Abstract
The RNA-binding motif protein 4 (RBM4) plays multiple roles in mRNA metabolism, including translation control. RBM4 suppresses cap-dependent translation but activates internal ribosome entry site-mediated translation. Because of its high expression level in muscle and heart, we investigated the function of RBM4 in myoblast cells. Here, we demonstrate that RBM4 is phosphorylated and translocates to the cytoplasm in mouse C2C12 cells upon cell differentiation. Notably, RBM4 is transiently deposited into cytoplasmic granules containing microtubule assembly factors as well as poly(A)+ RNAs. Moreover, RBM4 colocalizes with the components of micro-ribonucleoproteins, including the Argonaute2 (Ago2) protein, during muscle cell differentiation. RBM4 interacts directly with Ago2 and may recruit Ago2 to suppress translation of target mRNAs. Furthermore, RBM4 selectively associates with muscle cell-specific microRNAs and potentiates their translation repression activity by promoting micro-ribonucleoprotein association with target mRNAs. Altogether, our results suggest that RBM4 translocates to the cytoplasm and participates in translation suppression during muscle cell differentiation.
Introduction
RBM4 is a ubiquitous RNA-binding protein with relatively high abundance in skeletal muscle and heart. RBM4 is primarily localized in the nucleus and participates in alternative precursor mRNA splicing regulation (1, 2). After export to the cytoplasm, RBM4 may suppress translation of CU-rich elements containing mRNAs. Moreover, it can substantially activate internal ribosome entry site-mediated translation (3). The differential role of RBM4 in translation control is likely determined in response to cell stress cues.
In this study, we explore the function of RBM4 in muscle cells. We observed that RBM4 is colocalized with several centrosomal proteins and micro-RNP (miRNP)2 components in cytoplasmic granules of differentiated mouse C2C12 myoblasts (see “Results”). During myogenic differentiation, the microtubule cytoskeleton is reorganized in parallel with redistribution of several centrosomal proteins, including pericentrin and γ-tubulin to a few foci near the nuclear periphery (4, 5). Microtubules are nucleated from these perinuclear foci along the nuclear surface as well as through the cytoplasm. We have previously reported that RBM4 can translocate to cytoplasmic stress granules (SGs) when cells encounter environmental stress (3). SGs contain stalled translation initiation complexes (6). Moreover, cytoplasmic processing bodies (P-bodies) also contain translationally inert mRNAs. Unlike SGs, P-bodies lack the majority of translation initiation factors but contain mRNA degradation factors and miRNP components (7). miRNAs are a class of ∼21-nucleotide-long RNAs that typically base pair imperfectly to their target mRNAs and induce translation inhibition or promote RNA degradation (8, 9). The Argonaute (Ago) proteins, a central factor of miRNPs, contribute to translation inhibition likely via distinct mechanisms (9, 10).
Some miRNAs are expressed in a tissue- or time-specific manner and may regulate cell type-specific activity or cell differentiation (11). For example, a pair of muscle cell-specific miRNAs, miR-1 and miR-133, are particularly expressed during myogenesis and coordinately promote cell differentiation (12). Moreover, numerous lines of evidence have suggested an obligatory role of miRNAs for the development of skeletal and cardiac muscles (11). Also, emerging evidence indicates that miRNPs may function coordinately with some RNA-binding proteins in control of gene expression (13, 14); however, there is still much to learn about underlying mechanisms.
In this study, we explore the function of RBM4 during muscle cell differentiation. We found that RBM4 translocates to the cytoplasm most likely upon its phosphorylation, and it subsequently suppresses translation via binding to the CU-rich element(s) of target mRNAs and also acts as a cofactor of miRNPs to potentiate their activity in translation suppression during muscle cell differentiation.
EXPERIMENTAL PROCEDURES
Plasmid Construction
The pRL reporters were constructed by insertion of one (miR-1) or three copies (3×miR1) of the miR-1 artificial target sequence (12) or a 540-bp sequence of the mouse CCND1 3′-UTR (CCND1; nucleotides 1801–2340 of NM_007631) at two nucleotides downstream of the Renilla luciferase (RL) coding region of pRL-SV40 (Promega). The pRL-2×CU reporter was described previously (3). To express mouse miR-1 RNA, the PCR product coding for its primary sequence was cloned into a pcDNA-derived vector (Invitrogen). The mutant miR-1 was generated by a site-directed mutagenesis system (QuikChange, Stratagene). The expression vector for FLAG-tagged Ago2 was obtained by insertion of the PCR-amplified human Ago2 coding region in-frame with the FLAG tag in pcDNA3.1 (Invitrogen). The expression vectors for FLAG-tagged RBM4 and HA-tagged RBM4, TIA-1, eIF4A, and eIF4E were described previously (3). The Myc-Dcp1 expression vector was obtained from J. Lykke-Andersen (University of Colorado, Boulder).
Culture and Differentiation of Mouse Myoblasts
Mouse C2C12 myoblasts were maintained in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen). To induce C2C12 cell differentiation, the supplement was changed to 2% horse serum containing 20 μg/ml insulin; cells were cultured to ∼60–70% confluency and then used for transient transfection with Lipofectamine 2000 (Invitrogen).
Immunoblotting
Immunoblotting was conducted as recommended using the ECL system (Amersham Biosciences). Antibodies used included polyclonal anti-RBM4 and anti-pS309 that specifically recognize phosphorylated RBM4 (3) and monoclonal antibodies against myosin heavy chain (Abcam), α-tubulin (NeoMarkers), CCND1 (Santa Cruz Biotechnology), and Ago2 (Abnova).
Immunofluorescence
Indirect immunofluorescence was performed as described previously (3). Primary antibodies used were polyclonal antibodies against RBM4, Dcp1, Myc tag (2 μg/ml; Upstate), and pericentrin (2 μg/ml; Abcam) and monoclonal antibodies against FLAG tag (20 μg/ml; Sigma), HA epitope (2 μg/ml; Babco), Ago2 (2.5 μg/ml; Abnova), and GW182 (10 μg/ml; Abcam). Dcp1 antisera were raised by immunizing rabbits with recombinant protein corresponding to the N-terminal 251 residues of human Dcp1; antibodies were affinity-purified for use in immunofluorescence. Secondary antibodies were used as described (3). Fluorescence in situ hybridization for poly(A)+ RNA detection was performed according to Li et al. (20).
Immunoprecipitation
To investigate the interaction of RBM4 with endogenous miR-1, the FLAG-RBM4 expression vector was transfected into C2C12 cells. Cells were initially incubated in growth medium for 24 h and then transferred to differentiation medium for 48 h. Analogously, to examine the interaction of FLAG- or HA-tagged translation factors with the reporter mRNAs, each of their corresponding expression vectors was cotransfected with the reporter into HEK293 cells for 48 h. Cell lysates were prepared in RIPA buffer (3) containing 1% Nonidet P-40 and then subjected to immunoprecipitation by incubation with anti-FLAG- or anti-HA-conjugated beads at 4 °C for 2 h. The beads were extensively washed with RIPA buffer containing 0.5% Nonidet P-40. RNA was extracted from the immunoprecipitates after proteinase K treatment using phenol/chloroform/isoamyl alcohol and recovered for Northern blotting or RT-PCR.
Northern Blot Analysis
To detect miRNAs, RNA samples were fractionated on 8% denaturing polyacrylamide gels and transferred onto nylon membranes (Zeta-Probe, Bio-Rad). The membranes were preincubated in a mixture containing 0.2 m Na2PO4 (pH 7.0) and 7% SDS and subsequently hybridized with 1 × 106 cpm of γ-32P-labeled DNA oligonucleotide probe (2 × 106 cpm/pmol) in the same solution. The miRNA probes used are as follows: miR-1, 5′-TACATACTTCTTTACATTCCA; miR-133, 5′-ACAGCTGGTTGAAGGGGACCAA; miR-206, 5′-CCACACACTTCCTTACATTCCA; and miR-16; 5′-AATATTTACGTGCTGCTA. The probe (5′-AAAGCCTACAGCACCC) for 5 S rRNA was used as control.
In Vivo Translation Assay
RL reporter was cotransfected with the firefly luciferase vector pGL3-control (Promega) as reference into cells. RL activity was normalized to firefly luciferase activity in individual transfectants. For each set of the assays, the data were obtained from at least three independent experiments.
Electrophoretic Mobility Shift Assay
C2C12 cells were transiently transfected with the FLAG-Ago2 expression vector and maintained in growth or differentiation medium. The S100 extract was prepared according to Tarn and Steitz (21). Each 20-μg extract was incubated with 2 × 105 cpm of γ-32P-labeled transcript (134 nucleotides) containing three copies of the miR-1-binding site derived from the 3′-UTR of pRL-3×miR1 in a 20-μl reaction containing 10 mm HEPES (pH 7.9), 50 μm EDTA, 10% glycerol, 1 mm dithiothreitol, 5 mm MgCl2, 0.5 μg/ml bovine serum albumin, and 12.5 ng/ml tRNA for 15 min at room temperature. The mixture was subsequently incubated with 0.5 μg of purified anti-RBM4 (3) or anti-FLAG antibody (Zymed Laboratories Inc.) or mouse immunoglobulin (Sigma) in the presence of heparin (5 mg/ml) for an additional 15 min. The reactions were analyzed by electrophoresis on a 6% polyacrylamide nondenaturing gel in a buffer containing 45 mm Tris-HCl, 45 mm boric acid, and 1 mm EDTA (pH 8.0) (21).
RESULTS
RBM4 Is Phosphorylated during Cell Differentiation
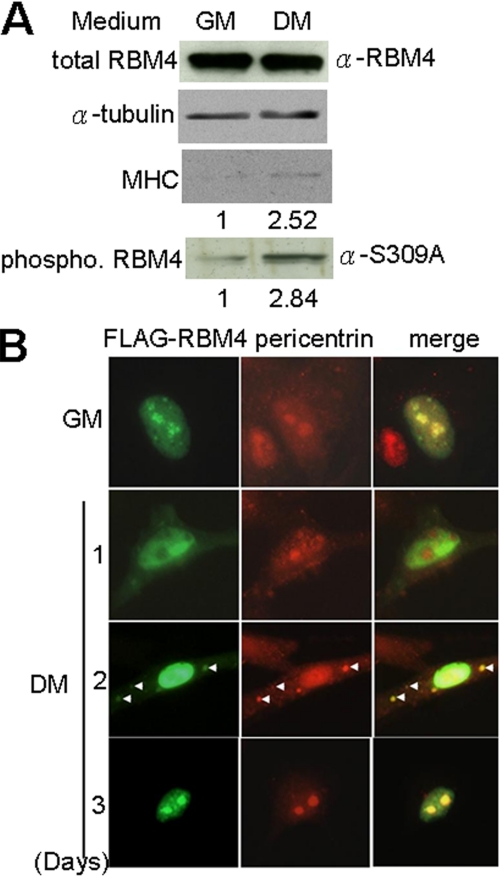
We took advantage of mouse C2C12 myoblasts to investigate the possible function of RBM4 in muscle cell mRNA metabolism. Upon depletion of serum, we observed that the expression level of MyoD and myogenin gradually increased (data not shown), and myosin heavy chain was also induced in C2C12 cells (Fig. 1A). Multiple nuclei and elongated cell shape could be detected 4–5 days after serum reduction (data not shown), indicating cell differentiation into myocytes. In differentiated cells, although the expression level of RBM4 remained unchanged, its phosphorylation level was enhanced by ∼3-fold as detected by using antibody that specifically recognizes phosphorylated RBM4 (3) (Fig. 1A). Thus, RBM4 phosphorylation at serine 309 could be induced by environmental stress (3) as well as by cell differentiation (this study).
FIGURE 1.
Phosphorylation and subcellular localization of RBM4 during myoblast differentiation. A, immunoblotting of cell lysates prepared from C2C12 cells that were cultured in growth (GM) or differentiation medium (DM) for 2 days. Relative level of myosin heavy chain (MHC) protein expression and RBM4 phosphorylation in GM versus DM is indicated below the gels. B, immunofluorescence of C2C12 cells that transiently expressed FLAG-RBM4 and were cultured in GM or DM for 1–3 days.
RBM4 Transiently Localizes in Cytoplasmic Granules
Using indirect immunofluorescence, we observed that overexpressed FLAG-tagged RBM4 primarily distributed in the nucleus with a higher concentration in nucleoli under cell growth conditions, but it was largely depleted from the nucleoli and meanwhile accumulated in the cytoplasm upon cell differentiation (Fig. 1B). Two days after differentiation initiation, FLAG-RBM4 appeared in cytoplasmic granules (Fig. 1B). We have also examined the effect of phosphorylation on cellular redistribution of RBM4. The nonphosphorylatable mutant had a lower tendency to locate in the cytoplasm and to form granules than the wild type, whereas the phosphomimetic mutant had a higher level in cytoplasmic localization and granule formation (supplemental Fig. 1). Therefore, the result suggests that cytoplasmic localization of RBM4 is perhaps a result of its phosphorylation. Interestingly, endogenous pericentrin, a marker of differentiation-induced muscle cell granules, changed its subcellular localization throughout differentiation similarly to FLAG-RBM4 (Fig. 1B). Moreover, RBM4 was also colocalized with γ-tubulin (data not shown). Therefore, RBM4 appeared to be a component of perinuclear granules in differentiating myocytes.
RBM4 Colocalizes with miRNP Components
Using in situ hybridization, we observed colocalization of poly(A)+ RNAs with RBM4 as well as pericentrin in differentiation-induced muscle cell granules (supplemental Fig. 2). This result prompted us to characterize whether these granules contain any other RNA metabolic factors. During cell differentiation, cytoplasmic FLAG-RBM4 was indeed colocalized with endogenous Dcp1, a representative component of P-bodies, in granules (Fig. 2A). Overexpression of Myc-tagged Dcp1 could induce relocalization of endogenous RBM4 to granules even under cell growth conditions (Fig. 2B). RBM4 was also colocalized substantially with Ago2 and GW182 during differentiation (Fig. 2C), although these two P-body marker proteins formed numerous granules in the cytoplasm of both proliferating (data not shown) and differentiating cells (Fig. 2C). However, unlike Dcp1, overexpression of HA-tagged SG marker protein TIA-1 did not induce RBM4-containing granules in proliferating C2C12 cells (Fig. 2D). Nevertheless, these two proteins were still colocalized in granules in differentiated cells. Collectively, the above results provide a hint that muscle cell-specific granules containing RBM4 may constitute sites for cytoplasmic mRNA metabolism during differentiation.
FIGURE 2.
Colocalization of RBM4 with the miRNP components. C2C12 cells that transiently expressed (A) FLAG-RBM4, (B) Myc-Dcp1, or (D) HA-TIA1 were cultured in GM for DM for 2 day, and then subjected to immunofluorescence using antibody specific to individual tag. C, immunofluorescence of nontransfected C2C12 cells using anti-RBM4, GW182, and Ago2. Representative arrowheads show granules with merged signals. White boxes show RBM4 granules that are colocalized with maker proteins.
RBM4 Recruits Ago2 for Translation Suppression
Colocalization of RBM4 with Ago2 prompted us to examine whether these two proteins interact with each other. As shown in Fig. 3A, overexpressed FLAG-RBM4 in C2C12 cells coprecipitated with Ago2 under both cell proliferation and differentiation conditions. The interaction between RBM4 and Ago2 was independent of RNA and was regardless of the phosphorylation status of RBM4 (supplemental Fig. 3). It should be noted that RBM4 was largely localized in the nucleus and that Ago2 was distributed throughout the whole cells during cell proliferation, but both proteins accumulated in the cytoplasm and colocalized in granules when cells were induced to differentiate (Fig. 2 and supplemental Fig. 4). Therefore, a slightly enhanced interaction between RBM4 and Ago2 in differentiating C2C12 cell extract (Fig. 3A) may be consistent with their better colocalization in the cytoplasm. Next, to examine whether RBM4 functions coordinately with Ago2 in translation control, we used the Renilla luciferase (RL) reporter containing duplicate CU-rich elements derived from α-tropomyosin in the 3′-UTR (3) and performed the assay in HEK293 cells. We observed that HA-tagged RBM4 inhibited translation of this pRL-2×CU reporter in a dose-dependent manner (Fig. 3B). Using immunoprecipitation followed by RT-PCR, we observed that RBM4 could considerably promote coexpressed FLAG-Ago2 association with the reporter transcript (Fig. 3D, lanes 1–3). Therefore, RBM4 might recruit Ago2 for translation regulation of CU-rich element-containing mRNAs. Next, using siRNA to suppress Ago2 expression to ∼30% of the control, we observed that the translation suppression effect of RBM4 was attenuated (Fig. 3C, lane 8). This effect was not observed with a control siRNA (Fig. 3C, lane 7). Therefore, Ago2 might be involved in RBM4-mediated translation control. To explore how RBM4 suppresses translation, we inspected the binding of two translation initiation factors to the RL-2×CU mRNA. Immunoprecipitation and RT-PCR showed that overexpression of RBM4 reduced the association of coexpressed eIF4E, but not eIF4A, with the reporter mRNA (Fig. 3D, lanes 4–9). Nevertheless, the detailed mechanism by which RBM4 might abolish eIF4E binding to the mRNA remains to be investigated.
FIGURE 3.
RMB4-mediated translation suppression involves Ago2. A, cell lysates were prepared from FLAG-RBM4-expressing or mock C2C12 cells grown in GM or DM and then subjected to anti-FLAG immunoprecipitation (IP) followed by immunoblotting. The luciferase assay in B and C was performed using the mock pRL or pRL-2×CU reporter in HEK293 cells and as described under “Experimental Procedures”; cotransfected vectors of effector proteins or siRNA are as indicated. Bar scales show relative RL activity (see under “Experimental Procedures”). Ago2 knockdown efficiency was assessed by using immunoblotting. D, pRL-2×CU was cotransfected with effector expression vector(s) as indicated. Immunoprecipitation was performed 48 h after transfection; coprecipitates were analyzed by RT-PCR using primers specific for the RL-2×CU mRNA.
RBM4 Enhances Ago2 Association with an miR-1 Target
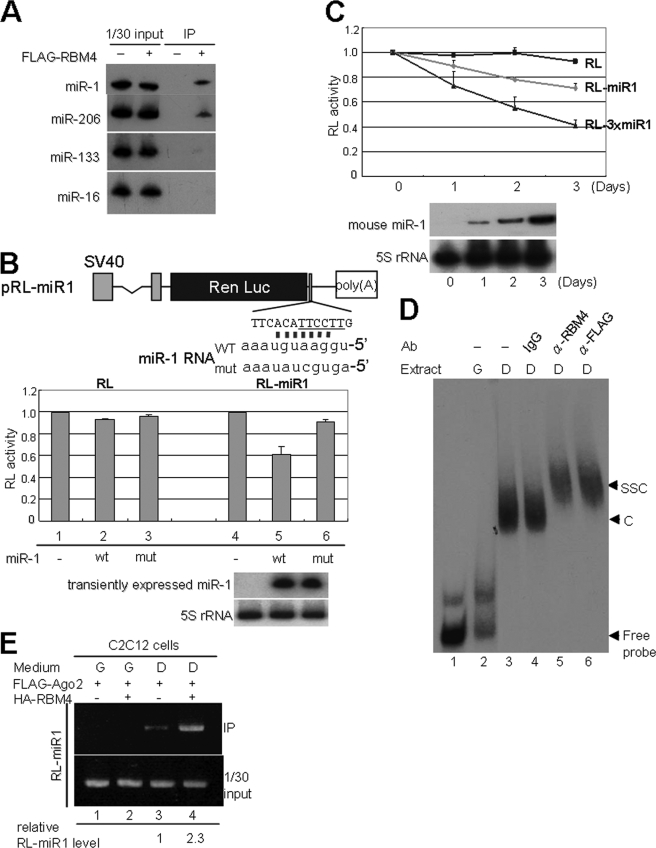
Because RBM4 colocalized and interacted with Ago2, we suspected that RBM4 may associate with miRNAs. Immunoprecipitation of FLAG-RBM4 from the lysates of differentiated C2C12 cells showed that it could associate with two muscle-specific miRNAs, miR-1 and miR-206, but had minimal or no interaction with another muscle miRNA, miR-133, and the ubiquitous miR-16 (Fig. 4A). The question of why RBM4 confers such selectivity for miRNAs needs future investigation. To test whether RBM4 is involved in miRNP-mediated gene expression regulation, we established the RL reporters containing one or three copies of an artificial miR-1 targeting site in the 3′-UTR (Fig. 4B). The transfection assay was performed in HEK293 cells because this cell line does not have endogenous miR-1 RNA but can properly express ectopic miR-1 upon transfection of an miR-1-expressing vector. Transient expression of the wild-type, but not mutant, miR-1 could suppress the expression of pRL-miR1 in HEK293 cells (Fig. 4B) without altering its mRNA level (Fig. 5A), indicating that translation of this reporter was regulated by miR-1 in a sequence-specific manner. Moreover, expression of RL-miR1 in C2C12 cells was gradually reduced throughout cell differentiation and was inversely proportional to the level of miR-1 (Fig. 4C). Enhanced repression was observed with the RL-3×miR1 reporter containing triplicate miR-1 target site, probably because of the binding of multiple miR-1 to the 3′-UTR (Fig. 4C).
FIGURE 4.
RBM4 selectively interacts with miRNAs and promotes Ago2 association with miRNA targets. A, FLAG-RBM4 was immunoprecipitated (IP) from differentiated C2C12 cells. Coprecipitates were analyzed by Northern blotting. B, diagram shows the pRL-miR1 reporter containing a synthetic miR-1 targeting sequence; the CU-rich sequence is underlined. The wild-type (WT) and mutant (mut) miR-1 RNAs are also depicted. The pRL or pRL-miR1 reporter was cotransfected with miR-1 expression vector in HEK293 cells. The reporter assay was essentially as in Fig. 3. Expression of miR-1 RNA was detected by Northern blotting in both B and C. C, reporter assay using pRL-miR1 or pRL-3×miR1 was performed in C2C12 cells that were cultured in DM for 0–3 days. D, EMSA was performed using 32P-labeled RL-3×miR1 3′-UTR RNA as probe and C2C12 cell cytoplasmic extract containing FLAG-Ago2. C and SSC represent RNA-protein complex and supershifted complex, respectively. E, immunoprecipitation-RT-PCR was as similar to Fig. 3D (lanes 1–3), except that transfection was performed in C2C12 cells. Relative amount of coprecipitated RL-miR1 RNA is indicated. G, GM; D, DM.
FIGURE 5.
RBM4 potentiates miRNA-mediated translation suppression. The RL reporter assay of A–C was performed as in Fig. 3; coexpressed effectors, miR-1, or short hairpin (sh) RNA are as indicated. Diagram in panel A shows the pRL-CCND1 reporter; the mouse cyclin D1 3′-UTR contains a putative miR-1 target site as depicted. Reporter mRNAs were examined by RT-PCR. Reporter assay was performed by transfection of an effector-expression vector alone (miR-1-minus) or together with miR-1 (miR-1-plus). The miR-1 suppression effect refers relative RL activity of each pair of miR-1-plus versus miR-1-minus transfectants. Scale bars show the translation suppression effect of miR-1 or/and effector-expressing transfectants relative to that of the mock. Rescue efficiency (rescue eff in panel C) refers to fold increase in relative RL activity upon RBM4 depletion; p value indicating the mean difference was obtained by using a paired t test. Immunoblotting shows the efficiency of RBM4 knockdown. D, immunoprecipitation (IP) and RT-PCR were similar to Fig. 3D (lanes 1–3).
Based on the above results, we speculated that RBM4 may participate in miRNA-mediated translation regulation. Therefore, we examined whether RBM4 is associated with miRNA targets. Gel mobility shift assay showed that a 32P-labeled RNA derived from the 3′-UTR of RL-3×miR1 formed a complex in the cytoplasmic extract prepared from differentiating C2C12 cells, which could be supershifted by purified anti-RBM4 or anti-FLAG antibody that recognizes transiently expressed FLAG-Ago2 (Fig. 4D, lanes 3, 5, and 6). No specific RNP complex was detected in growing C2C12 cells, perhaps due to nuclear localization of RBM4 and absence of miR-1 during this stage (Fig. 4D, lane 2). Therefore, RBM4, like Ago2, might associate with miR-1 target mRNPs in differentiated cells. Next, we examined whether RBM4 may act as a cofactor of Ago2-containing miRNPs to modulate their association with the target mRNAs. Using immunoprecipitation and RT-PCR, we observed that FLAG-Ago2 bound to the RL-miR1 transcript in differentiated cells, and this binding was enhanced ∼2.3-fold upon overexpression of HA-RBM4 (Fig. 4E). Therefore, RBM4 not only associated with the miR-1 target mRNAs but could also promote or stabilize the Ago2-target mRNA interaction.
RBM4 Potentiates the Activity of miRNAs
Next, we examined whether RBM4 is engaged in miR-1-mediated translation control. Transient expression of RBM4 alone down-regulated RL-miR1 translation in HEK293 cells (Fig. 5A, top panel, lane 7); this was perhaps due to the binding of RBM4 to the CU-rich miR-1 target site. Overexpression of RBM4 could further the effect of miR-1-mediated suppression (Fig. 5A, top panel, lane 8), albeit additively; however, this was not observed with the RBM4 mutant (Fig. 5B, lane 3). We analyzed another splicing and translation regulatory factor, the alternative splicing factor, as comparison. The result showed that the alternative splicing factor somewhat reversed the translation inhibitory effect of miR-1 (Fig. 5B, lane 4). Therefore, the effect of RBM4 in promoting miR-1-mediated translation suppression appeared to be specific. Furthermore, we examined a cellular miR-1 targeting element that resides within the mammalian cyclin D1 genes. A segment of the mouse cyclin D1 3′-UTR was introduced into the RL reporter (Fig. 5A, pRL-CCND1). RBM4 and miR-1 could individually reduce the expression of this reporter in HEK293 cells (Fig. 5A, lower panel, lanes 6 and 7). As observed with RL-miR1, overexpression of RBM4 augmented the suppressive effect of miR-1 on RL-CCND1 translation (Fig. 5A, lower panel, lane 8). We next evaluated whether the activity of miR-1 is affected upon depletion of RBM4. When RBM4 expression was reduced by ∼70%, the RL activity of the miR-1 target site-null reporter was lightly enhanced (Fig. 5C, lanes 1, 2, 5, and 6), which was consistent with our previous observation (3). Nevertheless, depletion of RBM4 could relive miR-1-mediated suppression of both reporters to a statistically significant level (Fig. 5C, lanes 3, 4, 7, and 8). Therefore, RBM4 might act, at least additively, in potentiation of miR-1 activity in translation inhibition. However, the possibility that RBM4 and miR-1 suppressed the translation of common target mRNAs in a separate manner cannot be completely excluded.
We finally examined whether RBM4 has any effect on miR-1 RNP association with its target mRNAs. In the absence of miR-1, transiently expressed FLAG-Ago2 minimally bound to the RL-miR1 mRNA in HEK293 cells (Fig. 5D, lane 2). Overexpression of RBM4 or miR-1 could slightly enhance Ago2 interaction with this mRNA (Fig. 5D, lanes 3 and 4), but their coexpression could yield a greater extent of such interaction (Fig. 5D, lane 5). Therefore, RBM4 might assist certain miRNAs in translation suppression by promoting association of Ago2-containing miRNPs with their cognate target mRNAs (Fig. 6).
FIGURE 6.
Model of RBM4-mediated translation suppression during muscle cell differentiation. In growing myoblasts, RBM4 is essentially localized in the nucleus, and miR-1 is not expressed. Therefore, translation of the mRNAs containing CU-rich miR-1 targeting sites is essentially active, unless RBM4 is overexpressed. Upon cell differentiation, RBM4 is phosphorylated and translocates to the cytoplasm, and miR-1 is expressed. Translation of such mRNAs may be suppressed by RBM4 (a) or miR-1 RNP (b) alone or in an RBM4 and miRNP cooperative manner (c). ORF, open reading frame.
DISCUSSION
Cell stress induces phosphorylation and cytoplasmic accumulation of RBM4 via the mitogen-activated protein kinase (MAPK)/p38 kinase pathway (3). Notably, this kinase cascade is also activated in myogenic differentiation (15, 16). We indeed observed that RBM4 became phosphorylated and translocated to the cytoplasm during muscle cell differentiation (Fig. 1). Perhaps activation of specific signaling pathways may yield a common effect on RBM4 in different cell types and circumstances. Moreover, both cell stress and differentiation could result in RBM4 colocalization with mRNA metabolic factors in cytoplasmic granules (Fig. 2) (3). Therefore, RBM4 may likely participate in certain cytoplasmic RNA metabolic activities, such as translational control, under specific cellular conditions.
Several key factors of P-bodies or SGs can be found in cell type-specific RNA granules. For example, the P-body protein Dcp1 locates in dendritic RNA transport granules, and together with the Ago family protein MIWI, it is also present in cytoplasmic chromatoid bodies of male germ cells (6, 17). Our detection of RBM4 as well as Dcp1 and miRNP components in muscle cell cytoplasmic granules reinforces that compositionally and perhaps functionally similar granules exist in various cell types. This observation is also in line with the expression of muscle-specific miRNAs during differentiation and suggests that such muscle cell granules may constitute sites of miRNA-mediated activities that modulate cell differentiation. Nevertheless, whether these granules host specific species of mRNAs/miRNAs or exhibit any specific activities must be examined in future experiments.
We have previously reported that RBM4 suppresses the cap-dependent translation via binding to CU-rich elements of mRNAs. On the other hand, RBM4 recruits eIF4A to activate encephalomyocarditis virus internal ribosome entry site-mediated translation (3). Here, we report that RBM4 suppresses translation perhaps by promoting Ago2 association with the mRNA, which may lead to dissociate eIF4E but not eIF4A from the mRNA (Fig. 3). Indeed, Ago2 may act through multiple mechanisms to negatively regulate translation (8, 9). For example, Ago proteins might deprive eIF4E from the mRNA cap or disrupt its interaction with eIF4G or even act at different steps of translation (10, 18). Therefore, whether Ago2 contributes to eIF4E dissociation in RBM4-mediated translation suppression remains to be clarified. Furthermore, our data revealed that RBM4 selectively interacted with miRNAs during muscle cell differentiation and might potentiate their activity in translation suppression (Figs. 4 and 5). This result is in line with a previous report that RBM4 association with an Ago2-containing mRNP post-transcriptionally suppresses gene expression (19). However, the question of whether RBM4 is a bona fide integral component of global or specific miRNPs in muscle and non-muscle cells still remains to be investigated. Together with our previous study (3), we provide evidence suggesting that RBM4 modulates different aspects of translation via multiple mechanisms. In particular, during muscle cell differentiation, RBM4 translocates to the cytoplasm and suppresses translation by itself or in conjunction with a set of miRNAs (Fig. 6).
Acknowledgments
We thank J. Lykke-Andersen for cDNA clones, W. Filipowicz (Friedrich Miescher Institute, Switzerland) and J. A. Steitz (Yale University) for critical comments on the manuscript, and W.-L. Chang for antibody preparation.
This work was supported by National Science Council Grant NSC 96-2628-B001-013.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
- miRNP
- micro-ribonucleoprotein
- HA
- hemagglutinin
- UTR
- untranslated region
- RT
- reverse transcription
- GM
- growth medium
- DM
- differentiation medium
- SG
- stress granule
- RL
- Renilla luciferase
- P-body
- processing bodies.
REFERENCES
- 1.Lai M. C., Kuo H. W., Chang W. C., Tarn W. Y. (2003) EMBO J. 22, 1359–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin J. C., Tarn W. Y. (2005) Mol. Cell. Biol. 25, 10111–10121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin J. C., Hsu M., Tarn W. Y. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 2235–2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bugnard E., Zaal K. J., Ralston E. (2005) Cell Motil. Cytoskeleton 60, 1–13 [DOI] [PubMed] [Google Scholar]
- 5.Srsen V., Fant X., Heald R., Rabouille C., Merdes A. (2009) BMC Cell Biol. 10, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson P., Kedersha N. (2006) J. Cell Biol. 172, 803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parker R., Sheth U. (2007) Mol. Cell 25, 635–646 [DOI] [PubMed] [Google Scholar]
- 8.Jackson R. J., Standart N. (2007) Sci. STKE 2007, 367:re1. [DOI] [PubMed] [Google Scholar]
- 9.Filipowicz W., Bhattacharyya S. N., Sonenberg N. (2008) Nat. Rev. Genet. 9, 102–114 [DOI] [PubMed] [Google Scholar]
- 10.Iwasaki S., Kawamata T., Tomari Y. (2009) Mol. Cell 34, 58–67 [DOI] [PubMed] [Google Scholar]
- 11.van Rooij E., Liu N., Olson E. N. (2008) Trends Genet. 24, 159–166 [DOI] [PubMed] [Google Scholar]
- 12.Chen J. F., Mandel E. M., Thomson J. M., Wu Q., Callis T. E., Hammond S. M., Conlon F. L., Wang D. Z. (2006) Nat. Genet. 38, 228–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammell C. M. (2008) RNA Biol. 5, 123–127 [DOI] [PubMed] [Google Scholar]
- 14.Kedde M., Agami R. (2008) Cell Cycle 7, 899–903 [DOI] [PubMed] [Google Scholar]
- 15.Keren A., Tamir Y., Bengal E. (2006) Mol. Cell. Endocrinol. 252, 224–230 [DOI] [PubMed] [Google Scholar]
- 16.Lluís F., Perdiguero E., Nebreda A. R., Muñoz-Cánoves P. (2006) Trends Cell Biol. 16, 36–44 [DOI] [PubMed] [Google Scholar]
- 17.Kotaja N., Sassone-Corsi P. (2007) Nat. Rev. Mol. Cell Biol. 8, 85–90 [DOI] [PubMed] [Google Scholar]
- 18.Kiriakidou M., Tan G. S., Lamprinaki S., De Planell-Saguer M., Nelson P. T., Mourelatos Z. (2007) Cell 129, 1141–1151 [DOI] [PubMed] [Google Scholar]
- 19.Höck J., Weinmann L., Ender C., Rüdel S., Kremmer E., Raabe M., Urlaub H., Meister G. (2007) EMBO Rep. 8, 1052–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li C., Lin R. I., Lai M. C., Ouyang P., Tarn W. Y. (2003) Mol. Cell. Biol. 23, 7363–7376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tarn W. Y., Steitz J. A. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 2504–2508 [DOI] [PMC free article] [PubMed] [Google Scholar]