Abstract
The recognition of bacteria, viruses, fungi, and other microbes is controlled by host immune cells, which are equipped with many innate immunity receptors, such as Toll-like receptors, C-type lectin receptors, and immunoglobulin-like receptors. Our studies indicate that the immune modulating properties of many herbal drugs, for instance, the medicinal fungus Reishi (Ganoderma lucidum) and Cordyceps sinensis, could be attributed to their polysaccharide components. These polysaccharides specifically interact with and activate surface receptors involved in innate immunity. However, due to the complexity of polysaccharides and their various sources from medicinal fungi, quantitative analysis of medicinal polysaccharide extracts with regard to their functions represents a major challenge. To profile carbohydrate-immune receptor interactions, the extracellular domains of 17 receptors were cloned as Fc-fusion proteins, such that their interactions with immobilized polysaccharides could be probed in an enzyme-linked immunosorbent assay. The results show that several innate immune receptors, including Dectin-1, DC-SIGN, Langerin, Kupffer cell receptor, macrophage mannose receptor, TLR2, and TLR4, interact with the polysaccharide extracts from G. lucidum (GLPS). This analysis revealed distinct polysaccharide profiles from different sources of medicinal fungi, and the innate immune receptor-based enzyme-linked immunosorbent assay described here can serve as a high-throughput profiling method for the characterization and quality control of medicinal polysaccharides. It also provides a means to dissect the molecular mechanism of medicinal polysaccharide-induced immunomodulation events.
Introduction
The immune system enables a host to discriminate self from non-self antigens so that invading pathogens can be recognized, captured, and eradicated. Adaptive immunity relies on highly polymorphic molecules, such as class I and class II major histocompatibility complexes, to present antigens to T and B cells and to initiate an immune response against foreign antigens. Whereas adaptive immunity implements highly polymorphic major histocompatibility complex molecules for presenting specific antigens, innate immunity recognizes a diverse class of antigens by “pattern recognition” (1). The identification of pathogen-associated molecular patterns (PAMPs)3 is controlled by pattern recognition receptors (PRRs), such as Toll-like receptors (TLRs) (2–4), C-type lectin receptors (CLRs) (5), immunoglobulin-like (Ig-like) receptors (e.g. TREMs and TREM-like transcripts known as TLTs) (6), NOD proteins (4), and others (7, 8). Notably, immune cells are capable of recognizing pathogens through specific carbohydrate antigens by non-TLRs. For example, macrophage mannose receptor (MMR) recognizes the N-linked high-mannose glycans expressed on the surface of pathogens (9), whereas Dectin-1 binds specifically to β-glucans, the major polysaccharide backbone of the fungal cell wall (10, 11). These findings indicate that unique carbohydrate structures associated with pathogens can serve as the PAMPs in innate immunity.
Among the innate immune receptors (IIRs), CLRs are vital carbohydrate-recognizing proteins that mediate pathogen recognition/clearance, immune cell interactions, and endothelium cell adhesion. These proteins are characterized by the presence of highly conserved carbohydrate recognition domains that are responsible for binding to a variety of glycoproteins and glycolipids via their carbohydrate portion of the molecule. CLRs are able to activate and modulate host immunity through protein-protein interactions and/or by using their carbohydrate recognition domains to interact with glycans (5, 12). For example, DC-SIGN, a typical CLR specifically expressed on dendritic cells and macrophages, participates in the immune response and pathogenesis by binding to viral pathogens (e.g. human immunodeficiency virus, cytomegalovirus, dengue virus, Ebola virus, and SARS corona virus), various bacterial pathogens (e.g. Mycobacterium and Helicobacter), and parasitic antigens (13–17). In addition to its carbohydrate-binding capability, DC-SIGN can interact with cell surface proteins such as intercellular adhesion molecule (ICAM)-2 and -3 to mediate cell trafficking and the formation of the immunological synapse between dendritic cells and T cells (18, 19). To better understand the underlying mechanisms of self versus non-self recognition in the immune system, it is important to study the binding characteristics of CLRs to their glycoconjugate ligands.
Ganoderma and Cordyceps are among the most popular fungi used for dietary supplement and/or herbal medicine in eastern Asia. The polysaccharide extracts from Ganoderma lucidum (Reishi or Ling-Zhi), abbreviated as GLPS, contain active anti-tumor and immune modulation activities (20–23). In addition, the polysaccharides extracted from Cordyceps (caterpillar fungus), also known as CPS, alter apoptotic homeostasis, improve respiratory, renal, and cardiovascular functions (24–26), and increase sensitivity to insulin (27). The major polysaccharide backbones of GLPS and CPS are β-glucans and α(1–4)-mannan, although the composition is highly variable due to strain diversity, culture conditions, and extraction methods. As such, the beneficial activities of GLPS and CPS are difficult to reproduce. Analytical methods such as high-performance liquid chromatography and proton-nuclear magnetic resonance techniques have been applied to monitor and investigate the components derived from medicinal herbs, but these methods are not suitable for analyzing large and complex molecules such as GLPS and CPS.
Here, we develop an enzyme-linked immunosorbent assay (EIA) based on Fc-tagged recombinant IIR fusion proteins to investigate the profiles of GLPS and CPS derived from various sources. This IIR-EIA analyzes the polysaccharides and glycoconjugates in natural products quantitatively and qualitatively. The method of IIR-EIA will be useful for resolving the biological functions of medicinal polysaccharides.
EXPERIMENTAL PROCEDURES
Cell Culture
The FreeStyleTM 293-F cells (Invitrogen) were cultured with FreeStyle 293 Expression Medium (Invitrogen) in Erlenmeyer flasks and incubated in a 37 °C incubator containing 8% CO2 on an orbital shaker rotating at 125 rpm.
Polysaccharides and Monosaccharides
Mannan (α-linked, from Saccharomyces cerevisiae), β-glucan (from barley), cellulose (carboxymethylated, with average Mr ∼90,000), dextran (Mr ∼100,000), laminarin, fucoidan, β(1–4)-mannan, and monosaccharides were purchased from Sigma; amylopectin, galactan, and arabinogalactan (from larch) were purchased from Megazyme (Wicklow, Ireland). Mannan, cellulose, dextran, laminarin, fucoidan, galactan, arabinogalactan, and all the monosaccharides were dissolved in pure water. Stock solutions of β(1–4)-mannan (5 mg/ml) and amylopectin (10 mg/ml) were made by dissolving in 0.75 and 0.5 n NaOH, respectively. For dissolving cellulose, 5 mg/ml of stock solution was heated at 90 °C with vigorous shaking for 30 min. For preparing β-glucan stock solution, 3 mg of β-glucan was first dissolved in 0.2 ml of DMSO by heating at 60 °C with shaking for 30 min, followed by mixing in 0.8 ml of pure water and another 30 min of 60 °C incubation with vigorous shaking.
Construction, Expression, and Purification of Fc-tagged Recombinant Immune Receptor Fusion Proteins
The DNA fragments encoding extracellular domains of CLRs, and TLRs were amplified by reverse transcriptase-PCR and subcloned into pcDNA3.1(+)-hIgG1 Fc or pSecTag2C-hIgG1 Fc (gifts from Hugh O. McDevitt, Stanford University, Palo Alto, CA) vectors to generate N- or C-terminal hIgG1 Fc-tagged fusion proteins, respectively. Primer pairs for amplifying extracellular domains of IIRs are listed in Table 1. The receptor-Fc proteins were overexpressed using the FreeStyle 293 Expression System (Invitrogen). Briefly, 3 × 107 293-F cells in 28 ml of culture medium were transfected with a mixture of 30 μg of plasmid DNA/40 μl of 293fectinTM in a total of 2 ml of Opti-MEM® I (Invitrogen). The culture supernatants were harvested at days 3 and 5, and the recombinant receptor-Fc proteins were purified by Protein A columns (GE Healthcare). Purified IIR-Fc proteins were examined for their sizes by SDS-PAGE with or without the addition of 5% 2-mercaptoethanol (supplemental Fig. S1).
TABLE 1.
Primer pairs for amplifying IIR extracellular domain fragments
| Innate immune receptor (IIR) | Other names | Forward primer | Reverse primer |
|---|---|---|---|
| AICL | CLEC2B, CLECSF2 | GGATCCTCTCAGAGTTTATGCCCC | GGATCCCCCCATTATCTTAGACAT |
| BDCA2 | CLEC4C, DLEC, HECL, CLECSF11, CLECSF7 | GGATCCTTTATGTATAGCAAAACTGTCAAG | GAATTCTTATATGTAGATCTTCTTCATCTT |
| CLEC2 | CLEC1B | GGATCCATGCAGCGCAATTACCTACAA | GAATTCTTAAGGTAGTTGGTCCACCTTG |
| CLEC6 | CLEC4D, Mpc1, MCL, CLECSF8 | GGATCCCATCACAACTTTTCACGCTGT | GAATTCCTAGTTCAATGTTGTTCCAGG |
| DCIR | CLEC4A, DDB27, CLECSF6 | GGATCCTTTCAAAAATATTCTCAGCTTCTT | GAATTCTCATAAGTGGATCTTCATCATC |
| DC-SIGN | CLEC4L, CD209 | GGATCCAAGGTCCCCAGCTCCATAAG | GAATTCCTACGCAGGAGGGGGGT |
| Dectin-1 | CLEC7A, β-glucan receptor, CLECSF12 | GGATCCACCATGGCTATTTGGAGATCC | GAATTCTTACATTGAAAACTTCTTCTCAC |
| Langerin | CLEC4K, CD207 | GGATCCCGGTTTATGGGCACCATA | GGATCCTCACGGTTCTGATGGGAC |
| NKG2D | KLRK1 | GGAGTGCTGTATTCCTAAAC | GAATTCCTGGCTTTTATTGAGATGG |
| MDL1 | CLEC5A, CLECSF5 | AGATCTAGTAACGATGGTTTCACCAC | GAATTCCTGTGATCATTTGGCATTCTT |
| Mouse KCR | Kupffer cell receptor | CAGCCTTGGAGACCTGAGT | CTAGCCTACTCTGGCCGC |
| DCAL1 | CLECL1 | GGATCCTACGCTGACATCAAAACTGTT | GAATTCTAAATGTTAAATCTCACCATAGC |
| DEC205 | CLEC13B, LY75, CD205 | GCCGGCCTGGCCGCGCAGCTAATGA | GCGGCCGCGGCACTTTGCAGACAACTC |
| Endo180 | CLEC13E, MRC2 | GATATCCGGAACCCAACATCTTCC | GCGGCCGCAGCGCTGATGGGGAGAAGCT |
| MMR | CLEC13D, MRC1 | GGTACCTACTGGACACCAGGCAATT | GCGGCCGCGAAGGGTCCATCTTCCTTG |
| TLR2 | TIL4, CD282 | GCGGATCCAGGAAGAATCCTCCAATCAG | GCGCGGCCGCGTCCTGTGACATTCCGACAC |
| TLR4 | hToll, CD284 | GCGGATCCAAAGCTGGGAGCCCTGCGT | GCGCGGCCGCTTATTCATCTGACAGGTGATA |
Standardized Isolation of the F3 Fraction of G. lucidum Polysaccharides (GLPS-F3)
The polysaccharide components of GLPS-F3 were prepared according to the procedures described previously (23). In brief, crude Reishi powders (broken spores and fruiting bodies) were stirred in boiling water for 2 h, and centrifuged to remove the insoluble materials. The resulting solution was concentrated, lyophilized, and dissolved in a small volume of 0.1 n Tris buffer (pH 7.0) containing 0.1 n sodium azide, and purified by gel filtration chromatography at 4 °C using a Sephacryl S-500 column (95 × 2.6 cm) to obtain GLPS-F3. Fractions were subjected to the phenol-sulfuric acid method to detect sugar concentration, concentrated, dialyzed with a 6000–8000-dalton MWCO membrane against water, and lyophilized to give GLPS-F3 powders. The estimated molecular weights of GLPS-F3 were larger than 1,000,000 according to the elution time shown in gel filtration chromatography.
Preparation of Biotinylated GLPS-F3
The biotinylation of GLPS-F3 was performed in a one-pot procedure. GLPS-F3 (100 mg) in 0.2 n NaHCO3/Na2CO3 (10 ml) was reacted with biotinamidohexanoyl-6-amino-hexanoic acid N-hydroxysuccinimide ester (biotin-XX-NHS, 1.0 mg) in N,N-dimethylformamide (1 ml) and stirred at room temperature for 12 h. The resulting solution was dialyzed against water (500 ml for five times) with a 6000–8000-dalton MWCO membrane at 4 °C within 48 h. After dialysis, the biotinylated F3 was lyophilized to give a brown powder of 90 mg (90%). The purity of biotinylated GLPS-F3 was monitored by high-pressure liquid chromatography and streptavidin-fluorescein isothiocyanate in binding assay.
Enzyme-linked Immunoassay
Samples of GLPS-F3 and biotinylated GLPS-F3 were weighed, dissolved, and diluted with 100 mm Tris buffer (pH 9.5) to give 0.2–20 μg/ml. Diluted GLPS-F3 was then immobilized in the wells of 96-well Nunc MaxiSorpTM plates (50 μl/well, Thermo Fisher Scientific, Waltham, MA). After incubation overnight at 4 °C, wells were washed twice with TBST (0.05% Tween 20/TBS), followed by blocking with 200 μl of blocking buffer (2% bovine serum albumin/TBST) for 1 h at room temperature. Peroxidase-conjugated streptavidin was diluted (1:5000) in blocking buffer (Vector Laboratories, Burlingame, CA) and added 100 μl per well to detect the immobilized biotinylated GLPS-F3 after incubation at room temperature for 30 min. Alternatively, for examining the interaction with GLPS-F3 that was immobilized and blocked as described above, 100 μl of IIR-Fc fusion protein (1 μg/ml in 2 mm MgCl2, 2 mm CaCl2, 1% bovine serum albumin/TBST) in the presence or absence of competitors was added into each well and incubated for 1 h at room temperature. After washing with TBST, each well was incubated with 100 μl of peroxidase-conjugated goat anti-human IgG Ab (Jackson ImmunoResearch Laboratories) in blocking buffer (1:5000) at room temperature for 30 min. Wells were incubated with 100 μl of tetramethylbenzidine substrate (Pierce) for 15 min after the TBST wash and read at 450 nm in a plate reader. For profiling of GLPS and the CPS mixture derived from different sources, samples were first measured for their sugar contents by phenol-sulfuric acid method, and the sugar concentrations were then used to calculate the amounts for immobilization on 96-well plates (250 ng/well). CPS samples were obtained from Wyntek Corporation (Taipei, Taiwan).
RESULTS
Expression of Recombinant IIR-Fc Fusion Proteins
The DNA fragments encoding the extracellular domain of IIRs including CLRs and TLRs were amplified by reverse transcriptase-PCR using the cDNA templates derived from various immune cells. These amplified DNA fragments were subcloned into mammalian expression vectors. The vectors contained an Fc fusion, consisting of a mutated human IgG1 Fc portion lacking the human Fc receptors binding capability (28). In addition, the hinge region of IgG1 Fc was preserved to produce dimeric fusion proteins that allow for divalent interactions. The extracellular domains of the type I and II transmembrane receptors were fused at the N terminus (receptor-Fc) and C terminus (Fc-receptor), respectively.
Glycan-binding Properties of IIR-Fc Fusion Proteins
The IIR-Fc proteins were submitted to the Consortium for the Functional Glycomics (CFG) for glycan array analysis to understand more about their detailed carbohydrate recognition capability. All of the glycan binding data for the IIRs are available through Functional Glycomics Gateway. Table 2 summarizes the IIR-Fc proteins studied by EIA along with their glycan-binding characteristics. Of these 17 IIRs, 12 were submitted for CFG glycan array analysis by us, and six IIRs among them (Fc-tagged AICL, BDCA2, CLEC2, DCIR, Langerin, and MMR) show binding to the glycans on this array. The individual glycan binding profiles for these recombinant IIR proteins we submitted for CFG glycan array analysis can be found in supplemental Figs. S2–S7. In addition to these six IIRs, recombinant DC-SIGN and Kupffer cell receptor (KCR) proteins from other groups have also been characterized for their sugar-binding properties via CFG glycan arrays (29, 30). The binding profiles of these two proteins are thus included in Table 2 as well. A high proportion of IIRs interact with sulfated lactose (Lac) or N-acetyllactosamine (LacNAc). CLEC2, DC-SIGN, Dectin-2, Langerin, and MMR bind to high mannose (Man) structures. In addition to the sulfated LacNAc/Lac, BDCA2 and DC-SIGN also bind biantennary N-glycans with or without core fucosylation and terminal sialylation, suggesting that they interact with heavily N-glycosylated proteins on the cell surface. AICL, CLEC2, DC-SIGN, and Langerin can bind fucose (Fuc)-containing antigens, such as H, Lewis, or sialyl Lewis antigens, indicating their ability to recognize Fuc. In addition, the results indicate that CLEC2 and MMR have a higher tendency to bind poly α2–8-linked sialic acid (NeuAc); notably, this polysialic acid has been shown to mediate cancer growth and invasion (31, 32). Overall, the results from glycan array analysis provide greater insight into the carbohydrate recognition capabilities of each IIR, which will likely help delineate the molecular characteristics of their natural glycoconjugate substrates.
TABLE 2.
Carbohydrate-binding characteristics of IIRs
| Innate immune receptor (IIR) | Other names | Data available in CFG glycan array database | Carbohydrate-binding characteristics (CFG glycan array data) |
|---|---|---|---|
| AICL | CLEC2B, CLECSF2 | Yes | 1. Sulfated LacNAca or Lac |
| 2. H antigen (type 3) | |||
| 3. GlcA | |||
| 4. Sialyl-LacNAc or -Lewis a | |||
| BDCA2 | CLEC4C, DLEC, HECL, CLECSF11, CLECSF7 | Yes | 1. Sulfated LacNAc or Lac |
| 2. Biantennary N-glycans | |||
| 3. Galβ4GalNAcβ3-H antigen | |||
| CLEC2 | CLEC1B | Yes | 1. Sulfated LacNAc or Lac |
| 2. Sialyl-LacNAc, -Lac or -Lewis a | |||
| 3. Poly α2–8-linked NeuAc | |||
| 4. High Man | |||
| CLEC6 | CLEC4D, Mpc1, MCL, CLECSF8 | Yes | No binding to CFG glycan array |
| DCIR | CLEC4A, DDB27, CLECSF6 | Yes | 1. Sulfated LacNAc or Lac |
| 2. Biantennary N-glycans | |||
| DC-SIGNb | CLEC4L, CD209 | Yes | 1. High Man |
| 2. Lewis a/b/x/y | |||
| 3. A and B antigens | |||
| 4. Fucosyl biantennary N-glycans | |||
| Dectin-1 | CLEC7A, β-glucan receptor, CLECSF12 | Yes | No binding to CFG glycan array |
| Langerin | CLEC4K, CD207 | Yes | 1. Sulfated LacNAc or Lac |
| 2. Sialyl-Lewis x or Lewis x/y | |||
| 3. H and B antigens | |||
| 4. GlcNAcβGal/GalNAc | |||
| 5. High Man | |||
| 6. Core 3/4 | |||
| NKG2D | KLRK1 | No | Not determined |
| MDL1 | CLEC5A, CLECSF5 | Yes | No binding to CFG glycan array |
| KCRc | Kupffer cell receptor | Yes (rat) | 1. Oligosaccharides with terminal Gal or GalNAc |
| 2. Biantennary N-glycans | |||
| DCAL1 | CLECL1 | Yes | No binding to CFG glycan array |
| DEC205 | CLEC13B, LY75, CD205 | Yes | No binding to CFG glycan array |
| Endo180 | CLEC13E, MRC2 | Yes | No binding to CFG glycan array |
| MMR | CLEC13D, MRC1 | Yes | 1. High Man |
| 2. Sulfated sugars | |||
| 3. Poly α2–8-linked NeuAc | |||
| TLR2 | TIL4, CD282 | Yes | No binding to CFG glycan array |
| TLR4 | hToll, CD284 | No | Not determined |
a Nomenclature: LacNAc, Galβ1–4GlcNAc; Lac, Galβ1–4Glc; H antigen (type 2), Fucα1–2Galβ1–4GlcNAc-R; H antigen (type 3), Fucα1–2Galβ1–3GalNAc-R; A antigen, GalNAcα1–3(Fucα1–2)Gal-R; B antigen, Galα1–3(Fucα1–2)Gal-R; Lewis a, Galβ1–3(Fucα1–4)GlcNAc-R; Lewis x, Galβ1–4(Fucα1–3)GlcNAc-R; Lewis y, Fucα1–2Galβ1–4(Fucα1–3)GlcNAc-R; sialyl Lewis a, NeuAcα2–3Galβ1–3(Fucα1–4)GlcNAc-R; sialyl Lewis x, NeuAcα2–3Galβ1–4(Fucα1–3)GlcNAc-R.
b Ref. 30.
c Ref. 29.
Identification of Receptors Capable of Interacting with GLPS-F3 by IIR-EIA
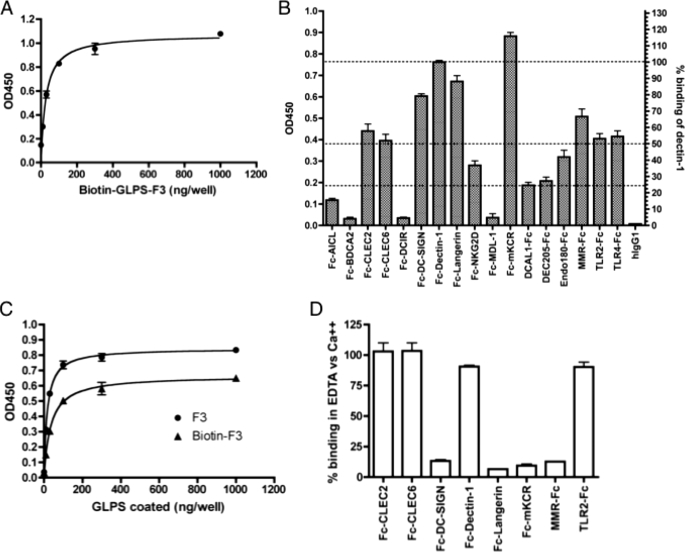
To demonstrate the possibility of using IIR for profiling unidentified glycan mixtures, all 17 IIR-Fc fusion proteins were tested for binding specificity and sensitivity to GLPS-F3. GLPS-F3 is the water-soluble polysaccharide fraction extracted from G. lucidum capable of inducing immune cell activation (23). According to the previous analysis (23), the carbohydrates in GLPS-F3 are composed of 58% glucose, 15% mannose, 14% galactose, 7% Fuc, 3% xylose, and 1% N-acetylglucosamine (GlcNAc), and 2% other monosaccharides, but no sialic acid. Protein accounts for ∼30% of GLPS-F3 in weight (data not shown), indicating that GLPS-F3 is a mixture of polysaccharides and proteins (or glycoproteins) with very high molecular masses (>1000 kDa).
The IIR-EIA of immobilized GLPS-F3 in microtiter plates was established to investigate the interaction between GLPS-F3 and IIR fusion proteins. To optimize the immobilization condition, various amounts of biotinylated-GLPS-F3 (biotin-GLPS-F3) ranging from 10 to 1000 ng/well were coated on microtiter plates through both hydrophilic and hydrophobic interactions. For read out, peroxidase-conjugated streptavidin and then chromogen substrate tetramethylbenzidine were subsequently added. As shown in Fig. 1A, the intensity of coloration reached a plateau at 100 ng/well; therefore this concentration was used for immobilization of un-biotinylated GLPS-F3 in the IIR-EIA. As expected, Fc-Dectin-1 (β-glucan receptor) bound strongly to GLPS-F3 (Fig. 1B), which contains β(1,3)-glucan as the major backbone (33). In addition, Fc-mKCR, Fc-Langerin, and Fc-DC-SIGN also bound to GLPS-F3 specifically (OD450 > 0.6), whereas weaker but positive binding was also observed between GLPS-F3 and TLR2, TLR4, MMR, CLEC2, and CLEC6 (0.4 < OD450 < 0.6). Notably, both TLR2 and TLR4 were reported to mediate GLPS-induced cell activation (34–37). Weaker binding (0.2 < OD450 < 0.4) was also observed in Fc-tagged NKG2D, DCAL1, DEC205, and Endo180. However, none of these receptors have been reported to play a role in GLPS-mediated cell activation. Other CLRs including Fc-tagged AICL, BDCA2, DCIR, and MDL-1 showed a minimal capability to bind to GLPS-F3, with the signal similar to that of human IgG1 control. Among the non-binders, Fc-AICL, Fc-BDCA2, and Fc-DCIR display binding capabilities to specific ligands on CFG glycan arrays, whereas Fc-MDL-1 (also known as CLEC5A) has been demonstrated to interact with dengue virus (38) although it shows no binding on the glycan array. Here, we provide evidence that these five recombinant IIR proteins do have binding abilities to various ligands but not GLPS-F3.
FIGURE 1.
Identification of human IIRs that interact with GLPS-F3. A, optimization of GLPS-F3 EIA. Biotinylated GLPS-F3 was immobilized at several dilutions onto 96-well EIA plates. The quantity of coated GLPS-F3 was then examined by peroxidase-conjugated streptavidin. B, detection of GLPS-F3-interacting IIRs by EIA. EIA was performed as described under ”Experimental Procedures.“ To compare the binding intensities of GLPS-F3 among IIR-Fc fusion proteins, OD450 readings of different IIR-Fc were normalized against that of Fc-Dectin-1, as shown in the right y axis. IIR-Fc that shows over 50% of binding is considered as a stronger binder. Data were calculated from five rounds of independent experiments. Dashed lines indicate 100%, 50%, and 25% binding of Dectin-1. C, comparison of the binding curves of both plate-coated biotin-GLPS-F3 and un-biotinylated GLPS-F3 to Fc-Dectin-1. D, calcium dependence of receptor-Fc binding to GLPS-F3. EIA was performed in binding buffer with 2 mm CaCl2 (binding in Ca2+) or with 10 mm EDTA and no CaCl2 (binding in EDTA). For A, C, and D, one representative data from three independent experiments is shown. Error bars represent standard error of the mean.
Considering that the biotinylation of GLPS-F3 may change the immobilization behavior of GLPS-F3 on EIA plates, we compared the binding curves of biotin-GLPS-F3 and un-biotinylated GLPS-F3 to Fc-Dectin-1. Results revealed a similar amount (100 ng/well) of coated biotin- and un-biotinylated GLPS-F3 to reach the plateau of Fc-Dectin-1 binding, although the binding intensity was higher in coated GLPS-F3 (Fig. 1C). This suggests that the optimal coating level on EIA plates evaluated by biotin-GLPS-F3 can be applied to GLPS.
We further examined whether the binding of each fusion protein to GLPS-F3 is calcium ion-dependent, a common feature among CLRs, to reflect their specific carbohydrate-interacting capabilities. To address calcium dependence, GLPS-F3-interacting IIR-Fc proteins were selected for IIR-EIA with either the standard lectin-binding buffer (with calcium ion) or EDTA-binding buffer (lectin-binding buffer with 10 mm EDTA, but without calcium ions). The results shown in Fig. 1D are consistent with previous reports that the presence of EDTA abolished the binding of DC-SIGN, Langerin, mKCR, and MMR to carbohydrates (39), indicating the requirement of calcium ions for their carbohydrate recognition. The results also revealed that in addition to TLR2, the interactions of Dectin-1, CLEC2, and CLEC6 with GLPS-F3 were calcium-independent. Besides Dectin-1, which interacts with β-glucan in a calcium-independent fashion (10), CLEC2 shows no association with calcium ions based on its crystal structure (40), supporting the observation of its calcium-independent binding. As for CLEC6, there is no evidence showing that it interacts with carbohydrates. Like CLEC2, the C-type lectin-like domain of CLEC6 lacks some of the conserved calcium- and sugar-binding residues for lectin activity. This may provide a possible explanation for their calcium-independent binding property.
Determination of Binding Specificity by Competition Assay
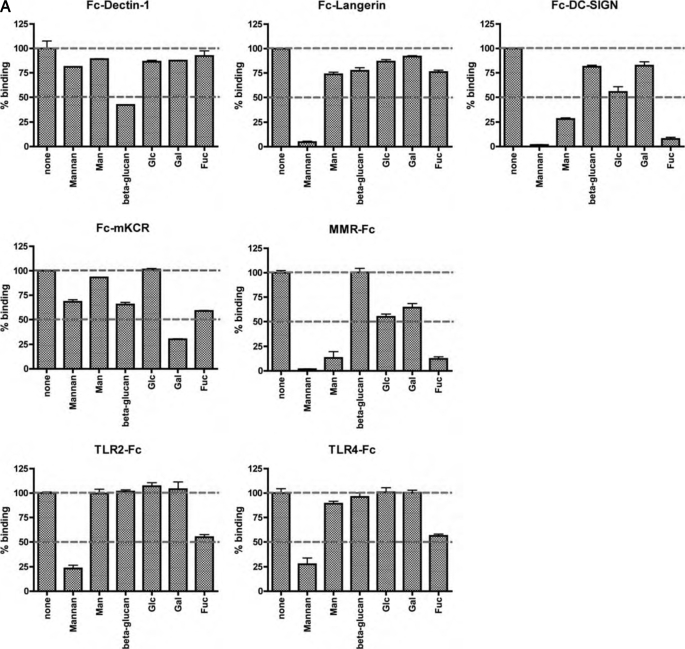
To understand the nature of IIR fusion protein binding to GLPS-F3, a competition study was performed with the polysaccharides mannan and β-glucan, and monosaccharides such as d-mannose (Man), d-glucose (Glc), d-galactose (Gal), and l-fucose (Fuc). The IIRs that showed higher binding to GLPS-F3, including Fc-tagged Dectin-1, Langerin, DC-SIGN, MMR, mKCR, TLR2, and TLR4 fusion proteins were examined. As shown in Fig. 2A, under 1 mg/ml of competitive sugars, the interaction between GLPS-F3 and Fc-Dectin-1 was inhibited significantly by β-glucan (58% inhibition), in accordance with previously published results (10, 41–43). The interaction between Fc-Langerin and GLPS-F3 was disrupted by mannan (95% inhibition), which are known sugar ligands for Langerin (44). The binding of Fc-DC-SIGN to GLPS-F3 was considerably blocked by mannan, Man, and Fuc (98, 72, and 92% inhibition), whereas Glc also had a weaker effect at 45% inhibition of binding. In agreement with the known binding properties of MMR, the interaction between GLPS-F3 and MMR was abolished by mannan, Man, and Fuc, at 98, 87, and 88% respective inhibition (45, 46). The binding of Fc-mKCR to GLPS-F3 was blocked in the presence of Gal and Fuc (70 and 41% inhibition, respectively), two known ligands of mKCR (47–49). Mannan and Fuc inhibited the interaction of GLPS-F3 with TLR2-Fc (77 and 45% inhibition, respectively) and TLR4-Fc (72 and 44% inhibition, respectively). Although the sugar ligands of TLR2 and TLR4 have not been reported yet, it has been demonstrated that mannan derived from yeasts triggers tumor necrosis factor-α secretion via TLR4 (50), implying the possibility of mannan-TLR4 interaction.
FIGURE 2.
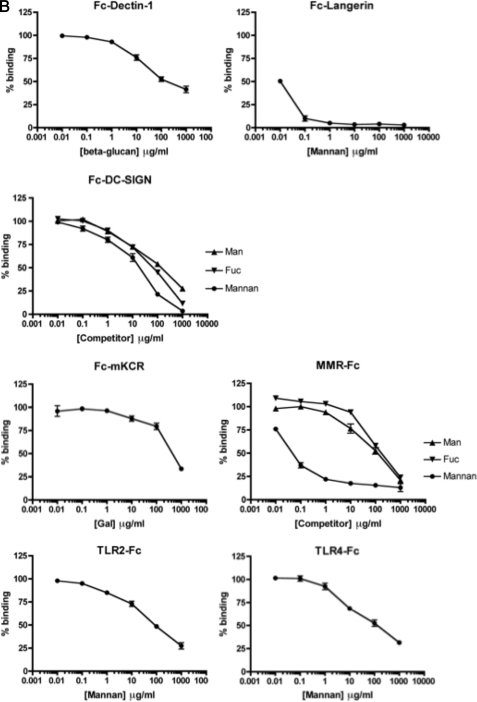
Competition assay for GLPS-F3-interacting IIRs. A, competitor sugars (1 mg/ml) and IIR-Fc in EIA were added simultaneously to immobilized GLPS. The percentage of binding was normalized against the sample without competitor in each receptor-Fc group. Shown are representative data from three independent experiments. The percentages of binding at 100% and 50% are indicated as dashed lines. B, dose-response curves of the sugars that inhibited >50% GLPS-F3-IIR interaction at 1 mg/ml. One representative data from three independent experiments is shown. Error bars represent standard error of the mean.
The sugars that showed over 50% competition at 1 mg/ml were further tested for their dose-response curves. As revealed in Fig. 2B, the IC50 of β-glucan for the Fc-Dectin-1-GLPS-F3 interaction was 100 μg/ml, whereas the IC50 of mannan for the binding of IIR-Fc to GLPS-F3 was lower: 10 ng/ml for Langerin, 17 μg/ml for DC-SIGN, 50 ng/ml for MMR, and 58 and 43 μg/ml for TLR2 and TLR4, respectively. Man gave a much higher IC50 for the MMR-GLPS-F3 interaction, when compared with mannan (87 μg/ml versus 50 ng/ml), however, gave higher but comparable IC50 values for the DC-SIGN-GLPS-F3 interaction compared with mannan (60 versus 17 μg/ml). The IC50 values of Fuc for the binding of DC-SIGN and MMR with GLPS-F3 were 54 and 138 μg/ml, and the IC50 of Gal in inhibiting the mKCR-GLPS-F3 interaction was 437 μg/ml. The results showed dose responsiveness of sugar competitors for IIR-Fc-GLPS-F3 binding. Additionally, the data also supports the concept that polyvalent sugar ligands (e.g. mannan versus Man) increase the binding affinity of lectins.
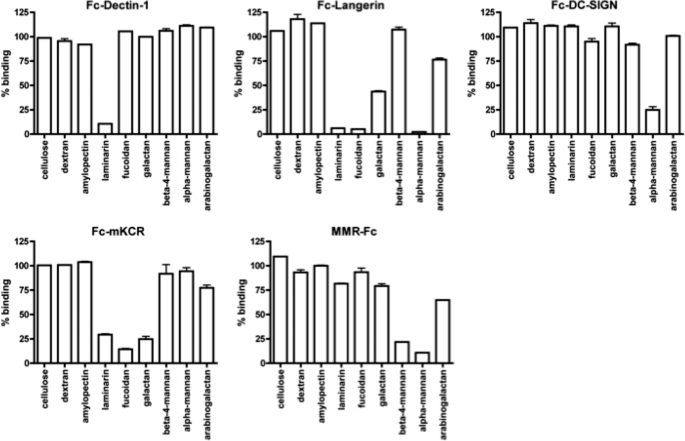
To further explore the binding specificities of IIR-Fc proteins to other polysaccharides, we added several polysaccharides to compete with the binding of individual IIR-Fc with GLPS-F3 (Fig. 3). Fc-Dectin-1, Fc-Langerin, Fc-DC-SIGN, Fc-mKCR, and MMR-Fc proteins were selected to examine their binding to cellulose (β(1–4)-glucan), dextran (α(1–6)-glucan with α(1–3)-linked Glc side chains), amylopectin (α(1–4)-glucan with α(1–6)-linked Glc side chains), laminarin (β(1–3)-glucan with β(1–6)-linked Glc side chain), fucoidan (composed predominated of sulfated Fuc), galactan (Gal polymer), β(1–4)-mannan, α-mannan, or arabinogalactan (Gal polymer with arabinose). Fc-Dectin-1 showed a specific binding toward laminarin among these polysaccharides, whereas Fc-Langerin showed binding to laminarin, fucoidan, galactan, and α-mannan. Fc-DC-SIGN had binding specificity only to α-mannan, but not β(1–4)-mannan, whereas MMR-Fc exhibited binding to both α- and β(1–4)-mannan, and Fc-mKCR showed binding to laminarin, fucoidan, and galactan. Notably, the results revealed in this competition assay indicate that in addition to binding to the reported sugar ligands, both Langerin and mKCR could interact with laminarin. In summary, the results indicate that Fc-tagged IIR proteins preserve their binding specificity, and GLPS may activate immune cells through interactions with multiple receptors.
FIGURE 3.
Competition of GLPS-F3-interacting IIRs by various polysaccharides with known structures. Competitor polysaccharides (1 mg/ml) and indicated IIRs were added to immobilized GLPS in microtiter plates. The percentage of binding was normalized against the sample without competitor in each group. Shown are representative data from three independent experiments. Error bars represent standard error of the mean from triplicates.
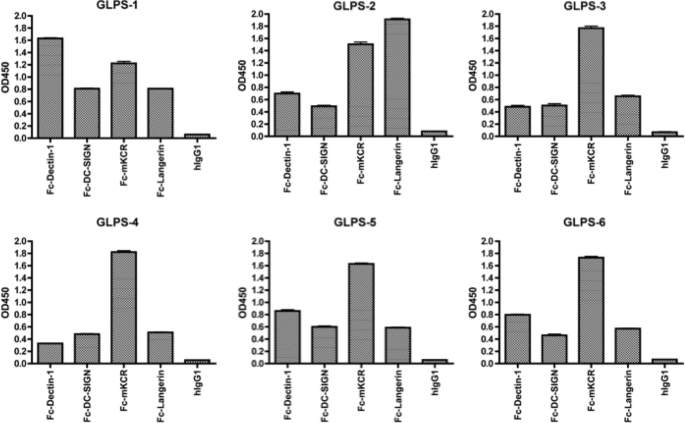
Profiling of Polysaccharides from Different Sources
IIR-EIA was applied to profiling commercially available GLPS prepared from different Reishi sources. To cover the maximal numbers of sugar ligands recognized by our receptor probes, Fc-Dectin-1, Fc-DC-SIGN, Fc-mKCR, and Fc-Langerin were used to establish the binding profiles of each polysaccharide sample by EIA (Fig. 4). Sample GLPS-1 consisted of the polysaccharides extracted from both mycelia and fruiting bodies, whereas GLPS-2 was derived from fruiting bodies; GLPS-3 and -4 were different batches of blended crude extracts from spores and fruiting bodies; GLPS-5 and -6 were batches of GLPS further extracted from GLPS-3/4. Compared with GLPS-2, Fc-Dectin-1 bound to GLPS-1 much stronger than GLPS-2, indicating that Reishi mycelia contain more β-glucan than Reishi fruiting bodies. Compared with the relatively crude GLPS-3/4, the further purified GLPS-5/6 showed increased Fc-Decitn-1 binding, suggesting that the purification process has further enriched Dectin-1-binding components, possibly glycan chains with more β-glucan. The binding of Fc-mKCR to all GLPS samples was relatively strong, except GLPS-1. The IIR-EIA results demonstrate that GLPS isolated from different sources can have distinct binding patterns.
FIGURE 4.
Profile of GLPS interacting with IIR-Fc. GLPS from different sources was immobilized on 96-well EIA plates (sugar concentrations is 250 ng/well). The binding of selected IIR-Fc to GLPS was examined by EIA. The selected data shown is representative of three independent experiments.
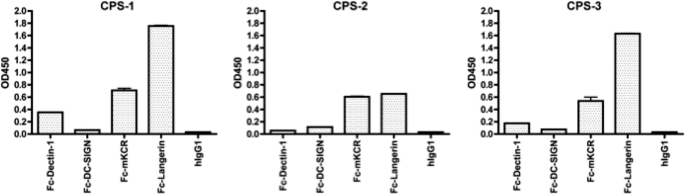
The binding fingerprints of polysaccharides extracted from three different strains of Cordyceps (CPS) were also tested using the fusion protein probes. As shown in Fig. 5, the binding patterns of CPS samples were very different from GLPS samples. In comparison to the binding profiles of GLPS samples, the CPS sample profiles showed weaker binding to Fc-mKCR. Significant differences between GLPS and CPS samples were also detected by probing with Fc-Langerin (binding was higher in two of three CPS samples). Moreover, the amounts of Dectin-1 and DC-SIGN-interacting components seemed to be much lower in CPS than in GLPS samples. These results conclusively demonstrate that IIR-EIA can be used to profile the polysaccharides extracts prepared from different strains, sources, or batches of Reishi and Cordyceps. Here we demonstrate that IIR-EIA is useful for understanding the receptor binding properties of polysaccharide-containing mixtures, it is a practical tool for profiling polysaccharide structures; and, more importantly, it provides a direct link between components/structures of polysaccharides to biological functions initiated through interactions with cell surface receptors.
FIGURE 5.
Profile of polysaccharides extracted from CPS interacting with IIR-Fc. CPS was extracted from different Cordyceps strains and immobilized in microtiter plates for EIA (sugar concentrations is 250 ng/well). CPS-1, -2, and -3 were extracted from Acremonium terricola, Paecilomyces hepiali, and Hirsutella sinensis. Representative data derived from two independent experiments is shown.
DISCUSSION
PRRs are innate immune receptors that serve as the front line in sensing and defending invading pathogens. PRRs induce cell phagocytosis and transmit signals to trigger and regulate immune responses after binding to PAMPs (39). To exploit the specific PAMP-recognizing properties of PRRs, and specifically the less well characterized carbohydrate-type PAMPs, we cloned and expressed PRRs as recombinant receptor-Fc fusion proteins for expression in mammalian cells to ensure proper folding and glycosylation. IIR-EIA demonstrates the use of PRRs as probes to reveal the constituents of polysaccharide-containing mixtures, which can be glycosylated natural products, a pool of glycoproteins, or the poly- or oligosaccharides from bacterial and viral pathogens. In addition to being a tool for carbohydrate profiling, IIR-EIA allows complex molecular saccharide structures to be directly coupled to their biological functions, providing invaluable information that is difficult to obtain by other means.
To demonstrate the power of the IIR-EIA platform, we analyzed a GLPS polysaccharide mixture with a molecular composition and biological function that had been partially characterized. GLPS contains β(1–3)-linked glucan backbones with variable attachment of β(1–2, 4, 6)-linked Glc (33) and α(1–4)-linked mannan backbones appended with xylose and Fuc (51). Several innate immune receptors bound to GLPS-F3 in accordance with their reported recognition properties. Dectin-1, which recognizes β-glucans (52) and has been determined to bind β(1–3)-linked glucose oligomers with at least 10- or 11-mer in length (42); and DC-SIGN, MMR, and Langerin, which recognize Fuc and mannan and/or Man (39) (Fig. 1B and supplemental data). A sugar competition assay confirmed that the interaction of GLPS-F3 with these Fc-tagged receptors was specific to such carbohydrate epitopes. In addition to CLRs, TLR2 and TLR4, two receptors reported to mediate GLPS-induced cell activation (34–37), showed binding to GLPS-F3 directly. In this case, competition experiments indicate that TLR2 and TLR4 interact with GLPS-F3 mannan and other structures linked to Fuc (Fig. 2). We observed that mKCR interacts with GLPS-F3 and that this interaction can be interrupted by the presence of Gal and Fuc, consistent with previous results that KCR binds to Gal, GalNAc, and Fuc (47–49). In addition, CLEC2 and CLEC6 bind to GLPS-F3, however, under our sugar competition conditions, no antagonism was observed. This suggests that other as yet unknown structures, for instance, sulfated sugars, and possibly the proteins in GLPS-F3 are involved in binding to CLEC2 and CLEC6. Another member of the mannose receptor family, Endo180 (53, 54), which showed negative binding in glycan array analysis in this study, has been reported to bind to Man, Fuc, and GlcNAc (55) and could also bind to GLPS-F3. This could be due to the mannan backbone in GLPS-F3, which provides multivalent sugar epitopes to enhance the binding of Endo180. However, the possibility that Endo180 binds to the protein portion of the GLPS/F3 mixture cannot be ruled out.
The results from the sugar competition assay (Fig. 2) showed that even at the same mass-volume concentration, the competitors bearing multiple carbohydrate determinants have higher competitive ability than the monosaccharide competitors (e.g. mannan versus Man). This supports the concept that multivalency of carbohydrate results in higher binding affinity. This also implies that in addition to terminal sugars, the internal sugars, which are the major contributors in generating multiple carbohydrate epitopes, can play a role in interacting with CLRs, even though the glycan array analysis indicates that the binding specificities of IIR-Fc proteins are mostly terminal sugars.
It has been reported that the terminal sialylation of the MMR protein is crucial for its mannose recognition capability and modulates its binding with sulfated sugars via regulating MMR self-association (56). Thus it would be interesting to understand whether the sialyl residues or the sulfate groups in oligosaccharides could also influence the binding affinities of other CLRs.
Because GLPS-F3 has been shown to stimulate a variety of cytokines including interleukin-1β, interleukin-6, interleukin-12, interferon-γ, tumor necrosis factor-α, granulocyte macrophage-colony stimulating factor, granulocyte-colony stimulating factor, and macrophage-colony stimulating factor in different immune cells (22, 23, 32, 33), it is highly possible that GLPS activates different immune cells with distinctive sets of cell surface IIRs to produce cell type-specific cytokine signatures. The correlations between GLPS-induced cytokine secretion and IIR expression in specific cell types, once established, will help understand the detailed mechanism of action for better purification methods and drug designs.
As demonstrated in Figs. 4 and 5, polysaccharide extracts prepared from different sources or strains have different fingerprints, which likely relate to their biological effects. For example, a recent study on GLPS-modulated maturation of dendritic cells suggests that the GLPS prepared from mycelia may be a better immunostimulator than the GLPS derived from spores (57). IIR-EIA can directly monitor the polysaccharide variability of extracts from the same source so that differences in culture conditions, chemical extraction methods, and/or preparation methods (e.g. enzymatic digestion) can be evaluated. By correlating the polysaccharide mixture binding patterns in IIR-EIA, it may be possible to optimize procedures for preparing herbal or natural products for particular treatments. Alternatively, IIRs can be implemented for affinity fractionation to enrich certain components of polysaccharide mixtures. The isolated fractions can be cross-examined by cell-based assays and in animal models so that their biological effects can be determined and the most medicinally beneficial fractions can be used.
By profiling the glycan-binding properties of IIRs, both their binding preferences and their carbohydrate ligand structures can be better understood. For instance, many CLRs recognize Lewis and sialyl Lewis antigens but with different binding properties: DC-SIGN tends to bind to Lewis but not sialyl Lewis antigens, Langerin binds to Lewis x/y but not Lewis a/b, and AICL and CLEC2 recognize sialyl Lewis a as well as sialyl LacNAc (Table 2). Thus, understanding the sugar-binding properties of IIRs is useful for establishing carbohydrate structures in mixtures of undefined saccharide contents. As newer versions of CFG glycan arrays (58) containing more carbohydrate structures become readily available, the specificity of carbohydrate-binding proteins like IIRs can continuously be updated. To further characterize the binding pattern in carbohydrate recognition, the polysaccharides identified by immune receptors can also be compared with those derived from plant lectin interaction. All of these tools, glycan arrays, profiling by plant lectins, and immune receptor fingerprinting, will add additional dimensions to the study of structure-function relationships that are instrumental in immuno- and glycobiology research. A particular strategic benefit of IIR-EIA is that carbohydrate recognition can subsequently be directly coupled to immunological function.
The IIR-EIA system established here is a useful tool for high-throughput profiling of polysaccharide mixtures derived from herbal drugs, natural products, and the oligosaccharides displayed on pathogen-associated glycoproteins. With this strategy, we have recently reported the interaction of dengue virus with CLEC5A (also known as MDL-1) that leads to further investigation of the critical role of CLEC5A in dengue virus-induced lethal syndromes (38), demonstrating the potential of the IIR-EIA technique in discovering novel interactions between carbohydrates and IIRs. IIR-EIA is currently being extended to glass slide arrays for profiling glycan-containing mixtures (59), and undoubtedly, the information gathered from specific poly- and oligosaccharide mixtures and/or pathogens that are recognized by IIRs will be important for understanding the underlying molecular mechanisms of glycan-immune cell interactions, including downstream signaling pathways, cross-talk, and receptor collaboration.
Acknowledgments
We thank Dr. Sarah R. Hanson for critically reading the manuscript. The glycan array resources were provided by the Consortium for the Functional Glycomics funded by National Institutes of Health NIGMS Grant GM62116.
This work was supported by National Sciences Council, Taiwan, Grant 95-2320-B-010-040-MY3, Taipei Veterans General Hospital, Taiwan, Grant V96S5-001 (to S. L. H.), and Taiwan Merit Scholarship TMS-094-1-B-003 (to T. L. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7.
- PAMP
- pathogen-associated molecular pattern
- CLR
- C-type lectin receptor
- CPS
- Cordyceps polysaccharides extracts
- EIA
- enzyme-linked immunosorbent assay
- Fuc
- fucose
- Gal
- galactose
- GalNAc
- N-acetylgalactosamine
- Glc
- glucose
- GlcA
- glucuronic acid
- GlcNAc
- N-acetylglucosamine
- GLPS
- G. lucidum polysaccharide extracts
- IIR
- innate immune receptors
- Lac
- lactose
- LacNAc
- N-acetyllactosamine
- Man
- mannose
- mKCR
- mouse Kupffer cell receptor
- MMR
- macrophage mannose receptor
- NeuAc
- N-acetylneuraminic acid or sialic acid
- PRR
- pattern recognition receptor
- TBS
- Tris-buffered saline.
REFERENCES
- 1.Janeway C. A., Jr. (1989) Cold Spring Harbor Symp. Quant. Biol. 54, 1–13 [DOI] [PubMed] [Google Scholar]
- 2.Aderem A., Ulevitch R. J. (2000) Nature 406, 782–787 [DOI] [PubMed] [Google Scholar]
- 3.Akira S., Takeda K. (2004) Nat. Rev. Immunol. 4, 499–511 [DOI] [PubMed] [Google Scholar]
- 4.Athman R., Philpott D. (2004) Curr. Opin. Microbiol. 7, 25–32 [DOI] [PubMed] [Google Scholar]
- 5.Cambi A., Figdor C. G. (2003) Curr. Opin. Cell Biol. 15, 539–546 [DOI] [PubMed] [Google Scholar]
- 6.Daws M. R., Sullam P. M., Niemi E. C., Chen T. T., Tchao N. K., Seaman W. E. (2003) J. Immunol. 171, 594–599 [DOI] [PubMed] [Google Scholar]
- 7.Liu C., Xu Z., Gupta D., Dziarski R. (2001) J. Biol. Chem. 276, 34686–34694 [DOI] [PubMed] [Google Scholar]
- 8.McDonald C., Inohara N., Nuñez G. (2005) J. Biol. Chem. 280, 20177–20180 [DOI] [PubMed] [Google Scholar]
- 9.Stahl P. D., Ezekowitz R. A. (1998) Curr. Opin. Immunol. 10, 50–55 [DOI] [PubMed] [Google Scholar]
- 10.Brown G. D., Gordon S. (2001) Nature 413, 36–37 [DOI] [PubMed] [Google Scholar]
- 11.Herre J., Gordon S., Brown G. D. (2004) Mol. Immunol. 40, 869–876 [DOI] [PubMed] [Google Scholar]
- 12.Figdor C. G., van Kooyk Y., Adema G. J. (2002) Nat. Rev. Immunol. 2, 77–84 [DOI] [PubMed] [Google Scholar]
- 13.Alvarez C. P., Lasala F., Carrillo J., Muñiz O., Corbí A. L., Delgado R. (2002) J. Virol. 76, 6841–6844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Appelmelk B. J., van Die I., van Vliet S. J., Vandenbroucke-Grauls C. M., Geijtenbeek T. B., van Kooyk Y. (2003) J. Immunol. 170, 1635–1639 [DOI] [PubMed] [Google Scholar]
- 15.Geijtenbeek T. B., Kwon D. S., Torensma R., van Vliet S. J., van Duijnhoven G. C., Middel J., Cornelissen I. L., Nottet H. S., KewalRamani V. N., Littman D. R., Figdor C. G., van Kooyk Y. (2000) Cell 100, 587–597 [DOI] [PubMed] [Google Scholar]
- 16.Halary F., Amara A., Lortat-Jacob H., Messerle M., Delaunay T., Houlès C., Fieschi F., Arenzana-Seisdedos F., Moreau J. F., Déchanet-Merville J. (2002) Immunity 17, 653–664 [DOI] [PubMed] [Google Scholar]
- 17.Tassaneetrithep B., Burgess T. H., Granelli-Piperno A., Trumpfheller C., Finke J., Sun W., Eller M. A., Pattanapanyasat K., Sarasombath S., Birx D. L., Steinman R. M., Schlesinger S., Marovich M. A. (2003) J. Exp. Med. 197, 823–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geijtenbeek T. B., Krooshoop D. J., Bleijs D. A., van Vliet S. J., van Duijnhoven G. C., Grabovsky V., Alon R., Figdor C. G., van Kooyk Y. (2000) Nat. Immunol. 1, 353–357 [DOI] [PubMed] [Google Scholar]
- 19.Geijtenbeek T. B., Torensma R., van Vliet S. J., van Duijnhoven G. C., Adema G. J., van Kooyk Y., Figdor C. G. (2000) Cell 100, 575–585 [DOI] [PubMed] [Google Scholar]
- 20.Lien E. J. (1990) Prog. Drug Res. 34, 395–420 [DOI] [PubMed] [Google Scholar]
- 21.Shiao M. S. (2003) Chem. Rec. 3, 172–180 [DOI] [PubMed] [Google Scholar]
- 22.Wang S. Y., Hsu M. L., Hsu H. C., Tzeng C. H., Lee S. S., Shiao M. S., Ho C. K. (1997) Int. J. Cancer 70, 699–705 [DOI] [PubMed] [Google Scholar]
- 23.Wang Y. Y., Khoo K. H., Chen S. T., Lin C. C., Wong C. H., Lin C. H. (2002) Bioorg. Med. Chem. 10, 1057–1062 [DOI] [PubMed] [Google Scholar]
- 24.Buenz E. J., Bauer B. A., Osmundson T. W., Motley T. J. (2005) J. Ethnopharmacol. 96, 19–29 [DOI] [PubMed] [Google Scholar]
- 25.Zhu J. S., Halpern G. M., Jones K. (1998) J. Altern. Complement Med. 4, 429–457 [DOI] [PubMed] [Google Scholar]
- 26.Zhu J. S., Halpern G. M., Jones K. (1998) J. Altern. Complement Med. 4, 289–303 [DOI] [PubMed] [Google Scholar]
- 27.Balon T. W., Jasman A. P., Zhu J. S. (2002) J. Altern. Complement Med. 8, 315–323 [DOI] [PubMed] [Google Scholar]
- 28.Ettinger R., Browning J. L., Michie S. A., van Ewijk W., McDevitt H. O. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 13102–13107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coombs P. J., Taylor M. E., Drickamer K. (2006) Glycobiology 16, 1C–7C [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo Y., Feinberg H., Conroy E., Mitchell D. A., Alvarez R., Blixt O., Taylor M. E., Weis W. I., Drickamer K. (2004) Nat. Struct. Mol. Biol. 11, 591–598 [DOI] [PubMed] [Google Scholar]
- 31.Seidenfaden R., Krauter A., Schertzinger F., Gerardy-Schahn R., Hildebrandt H. (2003) Mol. Cell. Biol. 23, 5908–5918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suzuki M., Suzuki M., Nakayama J., Suzuki A., Angata K., Chen S., Sakai K., Hagihara K., Yamaguchi Y., Fukuda M. (2005) Glycobiology 15, 887–894 [DOI] [PubMed] [Google Scholar]
- 33.Usui T., Iwasaki Y., Mizuno T., Tanaka M., Shinkai K., Arakawa M. (1983) Carbohydr. Res. 115, 273–280 [Google Scholar]
- 34.Chen H. S., Tsai Y. F., Lin S., Lin C. C., Khoo K. H., Lin C. H., Wong C. H. (2004) Bioorg. Med. Chem. 12, 5595–5601 [DOI] [PubMed] [Google Scholar]
- 35.Hsu H. Y., Hua K. F., Lin C. C., Lin C. H., Hsu J., Wong C. H. (2004) J. Immunol. 173, 5989–5999 [DOI] [PubMed] [Google Scholar]
- 36.Lin K. I., Kao Y. Y., Kuo H. K., Yang W. B., Chou A., Lin H. H., Yu A. L., Wong C. H. (2006) J. Biol. Chem. 281, 24111–24123 [DOI] [PubMed] [Google Scholar]
- 37.Shao B. M., Dai H., Xu W., Lin Z. B., Gao X. M. (2004) Biochem. Biophys. Res. Commun. 323, 133–141 [DOI] [PubMed] [Google Scholar]
- 38.Chen S. T., Lin Y. L., Huang M. T., Wu M. F., Cheng S. C., Lei H. Y., Lee C. K., Chiou T. W., Wong C. H., Hsieh S. L. (2008) Nature 453, 672–676 [DOI] [PubMed] [Google Scholar]
- 39.Robinson M. J., Sancho D., Slack E. C., LeibundGut-Landmann S., Reis e Sousa C. (2006) Nat. Immunol. 7, 1258–1265 [DOI] [PubMed] [Google Scholar]
- 40.Watson A. A., Brown J., Harlos K., Eble J. A., Walter T. S., O'Callaghan C. A. (2007) J. Biol. Chem. 282, 3165–3172 [DOI] [PubMed] [Google Scholar]
- 41.Brown G. D., Taylor P. R., Reid D. M., Willment J. A., Williams D. L., Martinez-Pomares L., Wong S. Y., Gordon S. (2002) J. Exp. Med. 196, 407–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Palma A. S., Feizi T., Zhang Y., Stoll M. S., Lawson A. M., Díaz-Rodríguez E., Campanero-Rhodes M. A., Costa J., Gordon S., Brown G. D., Chai W. (2006) J. Biol. Chem. 281, 5771–5779 [DOI] [PubMed] [Google Scholar]
- 43.Willment J. A., Gordon S., Brown G. D. (2001) J. Biol. Chem. 276, 43818–43823 [DOI] [PubMed] [Google Scholar]
- 44.Stambach N. S., Taylor M. E. (2003) Glycobiology 13, 401–410 [DOI] [PubMed] [Google Scholar]
- 45.Leteux C., Chai W., Loveless R. W., Yuen C. T., Uhlin-Hansen L., Combarnous Y., Jankovic M., Maric S. C., Misulovin Z., Nussenzweig M. C., Feizi T. (2000) J. Exp. Med. 191, 1117–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stahl P. D. (1990) Am. J. Respir. Cell Mol. Biol. 2, 317–318 [DOI] [PubMed] [Google Scholar]
- 47.Fadden A. J., Holt O. J., Drickamer K. (2003) Glycobiology 13, 529–537 [DOI] [PubMed] [Google Scholar]
- 48.Lehrman M. A., Haltiwanger R. S., Hill R. L. (1986) J. Biol. Chem. 261, 7426–7432 [PubMed] [Google Scholar]
- 49.Lehrman M. A., Pizzo S. V., Imber M. J., Hill R. L. (1986) J. Biol. Chem. 261, 7412–7418 [PubMed] [Google Scholar]
- 50.Tada H., Nemoto E., Shimauchi H., Watanabe T., Mikami T., Matsumoto T., Ohno N., Tamura H., Shibata K., Akashi S., Miyake K., Sugawara S., Takada H. (2002) Microbiol. Immunol. 46, 503–512 [DOI] [PubMed] [Google Scholar]
- 51.Miyazaki T., Nishijima M. (1982) Carbohydr. Res. 109, 290–294 [Google Scholar]
- 52.Brown G. D. (2006) Nat. Rev. Immunol. 6, 33–43 [DOI] [PubMed] [Google Scholar]
- 53.Sheikh H., Yarwood H., Ashworth A., Isacke C. M. (2000) J. Cell Sci. 113, 1021–1032 [DOI] [PubMed] [Google Scholar]
- 54.East L., Isacke C. M. (2002) Biochim. Biophys. Acta 1572, 364–386 [DOI] [PubMed] [Google Scholar]
- 55.East L., Rushton S., Taylor M. E., Isacke C. M. (2002) J. Biol. Chem. 277, 50469–50475 [DOI] [PubMed] [Google Scholar]
- 56.Su Y., Bakker T., Harris J., Tsang C., Brown G. D., Wormald M. R., Gordon S., Dwek R. A., Rudd P. M., Martinez-Pomares L. (2005) J. Biol. Chem. 280, 32811–32820 [DOI] [PubMed] [Google Scholar]
- 57.Chan W. K., Law H. K., Lin Z. B., Lau Y. L., Chan G. C. (2007) Int. Immunol. 19, 891–899 [DOI] [PubMed] [Google Scholar]
- 58.Blixt O., Head S., Mondala T., Scanlan C., Huflejt M. E., Alvarez R., Bryan M. C., Fazio F., Calarese D., Stevens J., Razi N., Stevens D. J., Skehel J. J., van Die I., Burton D. R., Wilson I. A., Cummings R., Bovin N., Wong C. H., Paulson J. C. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 17033–17038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen M. L., Adak A. K., Yeh N. C., Yang W. B., Chuang Y. J., Wong C. H., Hwang K. C., Hwu J. R., Hsieh S. L., Lin C. C. (2008) Angew. Chem. Int. Ed. Engl. 47, 8627–8630 [DOI] [PubMed] [Google Scholar]