Abstract
Loss of the RecQ DNA helicase WRN protein causes Werner syndrome, in which patients exhibit features of premature aging and increased cancer. WRN deficiency induces cellular defects in DNA replication, mitotic homologous recombination (HR), and telomere stability. In addition to DNA unwinding activity, WRN also possesses exonuclease, strand annealing, and branch migration activities. The single strand binding proteins replication protein A (RPA) and telomere-specific POT1 specifically stimulate WRN DNA unwinding activity. To determine whether RPA and POT1 also modulate WRN branch migration activity, we examined biologically relevant mobile D-loops that mimic structures in HR strand invasion and at telomere ends. Both RPA and POT1 block WRN exonuclease digestion of the invading strand by loading on the strand. However, only RPA robustly stimulates WRN branch migration activity and increases the percentage of D-loops that are disrupted. Our results are consistent with cellular data that support RPA enhancement of branch migration during HR repair and, conversely, POT1 limitation of inappropriate recombination and branch migration at telomeric ends. This is, to our knowledge, the first evidence that RPA can stimulate branch migration activity.
Introduction
The protein defective in Werner syndrome, WRN, is a member of the RecQ DNA helicase family (1, 2). Other human RecQ helicases include BLM, RECQL4, RECQ1, and RECQ5 (reviewed in Refs. 3 and 4). BLM mutations result in Bloom syndrome (5), whereas RECQ4 mutations cause Rothmund-Thomson or RAPADILINO syndromes (6, 7). Although clinically distinct, these disorders all exhibit genomic instability and increased cancer (reviewed in Ref. 4). There are no known human disorders caused by RECQ1 or RECQ5 defects, but loss of these proteins in transgenic mice causes genomic instability (8, 9).
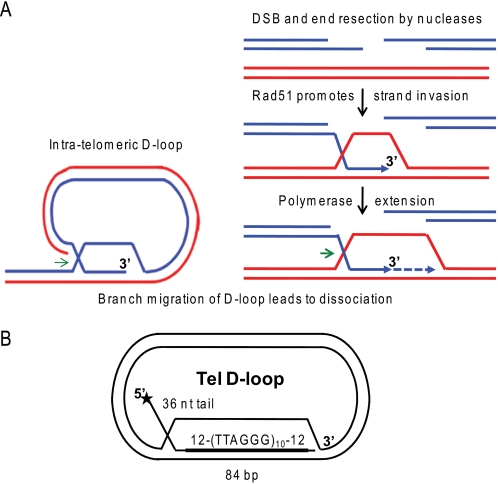
Previous studies indicate that RecQ helicases contribute to genome stability by regulating homologous recombination (HR)2 through various mechanisms. Inappropriate resolution of HR DNA intermediates can cause loss of heterozygosity, chromosome translocations, telomere loss, or toxic tangled DNA intermediates (10). HR functions in the repair of DNA double strand breaks (DSBs), restoration of collapsed or stalled replication forks, and the alternative lengthening of telomeres (ALT) pathways (10–13). HR initiates when RAD51 forms nucleoprotein filaments on protruding single-stranded DNA (ssDNA) tails (Fig. 1A). The RAD51 filaments catalyze invasion of the tail into homologous duplex DNA (double-stranded DNA; dsDNA) and promote strand exchange (14). The resulting joint molecule is a D-loop in which the invading strand is extended by a DNA polymerase (12, 15). Then the D-loops are dissociated, and repair proceeds by synthesis-dependent strand annealing, or they are processed into double Holliday junctions that are later resolved (reviewed in Refs. 10 and 11). Holliday junction resolution can lead to crossover recombinants, depending on the mechanism (11). Thus, D-loop processing represents a critical point in determining the cellular outcome of HR.
FIGURE 1.
Schematic of D-loops at telomeric ends and in homologous recombination. A, the 3′ ssDNA tails generated at a double strand break by nucleases or present at a collapsed replication fork are coated by Rad51 filaments. Then Rad51 promotes strand invasion and pairing with homologous sequence in duplex DNA. A polymerase can initiate DNA synthesis at the 3′-end of the invading strand. D-loops also exist at telomeric ends, whereby the 3′ tail invades homologous telomeric duplex DNA. Branch migration of the D-loops in the direction of the green arrows mediates D-loop disruption for replication fork progression at the telomeres or for completion of repair as in the synthesis-dependent strand-annealing pathway. B, schematic of 5′-tailed telomeric (Tel) mobile D-loop. The star denotes the 5′-end radiolabel. The invading strand base-pairs with the plasmid to form an 84-bp duplex with 10 TTAGGG repeats flanked by 12 bp of unique sequence with a protruding 5′ 36-nt single strand tail.
WRN loss causes defects in HR that is triggered to repair stalled or broken replication forks, whether fork demise is induced by lesions or depleted dNTPs (16, 17). The HR defect is rescued with a RAD51 dominant negative mutant, which prevents joint molecule formation (16). These data strongly suggest that WRN functions in the proper dissociation of HR intermediates during DNA replication. Consistent with this, WRN is required for normal replication fork elongation after DNA damage or fork arrest (18). WRN-deficient cells also exhibit features of abnormal HR at telomeric ends, including telomere loss, telomeric sister chromatid exchanges, and spontaneous extrachromosomal telomeric circles (19–22). These defects can result from aberrant cleavage of telomere structures, including the natural telomere D-loop/t-loop (23, 24). Telomeres contain a 3′ ssDNA tail that forms protective intratelomeric D-loops that stabilize the t-loop structure (25) (Fig. 1A). The precise role for WRN at stalled replication is unknown, but WRN is implicated in the dissociation of D-loops for HR repair (3, 4) and for replication fork progression at telomere ends (26, 27).
WRN is a 3′ to 5′ helicase and unwinds a variety of DNA substrates (28–30). Like other RecQ helicases, WRN is poorly processive during DNA unwinding and only unwinds short duplexes in vitro (∼30–50 bp) (3, 31–34). However, WRN possesses other catalytic activities that may be more relevant to processing HR intermediates and replication fork remodeling. WRN exhibits 3′ to 5′ exonuclease activity (35) and possesses single strand DNA annealing (36) and branch migration (BM) activities (37, 38). WRN, BLM, and RECQ1 all promote BM of mobile plasmid-based D-loops (Fig. 1B) to release the invading strand (33, 38–40). They can disrupt longer mobile D-loops compared with static oligomeric D-loops that cannot be branch-migrated but only unwound (38–41). Together, these data indicate important mechanistic differences between BM and DNA unwinding.
Single strand binding proteins (SSBs) are critical for HR repair (10), and previous reports indicate that RecQ helicases cooperate with SSBs. The human SSB, replication protein A (RPA), stimulates DNA unwinding by BLM, WRN, RECQ1, and RECQ5β, enabling them to unwind longer duplexes (31, 42–44). The only other SSB reported to stimulate WRN and BLM DNA unwinding is the human telomeric protein POT1 (protection of telomeres 1), but exclusively on telomeric duplexes (45, 46). POT1 binds telomeric ssDNA and preserves telomeric ends (24), whereas RPA is essential for DNA transactions throughout the genome (47). RPA also stimulates RECQ1 DNA unwinding but does not stimulate RECQ1 BM of mobile D-loops (40), perhaps due to mechanistic differences between BM and DNA unwinding. Given that BM activity is critical for HR repair, we asked whether WRN-interacting SSBs POT1 and RPA, which stimulate DNA unwinding, could also stimulate WRN BM activity.
Here we report that both RPA and POT1 loading on the invading strand of the mobile telomeric D-loops impedes WRN exonuclease progression. This favors the release of full-length ssDNA products. POT1 imparts a weak increase in the percentage of mobile telomeric D-loops disrupted by WRN BM activity. In contrast, RPA robustly stimulates WRN BM activity and significantly increases the percentage of total ssDNA product. These results are discussed in the context of WRN function in general HR repair as well as processing of D-loop structures at telomeric ends.
EXPERIMENTAL PROCEDURES
Proteins
Recombinant hexahistidine-tagged WRN protein and the exonuclease-dead E84A mutant (X-WRN) were purified as previously described (38, 46). The concentration of active protein was determined by a Bradford assay (Bio-Rad) and standard helicase activity assay with a 16-bp forked duplex. Recombinant human POT1 protein was purified using a baculovirus/insect cell expression system as described previously (46).
The construct for expression of hexahistidine Sumo-tagged TPP1 (amino acids 89–334) (48) was generously provided by Dr. Ming Lei (University of Michigan). Expression was induced with 0.1 mm isopropyl 1-thio-β-d-galactopyranoside in Escherichia coli BL21 (DE3) pLysS cells, and then cells were harvested by centrifugation at 4500 rpm for 20 min. Cell pellets were lysed in buffer (50 mm Tris, pH 8.0, 400 mm NaCl, 50 mm PO4, pH 8.0, 10% glycerol, 0.5% Igepal Ca-630, 1 mm DTT, and protease inhibitors (Roche Applied Science)) for 30 min at 4 °C. The lysate was then sonicated and clarified by centrifugation at 40,000 rpm for 30 min at 4 °C. The supernatant was loaded onto one HisTrap FF 1-ml column equilibrated in buffer A (50 mm Tris, pH 8.0, 400 mm NaCl, 50 mm PO4, pH 8.0, 10% glycerol, 2 μg/ml each of aprotinin, leupeptin, chymostatin, and pepstatin, 1 mm DTT, and 1 mm 4-(2-aminoethyl)-benzenesulfonyl fluoride). The column was washed with 15 mm imidazole, and protein was eluted with 250 mm imidazole. Fractions containing TPP1 were concentrated using a Centricon-10 device (Amicon) prior to loading onto a Superdex 75 10/300 GL column equilibrated in buffer S (25 mm Tris, pH 8.0, 150 mm NaCl, 2 μg/ml each of protease inhibitors, 1 mm DTT, and 1 mm 4-(2-aminoethyl)-benzenesulfonyl fluoride). Fractions containing TPP1 were pooled and concentrated. Protein concentrations were determined by a Bradford assay (Bio-Rad).
Recombinant hexahistidine-tagged RPA protein was expressed in E. coli BL21 (DE3) pLysS cells at 37 °C for 3 h. The protein was purified using nickel chromatography in buffer containing 50 mm Tris, pH 8.0, 50 mm NaCl, 5 mm β-mercaptoethanol, and a gradient of 10–300 mm imidazole. The protein was buffer-exchanged into 30 mm Hepes, pH 7.8, 1 mm dithiothreitol, 10 μm ZnCl2, 0.25% inositol, and 0.01% Nonidet P-40, passed through a Source Q column, and eluted over a five-column gradient with the same buffer containing 1 m NaCl. Protein concentration was determined by A280 nm measurements. Purity of all proteins was determined by SDS-PAGE and Coomassie staining.
RecA protein was from U.S. Biochemical Corp. (Cleveland, OH). T4 single strand binding protein was from New England Biolabs. E. coli RecQ helicase was generously provided by Dr. James Keck (University of Wisconsin).
DNA Substrates
All oligonucleotides used in this study were purchased from Integrated DNA Technologies (Coralville, IA) and were purified by PAGE or high pressure liquid chromatography by the manufacturer. Oligonucleotides were 5′-end-labeled with [γ-32P]ATP (3000 Ci/mmol) (PerkinElmer Life Sciences) using T4 polynucleotide kinase (New England BioLabs, Ipswich, MA), according to the manufacturer's protocol. The oligonucleotides for the three-stranded and forked duplex substrates contained three phosphorothioated nucleotides at the 3′-end to block WRN exonucuclease activity. The three-stranded branch migration substrate (Fig. 2A) was prepared by first annealing equal molar amounts of radiolabeled oligonucleotide a (a) and unlabeled oligonucleotide b (b) to form a forked intermediate (supplemental Table 1) in 1 mm LiCl. Oligonucleotide c (c) (48 nm) was incubated with the a/b forked duplex (32 nm) in reaction buffer (40 mm Tris-HCl, pH 8.0, 4 mm MgCl2, 5 mm DTT, 100 μg/μl bovine serum albumin, and 20 mm ATP) for 15 min at 37 °C and then diluted 1:3 immediately before the addition of WRN.
FIGURE 2.
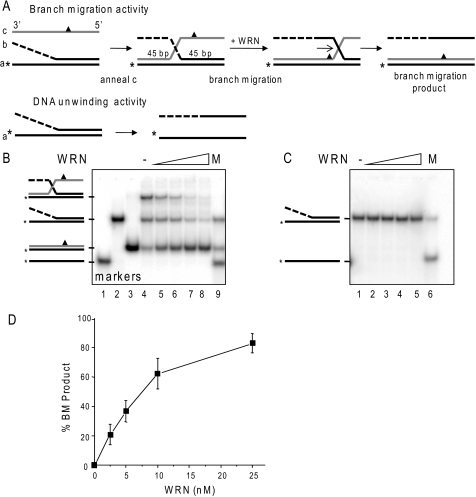
Comparison of WRN branch migration and DNA unwinding activities. A, schematic showing three-stranded branch migration and DNA unwinding of a forked duplex. The asterisk denotes a radiolabel, and the triangle indicates four mismatches used to limit spontaneous branch migration. The dashed line indicates ssDNA of non-complementary sequence. B, the three-stranded substrate-annealing reactions were incubated for 15 min at 37 °C, diluted 1:3 (10 nm labeled oligonucleotide), and immediately incubated with increasing WRN concentrations (0, 2.5, 5, 10, or 25 nm) (lanes 4–8, respectively) for 15 min under standard reaction conditions. The migration of DNA markers, ssDNA (lane 1), forked duplex (lane 2), BM duplex product (lane 3), or a mix of all three (M, lane 9) is shown. C, forked duplex (10 nm labeled oligonucleotide) was incubated with increasing WRN concentrations (0, 2.5, 5, 10, or 25 nm) for 15 min under standard reaction conditions. The migration of ssDNA is shown (M, lane 6). Reactions were run on an 8% native polyacrylamide gel and visualized by PhosphorImager analysis. D, the percentage of duplex BM product in B was calculated as described under “Experimental Procedures” and plotted against WRN concentration. Values represent the mean and S.D. from at least three independent experiments.
The plasmids used for making the 84-bp non-telomeric and telomeric D-loops were as previously described (38) and were purified by two rounds of ethidium bromide-saturated CsCl equilibrium gradient ultracentrifugation (Lofstrand Labs, Gaithersburg, MD). The plasmid-based D-loop substrates (Fig. 1B) were constructed as described previously (38). Briefly, RecA (4 μm) was incubated with the invading strand oligonucleotide (3.6 μm nucleotide) for 5 min at 37 °C, and then the supercoiled plasmid (300 μm nucleotides) was added and incubated for an additional 3 min. The reactions were terminated and deproteinized with proteinase K and SDS for 30 min, as described previously (38, 49). The D-loop constructs were purified by PAGE and electroelution and then concentrated and exchanged into storage buffer (10 mm Tris-HCl (pH 7.5), 10 mm MgCl2) using Micron-30 devices (Amicon). Purification quality and yields were determined by analysis on 4–20% native polyacrylamide gels, followed by visualization and quantitation with a Typhoon PhosphorImager and ImageQuant software (GE Healthcare).
Branch Migration and Exonuclease Reactions
The reactions were conducted in standard reaction buffer containing 40 mm Tris-HCl, pH 8.0, 4 mm MgCl2, 5 mm DTT, 100 μg/μl bovine serum albumin, and 2 mm ATP unless otherwise indicated. 14–28 ng/μl yeast tRNA was added to the reactions with mobile plasmid D-loops. The substrate and protein concentrations were as indicated in the figure legends. The substrates were preincubated with the various single strand binding proteins, and the reactions were initiated by adding wild type WRN or the X-WRN mutant protein, followed by incubation at 37 °C for 15 min, unless otherwise indicated. Reactions (10 μl) with the three-stranded and forked substrates were terminated with 3× stop dye (50) and run on 8% native polyacrylamide gels. Reactions (10 μl) with the plasmid mobile D-loops were terminated with 5 μl of 3× stop dye supplemented with 10 μg/ml proteinase K (50), deproteinized for 10 min at 37 °C, and separated on 4–20% native polyacrylamide gels. For analysis of the plasmid mobile D-loops reactions on 14% denaturing gels, the reactions were terminated with an equal volume of formamide stop dye (50). After drying the gels, the reactions were visualized using a Typhoon PhosphorImager and quantified using ImageQuant software (GE Healthcare).
For quantitation of total displaced ssDNA, the percentage of displaced products (full-length and shortened) was calculated as a function of the total radioactivity in the reaction lane (41). All values were corrected for background in the no enzyme control and heat-denatured substrate lanes.
RESULTS
WRN Catalyzes Three-stranded Branch Migration
RPA and POT1 are known to promote WRN DNA unwinding activity (42, 45). However, previous reports identified differences between DNA unwinding and BM activities (38–40). Such differences raise the possibility that RPA and POT1 may not necessarily stimulate WRN branch migration activity, as observed for RECQ1 and RPA (40). To test more directly for potential differences in these activities, we compared the ability of WRN to dissociate a 45-bp duplex by DNA unwinding activity versus branch migration activity in side by side reactions. For these studies, we prepared an oligomeric substrate similar to that used previously to demonstrate 3′ to 5′ BM activity of RAD54 and RECQ1 (40, 51). The branch-migrated duplex contains four mismatches (Fig. 2A, triangle), which minimize but do not eliminate spontaneous BM (Fig. 2B, lane 4), in agreement with previous reports (40). The annealing reaction time was kept to a minimum to prevent spontaneous BM (Fig. 2B, lane 4). Despite an excess of oligonucleotide c, not all of the forked duplex was driven into a three-stranded substrate during the annealing reaction (Fig. 2B, lane 4). Nevertheless, greater than 40% of the labeled oligonucleotide a was present in the three-stranded substrate. Under these conditions BM was monitored by conversion to duplex DNA in which oligonucleotides a and c were paired (Fig. 2, A and B, lane 3). In contrast, the product of unwinding the equivalent two-stranded forked duplex is ssDNA (Fig. 2, A and C).
WRN catalyzed BM in a dose-dependent manner and achieved nearly complete conversion of the three-stranded substrate to duplex DNA at the highest WRN concentration tested (Fig. 2, B and D). WRN BM activity was dependent on ATP hydrolysis (supplemental Fig. S1). In these reactions, we also noted a reduction in fork duplex at the higher WRN concentrations (Fig. 2B, lane 8). One possibility is that the annealing reaction of fork and oligonucleotide c was driven forward by WRN-mediated reduction in the three-stranded annealing product (Fig. 2A), thereby generating more three-stranded substrate that was then converted to duplex a/c DNA. In stark contrast, we observed no unwinding of the forked duplex even at the highest WRN concentrations (Fig. 2C) and no conversion of either the three-stranded substrate or two-stranded fork into unwound oligonucleotide a ssDNA product (Fig. 2, B and C). These data demonstrate that differences must exist in the mechanisms and product formation between WRN DNA unwinding and its branch migration activities.
RPA and POT1 Modulate WRN Processing of Telomeric Mobile D-loops
Although they are useful for examining BM, the three-stranded substrates are not suitable for testing RPA and POT1 promotion of BM activity. This substrate is non-telomeric and will not bind POT1 and contains an inherent forked region upstream of the BM region that is a known substrate for RPA stimulation of WRN unwinding (Fig. 2A). Therefore, we asked whether POT1 and RPA modulate WRN processing of more biologically relevant mobile D-loops.
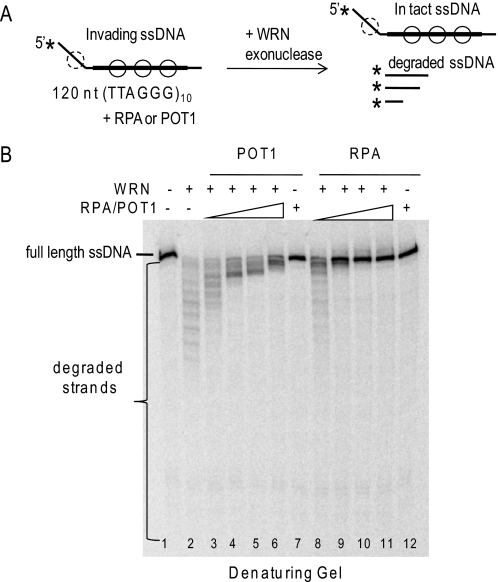
The assay used for these experiments involved RecA-catalyzed strand invasion of a 120-mer into negatively supercoiled plasmids to generate D-loops with a 5′ 36-nt ssDNA tail protruding from 84 bp of duplex DNA (Fig. 1B). The duplex region of the strand invasion contains the sequence (TTAGGG)10 flanked by 12 bp of unique sequence to ensure proper alignment of the repeats. RPA can preload on the 5′ ssDNA tail as well as the displaced ssDNA region of the plasmid. POT1 binds the consensus sequence (TTAGGGTTAG) (52, 53) and can preload only on the displaced ssDNA region of the plasmid (Fig. 3A).
FIGURE 3.
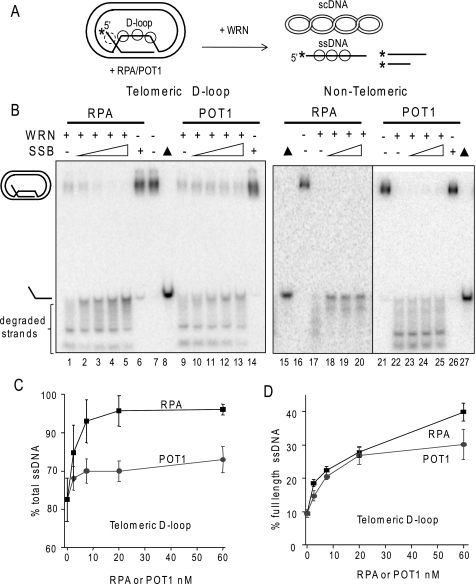
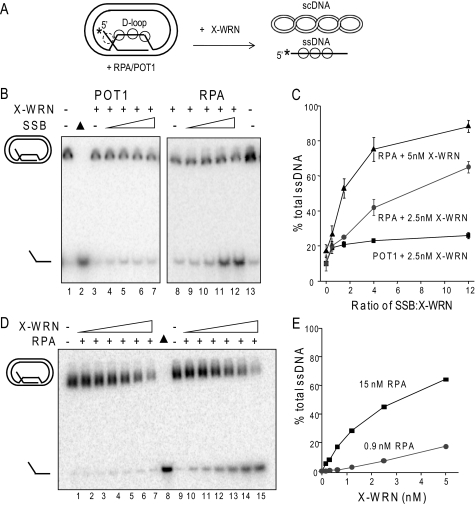
RPA and POT1 promote WRN release of full-length strands from mobile telomeric D-loops. A, a schematic is shown for the branch migration of mobile plasmid D-loops with 3′ invaded ssDNA and a 5′ ssDNA tail. The asterisk denotes a radiolabel, and scDNA indicates the supercoiled dsDNA product. Circles with solid lines indicate RPA or POT1 binding, and circles with dashed lines indicate only RPA binding for the telomeric D-loop (not to scale). B, the telomeric (lanes 1–14) or non-telomeric (lanes 15–27) mobile D-loops (50 pm) were incubated with 5 nm WRN and increasing concentrations (0, 2.5, 7.5, 20, or 60 nm) of either RPA or POT1 as indicated for 15 min under standard reaction conditions. Control reactions contained substrate with either 60 nm RPA (lane 6) or POT1 (lane 14). Reactions were run on a 4–20% native polyacrylamide gel and visualized by PhosphorImager analysis. ▲, heat denatured substrate. C, for the telomeric D-loop, the percentage of total displaced ssDNA was calculated as described under “Experimental Procedures” and plotted against RPA or POT1 concentration. Squares and black line, WRN and RPA; Circles and gray line, WRN and POT1. D, for the telomeric D-loop, the percentage of full-length ssDNA product was calculated as described under “Experimental Procedures” and plotted against RPA or POT1 concentrations. Filled squares and black line, WRN and RPA; filled circles and gray line, WRN and POT1. Values represent the mean and S.D. from at least three independent experiments.
WRN incubation with these mobile D-loops released invading strands of various lengths due to the combined action of WRN BM activity and WRN 3′ to 5′ exonucleolytic degradation of the invading strands (Fig. 3B, lanes 1 and 9) (38). Preloading the telomeric D-loops with either RPA or POT1 prior to the addition of WRN altered the product distribution in two ways. First, both RPA and POT1 increased the percentage of total D-loops disrupted by WRN (Fig. 3C). The maximal increase in total ssDNA displacement was slight for POT1 (12% increase) but was more pronounced for RPA (24% increase). Second, and more strikingly, preloading the D-loops with either RPA or POT1 led to a dose-dependent increase in the release of full-length strands and a decrease in degraded ssDNA products (Fig. 3, B and D). RPA increased the percentage of full-length ssDNA products by 76% (4.2-fold), whereas POT1 mediated a 68% increase (3.2-fold). RPA also strongly promoted WRN release of full-length strands from non-telomeric D-loops, whereas POT1 did not (Fig. 3B, lanes 15–27), confirming that POT1 must bind the substrate to modulate WRN exonuclease activity. These results indicate that RPA and POT1 limit the extent of WRN degradation of the invading strand of biologically relevant mobile D-loops.
Loading of RPA and POT1 on the Invading ssDNA Blocks Progression of the WRN Exonuclease
POT1 and RPA physical interaction with WRN does not alter its exonuclease catalytic activity (45, 46, 50). However, we showed previously that RPA and POT1 suppress WRN degradation of DNA strands in the context of oligomeric telomeric forks and static D-loops by promoting more rapid unwinding of the structures (41, 45, 50). Once unwound, these strands are no longer substrates for WRN digestion, because WRN does not effectively degrade short strands (<50 nt) and can only digest long strands (46, 54). However, the plasmid mobile D-loops differ from these oligomeric constructs in that 1) they are disrupted by branch migration, and 2) the invading strand is very long (120 nt), so it is effectively degraded by the WRN exonuclease activity after release from the D-loop (38). Therefore, RPA and POT1 may suppress WRN degradation of the invading strand in the mobile D-loops by two different mechanisms. First, RPA and POT1 loading on the ssDNA tails as it is being displaced might block the progression of the WRN exonuclease. Second, RPA and POT1 preloading on the bulging ssDNA in the “loop” region of the plasmid D-loop (Fig. 5A) might interfere with WRN digestion of the invading strand. POT1 could not preload on previously tested oligomeric static D-loops, which were short (33 nt) and non-telomeric (45).
FIGURE 5.
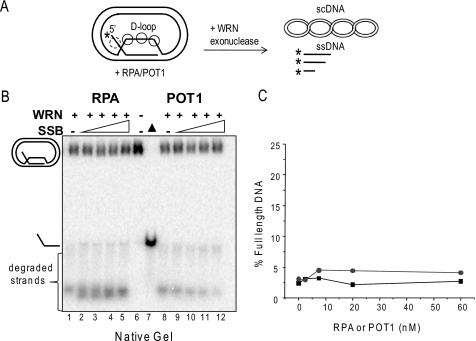
RPA and POT1 fail to alter WRN processing of mobile D-loops in the absence of branch migration activity. A, schematic of mobile D-loop disruption by WRN exonucleolytic degradation of the invading strand. The asterisk denotes a radiolabel, and scDNA indicates supercoiled dsDNA. Circles with solid lines represent either RPA or POT1 binding, and circles with dashed lines represent only RPA binding. B, the telomeric mobile D-loop (50 pm) was incubated with 5 nm WRN and increasing concentrations (0, 2.5, 7.5, 20, or 60 nm) of either RPA (lanes 1–5) or POT1 (lanes 8–12) for 15 min under standard reaction conditions except that 2 mm ATP was replaced with 2 mm ATPγS. Reactions were run on a 4–20% native polyacrylamide gel and visualized by PhosphorImager analysis. ▲, heat-denatured substrate. C, the percentage of full-length ssDNA products was calculated as described under “Experimental Procedures” and plotted against RPA or POT1 concentration. Squares and black lines, WRN and RPA; circles and gray line, WRN and POT1.
To test the first possibility, WRN was incubated with the 120-mer oligonucleotide containing 10 telomeric repeats prior to incorporation into the D-loop (Fig. 4A) (38). WRN degrades the 120-mer, as indicated by the appearance of shortened products on a denaturing gel (Fig. 4B, lane 2). Preloading with RPA or POT1 led to a dose-dependent inhibition of WRN degradation, as indicated by the loss of shortened fragments and increase in full-length oligonucleotides (Fig. 4B, lanes 3–12). POT1 is less effective than RPA in blocking the WRN exonuclease progression, because POT1 can only bind the 60-nt region of telomeric sequence in the oligonucleotide. These data indicate that coating of the released ssDNA strand by either RPA or POT1 protects against WRN digestion.
FIGURE 4.
WRN degradation of telomeric ssDNA is inhibited by RPA and POT1 loading. A, schematic of WRN exonuclease activity on the 120-nt ssDNA strand after release from the D-loop. The thick line represents the (TTAGGG)10 repeats, and the asterisk denotes the radiolabel. Circles with solid lines represent either RPA or POT1 binding, and circles with dashed lines represent only RPA binding. B, reactions contained 50 pm free telomeric ssDNA. The substrate was incubated with 5 nm WRN alone (lane 2) or with increasing concentrations (2.5, 7.5, 20, or 60 nm) of either POT1 (lanes 3–6) or RPA (lanes 8–11) for 15 min under standard reaction conditions. Control reactions contained substrate with either 60 nm POT1 (lane 7) or RPA (lane 12). Reactions were run on 14% polyacrylamide denaturing gels and visualized by PhosphorImager analysis.
Next we tested whether RPA and POT1 preloading on the bulging ssDNA in the “loop” region of the plasmid D-loop can interfere with WRN digestion of the invading strand (Fig. 5A). In other words, can RPA and POT1 inhibit WRN exonuclease in the absence of WRN-mediated BM of the mobile D-loop? To test this, the reactions were performed with APTγS, an ATP analog that is not efficiently hydrolyzed by WRN protein (55), because WRN BM activity requires ATP hydrolysis. Under these reaction conditions, WRN disrupts D-loops by degrading the invading strand duplex to thermally unstable lengths, thereby promoting release of very short strands (<48 nt) (Fig. 5, A and B, lanes 1 and 8) (38). The reactions were run on native gels to visualize whether the degraded ssDNA invading strands were released from the D-loop. Preloading the telomeric D-loops with either RPA or POT1 did not significantly increase the percentage of disrupted D-loops or the percentage of full-length ssDNA products in the absence of WRN BM activity (Fig. 5), in stark contrast to the reactions with WRN BM activity (Fig. 3B). Since the degraded ssDNA products are too short to be efficiently degraded by WRN (54), this precludes any protective effect of RPA or POT1 binding onto the released ssDNA (Fig. 5A). Similar results were observed with RPA and WRN on a non-telomeric D-loop of similar size (data not shown). The data confirm that POT1 and RPA do not alter WRN catalytic exonuclease activity and only suppress digestion of the invading strands when WRN BM is active.
RPA, but Not POT1, Robustly Stimulates WRN Branch Migration Activity
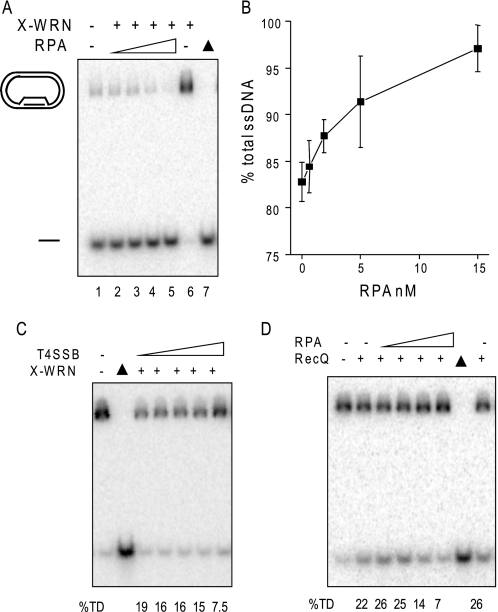
Although POT1 increased the percentage of full-length ssDNA products at the expense of degraded products, the percentage of total ssDNA product was only weakly increased, compared with RPA (Fig. 3C). To better examine POT1 and RPA modulation of WRN BM activity on telomeric mobile D-loops, the exonuclease was inactivated by mutating a single residue in the exonuclease domain (E84A, X-WRN) (35). Less D-loop is disrupted in the absence of exonuclease activity (Fig. 6A, lanes 3 and 8) (38). Preincubation of the D-loops with POT1 led to a slight increase in the percentage of D-loops disrupted by 2.5 nm X-WRN (up to 2.6-fold) that began to plateau at a 1.5-fold molar excess of POT1 over X-WRN (Fig. 6C). An N-terminal fragment of telomeric protein TPP1 (TPP1-N) was found to greatly enhance the ability of POT1 to increase telomerase processivity (48). However, TPP1-N did not alter POT1 effects on WRN BM or exonuclease activities on the telomeric mobile D-loop (supplemental Fig. S2).
FIGURE 6.
RPA robustly stimulates WRN branch migration activity. A, a schematic shows branch migration of the telomeric mobile D-loop with 3′ invaded ssDNA and a 5′ ssDNA tail in the absence of WRN exonuclease activity. The asterisk denotes a radiolabel, and scDNA indicates the supercoiled dsDNA product. Circles with solid lines indicate RPA or POT1 binding, and circles with dashed lines indicate RPA binding only to the telomeric D-loop (not to scale). B, the telomeric mobile D-loop was incubated with either 2.5 nm WRN exonuclease-dead variant (X-WRN) and increasing concentrations (1.3, 3.8, 10, or 30 nm) of either POT1 (lanes 4–7) or RPA (lanes 9–12). Reactions were conducted under standard conditions for 15 min and run on 4–20% polyacrylamide native gels. C, quantitation of reactions in A. The percentage of total ssDNA product was calculated as described under “Experimental Procedures” and plotted against the molar ratio of either POT1 or RPA to X-WRN. Squares and black line, 2.5 nm X-WRN and POT1; circles and gray line, 2.5 nm X-WRN and RPA; triangles and black line, 5 nm X-WRN and RPA. Values represent the mean and S.D. from at least three independent experiments. D, the telomeric mobile D-loop (50 pm) was incubated with either 0.9 nm RPA (lanes 2–7) or 15 nm RPA (lanes 11–16) and increasing concentrations of X-WRN (0.15, 0.31, 0.62, 1.2, 2.5, or 5 nm). Reactions were conducted under standard reaction conditions for 15 min and run on a 4–20% native polyacrylamide gel. ▲, heat-denatured substrate. E, the percentage of total ssDNA product was calculated as described under “Experimental Procedures” and plotted against WRN concentration. Black square, WRN and 15 nm RPA; gray circle, WRN and 0.9 nm RPA.
In contrast to the results with POT1, preincubating the D-loops with RPA led to a strong stimulation of X-WRN (2.5 nm) BM activity (up to a 6.5-fold increase) that was dose-dependent (Fig. 6C). Nearly complete disruption of the D-loops (88%) was achieved by increasing the amount of both X-WRN (5 nm) and RPA (up to 60 nm) (Fig. 7C). In contrast, higher amounts of X-WRN and POT1 did not significantly enhance D-loop disruption (data not shown). The percentage of ssDNA product was dependent on both X-WRN and RPA concentrations (Fig. 6, D and E). The addition of very low RPA concentrations (0.9 nm) did not significantly alter the percentage of ssDNA products from that achieved by X-WRN alone (average of 0.6, 1.6, 6.2, and 17% for 0.62, 1.2, 2.5, and 5 nm X-WRN (38)) (Fig. 6, D and E). However, higher levels of RPA (15 nm) promoted D-loop branch migration at all X-WRN concentrations tested, including very low X-WRN amounts (Figs. 6, D and E). For example, at 1.2 nm X-WRN, the addition of 15 nm RPA imparted a 28-fold increase in total ssDNA product.
FIGURE 7.
Specificity of RPA stimulation of WRN branch migration activity. A, RPA stimulation of WRN on D-loops lacking a tail. The 5′ no tail mobile non-telomeric D-loop (38) (50 nm) was incubated with 1.2 nm X-WRN and increasing concentrations of RPA (0, 0.62, 1.9, 5, or 5 nm) (lanes 1–5, respectively) for 15 min under standard reaction conditions. The reactions were run on a 4–20% native gel. B, quantitation of the reactions in A were calculated as described under “Experimental Procedures” and plotted against RPA concentration. Values represent the mean and S.D. from two or three independent reactions. C and D, the telomeric mobile D-loop (50 pm) was incubated with either 5 nm X-WRN and increasing concentrations of T4 single strand binding protein (0, 2.5, 7.5, 20, or 60 nm) (C) or 1.2 nm RecQ and increasing concentrations of RPA (0, 0.62, 1.9, 5, or 15 nm) (D). Reactions were conducted under standard conditions for 15 min, run on a 4–20% native polyacrylamide gel, and visualized by PhosphorImager analysis. ▲, heat-denatured substrate. The percentage of total ssDNA displacement (%TD) was calculated as described under “Experimental Procedures.”
Next, we examined the substrate and species specificity for RPA stimulation of WRN BM. We asked whether RPA binding to the protruding 5′ ssDNA tail of the telomeric D-loop was required to promote WRN BM of mobile D-loops, especially since POT1 cannot bind to this tail (Fig. 1B). For this, we tested a non-telomeric mobile D-loop that lacks protruding ssDNA tails, which is effectively disrupted by WRN BM, as shown previously (Fig. 7A, lane 1) (38). Preincubation of the no tail D-loop with RPA led to a dose-dependent increase in the percentage of ssDNA products generated by X-WRN BM activity that reached nearly 100% D-loop disruption (Fig. 7, A and B). To determine the specificity of the RPA stimulation of WRN BM activity, we tested other single strand DNA binding proteins. T4 phage single strand binding protein (T4 gene 32 protein, T4 SSB) did not stimulate X-WRN BM activity (Fig. 7C), and RPA did not stimulate E. coli RecQ helicase BM activity (Fig. 7D). X-WRN and RecQ activity were actually inhibited at the highest T4 SSB and RPA concentrations, respectively (Fig. 7, C and D). Thus, RPA, but not POT1, imparts a robust stimulation on WRN BM activity that is species-specific and not dependent on a protruding ssDNA tail from the D-loop.
DISCUSSION
Human RecQ helicases are crucial for maintaining genomic integrity, and cellular evidence indicates that they function in DNA replication and HR repair pathways (4, 11). Single strand binding proteins RPA and telomere-specific POT1 proteins interact with the WRN RecQ helicase in vivo and greatly enhance the ability of WRN to unwind duplex DNA in vitro (42, 45). However, we observed differences in WRN displacement of a duplex DNA by DNA unwinding versus branch migration activity (Fig. 2), raising the possibility that RPA and POT1 may not necessarily also stimulate WRN BM activity. To test this, BM activity was examined on biologically relevant mobile telomeric D-loops that are important intermediates in HR and at telomeric ends (Fig. 1). Both RPA and POT1protect the invading strand of mobile D-loops from excessive degradation by the WRN exonuclease, thereby favoring release of full-length strands at the expense of degraded products. We report the novel finding that RPA robustly stimulates WRN BM activity and increases the percentage of disrupted D-loops and total ssDNA product. To our knowledge, this is the first report that RPA can stimulate BM activity. In contrast, POT1 only imparted a weak increase in WRN BM activity that was not enhanced by the POT1 binding partner TPP1. Our results are consistent with cellular data that support roles for RPA and WRN in HR and, conversely, support roles for POT1 in limiting inappropriate recombination and branch migration at telomeric ends.
RPA stimulation of WRN BM activity is specific. RPA and T4 SSB did not stimulate branch migration activity of E. coli RecQ and WRN, respectively, but rather inhibited the activity of these helicases at high concentration (Fig. 7, C and D). In contrast, no inhibitory effect was detected with POT1 even when the binding partner TPP1 was added at high concentrations, although the stimulation of BM activity was weak (Figs. 6 and supplemental Fig. S2). However, POT1 is a modular protein that is predicted to exist in a variety of complexes at the telomeric end (56). Therefore, we cannot rule out the possibility that a POT1 complex with other telomeric proteins might further enhance or inhibit WRN BM of telomeric D-loops. For example, a telomere-specific RPA-like complex has been identified in Saccharomyces cerevisiae that consists of Cdc13 (POT1 homolog), Stn1, and Ten1 (57). The human homolog of Stn1 (OBFC1) has recently been identified and found to associate with POT1 partner TPP1 (58), raising the possibility that telomere-specific RPA-like trimers may also exist in humans. Further studies will be required to determine the protein specificity for the robust stimulation of WRN BM by RPA. For example, although RPA interacts with and stimulates DNA unwinding by RECQ1 (59), it does not stimulate RECQ1 BM of mobile D-loops (40). Other RecQ helicases remain to be tested. FANCM and RAD54 also branch-migrate plasmid-based mobile D-loops, and neither was stimulated by RPA preloading on the D-loop substrates (51, 60). Thus, the ability of RPA to stimulate BM activity of mammalian proteins may be confined to a select number.
We propose that RPA stimulates WRN DNA unwinding and WRN BM activity by different mechanisms. First, RPA activates WRN unwinding of long duplexes that cannot be unwound by WRN alone (as short as 42 bp) (42, 43), indicating that RPA greatly enhances the apparent helicase processivity of unwinding. However, WRN has a much higher apparent processivity during BM because it can disrupt an 84-bp mobile D-loop and can migrate a junction through >2700 bp in a recombination-like α structure (37, 38). Since WRN BM of long duplexes does not require RPA, it is unlikely that RPA stimulates WRN BM by increasing the apparent processivity. Second, earlier models (61, 62) suggested that RPA may partly stimulate WRN DNA unwinding by preventing the reannealing of partially unwound strands if WRN prematurely dissociates from the substrate. But for BM activity, SSBs are not needed to prevent strand reannealing. As WRN promotes BM through the mobile D-loop, the unpaired plasmid strands anneal and the D-loop “bubble” shrinks. Thus, if WRN dissociates prematurely, the plasmid strand is not available for reannealing with the partially separated invading strand. Hence, RPA stimulation of WRN BM activity may be through the recruitment of WRN to the mobile D-loop. An N-terminal region of the RPA large subunit (hRPA70) binds WRN and is sufficient to stimulate WRN helicase (61, 62). RPA binds to a motif of acidic amino acids in the WRN N terminus with high affinity, and a WRN protein fragment that lacks this motif displays reduced helicase activity compared with full-length WRN on long duplex substrates (50–100 bp) that require RPA for unwinding. This indicates that the high affinity physical interaction between RPA and WRN contributes to the stimulation of DNA unwinding and probably also contributes to the stimulation of BM activity. Thus, we propose that the primary mechanism for RPA stimulation of WRN BM activity is not by preventing strand reannealing but rather by physically interacting with WRN and helping to recruit WRN to the substrate.
Why is POT1 less effective in stimulating WRN BM compared with WRN DNA unwinding activity? Both RPA and POT1 were shown to stimulate X-WRN and BLM unwinding of telomeric oligomeric forks and D-loops (45). POT1 caused up to a 10-fold increase in the percentage of telomeric forks unwound by X-WRN in a dose-dependent manner (45). We showed previously that the mechanism for POT1 stimulation of WRN DNA unwinding is by POT1 binding the partially unwound strands to prevent strand reannealing and not by POT1 recruiting or retaining WRN on the substrate (46). In contrast, we observed only a weak stimulation of WRN BM activity by POT1 that was not dose-dependent (Figs. 3 and 6). As noted above, it is less critical for SSBs to hold partially separated strands apart during BM versus DNA unwinding, because the partially released invading ssDNA of the mobile D-loop is less likely to reanneal with the complementary plasmid strand. Given that POT1 does not recruit or retain WRN on forked substrates (46), we predict that POT1 is probably less effective at recruiting WRN to mobile telomeric D-loops than RPA, which may explain the lack of robust BM stimulation. Less is known about the physical interaction and binding affinity for WRN and POT1, compared with WRN and RPA. The WRN domain(s) that POT1 binds to remain to be mapped. Purified WRN binds to both full-length POT1 and a truncated variant that has the ssDNA binding domain but lacks the C terminus (45). However, the POT1 C terminus is required to bind WRN in vivo (45). Future studies that determine the affinity and map the WRN and POT1 interaction may help elucidate mechanistic differences when compared with the WRN and RPA physical and functional interaction.
Although RPA and POT1 differ in their ability to modulate WRN BM activity, both proteins protect mobile D-loops and ssDNA from excessive degradation by WRN exonuclease activity. POT1 and RPA favor WRN displacement of intact full-length ssDNA from mobile D-loops and decrease the percentage of degraded products (Fig. 3). In vivo POT1 protects the protruding 3′ ssDNA tail that occurs at telomere ends and prevents aberrant recombination at telomeres (63, 64). It is clear that the mechanism of WRN exonuclease inhibition requires BM activity and that RPA and POT1 physical interaction with WRN does not affect the catalytic exonuclease activity (Fig. 5). One possible mechanism of inhibition is that BM converts the D-loops to a product that is not a substrate for WRN exonuclease activity. For example, RPA and POT1 suppress WRN degradation of short forked duplexes by increasing the rate of duplex unwinding and conversion to unwound ssDNA products that are too short to be further degraded by WRN exonuclease (45, 50). However, the released invading ssDNA from the mobile D-loops are much longer (120 nt) and are effectively digested by WRN (38), so BM products are substrates for WRN digestion, making the above inhibition model unlikely. Another possibility involves BM generation of new ssDNA binding sites for RPA and POT1 on the partially and completely released invading strand of the mobile D-loop that are not present without BM activity. We show that RPA and POT1 loading block WRN digestion of the ssDNA product (Fig. 4), but given that WRN digests the invading strand both before and after complete release from the D-loop (38), it is also possible that these proteins impede WRN exonuclease progression on partially displaced invading strands. We propose that POT1 and RPA can preload on the plasmid ssDNA region of the telomeric D-loop (Fig. 3A) and may be transferred to the invading ssDNA as it is released by WRN BM activity, thereby impeding the exonuclease progression. Consistent with this, POT1 cannot bind non-telomeric D-loops and does not alter WRN degradation of this substrate (Fig. 3). Indeed, SV40 T antigen has been shown to actively load RPA on the unwound ssDNA strand as it is revealed (65). However, although the RPA and POT1 blockage of WRN exonuclease progression is interesting, it is not required for RPA stimulation of WRN BM activity (Fig. 6).
What is the biological relevance of the robust stimulation of WRN BM activity by RPA and the much weaker stimulation by POT1? RPA is an active and required component of the homologous recombination pathway (47). RPA physically interacts with RAD51, and this interaction promotes strand exchange, which drives the initial D-loop formation step of HR (Fig. 1) (66). Cellular evidence indicates that WRN protein functions in the recovery of stalled or collapsed replication forks by dissociating homologous recombination intermediates that are generated to restart the replication fork (16, 18, 67, 68). WRN has been shown to co-localize with RPA after the induction of replication fork arrest by dNTP depletion or DNA lesions that block replication forks (37, 69). Thus, RPA stimulation of WRN branch migration could promote the dissociation of the HR intermediates to complete repair. However, stimulation of BM at telomeric ends can have dire cellular consequences and lead to telomere loss (24). Improper processing of the natural telomere t-loop/D-loop by Holliday junction resolvases causes cleavage of the t-loop and telomere loss (23). In yeast, the absence of POT1 promotes telomere loss (70) and chromosome fusions by a synthesis-dependent strand-annealing pathway that is dependent on RPA and the RecQ helicase homolog (71). Thus, the lack of POT at telomeres may allow for more binding by RPA and the inappropriate stimulation of branch migration by WRN and possibly other RecQ helicases. Although POT1 may promote WRN helicase to unwind alternate DNA structures (45), we show here that POT1 also may act to prevent enhanced or untimely WRN branch migration at telomeric ends (Fig. 6). Although RPA may be generally more abundant in the cell, POT1 is enriched and more abundant at the telomeres due to an interaction with the shelterin complex (31). The biological consequences of preloading the telomeric tail with either RPA or POT1 will be influenced by how these proteins physically and functionally interact with DNA-processing enzymes, such as WRN protein.
Acknowledgments
We thank J. Keck for E. coli RecQ helicase protein, M. Lei for the TTP1 expression construct, and the Opresko laboratory for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants ES0515052 (to P. L. O.), GM065484 and CA092584 (to W. J. C.), and K99ES016758-01 (to H. W.). This work was also supported by a grant from the Ellison Medical Foundation (to P. L. O.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. S1 and S2.
- HR
- homologous recombination
- SSB
- single strand binding protein
- ssDNA
- single-stranded DNA
- dsDNA
- double-stranded DNA
- RPA
- replication protein A
- DSB
- double strand break
- ALT
- alternative lengthening of telomeres
- BM
- branch migration
- DTT
- dithiothreitol
- nt
- nucleotide
- APTγS
- adenosine 5′-O-(thiotriphosphate).
REFERENCES
- 1.Kudlow B. A., Kennedy B. K., Monnat R. J., Jr. (2007) Nat. Rev. Mol. Cell Biol. 8, 394–404 [DOI] [PubMed] [Google Scholar]
- 2.Yu C. E., Oshima J., Fu Y. H., Wijsman E. M., Hisama F., Alisch R., Matthews S., Nakura J., Miki T., Ouais S., Martin G. M., Mulligan J., Schellenberg G. D. (1996) Science 272, 258–262 [DOI] [PubMed] [Google Scholar]
- 3.Bachrati C. Z., Hickson I. D. (2003) Biochem. J. 374, 577–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brosh R. M., Jr., Bohr V. A. (2007) Nucleic Acids Res. 35, 7527–7544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ellis N. A., Groden J., Ye T. Z., Straughen J., Lennon D. J., Ciocci S., Proytcheva M., German J. (1995) Cell 83, 655–666 [DOI] [PubMed] [Google Scholar]
- 6.Kitao S., Shimamoto A., Goto M., Miller R. W., Smithson W. A., Lindor N. M., Furuichi Y. (1999) Nat. Genet. 22, 82–84 [DOI] [PubMed] [Google Scholar]
- 7.Siitonen H. A., Kopra O., Kääriäinen H., Haravuori H., Winter R. M., Säämänen A. M., Peltonen L., Kestilä M. (2003) Hum. Mol. Genet. 12, 2837–2844 [DOI] [PubMed] [Google Scholar]
- 8.Sharma S., Stumpo D. J., Balajee A. S., Bock C. B., Lansdorp P. M., Brosh R. M., Jr., Blackshear P. J. (2007) Mol. Cell. Biol. 27, 1784–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu Y., Raynard S., Sehorn M. G., Lu X., Bussen W., Zheng L., Stark J. M., Barnes E. L., Chi P., Janscak P., Jasin M., Vogel H., Sung P., Luo G. (2007) Genes Dev. 21, 3073–3084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sung P., Klein H. (2006) Nat. Rev. Mol. Cell Biol. 7, 739–750 [DOI] [PubMed] [Google Scholar]
- 11.Wu L., Hickson I. D. (2006) Annu. Rev. Genet. 40, 279–306 [DOI] [PubMed] [Google Scholar]
- 12.Heller R. C., Marians K. J. (2006) Nat. Rev. Mol. Cell Biol. 7, 932–943 [DOI] [PubMed] [Google Scholar]
- 13.Tarsounas M., West S. C. (2005) Cell Cycle 4, 672–674 [DOI] [PubMed] [Google Scholar]
- 14.Sung P. (1994) Science 265, 1241–1243 [DOI] [PubMed] [Google Scholar]
- 15.McIlwraith M. J., Vaisman A., Liu Y., Fanning E., Woodgate R., West S. C. (2005) Mol. Cell 20, 783–792 [DOI] [PubMed] [Google Scholar]
- 16.Saintigny Y., Makienko K., Swanson C., Emond M. J., Monnat R. J., Jr. (2002) Mol. Cell. Biol. 22, 6971–6978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dhillon K. K., Sidorova J., Saintigny Y., Poot M., Gollahon K., Rabinovitch P. S., Monnat R. J., Jr. (2007) Aging Cell 6, 53–61 [DOI] [PubMed] [Google Scholar]
- 18.Sidorova J. M., Li N., Folch A., Monnat R. J., Jr. (2008) Cell Cycle 7, 796–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crabbe L., Verdun R. E., Haggblom C. I., Karlseder J. (2004) Science 306, 1951–1953 [DOI] [PubMed] [Google Scholar]
- 20.Bai Y., Murnane J. P. (2003) Hum. Genet. 113, 337–347 [DOI] [PubMed] [Google Scholar]
- 21.Laud P. R., Multani A. S., Bailey S. M., Wu L., Ma J., Kingsley C., Lebel M., Pathak S., DePinho R. A., Chang S. (2005) Genes Dev. 19, 2560–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li B., Jog S. P., Reddy S., Comai L. (2008) Mol. Cell. Biol. 28, 1892–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang R. C., Smogorzewska A., de Lange T. (2004) Cell 119, 355–368 [DOI] [PubMed] [Google Scholar]
- 24.de Lange T. (2005) Genes Dev. 19, 2100–2110 [DOI] [PubMed] [Google Scholar]
- 25.Griffith J. D., Comeau L., Rosenfield S., Stansel R. M., Bianchi A., Moss H., de Lange T. (1999) Cell 97, 503–514 [DOI] [PubMed] [Google Scholar]
- 26.Opresko P. L. (2007) Mech. Ageing Dev. 128, 423–436 [DOI] [PubMed] [Google Scholar]
- 27.Gilson E., Géli V. (2007) Nat. Rev. Mol. Cell Biol. 8, 825–838 [DOI] [PubMed] [Google Scholar]
- 28.Machwe A., Xiao L., Theodore S., Orren D. K. (2002) J. Biol. Chem. 277, 4492–4504 [DOI] [PubMed] [Google Scholar]
- 29.Brosh R. M., Jr., Waheed J., Sommers J. A. (2002) J. Biol. Chem. 277, 23236–23245 [DOI] [PubMed] [Google Scholar]
- 30.Mohaghegh P., Karow J. K., Brosh R. M., Jr., Bohr V. A., Hickson I. D. (2001) Nucleic Acids Res. 29, 2843–2849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garcia P. L., Liu Y., Jiricny J., West S. C., Janscak P. (2004) EMBO J. 23, 2882–2891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharma S., Sommers J. A., Choudhary S., Faulkner J. K., Cui S., Andreoli L., Muzzolini L., Vindigni A., Brosh R. M., Jr. (2005) J. Biol. Chem. 280, 28072–28084 [DOI] [PubMed] [Google Scholar]
- 33.Popuri V., Bachrati C. Z., Muzzolini L., Mosedale G., Costantini S., Giacomini E., Hickson I. D., Vindigni A. (2008) J. Biol. Chem. 283, 17766–17776 [DOI] [PubMed] [Google Scholar]
- 34.Xu X., Liu Y. (2009) EMBO J. 28, 568–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang S., Li B., Gray M. D., Oshima J., Mian I. S., Campisi J. (1998) Nat. Genet. 20, 114–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Machwe A., Xiao L., Lloyd R. G., Bolt E., Orren D. K. (2007) Nucleic Acids Res. 35, 5729–5747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Constantinou A., Tarsounas M., Karow J. K., Brosh R. M., Bohr V. A., Hickson I. D., West S. C. (2000) EMBO Rep. 1, 80–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Opresko P. L., Sowd G., Wang H. (2009) PLoS ONE 4, e4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bachrati C. Z., Borts R. H., Hickson I. D. (2006) Nucleic Acids Res. 34, 2269–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bugreev D. V., Brosh R. M., Jr., Mazin A. V. (2008) J. Biol. Chem. 283, 20231–20242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Opresko P. L., Otterlei M., Graakjaer J., Bruheim P., Dawut L., Kølvraa S., May A., Seidman M. M., Bohr V. A. (2004) Mol. Cell 14, 763–774 [DOI] [PubMed] [Google Scholar]
- 42.Brosh R. M., Jr., Orren D. K., Nehlin J. O., Ravn P. H., Kenny M. K., Machwe A., Bohr V. A. (1999) J. Biol. Chem. 274, 18341–18350 [DOI] [PubMed] [Google Scholar]
- 43.Brosh R. M., Jr., Li J. L., Kenny M. K., Karow J. K., Cooper M. P., Kureekattil R. P., Hickson I. D., Bohr V. A. (2000) J. Biol. Chem. 275, 23500–23508 [DOI] [PubMed] [Google Scholar]
- 44.Cui S., Klima R., Ochem A., Arosio D., Falaschi A., Vindigni A. (2003) J. Biol. Chem. 278, 1424–1432 [DOI] [PubMed] [Google Scholar]
- 45.Opresko P. L., Mason P. A., Podell E. R., Lei M., Hickson I. D., Cech T. R., Bohr V. A. (2005) J. Biol. Chem. 280, 32069–32080 [DOI] [PubMed] [Google Scholar]
- 46.Sowd G., Lei M., Opresko P. L. (2008) Nucleic Acids Res. 36, 4242–4256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sakaguchi K., Ishibashi T., Uchiyama Y., Iwabata K. (2009) FEBS J. 276, 943–963 [DOI] [PubMed] [Google Scholar]
- 48.Wang F., Podell E. R., Zaug A. J., Yang Y., Baciu P., Cech T. R., Lei M. (2007) Nature 445, 506–510 [DOI] [PubMed] [Google Scholar]
- 49.Bachrati C. Z., Hickson I. D. (2006) Methods Enzymol. 409, 86–100 [DOI] [PubMed] [Google Scholar]
- 50.Opresko P. L., Laine J. P., Brosh R. M., Jr., Seidman M. M., Bohr V. A. (2001) J. Biol. Chem. 276, 44677–44687 [DOI] [PubMed] [Google Scholar]
- 51.Bugreev D. V., Hanaoka F., Mazin A. V. (2007) Nat. Struct. Mol. Biol. 14, 746–753 [DOI] [PubMed] [Google Scholar]
- 52.Lei M., Podell E. R., Cech T. R. (2004) Nat. Struct. Mol. Biol. 11, 1223–1229 [DOI] [PubMed] [Google Scholar]
- 53.Loayza D., Parsons H., Donigian J., Hoke K., de Lange T. (2004) J. Biol. Chem. 279, 13241–13248 [DOI] [PubMed] [Google Scholar]
- 54.Machwe A., Xiao L., Orren D. K. (2006) BMC Mol. Biol. 7, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kamath-Loeb A. S., Shen J. C., Loeb L. A., Fry M. (1998) J. Biol. Chem. 273, 34145–34150 [DOI] [PubMed] [Google Scholar]
- 56.Liu D., O'Connor M. S., Qin J., Songyang Z. (2004) J. Biol. Chem. 279, 51338–51342 [DOI] [PubMed] [Google Scholar]
- 57.Gao H., Cervantes R. B., Mandell E. K., Otero J. H., Lundblad V. (2007) Nat. Struct. Mol. Biol. 14, 208–214 [DOI] [PubMed] [Google Scholar]
- 58.Wan M., Qin J., Songyang Z., Liu D. (2009) J. Biol. Chem. 284, 26725–26731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cui S., Arosio D., Doherty K. M., Brosh R. M., Jr., Falaschi A., Vindigni A. (2004) Nucleic Acids Res. 32, 2158–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gari K., Décaillet C., Delannoy M., Wu L., Constantinou A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 16107–16112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Doherty K. M., Sommers J. A., Gray M. D., Lee J. W., von Kobbe C., Thoma N. H., Kureekattil R. P., Kenny M. K., Brosh R. M., Jr. (2005) J. Biol. Chem. 280, 29494–29505 [DOI] [PubMed] [Google Scholar]
- 62.Shen J. C., Lao Y., Kamath-Loeb A., Wold M. S., Loeb L. A. (2003) Mech. Ageing Dev. 124, 921–930 [DOI] [PubMed] [Google Scholar]
- 63.Denchi E. L., de Lange T. (2007) Nature 448, 1068–1071 [DOI] [PubMed] [Google Scholar]
- 64.Hockemeyer D., Daniels J. P., Takai H., de Lange T. (2006) Cell 126, 63–77 [DOI] [PubMed] [Google Scholar]
- 65.Jiang X., Klimovich V., Arunkumar A. I., Hysinger E. B., Wang Y., Ott R. D., Guler G. D., Weiner B., Chazin W. J., Fanning E. (2006) EMBO J. 25, 5516–5526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stauffer M. E., Chazin W. J. (2004) J. Biol. Chem. 279, 25638–25645 [DOI] [PubMed] [Google Scholar]
- 67.Franchitto A., Pirzio L. M., Prosperi E., Sapora O., Bignami M., Pichierri P. (2008) J. Cell Biol. 183, 241–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pirzio L. M., Pichierri P., Bignami M., Franchitto A. (2008) J. Cell Biol. 180, 305–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sakamoto S., Nishikawa K., Heo S. J., Goto M., Furuichi Y., Shimamoto A. (2001) Genes Cells 6, 421–430 [DOI] [PubMed] [Google Scholar]
- 70.Baumann P., Cech T. R. (2001) Science 292, 1171–1175 [DOI] [PubMed] [Google Scholar]
- 71.Wang X., Baumann P. (2008) Mol. Cell 31, 463–473 [DOI] [PubMed] [Google Scholar]