As I have written previously about my personal history and research journey since graduating from medical school (1), in this article, I wish to focus on the biochemistry of glutathione and glycosyltransferases, which, on the surface, would appear to be quite different subjects.
Early Training in Biochemistry and Studies of Glutathione Metabolism during Hepatocarcinogenesis
After obtaining an M.D. and finishing an internship at the university hospital in 1965, my early training in research was obtained in the Department of Biochemistry, Hokkaido University Graduate School of Medicine, where I received a Ph.D. degree in 1972. At that time, the department was chaired by Professor Hidematsu Hirai, and the focus of the departmental research effort was primarily on the biological and clinical significance of α-fetoprotein, an oncofetal protein that is synthesized by embryonic tissues and primary hepatomas but not by normal tissues (2). I had already developed an interest in enzymology, and I also became interested in an area that was very different from the mainstream program in the department. I decided to focus on the enzymes involved in glutathione metabolism during experimental hepatocarcinogenesis of rats that had been fed 0.06% 3′-methyl-4-dimethylaminoazobenzene under the guidance of Dr. Yutaka Tsukada. In these studies, rats were killed at weekly intervals, and the glutathione levels and glutathione-related enzymes were assayed spectrophotometrically. The experiments started early in the morning and usually ended at around midnight. I found that the activity of γ-glutamyl transpeptidase, one of the glutathione-degrading enzymes, in rats the fed azo dye underwent two biphasic changes, i.e. the activity was increased at an early stage of hepatocarcinogenesis, then decreased, and finally increased again at the late stage. Similar changes in the pattern for α-fetoprotein had been reported by a group in the same department. The actual level of glutathione decreased at the early stage, then increased, and again decreased at the late stage, mirroring the pattern observed for γ-glutamyl transpeptidase (3).
I hypothesized that γ-glutamyl transpeptidase might also be an oncofetal protein, analogous to α-fetoprotein. In fact, γ-glutamyl transpeptidase activity is high in fetal tissues, but the enzyme is also ubiquitously distributed and abundant in the kidney.
After receiving my Ph.D., I spent one year as a research fellow in the same laboratory and then moved to the Department of Hygiene and Preventive Medicine.
γ-Glutamyl Cycle and Alton Meister's Laboratory in New York
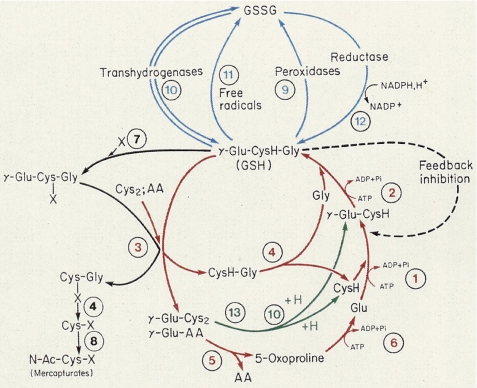
In 1974, Alton Meister, Professor and Chair of the Department of Biochemistry at the Cornell Medical School, presented a plenary lecture on the γ-glutamyl cycle at the annual meeting of the Japan Biochemical Society held in Okayama, Japan. He was agreeable to my working with him in New York, and I joined his group as a visiting associate professor on September 1, 1976. At that time, there were many postdoctoral fellows and faculty members in Meister's laboratory, and they published numerous excellent papers on glutathione metabolism, including the chemical synthesis of buthionine sulfoximine, a specific inhibitor of γ-glutamylcysteine synthetase; the chemical synthesis of glutathione esters that are able to enter the cell; the feedback inhibition of γ-glutamylcysteine synthetase; and the characterization of metabolic disorders of glutathione via the γ-glutamyl cycle, first thought to play a role in amino acid transport but later shown to be largely involved in the turnover of glutathione (Fig. 1) (4, 5). He was a heavy smoker, so I could soon recognize him by the smell of his cigars and pipes when he showed up in the lab and when I visited him to discuss research progress in his office on the first floor.
FIGURE 1.
γ-Glutamyl cycle proposed by Alton Meister (12). The cycle is involved primarily in the turnover of glutathione, i.e. glutathione synthesis and degradation. AA, amino acid.
Glutathione is synthesized via two enzymes, glutathione synthetase and γ-glutamylcysteine synthetase, both of which require ATP. Likewise, the degradation of glutathione is catalyzed by two enzymes, γ-glutamyl transpeptidase and γ-glutamylcyclotransferase. I was involved in the purification of γ-glutamylcyclotransferase from rat kidney, which was highly heterogeneous because of sulfhydryl modifications of the enzyme.
Hokkaido University and Cancer-associated Changes in Glycoproteins: A Saint's Maid Quotes Latin
I was supposed to become an associate professor in a newly launched department, i.e. the Graduate School of Environmental Science at Hokkaido University. I was obliged to return to Japan, but the research situation was not well organized, and I became interested in moving to another laboratory. Half a year later, Professor Akira Makita kindly offered me a position of associate professor in the Cancer Institute of the same university. His group discovered various cancer-associated changes in various enzymes such as arylsulfatases (6), β-glucuronidase, hexokinases, cathepsin D, and galactosyltransferase-1 and glycolipids such as sulfatides. Some of the work on sulfatides was subsequently carried out by Koichi Honke, who purified sulfatide synthase, cloned the cDNA and gene, produced null mice, and discovered many interesting phenotypes (7). He later joined our group as an associate professor and was a valuable member of our department. When I first joined the Makita laboratory, I was not familiar with glycoprotein or glycolipid research, and I was just like a saint's maid quoting Latin. Finally, however, I learned many techniques that subsequently proved to be useful in the area of glycobiology and published several articles under his guidance. I enjoyed my tenure as an associate professor in his group very much, as well as my interactions with the various graduate students in the group. I also published several papers on non-enzymatic glycosylation (glycation), the Maillard reaction, in relation to free radical research or diabetes, which subsequently became the other project of my research journey when I moved to Osaka University (8).
While there, I was able to restart experiments on γ-glutamyl transpeptidase. At that time, several glycolytic enzymes such as aldolase C, hexokinase, and pyruvate kinase had been reported to be activated in cancer and fetal and muscle tissues, and isozymic forms such as the muscle type were present in hepatoma tissues. Even though my preliminary experiments on γ-glutamyl transpeptidase suggested that isozymic forms were not present, I nevertheless continued to wonder if a specific isozymic form of γ-glutamyl transpeptidase was also activated in hepatomas. γ-Glutamyl transpeptidase is a glycoprotein and was first purified from bovine kidney by Meister's group. Therefore, I was interested in purifying the enzymes again from various rat tissues such as fetal, kidney, normal liver, and tumor tissues with the objective of comparing their biochemical properties as judged by kinetic properties, isoelectric focusing patterns, antigenicity, and amino acid analysis. The only difference we found was in the sugar composition, i.e. the levels of neutral sugars and sialic acid, as the isoelectric focusing patterns varied before and after treatment with sialidase (9, 10). Since then, I decided to focus on the sugar chains of the enzyme.
In collaboration with Professor Akira Kobata (Kobe University), an eminent glycobiology expert, we found that the enzyme purified from ascites hepatoma AH-66 cells contained bisecting GlcNAc, whereas the enzyme purified from normal liver did not (11).
Osaka University Medical School, Department of Biochemistry: From Protein Chemistry to Glycobiology (Purification of Glycosyltransferases) and from Gene Cloning to Functional Glycomics
In February 6, 1986, I received an unexpected phone call from the dean of the Osaka University Medical School, who told me that I was a candidate for the position of Professor and Chair of the Department of Biochemistry, one of the oldest and most prestigious biochemistry departments in Japan. The First Chair of the department was Professor Yashiro Kotake, who elucidated the chemistry of kynurenine and was an eminent biochemist and an expert in tryptophan metabolism. The Third Chair was Professor Osamu Hayaishi, a world-renowned and eminent biochemist famous for his pioneering work on oxygenase and prostaglandins. I was very happy and honored to accept this position and subsequently moved to Osaka on April 1, 1986. I decided to change my project from protein chemistry to glycobiology in this new department.
Purification of Glycosyltransferases and Gene Cloning
As 1988 was the 100th anniversary of the discovery of glutathione, I co-organized a meeting on the “Glutathione Centennial: Molecular Perspectives and Clinical Implications” with Dr. Yukiya Sakamoto and his group. Alton Meister also attended the conference as an organizer. At the meeting, he comprehensively reviewed the history of glutathione and its metabolism, designated as the γ-glutamyl cycle (12), as shown in Fig. 1. His group had been involved in studies in this area for the last 10 years or so. We reported on the purification and cDNA cloning of human γ-glutamyl transpeptidase and also reported on the significance of highly activated UDP-N-acetylglucosamine:β-d-mannoside β1,4-N-acetylglucosaminyltransferase III (GnT-III), which may modify γ-glutamyl transpeptidase in hepatoma tissues and hyperplastic nodules. In 1988, we also reported on a marked activation of GnT-III activity in primary hepatoma and ascites hepatoma, which could explain the unique changes in the sugar units attached to γ-glutamyl transpeptidase, i.e. the addition of a bisecting GlcNAc, a product of GnT-III.
In 1992, we organized the Sapporo Cancer Seminar, which was founded by Dr. Hiroshi Kobayashi, Professor Emeritus of Hokkaido University; this seminar is a Gordon Conference-like meeting that focuses on cancer research. I invited Alton Meister, Owen Griffith, Susumu Nishimura, Cecil Pickets, Harold Deutsch, and other distinguished speakers who were actively involved in glutathione research, redox regulation, and nitric oxide and reactive oxygen species (Fig. 2). Immediately after the meeting, we celebrated Alton's 70th birthday in Osaka, and many Japanese scientists, good friends of his or former colleagues, attended. Since Yoshitaka Ikeda in our group joined Alton's laboratory in 1993, we continued the work on the catalytic properties of γ-glutamyl transpeptidase in a collaborative effort, which focused mainly on studies of the catalytic mechanism of the enzyme by site-directed mutagenesis. Two years later, in 1995, Alton suffered a stroke during a trip to New Zealand, and after returning to New York, he passed away in December at the age of 72. The final paper that Alton actually handled was ours, which appeared in the Journal of Biological Chemistry in September 1995 (13); this article was the culmination of his work. Alton's death was a great loss not only for glutathione metabolism but also for biochemistry in the world. His enormous contributions to our understanding of glutathione metabolism through the concept of the γ-glutamyl cycle (Fig. 1) and amino acid metabolism will remain forever as his legacy. Our data on the catalytic mechanism of γ-glutamyl transpeptidase, including the work with Alton, were published (14).
FIGURE 2.
12th Sapporo Cancer Seminar in 1992. Shown are Alton Meister, to the left of me (front row, fourth from the right), and Hiroshi Kobayashi, to the left of Meister.
Starting in 1986, we began to purify the glycosyltransferase, which catalyzes the biosynthesis of the bisecting GlcNAc found in γ-glutamyl transpeptidase purified from ascites hepatoma AH-66 cells. At that time, the method used in assaying glycosyltransferases involved the incorporation of 14C- or 3H-labeled nucleotide sugars into the acceptor substrate, but this was a time-consuming and rather expensive method. Therefore, we attempted to develop a simpler, less costly method of analysis. The groups of Drs. Hase and Ikenaka at Osaka University developed a method involving the pyridylamination of the nonreducing end of glycans to analyze sugar chains. Atsushi Nishikawa and co-workers (15) successfully employed this technique to assay the activity of various glycosyltransferases such as GnT-III, -IV, -V, and -VI. Using this assay method, we measured the GnT-III activity of various hepatoma cells and azo dye-induced hepatomas and found that the enzyme activity was substantially elevated in most hepatomas. In some cases, the activity was 100-fold greater than that in a normal rat liver.
To obtain a cDNA clone, we attempted to purify GnT-III by affinity chromatography using biantennary sugar chains, the substrate for GnT-III. We first prepared a large amount of bisected biantennary sugar chains from human serum transferrin. Because GnT-III activity is very high in rat kidney, we collected a large number of kidneys from rats that had been killed during physiological experiments in other laboratories. We ultimately collected 10 kg of rat kidneys. The enzyme was purified >153,000-fold in 1.5% yield from a Triton X-100 extract of rat kidneys by fractionation procedures utilizing QAE-Sepharose, Cu2+-chelating Sepharose, and affinity chromatography on UDP-hexanolamine and substrate-conjugated Sepharose, and finally, the cDNA encoding GnT-III was successfully cloned (16). The enzyme is responsible for the formation of a bisecting glycan, as first published by Dr. Harry Schachter, who reported the presence of numerous N-acetylglucosaminyltransferases such as GnT-I, -II, -III, -IV, -V, and -VI. His group purified GnT-I and GnT-II for the first time and then cloned their genes (17). His contribution in this field has been enormous, and I met Harry for the first time in 1989, when Dr. Makita organized the Second Sapporo Cancer Seminar at a time when I was a new comer in this field. Since then, every time I met Harry, he gave me valuable suggestions and encouragement in our work on glycosyltransferases even though his lab has been our major competitor in this area because very few groups have attempted to purify and biochemically characterize the enzyme and then clone the gene (Fig. 3).
FIGURE 3.
Dr. Schachter's laboratory in Toronto (from left to right): Jim Dennis, Harry Schachter, Naoyuki Taniguchi, and Jeremy P. Carver.
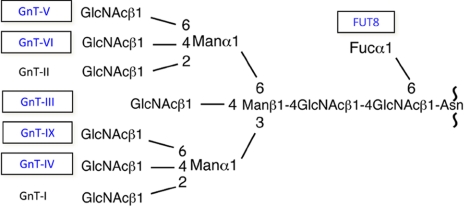
We further purified various glycosyltransferases involved in N-glycan branch biosynthesis one by one and cloned their genes; these included GnT-V (18) and GnT-IV, GnT-VI, synthetic enzyme for poly-N-acetyllactosamine, and α1,6-fucosyltransferase (FUT8) (19, 20). Our group also cloned other glycosyltransferases using molecular cloning techniques as depicted in Fig. 4. In 1989, I organized the first glycosyltransferase meeting in Osaka, and numerous eminent scientists in this field, including Robert Hill, Pamela Stanley, Jamey Marth, Gerald Hart, Ajit Varki, Jurgen Roth, Minoru Fukuda, Laurence Tabak, Andre Verbert, John Lowe, and many renowned Japanese glycobiologists, attended, and the significance of glycosyltransferases was discussed. Last year, the sixth meeting (now called Glyco T) was organized by Richard Cummings and Michael Pierce and held in Atlanta.
FIGURE 4.
Glycosyltransferases involved in N-glycan branching characterized by our group. The enzymes in boxes indicate the names of glycosyltransferases purified and their cDNAs cloned by our group. GnT-IX was cloned based on the genome sequence.
Functional Glycomics Approach and Lessons from GnT-III Transfectants
Using the cloned glycosyltransferase genes, we next initiated studies related to functional glycomics using sugar-remodeling techniques by gene transfections, knock-out and/or transgenic mice, and small interfering RNA. We were able to show various functional changes in glycans as the result of aberrant glycosylation (21–24) using the cloned glycosyltransferases. Moreover, to utilize a functional glycomics approach, we emphasized the importance of identifying the target protein(s) of the glycosyltransferase genes. At that time, some researchers asked, “What would happen if you were able to identify one or two target proteins among the numerous glycosylated proteins that could undergo glycosylation? This is analogous to pointing out one star among the universe.” However, we now all realize that, even though the knock-out of a certain glycosyltransferase gene may lead to various phenotypic changes due to a lack of glycan in various glycoproteins, in fact, phenotypic changes accompanied by functional changes in glycans are very limited. This indicates that the identification of target protein(s) is very important in characterizing glycan functions. In 2002, I co-edited and published a handbook in which most of the biochemical and molecular properties of glycosyltransferases and related genes are described (25). Our major glycosyltransferases of interest are depicted in Fig. 4.
As we observed previously, γ-glutamyl transpeptidase was one of the target proteins for GnT-III. To explore the other functions of the GnT-III gene and its target proteins, our group conducted experiments using cells that had been transfected by the GnT-III gene and found different types of likely target proteins as follows (21–24). K562 cells are usually killed by the immune systems in the spleen, but GnT-III-transfected K562 cells are not. This is due to a decreased sensitivity to natural killer cell cytotoxicity, which leads to increased spleen colonization by these cells in athymic mice. We identified target proteins that recognize bisecting GlcNAc, a product of GnT-III, as annexin V and apoprotein B.
Epidermal growth factor receptors (EGFRs) also contain N-glycans, and several researchers have proposed that they play a role in growth factor signaling. The deletion of Asn241 from the EGFR permits the molecule to dimerize spontaneously without phosphorylation (26). Erb-2 glycans play a key role in growth factor signaling because, without glycans, Erb-2 dimerizes without phosphorylation and enhances tumor development in vitro (27). In GnT-III-transfected glioma cells, EGF binding was blocked, and autophosphorylation of the EGFR occurred. In HeLa cells, GnT-III transfection inhibited the low affinity binding of EGF to the EGFR but enhanced the high affinity binding and the internalization rate of the EGFR. The transfection of GnT-III further resulted in 1) the inhibition of the growth factor response of tyrosine phosphorylation of the Trk/nerve growth factor receptor following the addition of nerve growth factor, 2) the inhibition of neurite growth induced by co-stimulation of EGF and integrins, and 3) the inhibition of EGFR-mediated extracellular signal-regulated kinase activation. The target proteins of GnT-III responsible for these effects were identified as the EGFR and integrin α3β1. CD44 obtained from GnT-III-transfected mouse melanoma cells exhibited increased adhesion to hyaluronate, which enhanced CD44-mediated tumor growth and metastasis in the spleen after subcutaneous inoculation into mice. The α1–3Gal epitope is a xenotransplantation antigen absent in humans. Thus, in the case of xenotransplantation from pigs to apes or humans, subacute rejection will occur because of the presence of an antibody against the αGal epitope in humans and apes. Interestingly, the transfection of GnT-III into pig cells or the generation of GnT-III transgenic pigs markedly reduced the αGal epitope by inhibiting α1–3Gal-transferase action, thus reducing its antigenicity.
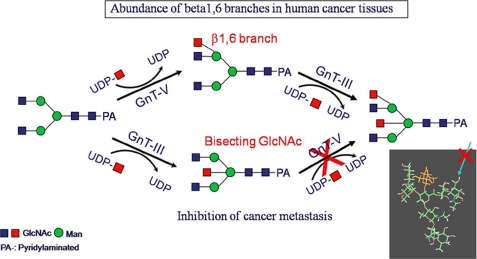
GnT-III and GnT-V Have Opposite Effects on Cancer Metastasis and Cell Adhesion
GnT-III serves as a STOP signal for other glycosyltransferases such as GnT-II, -IV, and -V, and once a bisecting GlcNAc is incorporated, those enzymes no longer act. Therefore, a high expression of GnT-III would be expected to inhibit the synthesis of multiantennary complex carbohydrates and to evoke various phenotypic alterations reflecting abolished glycan functions. Gu et al. (18) confirmed this substrate specificity using purified GnT-V and demonstrated that GnT-V does not act on a bisected biantennary chain. This catalytic function was originally reported by Schachter et al. (17) using a crude enzyme preparation. B16-hm cells, in which GnT-V is highly expressed, therefore leading to increased β1,6 branching, have a high metastatic potential. In contrast, lung metastasis in syngeneic and athymic mice after an intravenous injection of B16-hm cells transfected with the gene encoding GnT-III was dramatically suppressed (28). GnT-V catalyzes the addition of a β1–6 linkage to the Manα1–6 residue of GlcNAcβ1–2Manα1–6Man between the arm of the N-glycan core. In the GnT-III transfectants, the level of β1,6 branches is decreased due to competition for the substrate by intrinsic GnT-V and ectopically expressed GnT-III (Fig. 5). The underlying mechanism by which lung metastasis is suppressed is due to elevated E-cadherin levels on cell-cell borders. Upon overexpression of GnT-III in melanoma cells, E-cadherin remains on the cell-cell border and enhances the attachment of cancer cells to protect cancer metastasis. As E-cadherin·β-catenin complexes are quite stable, the increased E-cadherin levels result in the complete down-regulation of the tyrosine phosphorylation of β-catenin after EGF treatment (23, 24).
FIGURE 5.
Competitive reaction between GnT-III and GnT-V. Because GnT-III and GnT-V act on the same substrate but GnT-III acts on the substrate first and produces a bisecting GlcNAc structure, GnT-V is not able to act further. This leads to a suppression of the metastatic potential in vivo. Computer modeling of these reactions provided support for the this hypothesis.
Conversely, the disruption of E-cadherin-mediated cell adhesion appears to be a central event in the transition from noninvasive to invasive carcinomas. Interestingly, we recently found that E-cadherin-mediated cell-cell interaction up-regulated GnT-III expression, suggesting that the regulation of GnT-III and E-cadherin expression may represent a positive feedback loop and may be implicated in endothelial-mesenchymal transition (29). E-cadherin contains four N-glycan sites, and the role of glycans in terms of cell adhesion is now well recognized.
The increased GnT-V levels were attributed to the up-regulation of the Ets family of transcriptional activators, which increase GnT-V expression. Knock-out mice of Mgat5, encoding GnT-V, develop into apparently normal adults, but the incidences of polyoma virus middle T oncogene-induced mammary tumor growth and metastases are considerably lower in knock-out mice than in transgenic littermates. N-Glycans bearing the GnT-V branch amplify oncogene signaling of tumor growth in vivo (30).
Integrins, crucial molecules for cell adhesion, have been shown to be the major N-glycan-containing molecules on cell membranes. For example, integrin α5β1 contains 14 and 12 putative N-glycosylation sites on the α5 and β1 subunits, respectively. Introduction of a bisecting GlcNAc to N-glycans of integrin α5 by the overexpression of GnT-III diminished ligand binding, cell adhesion, and cell migration. Contrary to the functions of GnT-III, the overexpression of GnT-V promoted integrin α5β1-mediated cell migration (31). Indeed, the opposing effects of GnT-III and GnT-V have been observed for the same target protein, integrin α3β1. N-Glycosylation of the β-propeller domain on the α5 subunit (32) and the I-like domain on the β1 subunit is essential to both the heterodimerization and biological functions of the subunits. In addition, among many N-glycosylation sites of the α5 subunit, only N-glycosylation site-4 on the β-propeller was specifically modified by GnT-III, thereby regulating integrin α5β1-mediated cell spreading and cell migration.
Our group identified matriptase as a target protein for GnT-V and showed that the overexpression of GnT-V results in the increased expression of matriptase due to the resistance of matriptase to autodegradation. Matriptase is known to be associated with cancer invasion and metastasis, and its expression is enhanced in some types of tumor cells. Moreover, GnT-V may function as an inducer of angiogenesis, thereby promoting metastasis (22–24). TIMP-1 was identified as a target protein for GnT-V. The poly-N-acetyllactosamine chains on GnT-V-dependent N-glycan antennae serve as ligands for galectin-3, and it has been suggested that GnT-V strengthens the galectin-3/glycoprotein lattice on the T-cell receptor, thereby restricting the recruitment of T-cell receptors to the site of antigen presentation and impeding antigen-dependent receptor clustering and signal transduction (33).
Core Fucosylation of Glycoproteins Is Implicated in Growth, Lung Emphysema, Antibody Therapy, and a Cancer Biomarker
l-Fuc:N-acetyl-β-d-glucosaminide α1,6-fucosyltransferase (FUT8) catalyzes the transfer of a fucose residue from GDP-Fuc to position 6 of the innermost GlcNAc residue (core fucose residue) of an N-linked sugar hybrid and complex types of N-linked oligosaccharides on glycoproteins. The enzyme was first purified to homogeneity and cloned by our group from porcine brain as well as from human gastric cancer cells (19, 20). Deletion of the Fut8 gene resulted in marked phenotypic changes in mice, namely 60% of the mice died during the neonatal period, and the remainder died within 3 weeks. Using Fut8 knock-out mice, we found that the mRNA levels of MMP-1, -9, and -12 were specifically up-regulated among the various matrix metalloproteinases (MMPs) examined. MMP gene expression occurs specifically due to the deletion of the core fucose in the transforming growth factor-β (TGF-β) receptor, which usually suppresses MMP expression.
The up-regulation of MMPs results in the degradation of extracellular matrix proteins such as laminin and type IV collagen, which may facilitate the development of lung emphysema in mice and humans. In Fut8 knock-out mice, the phosphorylation of SMAD proteins was markedly down-regulated, suggesting an impairment in the TGF-β cascade. The core fucosylation of the TGF-β receptor was completely abolished in Fut8 knock-out mice, indicating that the binding affinity for the ligand, TGF-β, was impaired, and therefore, the negative regulation of TGF-β signaling was also suppressed (34). The Fut8 null mutation in mice also leads to the impairment of LRP-1 (low density lipoprotein receptor-related protein-1) (35) and an impaired pre-B-cell re-population due to dysfunction of VECAM-2 (vascular endothelial cell adhesion molecule-1) and integrin α4β1 (36).
Some natural killer cells contain receptors for the Fc domain of IgG and bind to the Fc portion of the IgG antibody on the surface of a target cell, such as tumor cells, and release cytolytic components that kill the target cell. This killing mechanism is referred to as antibody-dependent cell-mediated cytotoxicity.
It is well known that mouse monoclonal antibodies and humanized antibodies, including clinically effective agents such as trastuzumab (Herceptin®) and rituximab (Rituxan®), require both activation via FcγRIII and inhibition via FcγRIIB antibody receptors. The expression of antibodies with altered glycoforms, especially the addition of bisecting GlcNAc, leads to an increase in antibody-dependent cell-mediated cytotoxicity through a higher affinity for FCγRIII by up to 10–20-fold. Similar to the line of evidence described above, two groups reported that binding of fucose-deficient IgG1 to human FcγRIIIA was improved by up to 50–100-fold (37, 38). Core fucosylation also plays important roles in the adhesion of tumor cells via integrins (39). These findings strongly suggest that the α1,6-fucosylation of N-glycans modifies the function of the glycoproteins.
The Glycomics Approach Is One of the More Promising Approaches for the Discovery of Cancer Biomarkers
In 2003, the International Union of Biochemistry and Molecular Biology (IUBMB) Congress in Toronto was cancelled due to the SARS problem, and a joint meeting of the Human Proteome Organization (HUPO) with IUBMB was held in Montreal. I attended the council meeting of HUPO and was asked by several participants to launch a glycomics initiative because there was no official initiative on glycomics under HUPO. We finally launched the Human Disease Glycomics/Proteome Initiative (HGPI), with myself as the chair, along with many glycobiologists and biochemists who agreed to collaborate. Many experts, especially experts in mass spectrometry, joined this initiative and participated in the steering committee meeting. Among them was the Nobel Laureate Koichi Tanaka and many others. Since the launch of HGPI, two pilot studies of N-glycan (40) and O-glycan analyses have been performed with 26 participating laboratories. In 2006, with Jim Paulson, Sudhir Srivastava, Pamela Marino, and Ram Sasisekharan, I organized a joint meeting at the National Institutes of Health entitled “Frontiers in Glycomics: Bioinformatics and Biomarkers in Disease,” and on this occasion, we proposed a white paper emphasizing the importance of glycomics (41). In terms of identifying cancer biomarkers using glycomics, Dr. Sen-ichiro Hakomori has made many pioneering discoveries. The fucosylation of α-fetoprotein is markedly increased in patients with a primary hepatoma, and affinoelectrophoresis using lectin and antibody can be used to distinguish between patients with hepatocellular carcinoma and those with chronic hepatitis and liver cirrhosis as reported by Taketa et al. (42). In 2006, the Food and Drug Administration approved the use of fucosylated α-fetoprotein for the differential diagnosis of patients with liver cirrhosis and primary hepatoma. Glycan patterns of the α-fetoprotein L3 fraction and its enzymatic basis were reported by our group (43). Using a similar approach, fucosylated haptoglobin was identified as a possible marker for pancreatic cancer. Most fucosylated proteins are found in the bile duct under normal conditions, but in the case of cancer, polarity changes occur, which are possibly due to the expression of carrier protein(s) or the disorganization of tumor cells (44). The levels of GDP-fucose, as well as synthetic enzymes such as GDP-keto-6-deoxymannose 3,5-epimerase/4-reductase and GDP-mannose 4,6-dehydratase, play an important role in core fucosylation.
“Glycan Cycles” as Functional Units of Glycan Functions for Understanding the Systemic Approach: To Make Movies Rather Than to Take a Snapshot
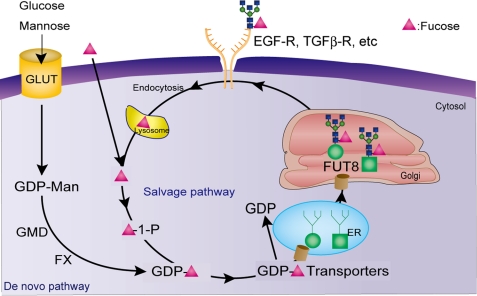
Here, I propose the concept of the “glycan cycle” as a functional unit of glycans that will permit the integration of glycan functions in analogy with the γ-glutamyl cycle for understanding glutathione metabolism (Fig. 1). Jim Dennis reported that Mgat5, a gene that encodes GnT-V, plays a key role in growth and arrest via the glycan structure of growth factor receptors and GLUT1 by elucidating the binding of galectin-3 (45). However, glycosylation is affected not only by a single glycosyltransferase such as GnT-V but also by various components such as the levels of mono-oligosaccharides, nucleotide sugars, nucleotide transporters, the localization of glycosyltransferases, nucleotide sugar levels in the Golgi, transcription factors, the pro-form and mature form of glycoproteins, and the structure of cell-surface glycoproteins (46). All of these components should be considered in detecting changes in more dynamic ways; namely, it is necessary not only to take a snapshot but also to make movies of the dynamic changes in glycan metabolism.
For example, glucose enters the cell via a glucose transporter and is converted to UDP-GlcNAc via the hexosamine pathway. UDP-GlcNAc plays a key role in the cytosol and is then incorporated into the Golgi via the UDP- GlcNAc transporter and serves as a donor substrate for various GlcNAc-transferases from GnT-I to GnT-VI. UDP-GlcNAc also serves as a donor for O-GlcNAc-transferase in the cytosol. These enzymes function to modify target proteins such as TGF-β, the EGFR, and the glucose transporter. These receptors are localized on the cell surface and bind to lectins such as galectin-3. When the affinity of lectin binding is decreased due to changes in receptor glycosylation, the receptors are endocytosed, and the free monosaccharide is probably recycled. This conceptual “functional unit of the glycan cycle,” such as just described for GlcNAc, is intended to help develop an understanding of the integrative and dynamic analysis of glycan functions. The same is true for the “fucose cycle,” in which GDP-fucose, GDP-mannose 4,6-dehydratase, GDP-keto-6-deoxymannose 3,5-epimerase/4-reductase, the GDP-fucose transporter, and fucosyltransferases are key components for core fucosylation as depicted in Fig. 6. Similarly, the “galactose cycle” and “sialic acid cycle,” etc., would be present.
FIGURE 6.
Schematic drawing of the glycan cycle (fucose cycle). Glycosylation is affected by various factors as described in text, and the pro-form is converted to the mature form of glycoproteins by glycosyltransferases and then localized on the cell surface, such as growth factor receptors by binding to lectins. The similar cycles such as the GlcNAc cycle, sialic acid cycle, etc., will be present. TGFβ-R; TGF-β receptor; GMD, GDP-mannose 4,6-dehydratase; FX, GDP-keto-6-deoxymannose 3,5-epimerase/4-reductase; ER, endoplasmic reticulum.
Among the components of the glycan cycle, nucleotide sugars play key roles in the function of glycans. Very recently, our group developed a method for the simultaneous assay of nucleotide sugars using ion-pair reversed-phase high pressure liquid chromatography and observed dramatic changes in nucleotide sugar levels in cells cultured under dense or sparse conditions (K. Nakajima, S. Kitazume, R. Fujinawa, E. Miyoshi, and N. Taniguchi, unpublished data). This indicates that nucleotide sugars may control glycan structures and function via the glycan cycle (Fig. 6). To clarify glycan functions in a more dynamic way, after I retired from the Osaka University Medical School in 2006 after reaching retirement age, I became an Endowed Chair Professor of the Department of Disease Glycomics in the same university, with support from Seikagaku Corp., and we subsequently launched a Systems Glycobiology Research Group at RIKEN (Wako, Japan). We expect that our understanding of glycan cycles will be enhanced considerably in the near future.
Acknowledgments
I thank my many colleagues who collaborated with me over three decades, many whose names also appear in the list of literature citations. During my stay at Osaka University, many eminent scientists came to visit as visiting professors and contributed to our research and education in our M.D./Ph.D. program as well. Among them, I thank Professors Dirk van den Eijnden, John Gutteridge, Kunihiko Suzuki, Harold F. Deutsch, Kosaku Uyeda, Takashi Yonetani, Peng George Wong, Mary Anderson, Owen W. Griffith, and William Lennarz, who spent from three weeks to one year as visiting professors and supported our research and the M.D./Ph.D. program in our faculty. I thank Drs. Vincent C. Hascall, Etorre Appella, Milton Feather, Jianguo Gu, and Eiji Miyoshi and Lisa Jenkins for reading this manuscript prior to submission and for valuable suggestions. I thank Dr. Kazuki Nakajima and Fumi Ota for help in preparing this manuscript. I acknowledge continued grant support from the Japan Society for the Promotion of Science and the Ministry of Education, Sports, Culture, Science, and Technology, Japan (from 1973 to the present).
REFERENCES
- 1.Taniguchi N. (2008) IUBMB Life 60, 703–705 [Google Scholar]
- 2.Hirai H., Nishi S., Watabe H., Tsukada Y. (1973) Gann Monogr. 14, 19–34 [Google Scholar]
- 3.Taniguchi N., Tsukada Y., Mukuo K., Hirai H. (1974) Gann 65, 381–387 [PubMed] [Google Scholar]
- 4.Meister A., Anderson M. E. (1983) Annu. Rev. Biochem. 52, 711–760 [DOI] [PubMed] [Google Scholar]
- 5.Meister A. (1986) in Molecular Perspectives and Clinical Implications ( Taniguchi N., Higashi T., Sakamoto Y., Meister A. eds) pp. 3–21, Academic Press, New York [Google Scholar]
- 6.Gasa S., Balbaa M., Nakamura M., Yonemori H., Makita A. (1987) J. Biol. Chem. 262, 1230–1238 [PubMed] [Google Scholar]
- 7.Honke K., Hirahara Y., Dupree J., Suzuki K., Popko B., Fukushima K., Fukushima J., Nagasawa T., Yoshida N., Wada Y., Taniguchi N. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 4227–4232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taniguchi N. (1992) Adv. Clin. Chem. 29, 1–59 [DOI] [PubMed] [Google Scholar]
- 9.Yokosawa N., Taniguchi N., Tsukada Y., Makita A. (1981) Oncodev. Biol. Med. 2, 165–177 [PubMed] [Google Scholar]
- 10.Taniguchi N., Yokosawa N., Iizuka S., Tukada Y., Sako F., Miyazawa N. (1983) Gann Monogr. 29, 263–272 [Google Scholar]
- 11.Yamashita K., Hitoi A., Taniguchi N., Yokosawa N., Tsukada Y., Kobata A. (1983) Cancer Res. 43, 5059–5063 [PubMed] [Google Scholar]
- 12.Meister A. (1998) in Glutathione Centennial ( Taniguchi N., Higashi T., Sakamoto Y., Meister A. eds) pp. 3–22, Academic Press, New York [Google Scholar]
- 13.Ikeda Y., Fujii J., Anderson M. E., Taniguchi N., Meister A. (1995) J. Biol. Chem. 270, 22223–22228 [DOI] [PubMed] [Google Scholar]
- 14.Taniguchi N., Ikeda Y. (1998) Adv. Enzymol. Relat. Areas Mol. Biol. 72, 239–278 [DOI] [PubMed] [Google Scholar]
- 15.Taniguchi N., Nishikawa A., Fujii S., Gu J. G. (1989) Methods Enzymol. 179, 397–408 [DOI] [PubMed] [Google Scholar]
- 16.Nishikawa A., Ihara Y., Hatakeyama M., Kangawa K., Taniguchi N. (1992) J. Biol. Chem. 267, 18199–18204 [PubMed] [Google Scholar]
- 17.Schachter H., Narasimhan S., Gleeson P., Vella G. (1983) Can. J. Biochem. Cell Biol. 61, 1049–1066 [DOI] [PubMed] [Google Scholar]
- 18.Gu J., Nishikawa A., Tsuruoka N., Ohno M., Yamaguchi N., Kangawa K., Taniguchi N. (1993) J. Biochem. 113, 614–619 [DOI] [PubMed] [Google Scholar]
- 19.Uozumi N., Yanagidani S., Miyoshi E., Ihara Y., Sakuma T., Gao C. X., Teshima T., Fujii S., Shiba T., Taniguchi N. (1996) J. Biol. Chem. 271, 27810–27817 [DOI] [PubMed] [Google Scholar]
- 20.Yanagidani S., Uozumi N., Ihara Y., Miyoshi E., Yamaguchi N., Taniguchi N. (1997) J. Biochem. 121, 626–632 [DOI] [PubMed] [Google Scholar]
- 21.Taniguchi N., Yoshimura M., Miyoshi E., Ihara Y., Nishikawa A., Fujii S. (1996) Glycobiology 6, 691–694 [DOI] [PubMed] [Google Scholar]
- 22.Taniguchi N., Ihara S., Saito T., Miyoshi E., Ikeda Y., Honke K. (2001) Glycoconj. J. 18, 859–865 [DOI] [PubMed] [Google Scholar]
- 23.Taniguchi N., Gu J., Takahashi M., Miyoshi E. (2004) Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 80, 82–91 [Google Scholar]
- 24.Taniguchi N., Miyoshi E., Gu J., Jianguo G., Honke K., Matsumoto A. (2006) Curr. Opin. Struct. Biol. 16, 561–566 [DOI] [PubMed] [Google Scholar]
- 25.Taniguchi N., Honke K., Fukuda M. (eds) (2002) Handbook of Glycosyltransferases and Related Genes, Springer, Tokyo [Google Scholar]
- 26.Tsuda T., Ikeda Y., Taniguchi N. (2000) J. Biol. Chem. 275, 21988–21994 [DOI] [PubMed] [Google Scholar]
- 27.Takahashi M., Yokoe S., Asahi M., Lee S. H., Li W., Osumi D., Miyoshi E., Taniguchi N. (2008) Biochim. Biophys. Acta 1780, 520–524 [DOI] [PubMed] [Google Scholar]
- 28.Yoshimura M., Nishikawa A., Ihara Y., Taniguchi S., Taniguchi N. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 8754–8758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akama R., Sato Y., Kariya Y., Isaji T., Fukuda T., Lu L., Taniguchi N., Ozawa M., Gu J. (2008) Proteomics 8, 3221–3228 [DOI] [PubMed] [Google Scholar]
- 30.Granovsky M., Fata J., Pawling J., Muller W. J., Khokha R., Dennis J. W. (2000) Nat. Med. 6, 306–312 [DOI] [PubMed] [Google Scholar]
- 31.Guo H. B., Lee I., Kamar M., Akiyama S. K., Pierce M. (2002) Cancer Res. 62, 6837–6845 [PubMed] [Google Scholar]
- 32.Zhao Y., Nakagawa T., Itoh S., Inamori K., Isaji T., Kariya Y., Kondo A., Miyoshi E., Miyazaki K., Kawasaki N., Taniguchi N., Gu J. (2006) J. Biol. Chem. 281, 32122–32130 [DOI] [PubMed] [Google Scholar]
- 33.Lagana A., Goetz J. G., Cheung P., Raz A., Dennis J. W., Nabi I. R. (2006) Mol. Cell. Biol. 26, 3181–3193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X., Inoue S., Gu J., Miyoshi E., Noda K., Li W., Mizuno-Horikawa Y., Nakano M., Asahi M., Takahashi M., Uozumi N., Ihara S., Lee S. H., Ikeda Y., Yamaguchi Y., Aze Y., Tomiyama Y., Fujii J., Suzuki K., Kondo A., Shapiro S. D., Lopez-Otin C., Kuwaki T., Okabe M., Honke K., Taniguchi N. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 15791–15796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee S. H., Takahashi M., Honke K., Miyoshi E., Osumi D., Sakiyama H., Ekuni A., Wang X., Inoue S., Gu J., Kadomatsu K., Taniguchi N. (2006) J. Biochem. 139, 391–398 [DOI] [PubMed] [Google Scholar]
- 36.Li-Nakagawa T., Miyoshi E., Yakushijin T., Hiramatsu N., Igura T., Hayashi N., Taniguchi N., Kondo A. (2008) J. Proteome Res. 7, 2222–2233 [DOI] [PubMed] [Google Scholar]
- 37.Shields R. L., Lai J., Keck R., O'Connell L. Y., Hong K., Meng Y. G., Weikert S. H., Presta L. G. (2002) J. Biol. Chem. 277, 26733–26740 [DOI] [PubMed] [Google Scholar]
- 38.Shinkawa T., Nakamura K., Yamane N., Shoji-Hosaka E., Kanda Y., Sakurada M., Uchida K., Anazawa H., Satoh M., Yamasaki M., Hanai N., Shitara K. (2003) J. Biol. Chem. 278, 3466–3473 [DOI] [PubMed] [Google Scholar]
- 39.Zhao Y., Itoh S., Wang X., Isaji T., Miyoshi E., Kariya Y., Miyazaki K., Kawasaki N., Taniguchi N., Gu J. (2006) J. Biol. Chem. 281, 38343–38350 [DOI] [PubMed] [Google Scholar]
- 40.Wada Y., Azadi P., Costello C. E., Dell A., Dwek R. A., Geyer H., Geyer R., Kakehi K., Karlsson N. G., Kato K., Kawasaki N., Khoo K. H., Kim S., Kondo A., Lattova E., Mechref Y., Miyoshi E., Nakamura K., Narimatsu H., Novotny M. V., Packer N. H., Perreault H., Peter-Katalinic J., Pohlentz G., Reinhold V. N., Rudd P. M., Suzuki A., Taniguchi N. (2007) Glycobiology 17, 411–422 [DOI] [PubMed] [Google Scholar]
- 41.Packer N. H., von der Lieth C. W., Aoki-Kinoshita K. F., Lebrilla C. B., Paulson J. C., Raman R., Rudd P., Sasisekharan R., Taniguchi N., York W. S. (2008) Proteomics 8, 8–20 [DOI] [PubMed] [Google Scholar]
- 42.Taketa K., Endo Y., Sekiya C., Tanikawa K., Koji T., Taga H., Satomura S., Matsuura S., Kawai T., Hirai H. (1993) Cancer Res. 53, 5419–5423 [PubMed] [Google Scholar]
- 43.Ohno M., Nishikawa A., Koketsu M., Taga H., Endo Y., Hada T., Higashino K., Taniguchi N. (1992) Int. J. Cancer 51, 315–317 [DOI] [PubMed] [Google Scholar]
- 44.Nakagawa T., Uozumi N., Nakano M., Mizuno-Horikawa Y., Okuyama N., Taguchi T., Gu J., Kondo A., Taniguchi N., Miyoshi E. (2006) J. Biol. Chem. 281, 29797–29806 [DOI] [PubMed] [Google Scholar]
- 45.Partridge E. A., Le Roy C., Di Guglielmo G. M., Pawling J., Cheung P., Granovsky M., Nabi I. R., Wrana J. L., Dennis J. W. (2004) Science 306, 120–124 [DOI] [PubMed] [Google Scholar]
- 46.Taniguchi N. (2007) Nat. Chem. Biol. 3, 307–309 [DOI] [PubMed] [Google Scholar]