Abstract
Human laeverin/aminopeptidase Q (LVRN/APQ) is a novel member of the M1 family of zinc aminopeptidases and is specifically expressed on the cell surface of human extravillous trophoblasts. Multiple sequence alignment of human M1 aminopeptidase revealed that the first Gly residue within the conserved exopeptidase motif of the M1 family, GXMEN motif, is uniquely substituted for His in human LVRN/APQ. In this study, we evaluated the roles of nonconserved His379, comprising the exopeptidase motif in the enzymatic properties of human LVRN/APQ. We revealed that the substitution of His379 with Gly caused significant changes in substrate specificity both toward fluorogenic substrates and natural peptide hormones. In addition, the susceptibilities of bestatin, a sensitive inhibitor for human LVRN/APQ, and natural inhibitory peptides were decreased in the H379G mutant. A molecular model suggested a conformational difference between wild-type and H379G human LVRN/APQs. These results indicate that His379 of the enzyme plays essential roles in its distinctive enzymatic properties and contributes to maintaining the appropriate structure of the catalytic cavity of the enzyme. Our data may bring new insight into the biological significance of the unique exopeptidase motif of LVRN/APQ obtained during the evolution of primates.
Introduction
The M1 family of mono-zinc aminopeptidases is widely distributed in bacteria, fungi, plants, and animals (1, 2). They hydrolyze peptide bonds linking to the N-terminal amino acids of peptide or protein substrates. The human M1 family consists of 11 enzymes that participate in many important physiological events, such as reproduction (3), angiogenesis (4–6), antigen presentation (7–10), blood pressure control (11–13), and memory retention (14), and thus play essential roles in the maintenance of homeostasis.
M1 aminopeptidases share two conserved domains: HEXXH(X)18E gluzincin motif and the exopeptidase motif, which is conserved as a GAMEN sequence in most members. The function of each conserved residue within these motifs has been elucidated using site-directed mutagenesis. For instance, two His residues and the second Glu within the HEXXH(X)18E motif coordinate the zinc atom essential for catalytic activity. The first Glu in the motif polarizes to the water molecule that coordinates the zinc atom and promotes nucleophilic attack of the carbonyl carbon of the peptide bond forming a tetrahedral intermediate in water (15). The conserved Glu within the exopeptidase motif, the GAMEN sequence, was shown to be involved in recognition of the α-amino group of substrates (16, 17).
Laeverin (LVRN)4 was originally identified as a cell-surface protein specifically expressed on human extravillous trophoblasts (18). The cDNA cloning of human LVRN revealed that it contains both consensus motifs and is a novel member of the family. Another group predicted the existence of the LVRN gene in the human genome through a genomic search and named it aminopeptidase Q (APQ) (19); however, its exopeptidase motif is uniquely composed of the HAMEN sequence, making its enzymatic activity uncertain. Recently, we established a large scale production system for a recombinant human LVRN/APQ and characterized its physicochemical and enzymatic properties (20). We found that the enzyme is indeed a novel leucine aminopeptidase with relatively high sensitivity to an aminopeptidase inhibitor, bestatin, and degraded several placenta-derived peptide hormones, such as angiotensin III, endokinin C, and kisspeptin-10. Considering the susceptibility of these peptides and their specific expression in the placenta, it was speculated that LVRN/APQ plays important roles in the maintenance of normal pregnancy in humans.
In this study, we investigated the catalytic mechanisms of human LVRN/APQ by site-directed mutagenesis to further evaluate its enzymatic properties. In the comparison of primary sequences between M1 aminopeptidases, we focused on the His379 residue uniquely substituted in the exopeptidase motif of human LVRN/APQ. It was shown that the substitution of His379 with Gly caused changes in substrate specificity and enzymatic properties of the enzyme. Construction of a three-dimensional model revealed a significant difference in the structure of the catalytic pocket between wild-type and mutant enzymes, suggesting an important role of His379 in the unique properties of the enzyme by the formation of an appropriate cavity structure.
EXPERIMENTAL PROCEDURES
Multiple Sequence Alignment and Phylogenetic Analysis of LVRN/APQ
The entire amino acid sequence of human LVRN/APQ was aligned with the whole sequences of other human M1 aminopeptidases and its orthologues using ClustalW2 software without taking known three-dimensional structural information into account.
For phylogenetic analyses, we used the nucleotide sequences encoding whole open reading frames of human M1 aminopeptidases and LVRN/APQ orthologue genes. Multiple sequence alignments of the nucleotide sequences of M1 aminopeptidase genes were carried out in a similar way for protein sequence alignments. From the alignment data, calculation of evolutionary distances and construction of phylogenetic trees were performed with the neighbor-joining method through ClustalW2 software. Nonrooted phylogenetic trees were displayed using TreeView software.
Site-directed Mutagenesis
To obtain a large amount of recombinant human LVRN/APQ, we slightly modified its cDNA by PCR. Briefly, to express the enzyme as a soluble protein, the coding sequences for the cytosolic and transmembrane region of the enzyme (Met1–Gln64) were replaced with that for human trypsin II signal peptide (i.e. MNLLLILTFVAAVAA) (21). Hexahistidine tag was added at its C-terminal end. Amplified cDNA was cloned into the BssHII-XhoI site of the baculovirus transfer vector pFastbac-1 (Invitrogen). The cDNAs encoding mutant human LVRN/APQs were generated using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The primers used to introduce nonsynonymous mutations (underlined nucleotides) into human LVRN/APQ cDNA were as follows: His379 to Gly sense primer, 5′-gtctagttttgacaacggtgcaatggaaaactgg-3′, and antisense primer, 5′-ccagttttccattgcaccgttgtcaaaactagac-3′; His379 to Phe sense primer, 5′-gtctagttttgacaactttgcaatggaaaactgg-3′, and antisense primer, 5′-ccagttttccattgcaaagttgtcaaaactagac-3′; His379 to Lys sense primer, 5′-gtctagttttgacaacaaggcaatggaaaactgg-3′, and antisense primer, 5′-ccagttttccattgccttgttgtcaaaactagac-3′; and His379 to Leu sense primer, 5′-gtctagttttgacaaccttgcaatggaaaactgg-3′, and antisense primer, 5′-ccagttttccattgcaaggttgtcaaaactagac-3′. The sequences of the products were confirmed by automated sequencing on an Applied Biosystems model 3730 (Foster City, CA). Replacement of Gly357 of human aminopeptidase A (APA) (11, 22), comprising the GAMEN motif, with His was carried out in a similar way using sense 5′-tccagattttggcactcatgccatggagaactgg-3′ and antisense 5′-ccagttctccatggcatgagtgccaaaatctgga-3′ primers. To generate recombinant bacmid DNA containing LVRN/APQ cDNAs, the generated transfer vectors were transformed to competent DH10Bac Escherichia coli cells harboring the baculovirus genome (bacmid) and a transposition helper vector (Invitrogen).
Expression and Purification of Recombinant LVRN/APQs in a Baculovirus System
Sf9 insect cells were transfected with bacmid DNA using Cellfectin reagent (Invitrogen), and after 72 h incubation, recombinant baculoviruses were harvested. For the expression of recombinant LVRN/APQs, Sf9 cells (2.0 × 106/ml) infected with the recombinant baculovirus (multiplicity of infection = ∼1–3) were cultured for 72 h in 3 liters of SFM-900 III medium (Invitrogen) at 27 °C supplied with 8.0 ppm of O2 (Cellmaster-1700; Wakenyaku, Kyoto, Japan).
The conditioned media containing recombinant LVRN/APQs were collected by centrifugation and then applied to a hydroxyapatite column (2.5 × 10 cm) (Nacalai Tesque, Kyoto, Japan) equilibrated in 50 mm Tris/HCl buffer (pH 7.5) and eluted with 100 mm sodium phosphate buffer (pH 7.5). The eluates were applied to a Ni2+-chelating Sepharose column (1.0 × 10 cm) (GE Healthcare) and then eluted with 200 mm imidazole. Production and purification of a soluble form of wild-type and G357H human APAs were also carried out in an essentially identical procedure except for using DEAE anion-exchange chromatography instead of hydroxyapatite column chromatography (11, 22). After purification, all enzymes gave a single band with a molecular mass of ∼120 kDa on SDS-PAGE (data not shown).
Measurement of Aminopeptidase Activity of LVRN/APQs
Aminopeptidase activities of LVRN/APQs were determined with various fluorogenic substrates, aminoacyl-4-methylcoumaryl-7-amides (aminoacyl-MCAs). The reaction mixture containing various concentrations of aminoacyl-MCA and the enzyme in 0.5 ml of 25 mm Tris/HCl buffer (pH 7.5) was incubated at 37 °C for 5 min. The amount of 7-amino-4-methylcoumarin released was measured by spectrofluorophotometry (F-2000; Hitachi, Tokyo, Japan) at an excitation wavelength of 360 nm and an emission wavelength of 460 nm. The kinetic parameters were calculated from Lineweaver-Burk plots. The results are represented by Km, kcat, and kcat/Km values. All measurements were performed in triplicate. The Ki values for aminopeptidase inhibitors and peptide hormones were measured using Leu-MCA (100 μm) as a substrate and calculated from Lineweaver-Burk plots.
Cleavage of Peptide Hormones by LVRN/APQs
Human peptide hormones (Peptide Institute, Osaka, Japan) (25 μm) were incubated with the enzyme (1 μg/ml) at 37 °C in 25 mm Tris/HCl buffer (pH 7.5). The reaction was terminated by adding 2.5% (v/v) formic acid. The reactants and products were separated on a reverse phase column, COSMOSIL (4.6 × 250 mm) (Nacalai Tesque, Kyoto, Japan), using an automated HPLC system (AT-10; Shimadzu, Kyoto, Japan). Peptides generated from angiotensin III and endokinin C were isocratically eluted with the following buffers at a flow rate of 0.5 ml/min: for peptides from angiotensin III, 19% acetonitrile containing 0.086% trifluoroacetic acid; for peptides from endokinin C, 30% acetonitrile containing 0.084% trifluoroacetic acid. Peptides generated from dynorphin-related peptides (Met-enkephalin and dynorphin A-(1–8), -(1–13), and -(1–17)) were loaded onto the column equilibrated in 10% acetonitrile containing 0.088% trifluoroacetic acid and eluted with a linear gradient of 10% acetonitrile containing 0.088% trifluoroacetic acid to 30% acetonitrile containing 0.084% trifluoroacetic acid in 20 min at a flow rate of 0.5 ml/min, and peptides from kisspeptin-10 were eluted with a linear gradient of 20% acetonitrile containing 0.086% trifluoroacetic acid to 40% acetonitrile containing 0.082% trifluoroacetic acid in 20 min at a flow rate of 0.5 ml/min. The molecular masses of peptides were determined by matrix-assisted laser desorption ionization-time of flight mass spectrometry with a REFLEX mass spectrometer (Bruker-Franzen Analytik) using α-cyano-4-hydroxycinnamic acid as the matrix.
Molecular Modeling of LVRN/ APQ
The recently published x-ray crystallographic structures of Thermoplasma acidophilum. tricorn interacting factor F3 (Protein Data Bank code 1Z5H) (23) was used as a template for modeling the catalytic pocket of human LVRN/APQ using the three-dimensional Jigsaw Protein Comparative Modeling Server. The structures were displayed using the CueMol program (R. Ishitani, CueMol, Molecular Visualization Framework).
Materials
All fluorogenic substrates and human peptide hormones, except for dynorphin A-(1–8), were purchased from the Peptide Institute (Osaka, Japan). Bestatin was obtained from Nacalai Tesque (Kyoto, Japan). Dynorphin A-(1–8) and amastatin were from Sigma. Endokinin C mutant K2A peptide was synthesized by the Research Resources Center (Brain Science Institute, RIKEN).
RESULTS
His379 in the Exopeptidase Motif of Human LVRN/APQ
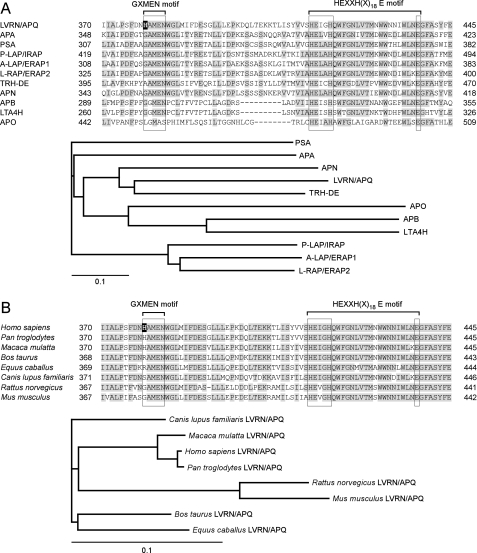
Multiple alignment of the M1 aminopeptidase family was performed to search for residues involved in the catalytic action of human LVRN/APQ (Fig. 1A). The exopeptidase motif, GXMEN, is well conserved among the family (2). Exceptionally, whereas aminopeptidase O completely lacks the motif, the first Gly in the motif of thyrotropin-releasing hormone-degrading enzyme is uniquely substituted with Ala (AAMEN). LVRN/APQ is also unique in that the first Gly in the motif is substituted with His, having a bulky side chain. Phylogenetic analysis of the human M1 family suggests that both LVRN/APQ and thyrotropin-releasing hormone-degrading enzyme having a substitution on the motif diverged from the ancestral aminopeptidase N (APN) gene. We next compared the exopeptidase motif among LVRN/APQ orthologues (Fig. 1B). GAMEN sequence substitutions were found in several mammalian species (i.e. humans and monkeys (HAMEN), dogs (SAMEN), and cattle and horses (RAMEN)) but not in mice and rats. It is noteworthy that the exopeptidase motif sequence is clearly conserved in LVRN/APQ orthologues among evolutionarily related species. We therefore focused in this study on the His379 residue within the exopeptidase motif of human LVRN/APQ and explored its roles in the enzymatic properties of the enzyme by site-directed mutagenesis.
FIGURE 1.
Molecular evolution of LVRN/APQ. Alignments of the human LVRN/APQ amino acid sequence with the sequences of other human M1 aminopeptidases (A, upper) and its orthologues (B, upper) are shown. Amino acid sequence alignments around the GXMEN and HEXXH(X)18E motifs of M1 aminopeptidases are shown. Gaps are inserted into the sequence to obtain optimal alignments. Residues conserved among more than six enzymes are shaded. His379 residue of human LVRN/APQ is boxed in black. Evolutionary relationships of human M1 aminopeptidases (A, lower panel) and LVRN/APQ orthologues (B, lower panel) are shown. The bar shown below the phylogenetic tree represents the genetic distance. The sequence data used for analyses are as follows: A, LVRN/APQ (NM_173800) (18); APA (NM_001977) (46, 47); PSA, puromycin-sensitive aminopeptidase (NM_006310) (48); P-LAP/IRAP, placental leucine aminopeptidase/oxytocinase/insulin-regulated aminopeptidase (NM_005575) (3); A-LAP/ERAP1, adipocyte-derived leucine aminopeptidase/endoplasmic reticulum aminopeptidase-1 (NM_001040458) (49); L-RAP/ERAP2, leukocyte-derived arginine aminopeptidase/endoplasmic reticulum aminopeptidase-2 (NM_022350) (9); TRH-DE, thyrotropin-releasing hormone-degrading enzyme (NM_013381) (50); APN (NM_001150) (51); APB, aminopeptidase B (NM_020216) (52); LTA4H, leukotriene A4 hydrolase (NM_000895) (53); and APO, aminopeptidase O (NM_032823) (54). B, human (Homo sapiens, NM_173800), monkeys (Pan troglodytes, XM_001149318, and Macaca mulatta, XM_001086413), cattle (Bos taurus, XM_001788658), horse (Equus caballus, XM_001918093), dog (Canis lupus familiaris, XM_538554), rat (Rattus norvegicus, XM_577617), and mouse (Mus musculus, XM_911283).
Aminopeptidase Activities of His379 Mutant LVRN/APQs
Kinetic studies of various synthetic substrates of wild-type and four mutant LVRN/APQs, i.e. H379G, H379F, H379K, and H379L LVRN/APQs, were initially performed (Table 1). In accordance with our previous study, wild-type LVRN/APQ showed maximum catalytic efficiency (kcat/Km) toward Leu-MCA, followed by Arg-Met-, Lys-, and Phe-MCA. The calculated Km values of H379G LVRN/APQ to neutral amino acids (Leu-, Met- and Phe-MCA) decreased to 41–60% of that of the wild-type enzyme, and turnover numbers decreased to 30–39%, resulting in a significant decrease in catalytic efficiency. Although the Km values of the mutant to Lys and Arg also decreased to ∼50%, catalytic efficiencies to both substrates were rather increased because of a moderate decrease in turnover numbers.
TABLE 1.
Kinetic parameters of wild-type and His379 mutant human LVRN/APQs toward various aminoacyl-MCA
Kinetic parameters were determined from Lineweaver-Burk plots. Reactions took place at 37 °C for 5 min.
| Substrate | LVRN/APQ | Kma | kcata | kcat/Km × 103a |
|---|---|---|---|---|
| μm | s−1 | μm−1·s−1 | ||
| Leu-MCA | Wild type | 98.6 ± 19 | 37.7 ± 4.8 | 385 ± 26 |
| H379G | 44.5 ± 15 | 11.3 ± 3.0 | 258 ± 19 | |
| H379F | 119 ± 21 | 49.8 ± 6.4 | 419 ± 24 | |
| H379K | 109 ± 9.6 | 33.7 ± 2.7 | 308 ± 3.2 | |
| H379L | 28.1 ± 2.7 | 4.85 ± 0.22 | 173 ± 11 | |
| Met-MCA | Wild type | 65.0 ± 5.0 | 10.9 ± 1.0 | 168 ± 4.3 |
| H379G | 26.4 ± 6.3 | 3.59 ± 0.66 | 137 ± 10 | |
| H379F | 62.8 ± 19 | 18.1 ± 3.9 | 292 ± 22 | |
| H379K | 61.5 ± 1.9 | 11.6 ± 2.7 | 189 ± 5.5 | |
| H379L | 9.26 ± 0.93 | 1.70 ± 0.066 | 184 ± 11 | |
| Arg-MCA | Wild type | 21.0 ± 5.1 | 4.48 ± 0.49 | 219 ± 33 |
| H379G | 11.1 ± 1.5 | 3.11 ± 0.23 | 282 ± 20 | |
| H379F | 21.5 ± 1.1 | 5.74 ± 0.17 | 267 ± 5.9 | |
| H379K | 18.9 ± 2.4 | 1.62 ± 0.12 | 84.9 ± 4.5 | |
| H379L | 9.85 ± 1.2 | 2.07 ± 0.17 | 210 ± 8.5 | |
| Lys-MCA | Wild type | 31.2 ± 1.6 | 4.83 ± 0.19 | 155 ± 2.0 |
| H379G | 15.5 ± 5.4 | 4.10 ± 1.2 | 269 ± 22 | |
| H379F | 31.3 ± 8.0 | 6.68 ± 1.5 | 215 ± 9.5 | |
| H379K | 27.5 ± 1.2 | 4.31 ± 0.14 | 157 ± 2.7 | |
| H379L | 5.82 ± 0.23 | 1.82 ± 0.044 | 313 ± 7.0 | |
| Phe-MCA | Wild type | 43.2 ± 4.5 | 2.19 ± 0.18 | 50.9 ± 1.1 |
| H379G | 26.1 ± 2.7 | 0.862 ± 0.062 | 33.2 ± 1.2 | |
| H379F | 44.7 ± 7.4 | 3.17 ± 0.56 | 71.0 ± 5.5 | |
| H379K | 44.2 ± 2.4 | 1.03 ± 0.11 | 23.1 ± 1.5 | |
| H379L | 18.2 ± 4.4 | 1.74 ± 0.22 | 97.7 ± 10 |
a Values are the mean ± S.D. (n = 3).
In the H379F mutant, affinities to all substrates tested were quite similar to those of the wild-type enzyme; however, because of a modest increase in the turnover number for each substrate, catalytic efficiency to all substrates was slightly increased (109–174%). In the H379K mutant, although a decrease was observed only in catalytic efficiencies toward Arg- and Phe-MCAs due to the significant reduction of kcat values, most parameters were similar to those of wild-type LVRN/APQ. In contrast, except for kcat toward Lys-MCA, a quite similar tendency was observed in Km and kcat values of H379L mutant to the values of H379G LVRN/APQ.
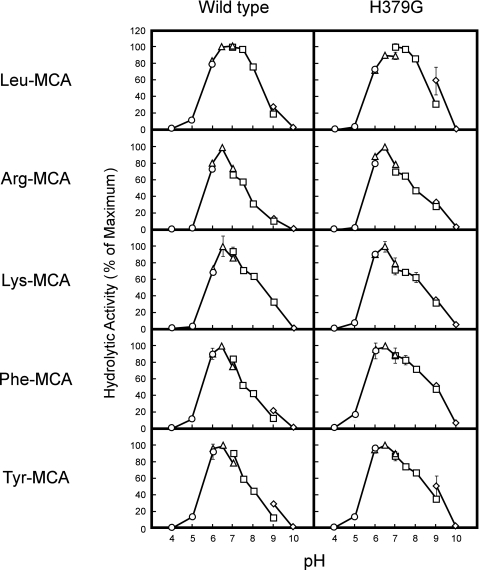
We next compared pH profiles between wild-type and H379G LVRN/APQs using five synthetic substrates (Fig. 2). Although the pH profiles were slightly different between each substrate, the optimal pH values and activity patterns over the pH range of both enzymes were almost identical, excluding the possibility that the hydration state of His379 affected cleavage of the substrates. Taken together, these results indicated that the substitution of His379 with Gly or Leu caused a significant change in the substrate specificity of LVRN/APQ, although substitution with Phe or Lys also partially affected the substrate specificity.
FIGURE 2.
pH profiles of wild-type and H379G LVRN/APQs toward various aminoacyl-MCAs. Enzymatic activity of recombinant LVRN/APQs (1 μg/ml) was measured in various buffers (pH 4.0–10.0) with 100 μm MCA-substrate at 37 °C for 5 min. ○, 25 mm sodium acetate; △, 25 mm PIPES/NaOH; □, 25 mm Tris/HCl; ◇, 25 mm glycine/NaOH. Each point represents the activity mean of triplicate determinations, and error bars indicate S.D. Similar results were obtained in four separate experiments. Fluorogenic substrates used for the assay are shown on the left side of the figure. The maximum activity toward each substrate was taken as 100%.
Susceptibility of Aminopeptidase Inhibitors to His379 Mutant LVRN/APQs
Unlike most other members of the M1 family, the aminopeptidase activity of human LVRN/APQ is inhibited by bestatin, a competitive aminopeptidase inhibitor, more efficiently than amastatin; therefore, we next compared the susceptibility of aminopeptidase inhibitors to His379 LVRN/APQ mutants (Table 2). Although bestatin inhibited the aminopeptidase activities of wild-type and H379F LVRN/APQ rather efficiently, H379G, H379K, and H379L LVRN/APQs were less susceptible to the inhibitor, suggesting that an aromatic ring of His379 is important for the bestatin sensitivity of wild-type LVRN/APQ. On the other hand, amastatin inhibited the aminopeptidase activity of H379G LVRN/APQ (Ki = 17.3 ± 5.4 μm) more effectively than adipocyte-derived leucine aminopeptidase/endoplasmic reticulum aminopeptidase-1 (Ki = 41.8 ± 10.3 μm), to which amastatin is much more effective than bestatin (24). These results indicated that His379 was responsible for the unique inhibitor profile of wild-type LVRN/APQ.
TABLE 2.
Ki values of aminopeptidase inhibitors for wild-type and His379 mutant LVRN/APQs
Kinetic parameters were determined from Lineweaver-Burk plots. Reactions took place at 37 °C for 5 min.
| LVRN/APQ | Bestatina | Amastatina |
|---|---|---|
| μm | μm | |
| Wild type | 3.81 ± 0.98 | 97.6 ± 10 |
| H379G | 12.2 ± 1.2 | 17.3 ± 5.4 |
| H379F | 4.19 ± 0.12 | 42.6 ± 12 |
| H379K | 22.5 ± 6.1 | 92.1 ± 17 |
| H379L | 8.37 ± 1.1 | 105 ± 13 |
a Values are the mean ± S.D. (n = 3).
Inhibition of the Aminopeptidase Activity of His379 Mutant LVRN/APQs by Natural Inhibitory Peptides
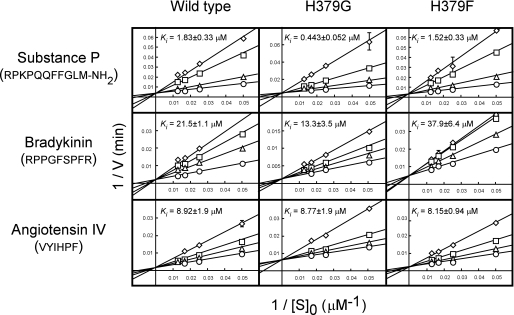
In our previous work, we showed that three peptide hormones, substance P, angiotensin IV, and bradykinin, known to be detected in the human placenta, were not degraded by LVRN/APQ and inhibited its aminopeptidase activity (20). We then compared the inhibitory effects of these human natural peptides on the aminopeptidase activities of His379 mutants. As shown in Fig. 3, substance P significantly inhibited the LVRN/APQ-mediated hydrolysis of Leu-MCA. Although substance P inhibited the aminopeptidase activity of the H379F mutant comparably with the wild type, the H379G mutant was much more sensitive to the hormone. Bradykinin, having the same N-terminal sequence as substance P (Arg-Pro), also inhibited the H379G mutant more than the wild-type and H379F enzymes. On the other hand, angiotensin IV, the N terminus of which is Val, inhibited all enzymes rather consistently. Because Lineweaver-Burk plots showed that all peptides inhibited the enzyme competitively, these results suggested that substitution of the His379 residue with Gly caused an increase in the affinity to two peptides with Arg at their N termini and thus suggested the involvement of His379 in the construction of the catalytic pocket of the enzyme.
FIGURE 3.
Competitive inhibition of the hydrolytic activity of LVRN/APQs by human natural peptides. The hydrolytic activities of wild-type and His379 mutant human LVRN/APQs (1 μg/ml) were measured at 37 °C for 5 min with Leu-MCA in the presence of various concentrations of substance P, bradykinin, or angiotensin IV. ○, control; △, 1 μm substance P (wild type and H379F), 0.3 μm substance P (H379G), 20 μm bradykinin (wild type and H379F), 5 μm bradykinin (H379G), 3 μm angiotensin IV; □, 5 μm substance P (wild type and H379F), 1 μm substance P (H379G), 40 μm bradykinin (wild type and H379F), 10 μm bradykinin (H379G), 10 μm angiotensin IV; and ◇, 10 μm substance P (wild type and H379F), 3 μm substance P (H379G), 60 μm bradykinin (wild type and H379F), 30 μm bradykinin (H379G), 20 μm angiotensin IV. Each point represents the activity mean of three separate experiments, and error bars indicate standard deviation. Ki values of peptide hormones for recombinant LVRN/APQs were determined from Lineweaver-Burk plots and are shown in each graph. Similar results were obtained in three separate experiments. Peptides used for inhibition are shown on the left side of the figure, and amino acid sequences are shown below.
Cleavage of Natural Peptide Hormones by His379 Mutant LVRN/APQs
We next compared the hydrolytic activities of wild-type, H379G, and H379F LVRN/APQs toward natural peptide hormones (Table 3 and supplemental Fig. S1). Wild-type LVRN/APQ cleaved about 30% of angiotensin III within 60 min (supplemental Fig. S1A). On the other hand, H379G LVRN/APQ converted angiotensin III into angiotensin IV more efficiently under the same conditions, and 60% was converted within this time. Kinetic study showed that the catalytic efficiency of the H379G mutant for angiotensin III was higher than those of wild-type and H379F LVRN/APQs, largely due to increased affinity (Table 3). These results showed that, in accordance with the hydrolytic efficiency toward synthetic substrates, H379G mutant degraded substrates with basic amino acid residues at their N termini more efficiently than wild-type enzyme.
TABLE 3.
Kinetic parameters of wild-type and H379G LVRN/APQs toward peptide substrates
Kinetic parameters were determined from Lineweaver-Burk plots. Reactions took place at 37 °C for 5 min.
| Substrate (sequence) | LVRN/APQ | Kma | kcata | kcat/Km × 103a |
|---|---|---|---|---|
| μm | s−1 | μm−1·s−1 | ||
| Angiotensin III | Wild type | 13.4 ± 2.1 | 1.24 ± 0.11 | 92.9 ± 7.2 |
| (RVYIHPF) | H379G | 4.30 ± 1.1 | 1.84 ± 0.22 | 440 ± 69 |
| H379F | 16.6 ± 2.4 | 1.86 ± 0.10 | 113 ± 10 | |
| Kisspeptin-10 | Wild type | 32.1 ± 8.9 | 3.62 ± 0.92 | 113 ± 5.2 |
| (YNWNSFGLRF-NH2) | H379G | 10.8 ± 4.1 | 0.501 ± 0.10 | 48.6 ± 8.1 |
| H379F | 16.7 ± 4.5 | 2.28 ± 0.45 | 138 ± 12 | |
| Dynorphin A-(1–8) | Wild type | 65.6 ± 15 | 22.3 ± 4.6 | 340 ± 7.3 |
| (YGGFLRRI) | H379G | 5.96 ± 1.2 | 0.748 ± 0.049 | 128 ± 18 |
| H379F | 11.3 ± 2.9 | 2.48 ± 0.55 | 219 ± 11 | |
| Endokinin C | Wild type | 38.4 ± 3.1 | 6.25 ± 0.86 | 162 ± 9.6 |
| (KKAYQLEHTFQGLL-NH2) | H379G | 22.3 ± 7.7 | 1.75 ± 0.41 | 80.3 ± 9.4 |
| H379F | 19.6 ± 1.3 | 3.25 ± 0.63 | 164 ± 20 | |
| Endokinin C K2A | Wild type | 180 ± 25 | 10.8 ± 3.5 | 59.5 ± 14 |
| (KAAYQLEHTFQGLL-NH2) | H379G | 24.4 ± 6.8 | 1.11 ± 0.31 | 45.3 ± 2.2 |
| H379F | 40.7 ± 3.8 | 2.58 ± 0.15 | 63.5 ± 2.2 |
a Values are the mean ± S.D. (n = 3).
We next tested the cleavage of peptides having the Tyr residue with N termini by H379G LVRN/APQ, because they were good substrates for the enzyme (20). Degradation of kisspeptin-10 to de-[Tyr]kisspeptin-10 by the H379G mutant (15% cleavage) was much slower than that by wild-type human LVRN/APQ (45% cleavage), in accordance with the hydrolysis of Phe-MCA (supplemental Fig. S1B). It is noteworthy that the pH profiles to angiotensin III and kisspeptin-10 were quite similar between wild-type and H379G LVRN/APQs, as in the case of fluorogenic substrates (supplemental Fig. S2).
Furthermore, although dynorphin A-(1–8) was efficiently digested by the wild type (90% cleavage) within 30 min, as expected, only 30% was digested after incubation with H379G mutant (supplemental Fig. S1C). Likewise, in kinetic parameters toward Phe-MCA, although the affinity of H379G LVRN/APQ for kisspeptin-10 and dynorphin-A-(1–8) was rather increased, the catalytic efficiency decreased due to marked falls in turnover numbers (Table 3). Meanwhile, Km and kcat values of the H379F mutant for both peptides were moderately decreased. Taken together, His379 of LVRN/APQ affects the hydrolytic efficiency of the enzyme toward various substrate peptides.
Endokinin C, a novel peptide hormone highly expressed in the human placenta, was shown to be a good substrate of human LVRN/APQ (20); however, endokinin C was unexpectedly less susceptible to H379G LVRN/APQ than the wild type, despite having a Lys residue at the N terminus of the peptide (supplemental Fig. S3A). The affinities of H379G and H379F mutants for the hormone were higher than the wild type in accordance with other peptide substrates; however, the turnover numbers of the mutants for endokinin C were lower than that of the wild type, resulting in decreased catalytic efficiency of the H379G mutant for the hormone (Table 3). Because the crystal structure of human leukotriene A4 hydrolase, a member of the M1 family, indicated that Gly268, which corresponded to His379 of LVRN/APQ, might interact with the P1′ residue of the substrate (25), we also examined the role of the P1′ residue of endokinin C in its susceptibility to LVRN/APQ by testing the hydrolysis of endokinin C mutant K2A, whose N-terminal second Lys was substituted with Ala. Wild-type LVRN/APQ-mediated degradation of endokinin C mutant K2A was apparently less than that of endokinin C, which was coincident with our previous observation that a degradation product of the hormone by LVRN/APQ (i.e. de-[Lys]endokinin C, KAYQLEHTFQGLL-NH2) was less susceptible to the enzyme than the hormone (supplemental Fig. S3B). Degradation of endokinin C mutant K2A was also impaired in the H379G mutant to a similar level of endokinin C mutant K2A degradation by the wild type. It should be noted that the decreased catalytic efficiency of wild-type LVRN/APQ for endokinin C mutant K2A was largely due to a marked increase in the Km value in comparison with that for endokinin C (Table 3). These results indicated that His379 played a significant role in not only recognition of the P1 residue but also interaction with the P1′ residue of the substrate or maintenance of the optimal cavity structure of the enzyme.
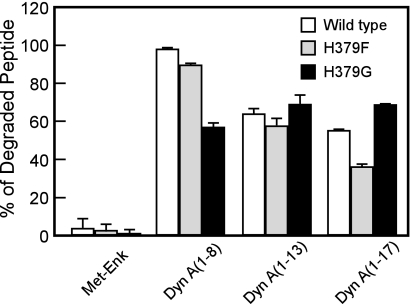
As we reported (20), human LVRN/APQ digested dynorphin A-(1–8) much more efficiently than the other three dynorphin-related peptides sharing the N-terminal five amino acid sequence, i.e. Met-enkephalin, dynorphin A-(1–13), and dynorphin A-(1–17). Thus, we next tested the hydrolysis of these peptides by the H379G mutant (Fig. 4). As in the wild-type enzyme, the H379G mutant did not digest Met-enkephalin at all, even when 2.5-fold more of the enzyme was employed. Strikingly, the H379G mutant cleaved dynorphin A-(1–8) less effectively than the wild-type enzyme even when it cleaved dynorphin A-(1–13) and dynorphin A-(1–17) comparably with the wild type. Another mutant enzyme, H379F LVRN/APQ, cleaved the shorter peptide more efficiently than the longer peptide, indicating the unique roles of His379 in the construction of the global structure of the catalytic cavity of the enzyme.
FIGURE 4.
Cleavage of dynorphin-related peptide hormones by wild-type and mutant LVRN/APQs. Twenty five micromolars of Met-enkephalin, dynorphin A-(1–8), -(1–13), and -(1–17) were incubated with wild-type (2 μg/ml), H379F (2 μg/ml), or H379G (5 μg/ml) LVRN/APQ at 37 °C for 10 min. After separating the generated peptides by reversed phase HPLC, the digested substrates were measured. Similar results were obtained in at least three separate experiments. Met-Enk, Met-enkephalin; Dyn A, dynorphin A.
Characterization of Enzymatic Properties of Gly357 Mutant Human APA
Finally, to evaluate the role of the first amino acid in the exopeptidase motif, we introduced a point mutation in the motif of human APA, the substrate specificity of which is readily distinguishable from that of LVRN/APQ.
As shown in Table 4, in the absence of CaCl2, the point mutation markedly altered Km values for four substrates tested (Km values for Glu-, Asp-, and Gln-MCAs decreased with different levels; however, the values for Ala-MCA increased). As for kcat, the mutation led to increased values for all substrates to 170–450%. Consequently, kcat/Km values for all substrates were higher in G357H APA than the wild type. In particular, catalytic efficiency of G357H APA for Glu-MCA increased to a similar level as that of Ca2+-activated wild-type APA. These results indicated changes in the substrate specificity of APA by the point mutation of Gly357. Even in the presence of 1 mm Ca2+, Km and kcat values toward each substrate were considerably different between two enzymes, resulting in the difference in kcat/Km values between wild-type and G357H APAs. In addition, as for wild-type APA, which shows a clear preference for acidic amino acids in the presence of Ca2+, G357H APA also showed an acidic amino acid preference in response to the addition of Ca2+. Furthermore, it should be noted that G357H APA was more active than the wild type toward all substrates both in the presence and absence of Ca2+.
TABLE 4.
Kinetic parameters of wild-type and Gly357 mutant human APAs toward various aminoacyl-MCA
Wild-type or G357H APAs (50 ng/ml) were incubated with aminoacyl-MCAs at 37 °C for 5 min with or without 1 mm CaCl2. Kinetic parameters were determined from Lineweaver-Burk plots.
| Substrate | APA | CaCl2 | Kma | kcata | kcat/Km × 103s |
|---|---|---|---|---|---|
| 1 mm | μm | s−1 | μm−1·s−1 | ||
| Glu-MCA | Wild type | − | 808 ± 68 | 6.89 ± 0.38 | 8.53 ± 0.28 |
| G357H | − | 148 ± 7.6 | 11.6 ± 0.75 | 78.0 ± 6.0 | |
| Asp-MCA | Wild type | − | 3040 ± 290 | 5.61 ± 0.33 | 1.85 ± 0.080 |
| G357H | − | 933 ± 90 | 9.49 ± 1.1 | 10.2 ± 0.28 | |
| Gln-MCA | Wild type | − | 852 ± 67 | 4.25 ± 0.23 | 4.98 ± 0.21 |
| G357H | − | 338 ± 18 | 11.0 ± 0.39 | 32.7 ± 0.72 | |
| Ala-MCA | Wild type | − | 234 ± 31 | 1.64 ± 0.18 | 7.00 ± 0.39 |
| G357H | − | 461 ± 27 | 7.37 ± 0.30 | 16.0 ± 1.2 | |
| Glu-MCA | Wild type | + | 201 ± 23 | 18.7 ± 0.29 | 90.3 ± 1.3 |
| G357H | + | 90.5 ± 6.6 | 34.2 ± 2.7 | 378 ± 22 | |
| Asp-MCA | Wild type | + | 1275 ± 69 | 10.8 ± 0.72 | 8.45 ± 0.63 |
| G357H | + | 292 ± 22 | 16.5 ± 0.56 | 56.5 ± 5.7 | |
| Gln-MCA | Wild type | + | 1288 ± 69 | 4.00 ± 0.22 | 3.10 ± 0.058 |
| G357H | + | 457 ± 55 | 5.17 ± 0.35 | 11.3 ± 0.65 | |
| Ala-MCA | Wild type | + | 930 ± 150 | 1.54 ± 0.20 | 1.65 ± 0.070 |
| G357H | + | 1159 ± 120 | 3.01 ± 0.26 | 2.60 ± 0.042 |
a Values are the mean ± S.D. (n = 4).
We next examined the susceptibility of G357H APA toward bestatin and amastatin (Table 5). Amastatin is quite a strong inhibitor of wild-type APA (Ki = 40 nm). The Ki value of G357H mutant increased to a similar level to the value of the wild type under 1 mm Ca2+. The Ki value of bestatin, a weaker inhibitor of APA, also increased with the point mutation. In the presence of Ca2+, Ki to amastatin of G357H mutant further increased, and bestatin became invalid up to 1 mm. These results suggested that replacement of Gly357 with His modified substrate specificity, Ca2+ response, and inhibitor susceptibility of human APA.
TABLE 5.
Ki values of aminopeptidase inhibitors for wild-type and Gly357 mutant human APAs
Wild-type or G357H APAs (250 ng/ml) were incubated with Glu-MCA at 37 °C for 5 min with or without 1 mm CaCl2. Ki values were calculated from Lineweaver-Burk plots.
| APA | CaCl2 | Bestatina | Amastatina |
|---|---|---|---|
| 1 mm | μm | μm | |
| Wild type | − | 31.4 ± 7.3 | 0.0399 ± 0.00096 |
| G357H | − | 275 ± 50 | 0.504 ± 0.080 |
| Wild type | + | 375 ± 58 | 0.303 ± 0.039 |
| G357H | + | No inhibition (up to 1 mm) | 11.9 ± 1.2 |
a Values are the mean ± S.D. (n = 3).
Taken together, our data strongly suggest that the first amino acid of the exopeptidase motif ((G/H)AMEN) might be involved as a determinant of the enzymatic properties of the M1 family of aminopeptidases.
DISCUSSION
In this study, we assessed how distinctive enzymatic properties of human LVRN/APQ were obtained. We first explored the significance of His379 of human LVRN/APQ in its enzymatic properties by performing a kinetic study of fluorogenic substrates using His379 mutant LVRN/APQs. When using Leu-, Met-, and Phe-MCAs as substrates, the H379G mutation caused significant reductions of Km values (41–60% of wild-type) and kcat (30–39% of wild-type) values, resulting in a significant decrease in catalytic efficiency (kcat/Km). In accordance with the kinetic parameters toward synthetic substrates, marked reductions in the release of Tyr residues from the N termini of dynorphin A-(1–8) and kisspeptin-10 were observed in H379G LVRN/APQ. Furthermore, the inhibitory activity of bestatin, an imitation di-peptide composed of a phenylalanine analogue and a leucine, was strikingly impaired by the substitution of His379 with Gly. These observations indicate that His379 is favorable for the binding and release of hydrophobic amino acids at N termini more than Gly, suggesting specific roles of human LVRN/APQ in the metabolism of peptides with an N-terminal hydrophobic amino acid.
By contrast, for basic synthetic substrates, no apparent reduction was observed in the turnover number of H379G mutant, despite marked falls in Km values for Arg- and Lys-MCA (53 and 50% reduction), resulting in increased catalytic efficiency (to 129 and 174%, respectively). In addition, the release of Arg residue from the N terminus of angiotensin III by the mutant was significantly faster than the wild type. Moreover, substance P and bradykinin inhibited the H379G mutant more effectively than wild-type LVRN/APQ, strongly suggesting that the H379G mutant prefers N-terminal basic amino acids to the wild-type enzyme.
Unlike the H379G mutant, all kinetic parameters for synthetic substrates and the inhibitor sensitivity of the H379F mutant were comparable with those of wild-type LVRN/APQ. Furthermore, in the H379L mutant, Km and kcat values for all synthetic substrates tested decreased and bestatin susceptibility was changed, as in the case of the H379G mutant. These results revealed the potential importance of an aromatic ring of the side chain of His379 for the unique enzymatic properties of human LVRN/APQ. In contrast, it seems unlikely that the hydrogen-bonding potential of the imidazole ring of His379 involves enzyme action such as substrate recognition, because pH profiles of H379G LVRN/APQs were almost identical to those of the wild-type enzyme (Fig. 2 and supplemental Fig. S2). In addition, H379K LVRN/APQ, to which an acyl chain with positive charge was introduced, also showed similar kinetic values to certain substrates when compared with the wild-type enzyme, whereas H379G and H379L mutants showed apparently different values. These results suggest that His at position 379 is required for optimum activity of human LVRN/APQ. The bulkiness of the side chain of the first residue of the exopeptidase motif might be an important factor for the hydrolytic action of LVRN/APQ.
Several studies on the catalytic mechanism of M1 aminopeptidases have demonstrated the roles of the conserved amino acid residue in the exopeptidase motif. It was shown that Glu352 and Asn353 in the motif play roles in the exopeptidase specificity of murine APA through interaction with the N-terminal amino acid of the substrate (16, 26). The significance of the Gly residue at position 1 of the exopeptidase motif was also elucidated in placental leucine aminopeptidase (P-LAP). Mutant P-LAP with Gly428 substituted with Ala retains comparable activity toward the synthetic substrate with the wild-type enzyme (27). Likewise, P-LAP mutants substituted with bulky amino acids, i.e. G428E, G428D, and G428K P-LAPs, also retained activity toward the synthetic substrates (28); however, all three mutants with a bulky amino acid residue were unable to degrade Leu-enkephalin, and the Arg-vasopressin-degrading activity of two mutants (G428E and G428D) was impaired. In addition, these mutations attenuated the susceptibility to peptide inhibitors, such as angiotensin IV and Leu-Val-Val-hemorphin-7. These results suggested that the Gly residue in the exopeptidase motif plays an important role in exerting the maximum activity of P-LAP. In contrast, as shown in this study, the Gly residue in the exopeptidase motif of human LVRN/APQ might be inadequate to achieve optimal enzymatic activity toward several synthetic and natural substrates.
The crystal structures of T. acidophilum tricorn interacting factor F3, human leukotriene A4 hydrolase, and E. coli APN were recently resolved (23, 25, 29, 30). The five amino acids comprising the exopeptidase motif form a short β-strand (β19 of T. acidophilum tricorn interacting factor F3) located in the catalytic pocket of the enzyme. In human leukotriene A4 hydrolase, it was also shown that Glu271 at position 4 within the motif formed a hydrogen bond with the N-terminal group on the substrate or an aminopeptidase inhibitor bestatin. In addition, the possibility that the main-chain amide of position 1 Gly (Gly268) within the motif directly formed a hydrogen bond with the backbone carbonyl group of bestatin or the P1′ residue of substrate peptide was also revealed (25). Moreover, in E. coli APN, a theoretical model was proposed that Met260 at position −1 from the exopeptidase motif might be involved in substrate recognition (30), i.e. a conformational change of Met260 causes volumetric change of the S1 pocket to make it possible to accept substrates of different N-terminal residue sizes. This cushion residue model well elucidates the broad substrate preference of the M1 aminopeptidases. Recently, it was also reported that Thr348 at position −1 from the exopeptidase motif of mouse APA plays an essential role in its substrate specificity through involvement of the configuration of the substrate and catalytic water molecules for optimal catalysis (31).
Taken together, it was suggested that if the Gly residue within the exopeptidase motif was replaced with another amino acid, especially with a bulky amino acid, it might cause some conformational changes of the side chain of the proximal residue and thus affect the enzymatic properties of M1 aminopeptidase.
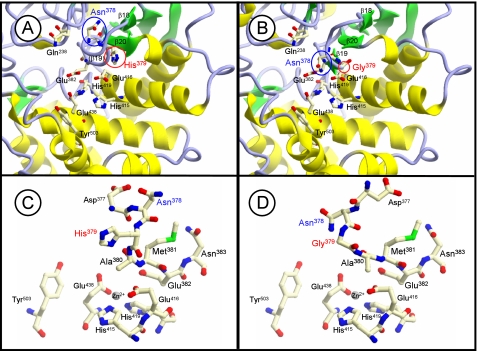
To evaluate this hypothesis, we simulated a molecular model of the catalytic pocket, which is well conserved among the M1 family, of wild-type and H379G human LVRN/APQs based on the crystal structure of T. acidophilum tricorn interacting factor F3, because of the highest similarity (>30%) around the region among the above three enzymes (Fig. 5). This model suggested a significant conformational difference in the S1 site surrounding the β19 strand composed of the exopeptidase motif between the enzymes (a subdomain corresponding to β18–β20 of T. acidophilum tricorn interacting factor F3) (Fig. 5, A and B). It is also conceivable that the mutation alters the side-chain conformation of other exopeptidase motif residues (Fig. 5, C and D). It should be noted that the position of the side chain of Asn378 at position −1 from the exopeptidase motif of LVRN/APQ is different between wild-type and H379G LVRN/APQs. Therefore, it is tempting to speculate that the substitution of Gly in the exopeptidase motif with His (and vice versa) might affect the substrate specificity of LVRN/APQ by changing the structure of the S1 pocket. This possibility can be supported by changes in the substrate specificity and Ca2+ response of APA by Gly357 mutation into His, because Ca2+ is a key factor to determine the substrate specificity of APA and binds to the S1 site of the enzyme (11, 22, 32).
FIGURE 5.
Molecular modeling of catalytic cavity of LVRN/APQ. Three-dimensional structural models of catalytic cavities of wild-type (A) and H379G (B) LVRN/APQs are shown. Models were generated as a template of the T. acidophilum tricorn interacting factor F3 x-ray structure. Asn378 and His379 around the exopeptidase motif, His415, Glu416, His419, and Glu438 in the HEXXH(X)18E motif (15), Gln238 (24), Glu382 (16, 17), and Tyr503 (55) are shown as residues in the catalytic cavity of LVRN/APQ. The β-strands, which correspond to β18–β20 of T. acidophilum tricorn interacting factor F3, are numbered. Side-chain conformation is around the exopeptidase motif. C and D, side-chain conformations around the exopeptidase motif of wild-type LVRN/APQ and H379G LVRN/APQ are shown, respectively.
Furthermore, we revealed the significance of the amino acid residue at position 379 of human LVRN/APQ for recognition of the P1′ residue of endokinin C (supplemental Fig. S3 and Table 3) and the chain length of dynorphin-related peptides by the enzyme (Fig. 4). We also found that a point mutation replacing Gly357 of human APA with His altered not only the affinity to substrate (Km) but also the turnover numbers (kcat) (Table 4). The molecular model suggests that the substitution of His379 of LVRN/APQ with Gly may also cause significant changes in side-chain conformation of other exopeptidase motif residues (Fig. 5, C and D). It is suggested from the E. coli APN three-dimensional structure that Ala380 and Glu382 of LVRN/APQ (corresponding to Ala262 and Glu264 of E. coli APN) interact with the carbonyl oxygen of the scissile peptide bond and α-amino group of the substrate N terminus, respectively (30). In addition, introduction of a point mutation into Thr348 of APA made changes in turnover numbers (31). The substitution of His379 may also affect amino-docking efficiency and/or the catalysis of LVRN/APQ.
Taken together, these results suggested that His379 may be involved in the formation of the optimal global structure of the catalytic cavity for the unique enzymatic action of LVRN/APQ. X-ray structural analysis of LVRN/APQ is required to elucidate the mode of its characteristic catalytic action and the difference among other M1 aminopeptidases.
Tissue distribution of the M1 aminopeptidase family is usually ubiquitous; however, the expression of LVRN/APQ is shown to be restricted. Therefore, we reported that the transcript of human LVRN/APQ gene can be detected only in the placenta (18). In agreement with the result of the Northern blot analysis, by using the immunohistochemical technique, it was revealed that LVRN/APQ was specifically expressed in the human placenta, only on the cell surface of extravillous trophoblasts in chorion laeve (18), but not in other M1 aminopeptidase-expressing tissues, such as kidney, liver, ovary, spleen, and uterine tube.5 Furthermore, when searched for human sequences similar to human LVRN/APQ cDNA (accession number NM_173800) in the expressed sequence tag data base, no more than 65 sequences were retrieved (E-value < e-100). Among 53 sequences, each source tissue of which is clear, 39 sequences (73.5%) were derived from the placenta. These findings indicated that LVRN/APQ might not be abundant in our body, and placenta is one of the major tissues expressing LVRN/APQ. Moreover, it is notable that LVRN/APQ is highly expressed in the placenta of pre-eclampsia patients and the synovial tissues of rheumatoid arthritis patients (33, 34). In the placenta, several M1 aminopeptidases are known to be widely distributed. P-LAP, APA, and adipocyte-derived leucine aminopeptidase/endoplasmic reticulum aminopeptidase-1 are expressed in not only invasive extravillous trophoblasts but also proliferative extravillous trophoblasts and villous trophoblasts (35–37). In contrast, human APN is specifically expressed on the maternal endometrial stromal cells, i.e. decidual cells that receive trophoblast invasion (38). On the other hand, human LVRN/APQ is specifically expressed on the cell surface of invasive extravillous trophoblasts in maternal decidua (18). This expression profile of M1 aminopeptidases in the human placenta was confirmed again by immunohistochemistry (supplemental Fig. S4).
It is well known that the M1 aminopeptidases play important roles in the metabolism of vasoactive peptide hormones, such as angiotensins and kallidin in the placenta (39). In brief, APA degrades angiotensin II to angiotensin III, and then APN, P-LAP, and LVRN/APQ can generate angiotensin IV from angiotensin III and bradykinin from kallidin. Thus, these aminopeptidases are functionally linked and disorders of their activities might be associated with pre-eclampsia, a hypertension peculiar to pregnancy. Judging from subcellular localization, the enzymatic property (substrate specificity and inhibitor profile), and sequence similarity, APN and P-LAP could share substrates, e.g. angiotensin III, and pathophysiological functions with LVRN/APQ in the placenta. For example, kisspeptins, Kiss-1 gene products, are expressed in syncytiotrophoblasts in the placenta and their cognate receptor, GPR54, is expressed in not only syncytiotrophoblasts but also extravillous and villous trophoblasts (40, 41). Because kisspeptin-10 suppresses trophoblast invasion and LVRN/APQ inactivates kisspeptin-10, it is tempting to speculate that human LVRN/APQ and APN are involved in trophoblast functions, such as invasion into the uterine arteries and subsequent vascular remodeling through adjacent invalidation of the hormone. In particular, as it was recently reported that the gene expression of LVRN/APQ, but not other M1 aminopeptidases, was remarkably enhanced in the placenta of patients with severe pre-eclampsia, it is strongly suggested that pathophysiological functions of LVRN/APQ are associated with trophoblast functions in pregnancy (34).
Bestatin is an aminopeptidase inhibitor with multiple pharmacological activities, such as inhibition of the proliferation of tumor cells, including trophoblastoma (42) and interleukin-8-induced apoptosis of leukemic cells (43), modulation of inflammatory cytokine production from activated monocytes and macrophages (44), and enhancement of hormone secretion from granulosa cells (45). Effective dosages of bestatin in the first two studies were around 3 μm (1 μg/ml). Because the Ki value of the wild type and H379G LVRN/APQ are 3.81 and 12.2 μm, respectively, wild-type LVRN/APQ but not H379G mutant, e.g. murine LVRN/APQ, could be involved in various pharmacological activities of bestatin.
In this study, we focused on His379 of human LVRN/APQ, comprising the exopeptidase motif, and addressed its roles in the catalytic mechanism of the enzyme. We revealed that this characteristic His379 might contribute to maintain the appropriate structure of catalytic cavity, which in turn gives human LVRN/APQ distinct enzymatic properties from other M1 aminopeptidases and orthologues. It is speculated that the gene evolution of LVRN/APQ through the replacement of Gly with His may be associated with the evolutionarily different function of the placenta between primates and rodents.
Acknowledgments
We are grateful to Dr. Masayuki Nakanishi of Matsuyama University for fruitful discussion and Dr. Hiroshi Nakayama of RIKEN Biomolecular Characterization Team for the assistance in kinetic study. We also thank Drs. Masashi Miyano and Mitsuaki Sugahara of RIKEN SPring-8 Center, Harima Institute, for helpful discussions.
This work was supported in part by grants-in-aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and a grant for the “Chemical Biology Research Program” from RIKEN.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
H. Fujiwara, unpublished data.
- LVRN
- laeverin
- APQ
- aminopeptidase Q
- MCA
- 4-methylcoumaryl-7-amide
- P-LAP
- placental leucine aminopeptidase
- APA
- aminopeptidase A
- APN
- aminopeptidase N
- HPLC
- high performance liquid chromatography
- PIPES
- 1,4-piperazinediethanesulfonic acid.
REFERENCES
- 1.Hooper N. M. (1994) FEBS Lett. 354, 1–6 [DOI] [PubMed] [Google Scholar]
- 2.Tsujimoto M., Hattori A. (2005) Biochim. Biophys. Acta 1751, 9–18 [DOI] [PubMed] [Google Scholar]
- 3.Rogi T., Tsujimoto M., Nakazato H., Mizutani S., Tomoda Y. (1996) J. Biol. Chem. 271, 56–61 [DOI] [PubMed] [Google Scholar]
- 4.Rangel R., Sun Y., Guzman-Rojas L., Ozawa M. G., Sun J., Giordano R. J., Van Pelt C. S., Tinkey P. T., Behringer R. R., Sidman R. L., Arap W., Pasqualini R. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 4588–4593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marchiò S., Lahdenranta J., Schlingemann R. O., Valdembri D., Wesseling P., Arap M. A., Hajitou A., Ozawa M. G., Trepel M., Giordano R. J., Nanus D. M., Dijkman H. B., Oosterwijk E., Sidman R. L., Cooper M. D., Bussolino F., Pasqualini R., Arap W. (2004) Cancer Cell 5, 151–162 [DOI] [PubMed] [Google Scholar]
- 6.Sato Y. (2004) Biol. Pharm. Bull. 27, 772–776 [DOI] [PubMed] [Google Scholar]
- 7.Serwold T., Gonzalez F., Kim J., Jacob R., Shastri N. (2002) Nature 419, 480–483 [DOI] [PubMed] [Google Scholar]
- 8.Saric T., Chang S. C., Hattori A., York I. A., Markant S., Rock K. L., Tsujimoto M., Goldberg A. L. (2002) Nat. Immunol. 3, 1169–1176 [DOI] [PubMed] [Google Scholar]
- 9.Tanioka T., Hattori A., Masuda S., Nomura Y., Nakayama H., Mizutani S., Tsujimoto M. (2003) J. Biol. Chem. 278, 32275–32283 [DOI] [PubMed] [Google Scholar]
- 10.Hattori A., Tsujimoto M. (2004) Biol. Pharm. Bull. 27, 777–780 [DOI] [PubMed] [Google Scholar]
- 11.Goto Y., Hattori A., Ishii Y., Mizutani S., Tsujimoto M. (2006) J. Biol. Chem. 281, 23503–23513 [DOI] [PubMed] [Google Scholar]
- 12.Goto Y., Hattori A., Ishii Y., Tsujimoto M. (2006) FEBS Lett. 580, 1833–1838 [DOI] [PubMed] [Google Scholar]
- 13.Mizutani S., Ishii M., Hattori A., Nomura S., Numaguchi Y., Tsujimoto M., Kobayshi H., Murohara T., Wright J. W. (2008) Heart Fail. Rev. 13, 273–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Albiston A. L., McDowall S. G., Matsacos D., Sim P., Clune E., Mustafa T., Lee J., Mendelsohn F. A., Simpson R. J., Connolly L. M., Chai S. Y. (2001) J. Biol. Chem. 276, 48623–48626 [DOI] [PubMed] [Google Scholar]
- 15.Vazeux G., Wang J., Corvol P., Llorens-Cortès C. (1996) J. Biol. Chem. 271, 9069–9074 [DOI] [PubMed] [Google Scholar]
- 16.Vazeux G., Iturrioz X., Corvol P., Llorens-Cortes C. (1998) Biochem. J. 334, 407–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luciani N., Marie-Claire C., Ruffet E., Beaumont A., Roques B. P., Fournié-Zaluski M. C. (1998) Biochemistry 37, 686–692 [DOI] [PubMed] [Google Scholar]
- 18.Fujiwara H., Higuchi T., Yamada S., Hirano T., Sato Y., Nishioka Y., Yoshioka S., Tatsumi K., Ueda M., Maeda M., Fujii S. (2004) Biochem. Biophys. Res. Commun. 313, 962–968 [DOI] [PubMed] [Google Scholar]
- 19.Puente X. S., Sánchez L. M., Overall C. M., López-Otín C. (2003) Nat. Rev. Genet. 4, 544–558 [DOI] [PubMed] [Google Scholar]
- 20.Maruyama M., Hattori A., Goto Y., Ueda M., Maeda M., Fujiwara H., Tsujimoto M. (2007) J. Biol. Chem. 282, 20088–20096 [DOI] [PubMed] [Google Scholar]
- 21.Matsumoto H., Rogi T., Yamashiro K., Kodama S., Tsuruoka N., Hattori A., Takio K., Mizutani S., Tsujimoto M. (2000) Eur. J. Biochem. 267, 46–52 [DOI] [PubMed] [Google Scholar]
- 22.Goto Y., Hattori A., Mizutani S., Tsujimoto M. (2007) J. Biol. Chem. 282, 37074–37081 [DOI] [PubMed] [Google Scholar]
- 23.Kyrieleis O. J., Goettig P., Kiefersauer R., Huber R., Brandstetter H. (2005) J. Mol. Biol. 349, 787–800 [DOI] [PubMed] [Google Scholar]
- 24.Goto Y., Tanji H., Hattori A., Tsujimoto M. (2008) Biochem. J. 416, 109–116 [DOI] [PubMed] [Google Scholar]
- 25.Thunnissen M. M., Nordlund P., Haeggström J. Z. (2001) Nat. Struct. Biol. 8, 131–135 [DOI] [PubMed] [Google Scholar]
- 26.Iturrioz X., Rozenfeld R., Michaud A., Corvol P., Llorens-Cortes C. (2001) Biochemistry 40, 14440–14448 [DOI] [PubMed] [Google Scholar]
- 27.Laustsen P. G., Vang S., Kristensen T. (2001) Eur. J. Biochem. 268, 98–104 [DOI] [PubMed] [Google Scholar]
- 28.Ye S., Chai S. Y., Lew R. A., Albiston A. L. (2007) Biol. Chem. 388, 399–403 [DOI] [PubMed] [Google Scholar]
- 29.Addlagatta A., Gay L., Matthews B. W. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 13339–13344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ito K., Nakajima Y., Onohara Y., Takeo M., Nakashima K., Matsubara F., Ito T., Yoshimoto T. (2006) J. Biol. Chem. 281, 33664–33676 [DOI] [PubMed] [Google Scholar]
- 31.Claperon C., Banegas-Font I., Iturrioz X., Rozenfeld R., Maigret B., Llorens-Cortes C. (2009) J. Biol. Chem. 284, 10618–10626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Claperon C., Rozenfeld R., Iturrioz X., Inguimbert N., Okada M., Roques B., Maigret B., Llorens-Cortes C. (2008) Biochem. J. 416, 37–46 [DOI] [PubMed] [Google Scholar]
- 33.Haas C. S., Creighton C. J., Pi X., Maine I., Koch A. E., Haines G. K., Ling S., Chinnaiyan A. M., Holoshitz J. (2006) Arthritis Rheum. 54, 2047–2060 [DOI] [PubMed] [Google Scholar]
- 34.Sitras V., Paulssen R. H., Grønaas H., Leirvik J., Hanssen T. A., Vårtun A., Acharya G. (2009) Placenta 30, 424–433 [DOI] [PubMed] [Google Scholar]
- 35.Ino K., Nagasaka T., Okamoto T., Uehara C., Nakazato H., Nakashima N., Mizutani S. (2000) Placenta 21, 63–72 [DOI] [PubMed] [Google Scholar]
- 36.Ino K., Kikkawa F., Suzuki T., Kajiyama H., Shibata K., Nomura S., Itakura A., Ito M., Nagasaka T., Hattori A., Tsujimoto M., Mizutani S. (2003) Lab. Invest. 83, 1799–1809 [DOI] [PubMed] [Google Scholar]
- 37.Nomura M., Tsukahara S., Ando H., Katsumata Y., Okada M., Itakura A., Nomura S., Kikkawa F., Nagasaka T., Mizutani S. (2002) Placenta 23, 631–639 [DOI] [PubMed] [Google Scholar]
- 38.Imai K., Maeda M., Fujiwara H., Okamoto N., Kariya M., Emi N., Takakura K., Kanzaki H., Mori T. (1992) Biol. Reprod. 46, 328–334 [DOI] [PubMed] [Google Scholar]
- 39.Mitsui T., Nomura S., Itakura A., Mizutani S. (2004) Biol. Pharm. Bull. 27, 768–771 [DOI] [PubMed] [Google Scholar]
- 40.Bilban M., Ghaffari-Tabrizi N., Hintermann E., Bauer S., Molzer S., Zoratti C., Malli R., Sharabi A., Hiden U., Graier W., Knöfler M., Andreae F., Wagner O., Quaranta V., Desoye G. (2004) J. Cell Sci. 117, 1319–1328 [DOI] [PubMed] [Google Scholar]
- 41.Janneau J. L., Maldonado-Estrada J., Tachdjian G., Miran I., Motté N., Saulnier P., Sabourin J. C., Coté J. F., Simon B., Frydman R., Chaouat G., Bellet D. (2002) J. Clin. Endocrinol. Metab. 87, 5336–5339 [DOI] [PubMed] [Google Scholar]
- 42.Ino K., Goto S., Okamoto T., Nomura S., Nawa A., Isobe K., Mizutani S., Tomoda Y. (1994) Jpn. J. Cancer Res. 85, 927–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mishima Y., Matsumoto-Mishima Y., Terui Y., Katsuyama M., Yamada M., Mori M., Ishizaka Y., Ikeda K., Watanabe J., Mizunuma N., Hayasawa H., Hatake K. (2002) J. Natl. Cancer Inst. 94, 1020–1028 [DOI] [PubMed] [Google Scholar]
- 44.Lkhagvaa B., Tani K., Sato K., Toyoda Y., Suzuka C., Sone S. (2008) Cytokine 44, 386–391 [DOI] [PubMed] [Google Scholar]
- 45.Tachibana T., Fujiwara H., Suginami H., Nakamura K., Honda T., Yamada S., Maeda M., Mori T. (1996) Hum. Reprod. 11, 497–502 [DOI] [PubMed] [Google Scholar]
- 46.Li L., Wang J., Cooper M. D. (1993) Genomics 17, 657–664 [DOI] [PubMed] [Google Scholar]
- 47.Nanus D. M., Engelstein D., Gastl G. A., Gluck L., Vidal M. J., Morrison M., Finstad C. L., Bander N. H., Albino A. P. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 7069–7073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Constam D. B., Tobler A. R., Rensing-Ehl A., Kemler I., Hersh L. B., Fontana A. (1995) J. Biol. Chem. 270, 26931–26939 [DOI] [PubMed] [Google Scholar]
- 49.Hattori A., Matsumoto H., Mizutani S., Tsujimoto M. (1999) J. Biochem. 125, 931–938 [DOI] [PubMed] [Google Scholar]
- 50.Schomburg L., Turwitt S., Prescher G., Lohmann D., Horsthemke B., Bauer K. (1999) Eur. J. Biochem. 265, 415–422 [DOI] [PubMed] [Google Scholar]
- 51.Look A. T., Peiper S. C., Rebentisch M. B., Ashmun R. A., Roussel M. F., Lemons R. S., Le Beau M. M., Rubin C. M., Sherr C. J. (1986) J. Clin. Invest. 78, 914–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fukasawa K. M., Fukasawa K., Kanai M., Fujii S., Harada M. (1996) J. Biol. Chem. 271, 30731–30735 [DOI] [PubMed] [Google Scholar]
- 53.Minami M., Ohno S., Kawasaki H., Rådmark O., Samuelsson B., Jörnvall H., Shimizu T., Seyama Y., Suzuki K. (1987) J. Biol. Chem. 262, 13873–13876 [PubMed] [Google Scholar]
- 54.Díaz-Perales A., Quesada V., Sánchez L. M., Ugalde A. P., Suárez M. F., Fueyo A., López-Otín C. (2005) J. Biol. Chem. 280, 14310–14317 [DOI] [PubMed] [Google Scholar]
- 55.Vazeux G., Iturrioz X., Corvol P., Llorens-Cortès C. (1997) Biochem. J. 327, 883–889 [DOI] [PMC free article] [PubMed] [Google Scholar]