Abstract
It has been shown previously that sub-complexes of the 26 S proteasome can regulate gene expression via non-proteolytic mechanisms. One such mechanism is the disruption of activator·promoter complexes in an ATP-dependent fashion, which was discovered in the context of the yeast Gal4 system. This activity strongly inhibits Gal4-driven gene expression unless the activator is mono-ubiquitylated, which protects it from the ATPases. To address whether this paradigm is also applicable to medically important mammalian transcriptional activators we report here a study of the interaction of the proteasomal ATPases with p53. It is shown that p53 binds directly to the ATPases via its C-terminal tetramerization and regulatory domain and that p53·promoter complexes are indeed vulnerable to ATPase-dependent disruption by the ATPase complex in vitro. Knockdown of one of the ATPases, Rpt6, in living cells results in increased occupancy of the p21waf1 promoter by p53 and increased expression of the gene, consistent with the idea that the proteasomal ATPases negatively regulate p53 function in a non-proteolytic fashion.
Introduction
The ubiquitin-proteasome system (UPS)2 is known to regulate the transcription of many genes via both proteolytic and non-proteolytic activities (1, 2). The 26 S proteasome consists of two major pieces, the 20 S core particle (CP) and the 19 S regulatory particle (RP) (3). The three proteolytic active sites in the proteasome are sequestered within the barrel-like 20 S CP (4). Substrate entry is regulated by the 19 S RP, which caps the top and/or bottom of the 20 S CP. Protein substrates modified with a chain of at least four Lys-48-linked ubiquitin molecules (5) bind to the 19 S RP and are then unfolded in an ATP-dependent fashion through the action of six triple AAA class ATPases (Rpts 1–6) that sit atop the opening to the 20 S CP cavity. It has been long known that this proteolytic function of the proteasome controls transcription by degrading key regulatory proteins (6). Proteasome-mediated proteolysis is also required for the function of some activators, thus acting in a positive fashion (7–9). Finally, the proteolytic activity of the proteasome is involved in the termination of RNA polymerase II-mediated transcription (10).
A part of the 19 S RP that includes the six ATPases, Rpn1 and Rpn2 (which we have termed APIS (11)), has been shown to regulate transcription in a non-proteolytic fashion. Studies of Gal4-dependent GAL gene transcription in yeast showed that the activator recruited these proteins, but not the 20 S CP or the lid subunit of the 19 S RP, to activated promoters in vivo (11). In vitro studies suggest that the Rpt proteins are critical for efficient promoter escape and elongation (12).
Analysis of certain Gal4 mutants (13, 14) also revealed a cryptic repressive activity of the APIS complex on GAL transcription, which resulted from the ATP-dependent destabilization of Gal4·promoter complexes by the APIS complex (15, 16). This activity requires direct interactions between the APIS complex and the activation domain of the Gal4 protein (15). APIS-mediated disruption of Gal4·DNA complexes occurs only when Gal4 is not mono-ubiquitylated within the DNA-binding domain (15, 17) revealing at least one function of this post-translational modification (18) to be protection against the destabilizing activity of the proteasomal ATPases. This protective effect involves direct interaction of the mono-ubiquitin moieties with Rpn1 and Rpt1 in the APIS complex, an interaction that disrupts the binding of Rpt4 and Rpt6 to the activation domains of the Gal4 dimer, thus preventing the APIS complex from using the activator as a substrate for its unwinding activity (17).
Although Gal4 is generally considered a paradigm of transactivators in general, studies of the applicability of these findings to important mammalian transactivators are in their infancy. There have been a few reports that the proteasomal ATPases stimulate transactivation of certain mammalian genes, apparently through a mechanism analogous to that worked out in the GAL system (19–21). It has also been found in a few cases that activator mono-ubiquitylation can stimulate transcription of the target genes (22, 23), although it is clear that mono-ubiquitylation can also inhibit transactivator activity by promoting nuclear exclusion (24), a dichotomy presumably explainable by context dependence. Furthermore, the Rpt6 protein (also known as Sug1 in yeast and Trip1 in mammals) has been found to associate with a number of mammalian activators and to be localized on some mammalian promoters by ChIP (19–21, 23, 25). However, to the best of our knowledge, no clear example of a repressive function of the APIS complex on mammalian transcription due to destabilization of activator·promoter complexes has been noted.
Among the promoters on which Rpt6 has been localized is that of p21waf1, a gene regulated by the p53 protein (25). There have been several previous studies about how UPS regulates the stability and activity of p53, and transcription of p53-dependent target genes. p53 has been shown to be degraded by both ubiquitin- dependent and ubiquitin-independent mechanisms (26). Proteasome inhibition by MG132 attenuated p53-mediated transcription and increased the occupancy of p53 onto p21waf1 promoter, suggesting positive regulation of p53-dependent transcription at p21waf1 promoter due to p53 turnover by UPS. Additionally, it was shown that the Sug1/Rpt6 subunit of the 19 S proteasome interacts with p53 in vitro and in vivo. ChIP analysis revealed that the Sug1 and S1 components of the 19 S regulatory particle are recruited to p21waf1 promoter by UV-induced DNA damage in MCF-7 cells (25). Finally, p53 is known to be ubiquitylated by several different E3 ligases, including Hdm2, E6-AP, Pirh2, Topors, COP1, ARF-BP1, BRCA1-BARD1, and E4F1, and some of these modifications have been shown to be stimulatory in nature (27, 28).
These hints of parallel behaviors of the Gal4 and p53 systems led us to probe whether the ability of the APIS complex to destabilize activator·promoter complexes might be relevant to p53 and the non-proteolytic regulation of p53-dependent transcription by APIS. We present here evidence that p53 indeed interacts physically with the APIS complex and that this interaction results in potent ATP-dependent disruption of p53·DNA complexes. This “stripping” of the p53 protein from DNA in vitro by the proteasomal ATPases would be predicted to repress p53 activity in cells. Indeed, we demonstrate that knockdown of the Rpt6 protein results in increased occupancy of the p21waf1 promoter by p53 and increased transcription of the gene. In striking contrast, the proteolytic activity of the proteasome is important for high levels of p53-mediated gene expression, as evidenced by the inhibition of p21waf1 expression in response to MG132, a proteasome inhibitor. These data argue that the proteasomal ATPases indeed act as repressors of p53-mediated p21waf1 gene expression by mediating a non-proteolytic destabilization process that is physically and functionally distinct from the interaction of the p53 protein with the proteolytic fragment of the proteasome.
EXPERIMENTAL PROCEDURES
Materials
A biotinylated DNA fragment (∼560 bp), containing the p53 binding site from the human p21 gene, major late promoter and the entire G-less cassette derived from pWAFMLT, was prepared by PCR with an upstream primer (5′-AACTCGACTGCAGCATATGTATCATACACATATACG-3′) and a biotinylated downstream primer (5′-biotin-CGATTCATTAATGCAGCTGG-3′), annealing to the flanking vector sequences. A nonspecific biotinylated DNA fragment that does not contain the p53 binding site was prepared by XbaI digestion of the above fragment. Yeast 26 S and 19 S proteasomes were purified using a FLAG affinity tag as described previously (29) with modifications (12). Mammalian PA700 (19 S) proteasome (30), which was purified from bovine blood, was kindly provided by G. DeMartino (University of Texas Southwestern Medical Center). Antibodies against p53 (1C12, Cell Signaling, and DO-1, Calbiochem), PentaHis (Qiagen), Ubiquitin (DakoCytomation), GST (Santa Cruz Biotechnology), and horseradish peroxidase-conjugated secondary antibodies (Bio-Rad) were used according to manufacturer's instructions. The yeast α-Rpt6 and α-20 S antibodies were produced in rabbit. The mammalian α-Rpt6 and α-Rpt4 antibodies were purchased from BIOMOL International and Abcam, respectively. The RIP-1 peptoid was synthesized according to a published protocol (31).
Recombinant Proteins
Purification of bacterially expressed FLAG-tagged p53 was described previously (32). His6-tagged p53 DNA binding domain (DBD, amino acids 102–292) was expressed in a BL21 (DE3) strain and purified by nickel-nitrilotriacetic acid chromatography (Qiagen). Recombinant GST-tagged p53 domains (NTD-(1–101), DBD-(102–292), and TDRD-(293–393)) were induced in BL21 (DE3) strain, and proteins were extracted with BC500 buffer (20 mm Tris-HCl (pH 8.0), 500 mm KCl, 1 mm dithiothreitol, 10% glycerol, 0.5 mm EDTA, 1 mm phenylmethylsulfonyl fluoride) containing 0.1% Nonidet P-40, and purified using glutathione-Sepharose bead (Amersham Biosciences).
Destabilization of Activator·DNA Complexes by the Proteasome
The destabilization assay has been described previously (15). Briefly, the p53·p21waf1 promoter complex formed by biotinylated DNA to the Dynal streptavidin magnetic bead (Invitrogen) was added at a 140 nm final concentration of activator·promoter complex to a reaction mixture containing 20 nm proteasome and 4 mm ATP in TR reaction buffer (10 mm HEPES (pH 8.0), 50 mm KCl, 6.25 mm MgCl2, 0.1 mm EDTA, 10% glycerol, 1 mm dithiothreitol, and 10 μm ZnCl2) for 15 min. After this incubation, activator·DNA complex was pulled down by the Dynal magnet and washed two times with TR buffer. The final bead complex was resuspended with TR buffer and SDS loading buffer and analyzed by 7.5% SDS-PAGE and Western blotting by ECL detection. The percent bound values were determined by quantitating each band by using ImageJ software. Competitor DNA, which has the same sequence with biotinylated DNA (Fig. 1A) except biotin label, was employed 10-fold excess compared with biotinylated DNA when used as a trap for dissociated activator. MG132 was employed at a concentration of 10 μm when used. Inhibition of the destabilization activity of the proteasome was done in the presence of the indicated amount of a pharmacological inhibitor of Rpt4 (RIP-1). All assays were repeated at least three times with at least two separate proteasome preparations for confirmation, and the representative blot was presented in each different set of experiments.
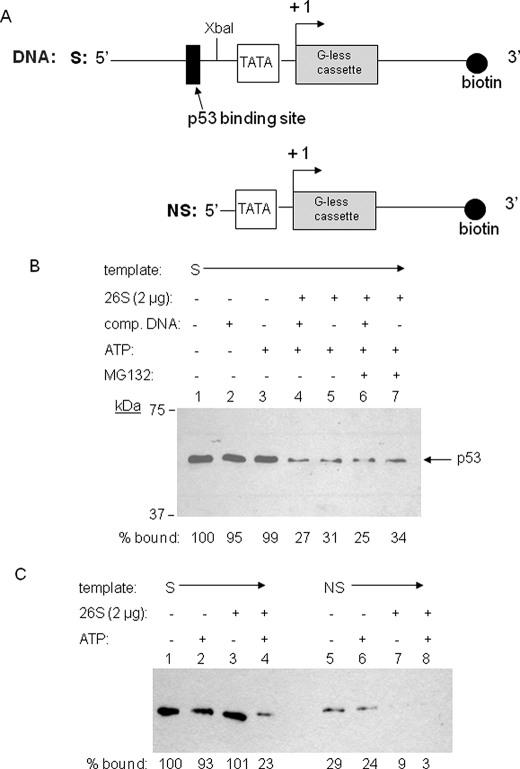
FIGURE 1.
A p53·p21waf1 promoter complex is destabilized by the yeast 26 S proteasome. A, schematic diagram of the biotinylated DNA fragments containing the p53 binding site (S) derived from pWAFMLT (i.e. the distal p53 binding site originally from the human p21waf1 gene) and containing no p53 binding site (NS), which was prepared by XbaI digestion of above fragment. The p53 binding site, TATA box, and transcription start site are indicated. B, Western blot using anti-p53 antibody that measures the amount of p53 protein that remains bound to the immobilized DNA after incubation for 15 min at 30 °C in the presence of the reagents indicated. The values shown at the bottom, which represent the percentage of p53 that remained bound to the immobilized DNA relative to the control lacking the proteasome and ATP (lane 1), were determined by quantitating each band using ImageJ software. C, the comparison of the destabilization effect of proteasomal ATPases on sequence-specific versus non-sequence specific p53 binding to the DNA.
ChIP Assay
Cell lines H460, HCT116 (p53 WT), and HCT116 (p53−/−) were kindly provided by the J. Minna laboratory (University of Texas Southwestern Medical Center), and these cells were maintained using RPMI 1640 media with l-glutamine (Mediatech Inc., Herndon, VA) and 10% FBS for H460, and using Dulbecco's modified Eagle's medium (high glucose) with l-glutamine (Invitrogen, Carlsbad, CA) and 10% FBS for HCT116. These cells were treated with 750 nm doxorubicin to induce genotoxic stress. Cells were harvested at the indicated times. Cells were cross-linked in 1% formaldehyde for 15 min before the reaction was quenched with glycine (0.125 m) for 5 min. The cells were washed with cold PBS twice and then collected and pelleted. The cells were resuspended in SDS lysis buffer (50 mm Tris-HCl (pH 8.1), 10 mm EDTA, 1% SDS) plus protease inhibitor mixture (2 × 107 cells/ml lysis buffer), and sonicated. After clearance of the precipitate, the soluble chromatin can be stored at −80 °C. 100 μl of the chromatin was diluted 10-fold with dilution buffer (16.7 mm Tris-HCl (pH 8.1), 167 mm NaCl, 1.2 mm EDTA, 0.01% SDS, 1.1% Triton X-100) and was pre-cleared by protein G plus beads. The sample was incubated with the antibody overnight at 4 °C before the protein G plus beads were added. After incubation with the protein G plus beads for 1 h at 4 °C, the solution was removed and the beads were subjected to several steps of washing, including one wash with the low salt buffer (20 mm Tris-HCl (pH 8.1), 150 mm NaCl, 2 mm EDTA, 0.1% SDS, 1% Triton X-100), one wash with the high salt buffer (20 mm Tris-HCl (pH 8.1), 500 mm NaCl, 2 mm EDTA, 0.1% SDS, 1% Triton X-100), one wash with LiCl wash buffer (10 mm Tris-HCl (pH 8.1), 1 mm EDTA, 0.25 m LiCl, 1% Nonidet P-40, 1% deoxycholic acid), and two washes with TE buffer (10 mm Tris-HCl (pH 8.0), 1 mm EDTA). The beads were then incubated with the elution buffer (1% SDS, 0.1 m NaHCO3) for 15 min twice before the eluates were collected by centrifugation. The cross-links were reversed by incubation at 65 °C overnight under high salt conditions. The eluates were then treated with RNase A and proteinase K sequentially before the DNAs in the eluates were purified by following the Qiagen PCR product purification protocol. The purified DNAs were used as templates for PCR amplification of the regions investigated. Quantitative real-time PCR of the purified DNAs was performed on an iCycler Thermal Cycler by using the IQTM SYBR Green Supermix (Bio-Rad). Relative enrichment of specific DNA was derived from comparing products amplified from the primers against the region of interest in the samples and the total input DNA, and considering a negative control sample. DNA of p21waf1 promoter regions was amplified using the following primers: forward, 5′-CTGGACTGGGCACTCTTGTC-3′; reverse, 5′-CTCCTACCATCCCCTTCCTC-3′. DNA of p21waf1 coding regions was amplified using the following primers: forward, 5′-TCTGTCTCGGCAGCTGACAT-3′; reverse, 5′-ACCACAAAAGATCAAGGTGAG-3′.
GST Pulldown Assay
The GST fusion proteins and proteasome pulldown assays were performed by mixing 1.2 μm GST fusion proteins with 50 nm 26 S proteasome in TR reaction buffer with additional 0.1% Nonidet P-40. After a 30-min incubation at 4 °C, glutathione-Sepharose beads (Amersham Biosciences) were added, and the mixture was incubated further for 30 min. The beads were pulled down, washed with TR buffer three times, and resuspended in 2× SDS loading buffer for SDS-PAGE and Western blot analysis.
siRNA Knockdown of Rpt6 19 S Proteasomal Subunit
Double-stranded RNA oligonucleotides directed against target sequences in Rpt6 (5′-AAGGTACATCCTGAAGGTAAA-3′), which was confirmed for specific knockdown efficiency in previous study (21), were obtained from Dharmacon (Boulder, CO). The negative control was a proven non-targeting siRNA provided from Dharmacon. The H460 cells were plated, grown to 30–50% confluency, and transfected with 20 nm (final) siRNA using Lipofentamine RNAiMAX (Invitrogen). 40 h after transfections, cells were treated with doxorubicin or not, and 8 h later cells were harvested. Half of the harvested cells were lysed using SDS sample buffer and were analyzed by Western blot to check the knockdown. The remaining cells were subjected to RNA extraction for RNA expression analysis. For ChIP analysis with siRNA knockdown samples, after 48 h of transfection of siRNA (with final 8 h doxorubicin treatment or not), cells were fixed and processed by following described ChIP procedure above.
RNA Expression
Harvested H460 cells were washed with cold phosphate-buffered saline twice, and total RNA were isolated with an RNeasy kit (Qiagen), according to the manufacturer's protocol. Reverse transcription was done using the iScript cDNA synthesis kit (Bio-Rad) in a 20-μl reaction volume following the manufacturer's instruction. For PCR detection, 1 μg of cDNA from each processed sample was subjected to real-time quantitative PCR and standard PCR using specific primers. The DNA sequences of the specific primers were as follows: p21waf1, forward (5′-CCTCAAATCGTCCAGCGACCTT-3′) and reverse (5′-CATTGTGGGAGGAGCTGTGAAA-3′); glyceraldehyde-3-phosphate dehydrogenase, forward (5′-GAAGGTGAAGGTCGGAGT-3′) and reverse (5′-GAAGATGGTGATGGGATTTC-3′).
RESULTS
Destabilization of p53·p21waf1 Promoter Complex by the Proteasomal ATPases in Vitro
To determine if the proteasomal ATPases are capable of destabilizing p53·DNA complexes, recombinant FLAG-tagged p53 was bound to an immobilized biotinylated double-stranded DNA containing one p53 binding site from the p21waf1 promoter (Fig. 1A). After addition of purified 26 S proteasome in the presence or absence of ATP, the amount of bound protein remaining on the DNA was determined. As shown in Fig. 1B, ∼70% of the bound p53 protein was stripped off of the immobilized DNA by the proteasome relative to the amount of p53 remaining on the promoter in the absence of the proteasome (compare lane 5 to lane 1). The presence of soluble competitor promoter DNA was not required to observe this net loss of p53 from the immobilized DNA (compare lanes 4 and 5), indicating that the disassembly process is irreversible. The addition of the proteolysis inhibitor MG132 did not inhibit the stripping activity (compare lanes 4 and 6 or 5 and 7), arguing that this is not a proteolytic reaction, as was demonstrated conclusively in the previous Gal4 studies (15). The reaction was completely dependent on the presence of ATP and proteasome. Elimination of either from the reaction resulted in an almost complete loss of the destabilization activity (Fig. 1B).
To examine whether this destabilization activity by proteasomal ATPases targets exclusively the sequence-specific DNA-binding activity of p53, we compared two DNAs with and without p53 binding site (Fig. 1A) as templates for destabilization activity. As shown in Fig. 1C, some extent of non-sequence-specific DNA binding of p53 was detected (∼25% compared with sequence specific binding), and this nonspecifically bound p53 was also affected for destabilization by the proteasomal ATPases, although the relative level of destabilized p53 was low compared with that by specifically bound p53.
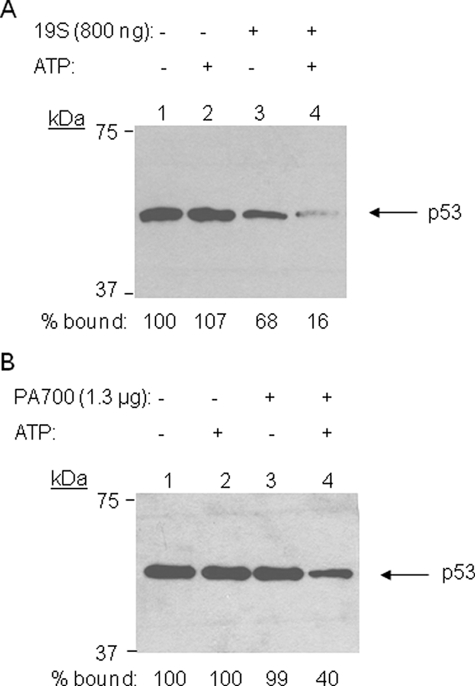
We also tested in this assay the activity of the 19 S RP (also known as PA700) purified from either yeast or mammalian (bovine) cells. Both were able to disrupt the p53·DNA complex in the presence of ATP, although the yeast 19 S RP was somewhat more active (Fig. 2, A and B). These 19 S RP preparations are free of immunologically detectable 20 S complex (12, 30) and thus further support the contention that this is a non-proteolytic process.
FIGURE 2.
Effect of the yeast and mammalian 19 S RPs on the stability of the p53·p21waf1 promoter complex. The figure shows an anti-p53 Western blot that measures the amount of protein that remained bound to the immobilized DNA (see Fig. 1) after incubation with the reagents indicated, including 19 S RPs from yeast (A) and cow red blood cells (B).
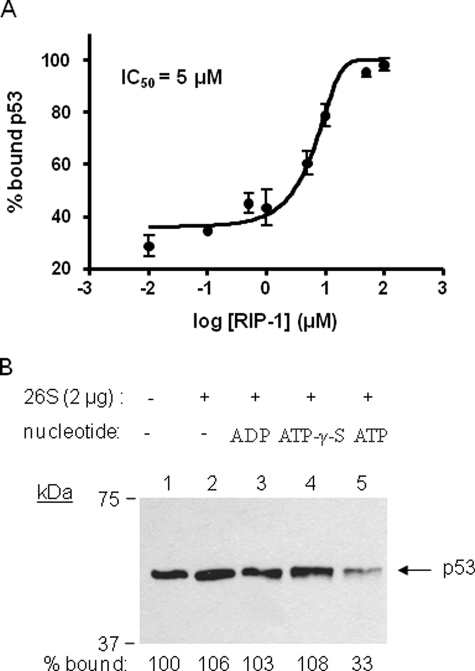
To ensure that the stripping activity observed in the above assays is due to an activity of the 19 S RP and not some putative contaminant in the preparations, we tested the effect of a pharmacological inhibitor of Rpt4 (33) called RIP-1. This reagent is highly specific for its target and has been shown previously to block destabilization of Gal4·VP16 complexes (15, 31). As shown in Fig. 3A, the RIP-1 blocked loss of p53 from the DNA in a dose-dependent fashion with an IC50 of ∼5 μm. We conclude that the active destabilization of the p53·DNA complex requires the activity of Rpt4 and, presumably, the other proteasomal ATPases.
FIGURE 3.
Inhibition of proteasomal ATPase destabilization of p53·p21waf1 promoter complexes by the pharmacological inhibitor of Rpt4 (RIP-1) and requirement of ATP hydrolysis for the destabilization activity of the proteasome. A, destabilization assay was carried out three times as described in Fig. 1 but with the addition of the indicated concentration of RIP-1, a synthetic inhibitor of Rpt4, and the resulting percent bound p53 values versus log concentration of RIP-1 were plotted and fitted into the equation from GraphPad software (log [inhibitor] versus normalized response). The determined IC50 value is displayed. B, destabilization assay was carried out as described in Fig. 1 in the absence of nucleotide or the presence of ADP, ATPγS, and ATP as noted.
Having established that the proteasomal ATPases are capable of destabilizing the p53·DNA complex, we next asked if ATP hydrolysis or simply ATP binding was necessary for this reaction. As shown in Fig. 3B, the 26 S proteasome did not destabilize p53·DNA complex in the presence of ADP nor ATPγS. Only in the presence of ATP, the 26 S proteasome destabilized the complex, indicating that ATP hydrolysis is essential for stripping the activator off of the DNA.
Destabilization of the p53·DNA Complex Requires Direct Interactions between the Proteasome and the Activator
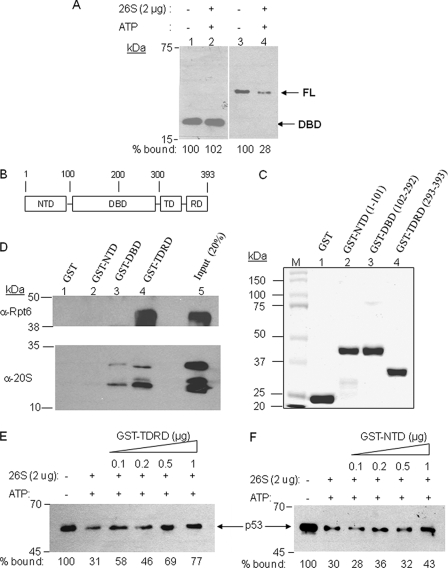
APIS-mediated destabilization of Gal4·DNA complexes absolutely required the presence of an activation domain (15), the region of the protein that binds the proteasomal ATPases (11, 34, 35). To determine if direct activator-APIS interactions are required for the destabilization of p53·DNA complexes, we compared the stability of the full-length p53·DNA complex with that of the p53 DNA-binding domain (DBD)·DNA complex in the presence of the proteasome and ATP. As shown in Fig. 4A, the p53 DBD itself was not stripped from the immobilized DNA by the 26 S proteasome, whereas the full-length protein was, consistent with the notion that a domain of p53 outside the DBD interacts with the APIS complex and renders p53 a substrate for the stripping activity.
FIGURE 4.
Identification of the p53 domain responsible for physical and functional interaction with the 19 S proteasome. A, the destabilization assay was carried out as described in Fig. 1 using either the isolated DNA-binding domain or full-length p53. B, domain map of p53. C, purification of GST-fused recombinant proteins of p53 domains. Each of the three domains, N-terminal domain (NTD), DNA-binding domain (DBD), and C-terminal tetramerization and regulatory domain (TDRD) and the number of amino acids are indicated in the SDS-PAGE of purified proteins. D, Western blots using either anti-Rpt6 or anti-20 S antibodies show the amount of protein retained by the p53 domain indicated after incubation of the GST-p53 domain fusion protein with intact 26 S proteasome. E and F, competition assay to examine the ability of the p53 TDRD (E) and NTD (F) regions to inhibit proteasome-mediated destabilization of immobilized p53·DNA complex. The assay was carried out as described in Fig. 1, except the indicated amounts of the GST fusion proteins were included as competitors for binding to the proteasomal ATPases. The amount of p53 retained on the DNA is shown.
We next attempted to identify the one or more domains of p53 that physically interact with the proteasomal ATPases. We prepared GST fusion proteins of each of three domains of p53, the N-terminal activation domain (NTD), the DNA-binding domain (DBD), and the tetramerization and regulatory domain (TDRD)) (Fig. 4, B and C). These constructs were incubated with the 26 S proteasome and then precipitated with glutathione-agarose beads. Retention of the Rpt6/Sug1 subunit as well as 20 S CP proteins by the GST fusion proteins was analyzed by SDS-PAGE and Western blotting. As shown in Fig. 4D, only GST-TDRD retained Rpt6, indicating that the C-terminal regulatory domain of p53 is responsible for interacting with the proteasomal ATPases. Some extent of 20 S CP binding was also detected by this domain and DBD, even after washing, though Rpt6 was highly enriched relative to the 20 S CP in the GST-TDRD- and GST-DBD-bound fraction (compare the ratios of Rpt6 and 20 S subunits in lanes 4 and 5). It is not clear if this 20 S CP retention is largely nonspecific or reflects some ability of the domain to retain the entire 26 S proteasome, which might indicate possible proteolytic role of the 20 S CP.
To determine if this physical interaction was of functional relevance in the destabilization reaction, we asked if the GST-TDRD fusion protein could act as a dominant negative inhibitor of the stripping reaction. As shown in Fig. 4E, the addition of GST-TDRD to the destabilization assay conditions inhibited loss of full-length p53 from the immobilized DNA in dose-dependent manner. Under the same conditions, the GST-NTD fusion protein, which does not interact with the ATPases, had little or no effect (Fig. 4F).
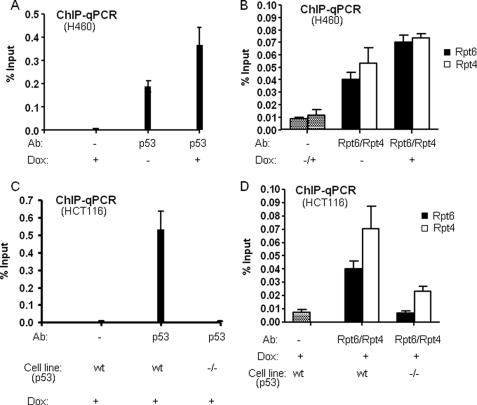
The Proteasomal ATPases Are Recruited to the p21waf1 Promoter in Vivo
If the destabilization of the p53·p21waf1 promoter complex observed in vitro is physiologically relevant, then it seems likely that the proteasomal ATPases would be physically resident on the promoter in living cells. To address this point, ChIP assays were carried out in a non-small cell lung cancer cell line, H460, where wild-type p53 activity was induced with doxorubicin. As shown in Fig. 5(A and B), clearly there is significant occupancy of the promoter by p53 and the ATPases under non-induced conditions in this cell type. However, induction with doxorubicin increased the association of the Rpt6 and Rpt4 subunits of the 19 S proteasome associate with the p21waf1 promoter region in concert with the increase in p53 occupancy. Therefore, their binding to the promoter seems to reflect that of p53, consistent with the activator recruiting these proteins to this locus. We did not observe significant ChIP signals using antibodies against the 20 S CP (data not shown), suggesting that it is not recruited to the promoter by p53, but we cannot rule out the possibility that this negative result is due to some technical issue. These results parallel those obtained in previously published ChIP experiments carried out in a different cell type (25).
FIGURE 5.
Association of p53 and proteasomal ATPase subunits with the p21waf1 promoter in vivo. A and B, promoter occupancy of p53 (A) and 19 S proteasomal subunits (Rpt4 and Rpt6) (B) were analyzed by ChIP assay using H460 cells with and without doxorubicin (Dox) induction. 8 h after doxorubicin treatment, cells were harvested for ChIP processing, and the antibodies used for precipitation and Dox treatments are indicated. C and D, promoter occupancy of p53 (C) and19 S proteasomal subunits (Rpt4 and Rpt6) (D) were analyzed by ChIP assay using HCT 116 p53 wt and null (−/−) cells with doxorubicin (Dox) induction. 8 h after doxorubicin treatment, cells were harvested for ChIP processing, and the antibodies used for precipitation and cells used are indicated.
Interestingly, when the same anti-Rpt6 or anti-Rpt4 chromatin samples were probed with primers complementary to the coding region of the gene, no evidence for Rpt6 or Rpt4 occupancy was observed (data not shown). This is in contrast to the situation in yeast, where strong ChIP signals for the ATPases were observed throughput the gene (11).
To further confirm that Rpt6 and Rpt4 recruitment to the p21waf1 promoter region is due to interaction with p53, we used different cells, HCT116 (p53WT) and HCT116 (p53−/−) for ChIP assay. As shown in Fig. 5 (C and D), no significant ChIP signals were detected in HCT116 (p53−/−) cells compared with HCT116 (p53WT), indicating that Rpt6 and Rpt4 are recruited to the p21waf1 promoter through specific binding to p53.
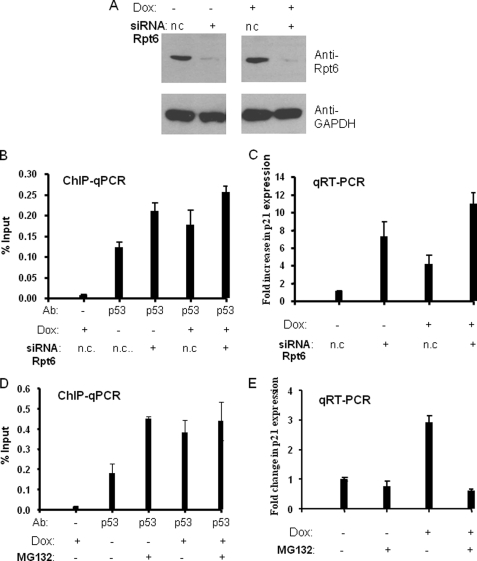
Rpt6 Antagonizes p53 Binding to the p21waf1 Promoter and Transcription of the Gene
The destabilization of the p53· p21waf1 promoter observed in vitro would predict that the proteasomal ATPases antagonize p53-mediated gene activation in living cells. To probe this point, we performed siRNA-mediated knockdown of Rpt6 (Sug1) and assessed the effect on p21waf1 promoter occupancy by p53 and p21waf1 RNA expression levels. siRNA-mediated knockdown of Rpt6 resulted in a decrease of ∼90–95% in the Rpt6 protein level in H460 cells with and without doxorubicin induction (Fig. 6A). It did not affect the level of glyceraldehyde-3-phosphate dehydrogenase, which was used as a control. We also attempted siRNA-mediated knockdown of two 20 S proteasome subunits (α4 and β5) separately in H460 cells using two different double-stranded RNA oligonucleotides provided by Dharmacon, which target α4 and β5, respectively. Unfortunately, knockdown was quite inefficient (only 10–20%, data not shown), and, therefore, these cells were not used in subsequent experiments.
FIGURE 6.
The effect of proteasome inhibition on p53 promoter occupancy and p21waf1 RNA transcription. A, H460 cells were transfected with nonspecific siRNA control (n.c.) or Rpt6-specific siRNA duplexes, and treated with doxorubicin (+) or not (−) at 40 h after transfection. After an additional 8 h, cells were harvested or processed for ChIP and RNA expression analysis. Western blots showed ∼90% knockdown of Rpt6 protein expression both in the absence (−) and presence (+) of Dox induction. B, ChIP analysis of p53 occupancy on the p21waf1 promoter with and without Dox induction. Negative control and specific siRNA of Rpt6 are indicated in each lane. Real-time PCR values were normalized to the input (input values represent 1% of the total lysate), and data are presented as % input. C, real-time RT-PCR analysis of p21waf1 expression with and without Dox induction. Real-time PCR values were normalized to the glyceraldehyde-3-phosphate dehydrogenase control and calibrated to the negative control siRNA and without Dox induction samples. Data were presented as -fold increase of p21waf1 gene expression. D, the effect of MG132 treatment on p53 occupancy on the p21waf1 promoter. H460 cells were induced by Dox (+) or not (−) when cells were grown to ∼70% confluency. After 8 h, cells were treated with MG132 (+) (5 μm) or not (−) and grown for an additional 4 h. After that, cells were harvested or processed for ChIP, and data were analyzed in the same way above. E, the effect of MG132 treatment on p21waf1 expression. Real-time RT-PCR analysis of p21waf1 expression was done in the same way above.
ChIP analysis showed that knockdown of Rpt6 increased the occupancy of p53 on the p21waf1 promoter by ∼2-fold compared with non-targeting negative control siRNA both in the absence and presence of doxorubicin induction (Fig. 6B). Knockdown of Rpt6 also resulted in up-regulation of p21waf1 gene expression as shown in Fig. 6C. In the absence of doxorubicin induction, this increase was quite large (∼7-fold; Fig. 6C, compare lanes 1 and 2). Under inducing conditions, knockdown of Rpt6 resulted in an increase of ∼2.5-fold relative to the cells containing the control siRNA. These data show that Rpt6, presumably in concert with the other proteasomal ATPases, acts as an antagonist of p53-mediated p21waf1 gene transcription and that it does so at the level of promoter occupancy.
Because siRNA-mediated knockdown of 20 S subunits was ineffective in our hands, we also examined the effect of MG132 in this cell line. As shown in Fig. 6 (D and E), treatment with MG132 increased the occupancy of the p21waf1 promoter by p53 but, conversely, decreased the level of p21waf1 mRNA production. Thus, inhibition of the proteasomal ATPases (via Rpt6 knockdown) and one of the 20 S CP proteolytic activities (with MG132) have opposite effects on p53-mediated p21waf1 gene expression, although both treatments increase promoter occupancy.
DISCUSSION
As reviewed briefly in the introduction, the UPS can modulate eukaryotic gene transcription in a number of ways. The proteolytic activity of the proteasome has been shown to either stimulate or suppress the transcription of many genes. p53 is one of a large class of activators that have overlapping degron and activation sequences (36), and it has been shown recently that proteasome-mediated turnover of the activator is important for its full activity (25), as is also the case for several other mammalian activators. Using a different cell line and a different inducer of p53-mediated transcription, we have observed the same result, as evidenced by the inhibition of p21waf1 transcription by the proteasome inhibitor MG132 (see Fig. 6E). Thus, the available data in the p53 system suggest that the UPS plays a dual role in acting to keep activator levels low under non-inducing conditions, thus presumably helping to repress p53 in the absence of genotoxic stress, as well as stimulating p53 activity through turnover of the activator under inducing conditions.
Studies of the Gal4 system and a few other yeast genes (37, 38) have revealed a different mechanism for regulation of transcription by elements of the proteasome that does not depend on the proteolytic activity of the UPS. A sub-complex of the 19 S RP, which we have termed APIS, that includes the base of the 19 S (the six ATPases (Rpts1–6), Rpn1, and Rpn2) and perhaps other proteins (20), has been shown to have two different effects on GAL transcription. One is a stimulatory role that is thought to result from more promoter escape and elongation due to APIS activity, although the mechanistic details are unclear. The other is a repressive function that stems from the ability of the APIS complex to potently dissociate Gal4·DNA complexes (15). This activity absolutely requires direct activation domain-Rpt6/Rpt4 interactions, presumably reflecting the need for APIS to engage the activator as a substrate for unfolding. This “stripping” activity is only manifest in the context of certain Gal4 mutants in vivo. The wild-type protein is immune. We have recently discovered that this is due to the ubiquitylation state of the activator. When Gal4 is mono-ubiquitylated somewhere in its DBD, it is insensitive to the stripping activity (15). The mutations that render Gal4 sensitive to this activity have been found to compromise its mono-ubiquitylation (15, 16). The detailed mechanism by which ubiquitylation protects Gal4 from the APIS complex has been worked out recently (17).
Given the many parallels between the mechanism of action of Gal4 and many other activators, we were interested in determining whether this repressive stripping activity was relevant to the regulation of mammalian transcription factors such as p53. We present here evidence that this is indeed the case. Biochemical experiments using purified p53 bound to DNA from the p21waf1 promoter demonstrate that the complex is disrupted by either the 19 S RP or the full 26 S proteasome (Figs. 1 and 2). This activity is dependent on ATP hydrolysis by the proteasomal ATPases (Fig. 3). Furthermore, as was the case for Gal4, direct binding of the ATPase complex to the activator appears to be essential, as indicated by the insensitivity of the p53 DBD·DNA complex to the stripping activity (Fig. 4). Experiments using GST fusion proteins containing fragments of the p53 protein identified the C-terminal TDRD segment to be the region of the protein that binds the ATPases (Fig. 4). Although some amount of 20 S CP is also retained by this fragment, Rpt6 is highly enriched in the TDRD-associated fraction relative to the 26 S input, suggesting that, like Gal4, p53 associates primarily with the 19 S RP or a fragment thereof.
The only biochemical difference noted so far in the studies of Gal4 and p53 in this reaction is the fact that disruption of the Gal4·DNA complexes was highly reversible and net loss of the protein from the immobilized DNA was observed only in the presence of excess soluble competitor DNA (15). This was not the case in this study. The presence or absence of soluble p21waf1 promoter DNA had no effect, suggesting that, when the ATPases unwind the p53 protein, they do so in an irreversible fashion. In summary, the biochemical experiments show clearly that a p53·DNA complex is a substrate for APIS-mediated disruption.
To gauge the physiological relevance of these in vitro observations, several experiments were performed in cultured H460 and HCT116 cancer cells. We showed that Rpt6 and Rpt4 are recruited to the p21waf1 promoter in an inducible fashion upon induction of cells with doxorubicin. Recruitment of the 20 S CP to the same promoter could not be detected by ChIP, but we cannot rule out that this negative result simply reflects a technical problem with the antibody or some such matter. These results parallel ChIP experiments published recently by Zhu et al. who showed recruitment of Rpt6 and Rpn2 to the p21waf1 promoter in MCF-7 cells after UV irradiation (25). These workers also showed that Rpt6 co-immunoprecipitates with p53 and p53 turnover by the proteasome positively regulates p53-mediated transcription at p21waf1 promoter.
We provide evidence here for the first time that the proteasomal ATPases regulate p53-mediated transcription by a non-proteolytic mechanism in vitro and in cells. This activity is repressive in nature, in contrast to the positive effect of the proteolytic activity of the proteasome on p21waf1 gene expression. siRNA-mediated knockdown of Rpt6 increases p53 occupancy of the p21waf1 promoter and stimulates p53-mediated p21waf1 gene expression (Fig. 6). Blocking proteasome-mediated proteolysis with MG132 also increased occupancy of the promoter by p53, but results in inhibition of p21waf1 mRNA production (Fig. 6), consistent with previous reports (25). The opposite effects of Rpt6 knockdown and MG132 on gene expression argue strongly that Rpt6 regulates p53 activity in some way that is different from merely contributing to efficient activator proteolysis. The observed effects of Rpt6 knockdown are consistent with the idea that this ablates the ability of the APIS complex to destabilize p53·p21waf1 promoter interactions in vivo, thus increasing the promoter occupancy and allowing increased gene expression. It is interesting that Rpt6 knockdown has a particularly dramatic stimulatory effect on p21waf1 gene expression in the absence of induction with doxorubicin, consistent with the idea that the stripping reaction may help to restrict p53 from binding stably to the promoter in the uninduced state and thus repress inappropriate basal transcription. It should be noted that the quantitative difference in the levels of promoter occupancy by p53 in cells treated with the control siRNA or the Rpt6-targeted siRNA are not as dramatically different as the differences in p21waf1 gene expression in the same cells (Fig. 6, compare B and C). This could simply reflect technical issues inherent in quantitative comparisons of different types of assays. Alternatively, it could be speculated that, although disruption of p53·promoter complexes by APIS is irreversible in vitro, it may be reversible in vivo in the presence of molecular chaperones. Because standard ChIP assays do not distinguish stable and kinetically labile binding events (39), but rather simply measure steady-state occupancy, it is possible that the half-life of the p53·promoter complexes is reduced by APIS in uninduced cells. A few studies have appeared showing that activator·promoter complexes with high off-rates drive lower levels of transcription than complexes with identical KD values, but lower off-rates (40, 41). Thus, by limiting the lifetime of the p53·promoter complex in vivo, it is possible that APIS could antagonize gene expression to a greater degree than would be predicted from simple ChIP data. What influence the various ubiquitylation events known to occur on p53 might have on its sensitivity to APIS-mediated stripping is unclear and will be the subject of future investigation.
Finally, the fact that knockdown of Rpt6 stimulates p53-mediated p21waf1 transcription suggests that the proteasomal ATPases are not critical for efficient elongation. This is also consistent with our inability to detect Rpt6 on the coding sequence of the gene, although it was clearly present on the promoter. In the case of the yeast GAL and heat shock genes, the ATPases do function to stimulate elongation and are clearly present throughout the gene (10, 11, 38). Thus, the p21waf1 gene would appear to represent a case in which only the inhibitory non-proteolytic function of the APIS complex is operative. Whether this will also be true of other p53-regulated genes remains to be elucidated.
In summary, we have demonstrated that the proteasomal ATPases negatively regulate p53 function via a non-proteolytic mechanism that involves active dissociation of p53·DNA complexes. Although a few instances of stimulatory, non-proteolytic effects of these ATPases in mammalian cells have been noted that parallel earlier observations in yeast, this is, to our knowledge, the first example of an inhibitory non-proteolytic action of proteasomal ATPases in mammalian cells.
This work was supported, in whole or in part, by National Institutes of Health Grants GM71833 (to T. K.) and CA103867 and CA124760 (to C. M. C.).
- UPS
- ubiquitin-proteasome system
- ChIP
- chromatin immunoprecipitation
- siRNA
- small interfering RNA
- 19 S RP
- 19 S regulatory particle
- 20 S CP
- 20 S core particle
- APIS
- AAA protein independent of the 20 S
- NTD
- N-terminal activation domain
- DBD
- DNA-binding domain
- TDRD
- tetramerization and regulatory domain.
REFERENCES
- 1.Collins G. A., Tansey W. P. (2006) Curr. Opin. Genet. Dev. 16, 197–202 [DOI] [PubMed] [Google Scholar]
- 2.Kodadek T., Sikder D., Nalley K. (2006) Cell 127, 261–264 [DOI] [PubMed] [Google Scholar]
- 3.Baumeister W., Walz J., Zühl F., Seemüller E. (1998) Cell 92, 367–380 [DOI] [PubMed] [Google Scholar]
- 4.Groll M., Ditzel L., Löwe J., Stock D., Bochtier M., Bartunik H. D., Huber R. (1997) Nature 386, 463–471 [DOI] [PubMed] [Google Scholar]
- 5.Thrower J. S., Hoffman L., Rechsteiner M., Pickart C. M. (2000) EMBO J. 19, 94–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lipford J. R., Deshaies R. J. (2003) Nat. Cell Biol. 5, 845–850 [DOI] [PubMed] [Google Scholar]
- 7.Reid G., Hübner M. R., Métivier R., Brand H., Denger S., Manu D., Beaudouin J., Ellenberg J., Gannon F. (2003) Mol. Cell 11, 695–707 [DOI] [PubMed] [Google Scholar]
- 8.Nawaz Z., O'Malley B. W. (2004) Mol. Endocrinol. 18, 493–499 [DOI] [PubMed] [Google Scholar]
- 9.Lipford J. R., Smith G. T., Chi Y., Deshaies R. J. (2005) Nature 438, 113–116 [DOI] [PubMed] [Google Scholar]
- 10.Gillette T. G., Gonzalez F., Delahodde A., Johnston S. A., Kodadek T. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 5904–5909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gonzalez F., Delahodde A., Kodadek T., Johnston S. A. (2002) Science 296, 548–550 [DOI] [PubMed] [Google Scholar]
- 12.Ferdous A., Gonzalez F., Sun L., Kodadek T., Johnston S. A. (2001) Mol. Cell 7, 981–991 [DOI] [PubMed] [Google Scholar]
- 13.Swaffield J. C., Melcher K., Johnston S. A. (1995) Nature 374, 88–91 [DOI] [PubMed] [Google Scholar]
- 14.Corton J. C., Moreno E., Johnston S. A. (1998) J. Biol. Chem. 273, 13776–13780 [DOI] [PubMed] [Google Scholar]
- 15.Ferdous A., Sikder D., Gillette T. G., Nalley K., Kodadek T., Johnston S. A. (2007) Genes Dev. 20, 112–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Archer C. T., Delahodde A., Gonzalez F., Johnston S. A., Kodadek T. (2008) J. Biol. Chem. 283, 12614–12623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Archer C. T., Burdine L., Liu B., Ferdous A., Johnston S. A., Kodadek T. (2008) J. Biol. Chem. 283, 21789–21798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salghetti S. E., Caudy A. A., Chenoweth J. G., Tansey W. P. (2001) Science 293, 1651–1653 [DOI] [PubMed] [Google Scholar]
- 19.Rasti M., Grand R. J., Yousef A. F., Shuen M., Mymryk J. S., Gallimore P. H., Turnell A. S. (2006) EMBO J. 25, 2710–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lassot I., Latreille D., Rousset E., Sourisseau M., Linares L. K., Chable-Bessia C., Coux O., Benkirane M., Kiernan R. E. (2007) Mol. Cell 25, 369–383 [DOI] [PubMed] [Google Scholar]
- 21.Bhat K. P., Turner J. D., Myers S. E., Cape A. D., Ting J. P., Greer S. F. (2008) Mol. Immunol. 45, 2214–2224 [DOI] [PubMed] [Google Scholar]
- 22.Greer S. F., Zika E., Conti B., Zhu X. S., Ting J. P. (2003) Nat. Immunol. 4, 1074–1082 [DOI] [PubMed] [Google Scholar]
- 23.Brès V., Kiernan R. E., Linares L. K., Chable-Bessia C., Plechakova O., Tréand C., Emiliani S., Peloponese J. M., Jeang K. T., Coux O., Scheffner M., Benkirane M. (2003) Nat. Cell Biol. 5, 754–761 [DOI] [PubMed] [Google Scholar]
- 24.Brooks C. L., Li M., Gu W. (2004) Cell Cycle 3, 436–438 [PubMed] [Google Scholar]
- 25.Zhu Q., Wani G., Yao J., Patnaik S., Wang Q. E., El-Mahdy M. A., Praetorius-Ibba M., Wani A. A. (2007) Oncogene 26, 4199–4208 [DOI] [PubMed] [Google Scholar]
- 26.Asher G., Shaul Y. (2005) Cell Cycle 4, 1015–1018 [DOI] [PubMed] [Google Scholar]
- 27.Le Cam L., Linares L. K., Paul C., Julien E., Lacroix M., Hatchi E., Triboulet R., Bossis G., Shmueli A., Rodriguez M. S., Coux O., Sardet C. (2006) Cell 127, 775–788 [DOI] [PubMed] [Google Scholar]
- 28.Lin L., Ozaki T., Takada Y., Kageyama H., Nakamura Y., Hata A., Zhang J. H., Simonds W. F., Nakagawara A., Koseki H. (2005) Oncogene 24, 3385–3396 [DOI] [PubMed] [Google Scholar]
- 29.Verma R., Chen S., Feldman R., Schieltz D., Yates J., Dohmen J., Deshaies R. J. (2000) Mol. Biol. Cell 11, 3425–3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strickland E., Hakala K., Thomas P. J., DeMartino G. N. (2000) J. Biol. Chem. 275, 5565–5572 [DOI] [PubMed] [Google Scholar]
- 31.Lim H. S., Archer C. T., Kodadek T. (2007) J. Am. Chem. Soc. 129, 7750–7751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomas M. C., Chiang C. M. (2005) Mol. Cell 17, 251–264 [DOI] [PubMed] [Google Scholar]
- 33.Lim H. S., Cai D., Archer C., Kodadek T. (2007) J. Am. Chem. Soc. 129, 12936–12937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Melcher K., Johnston S. (1995) Mol. Cell. Biol. 15, 2839–2848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Archer C., Burdine L., Kodadek T. (2005) Mol. BioSystems 1, 366–372 [DOI] [PubMed] [Google Scholar]
- 36.Tansey W. P. (2001) Genes Dev. 15, 1045–1050 [DOI] [PubMed] [Google Scholar]
- 37.Morris M. C., Kaiser P., Rudyak S., Baskerville C., Watson M. H., Reed S. I. (2003) Nature 423, 1009–1013 [DOI] [PubMed] [Google Scholar]
- 38.Sulahian R., Sikder D., Johnston S. A., Kodadek T. (2006) Nucleic Acids Res. 34, 1351–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nalley K., Johnston S. A., Kodadek T. (2006) Nature 442, 1054–1057 [DOI] [PubMed] [Google Scholar]
- 40.Yang E., Henriksen M. A., Sachaefer O., Zakharova N., Darnell J. E., Jr. (2002) J. Biol. Chem. 277, 13455–13462 [DOI] [PubMed] [Google Scholar]
- 41.Brady K. L., Ponnampalam S. N., Bumbulis M. J., Setzer D. R. (2005) J. Biol. Chem. 280, 26743–26750 [DOI] [PubMed] [Google Scholar]