Eyre S, Hinks A, Bowes J, Donn R, Symmons D, Hider S, Bruce IN, Wilson AG, Marinou I, Morgan A, Emery P, Carter A, Steer S, Hocking L, Reid DM, Wordsworth P, Harrison P, Strachan D, Worthington J. Rheumatoid arthritis association at 6q23. Nat Genet. 2007;39:1431–1433. doi: 10.1038/ng.2007.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remmers EF, Plenge RM, Lee AT, Graham RR, Hom G, Behrens TW, de Bakker PI, Le JM, Lee HS, Batliwalla F, Li W, Masters SL, Booty MG, JP Carulli, Padyukov L, Alfredsson L, Klareskog L, Chen WV, Amos CI, Criswell LA, Seldin MF, Kastner DL, Gregersen PK. STAT4 and the risk of rheumatoid arthritis and systemic lupus erythematosus. N Engl J Med. 2007;357:977–986. doi: 10.1056/NEJMoa073003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton A, Thomson W, Ke X, Eyre S, Hinks A, Bowes J, Gibbons L, Plant D, Wilson AG, Marinou I, Morgan A, Emery P, Steer S, Hocking L, Reid DM, Wordsworth P, Harrison P, Worthington J. Re-evaluation of putative rheumatoid arthritis susceptibility genes in the post-genome wide association study era and hypothesis of a key pathway underlying susceptibility. Hum Mol Genet. 2008;17:2274–2279. doi: 10.1093/hmg/ddn128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M, Rowland CM, Garcia VE, Schrodi SJ, Catanese JJ, Helm-van Mil AH van der, Ardlie KG, Amos CI, Criswell LA, Kastner DL, Gregersen PK, Kurreeman FA, Toes RE, Huizinga TW, Seldin MF, Begovich AB. A large-scale rheumatoid arthritis genetic study identifies association at chromosome 9q33.2. PLoS Genet. 2008;4:e1000107. doi: 10.1371/journal.pgen.1000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S, Remmers EF, Lee AT, Hackett R, Guiducci C, Burtt NP, Gianniny L, Korman BD, Padyukov L, Kurreeman FA, Chang M, Catanese JJ, Ding B, Wong S, Helm-van Mil AH van der, Neale BM, Coblyn J, Cui J, Tak PP, Wolbink GJ, Crusius JB, Horst-Bruinsma IE van der, Criswell LA, Amos CI, Seldin MF, Kastner DL, Ardlie KG, Alfredsson L, Costenbader KH, Altshuler D. Common variants at CD40 and other loci confer risk of rheumatoid arthritis. Nat Genet. 2008;40:1216–1223. doi: 10.1038/ng.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton A, Thomson W, Ke X, Eyre S, Hinks A, Bowes J, Plant D, Gibbons LJ. Wellcome Trust Case Control Consortium ; YEAR Consortium; BIRAC Consortium. Wilson AG, Bax DE, Morgan AW, Emery P, Steer S, Hocking L, Reid DM, Wordsworth P, Harrison P, Worthington J. Rheumatoid arthritis susceptibility loci at chromosomes 10p15, 12q13 and 22q13. Nat Genet. 2008;40:1156–1159. doi: 10.1038/ng.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimball ES, Gross JL. Angiogenesis in pannus formation. Agents Actions. 1991;34:329–331. doi: 10.1007/BF01988724. [DOI] [PubMed] [Google Scholar]
- Stupack DG, Storgard CM, Cheresh DA. A role for angiogenesis in rheumatoid arthritis. Braz J Med Biol Res. 1999;32:573–581. doi: 10.1590/S0100-879X1999000500011. [DOI] [PubMed] [Google Scholar]
- Clavel G, Bessis N, Boissier MC. Recent data on the role for angiogenesis in rheumatoid arthritis. Joint Bone Spine. 2003;70:321–326. doi: 10.1016/S1297-319X(03)00088-5. [DOI] [PubMed] [Google Scholar]
- Pandya NM, Dhalla NS, Santani DD. Angiogenesis--a new target for future therapy. Vascul Pharmacol. 2006;44:265–274. doi: 10.1016/j.vph.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Storgard CM, Stupack DG, Jonczyk A, Goodman SL, Fox RI, Cheresh DA. Decreased angiogenesis and arthritic disease in rabbits treated with an alphavbeta3 antagonist. J Clin Invest. 1999;103:47–54. doi: 10.1172/JCI3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio-Fortea YJ, Bukulmez H, Petit-Teixeira E, Michou L, Pierlot C, Cailleau-Moindrault S, Lemaire I, Lasbleiz S, Alibert O, Quillet P, Bardin T, Prum B, Olson JM, Cornelis F. Dense genome-wide linkage analysis of rheumatoid arthritis, including covariates. Arthritis Rheum. 2004;50:2757–2765. doi: 10.1002/art.20458. [DOI] [PubMed] [Google Scholar]
- Jacq L, Garnier S, Dieude P, Michou L, Pierlot C, Migliorini P, Balsa A, Westhovens R, Barrera P, Alves H, Vaz C, Fernandes M, Pascual-Salcedo D, Bombardieri S, Dequeker J, Radstake TR, Van Riel P, Putte L van de, Lopes-Vaz A, Glikmans E, Barbet S, Lasbleiz S, Lemaire I, Quillet P, Hilliquin P, Teixeira VH, Petit-Teixeira E, Mbarek H, Prum B, Bardin T, Cornelis F. The ITGAV rs3738919-C allele is associated with rheumatoid arthritis in the European Caucasian population: a family-based study. Arthritis Res Ther. 2007;9:R63. doi: 10.1186/ar2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahnert P, Kirsten H. Association of ITGAV supports a role of angiogenesis in rheumatoid arthritis. Arthritis Res Ther. 2007;9:108. doi: 10.1186/ar2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, Healey LA, Kaplan SR, Liang MH, Luthra HS, Medgser TA, Mitchell DM, Neustadt DH, Pinals RS, Schaller JG, Sharp JT, Wilder RL, Hunde GG. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;31:315–324. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15:97–98. doi: 10.1038/sj.cr.7290286. [DOI] [PubMed] [Google Scholar]
- The Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The R Project for Statistical Computing. http://www.r-project.org
- International HapMap Project. http://hapmap.ncbi.nlm.nih.gov
- Iikuni N, Kobayashi S, Ikari K, Tomatsu T, Hara M, Yamanaka H, Kamatani N, Momohara S. ITGAV polymorphism and disease susceptibility in a Japanese rheumatoid arthritis population. Arthritis Res Ther. 2007;9:405. doi: 10.1186/ar2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi F, Nakano K, Matsuta K, Nabeta H, Bannai M, Tanimoto K, Ito K. Positive and negative association of HLA-DR genotypes with Japanese rheumatoid arthritis. Clin Exp Rheumatol. 1996;14:17–22. [PubMed] [Google Scholar]
- MacGregor A, Ollier W, Thomson W, Jawaheer D, Silman A. HLA-DRB1*0401/0404 genotype and rheumatoid arthritis: increased association in men, young age at onset, and disease severity. J Rheumatol. 1995;22:1032–1036. [PubMed] [Google Scholar]
- Ikari K, Momohara S, Inoue E, Tomatsu T, Hara M, Yamanaka H, Kamatani N. Haplotype analysis revealed no association between the PTPN22 gene and RA in a Japanese population. Rheumatology (Oxford) 2006;45:1345–1348. doi: 10.1093/rheumatology/kel169. [DOI] [PubMed] [Google Scholar]
- Matsushita M, Tsuchiya N, Shiota M, Komata T, Matsuta K, Zama K, Oka T, Juji T, Yamane A, Tokunaga K. Lack of a strong association of CTLA-4 exon 1 polymorphism with the susceptibility to rheumatoid arthritis and systemic lupus erythematosus in Japanese: an association study using a novel variation screening method. Tissue Antigens. 1999;54:578–584. doi: 10.1034/j.1399-0039.1999.540607.x. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Yamada R, Chang X, Tokuhiro S, Sawada T, Suzuki M, Nagasaki M, Nakayama-Hamada M, Kawaida R, Ono M, Ohtsuki M, Furukawa H, Yoshino S, Yukioka M, Tohma S, Matsubara T, Wakitani S, Teshima R, Nishioka Y, Sekine A, Iida A, Takahashi A, Tsunoda T, Nakamura Y, Yamamoto K. Functional haplotypes of PADI4, encoding citrullinating enzyme peptidylarginine deiminase 4, are associated with rheumatoid arthritis. Nat Genet. 2003;34:395–402. doi: 10.1038/ng1206. [DOI] [PubMed] [Google Scholar]
- Ikari K, Kuwahara M, Nakamura T, Momohara S, Hara M, Yamanaka H, Tomatsu T, Kamatani N. Association between PADI4 and rheumatoid arthritis: a replication study. Arthritis Rheum. 2005;52:3054–3057. doi: 10.1002/art.21309. [DOI] [PubMed] [Google Scholar]
- Barton A, Bowes J, Eyre S, Spreckley K, Hinks A, John S, Worthington J. A functional haplotype of the PADI4 gene associated with rheumatoid arthritis in a Japanese population is not associated in a United Kingdom population. Arthritis Rheum. 2004;50:1117–1121. doi: 10.1002/art.20169. [DOI] [PubMed] [Google Scholar]
- Caponi L, Petit-Teixeira E, Sebbag M, Bongiorni F, Moscato S, Pratesi F, Pierlot C, Osorio J, Chapuy-Regaud S, Guerrin M, Cornelis F, Serre G, Migliorini P. A family based study shows no association between rheumatoid arthritis and the PADI4 gene in a white French population. Ann Rheum Dis. 2005;64:587–593. doi: 10.1136/ard.2004.026831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochi Y, Yamada R, Suzuki A, Harley JB, Shirasawa S, Sawada T, Bae SC, Tokuhiro S, Chang X, Sekine A, Takahashi A, Tsunoda T, Ohnishi Y, Kaufman KM, Kang CP, Kang C, Otsubo S, Yumura W, Mimori A, Koike T, Nakamura Y, Sasazuki T, Yamamoto K. A functional variant in FCRL3, encoding Fc receptor-like 3, is associated with rheumatoid arthritis and several autoimmunities. Nat Genet. 2005;37:478–485. doi: 10.1038/ng1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikari K, Momohara S, Nakamura T, Hara M, Yamanaka H, Tomatsu T, Kamatani N. Supportive evidence for a genetic association of the FCRL3 promoter polymorphism with rheumatoid arthritis. Ann Rheum Dis. 2006;65:671–673. doi: 10.1136/ard.2005.043489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thabet MM, Wesoly J, Slagboom PE, Toes RE, Huizinga TW. FCRL3 promoter 169 CC homozygosity is associated with susceptibility to rheumatoid arthritis in Dutch Caucasians. Ann Rheum Dis. 2007;66:803–806. doi: 10.1136/ard.2006.064949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre S, Bowes J, Potter C, Worthington J, Barton A. Association of the FCRL3 gene with rheumatoid arthritis: a further example of population specificity? Arthritis Res Ther. 2006;8:R117. doi: 10.1186/ar2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen CJ, Kelly H, Eden JA, Merriman ME, Pearce SH, Merriman TR. Analysis of the Fc receptor-like-3 (FCRL3) locus in Caucasians with autoimmune disorders suggests a complex pattern of disease association. J Clin Endocrinol Metab. 2007;92:1106–1111. doi: 10.1210/jc.2006-2183. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Allele and genotype distribution of rs10174098 and rs3911238. OR, odds ratio; CI, confidence interval; HWE, Hardy-Weinberg equilibrium; UK, United Kingdom; WTCCC, Wellcome Trust Case Control Consortium.