Abstract
Microplusin, a Rhipicephalus (Boophilus) microplus antimicrobial peptide (AMP) is the first fully characterized member of a new family of cysteine-rich AMPs with histidine-rich regions at the N and C termini. In the tick, microplusin belongs to the arsenal of innate defense molecules active against bacteria and fungi. Here we describe the NMR solution structure of microplusin and demonstrate that the protein binds copper II and iron II. Structured as a single α-helical globular domain, microplusin consists of five α-helices: α1 (residues Gly-9 to Arg-21), α2 (residues Glu-27 to Asn-40), α3 (residues Arg-44 to Thr-54), α4 (residues Leu-57 to Tyr-64), and α5 (residues Asn-67 to Cys-80). The N and C termini are disordered. This structure is unlike any other AMP structures described to date. We also used NMR spectroscopy to map the copper binding region on microplusin. Finally, using the Gram-positive bacteria Micrococcus luteus as a model, we studied of mode of action of microplusin. Microplusin has a bacteriostatic effect and does not permeabilize the bacterial membrane. Because microplusin binds metals, we tested whether this was related to its antimicrobial activity. We found that the bacteriostatic effect of microplusin was fully reversed by supplementation of culture media with copper II but not iron II. We also demonstrated that microplusin affects M. luteus respiration, a copper-dependent process. Thus, we conclude that the antibacterial effect of microplusin is due to its ability to bind and sequester copper II.
Introduction
Antimicrobial peptides (AMPs)3 are widely distributed in nature and represent essential components of the first line of defense against infections. In mammals, AMPs are an important part of the innate immune system and may act either directly on microorganisms or by modulating the immune response (innate and adaptative) through (i) enhancement of phagocytosis, (ii) recruitment of immune cells to infection sites, (iii) induction of cytokines, and (iv) neutralization of the septic effects of lipopolysaccharide. In invertebrates, which have only innate immunity, AMPs are extremely effective and work as powerful weapons against bacteria, fungi, and protozoa (1–3).
AMPs are often short cationic molecules (12–100 amino acids and with a net charge ranging between +2 and +10) with an amphipathic structure. Based on their structures, they are classified as linear, forming α-helices, or cysteine-containing β-sheet(s); they are also classified based on their richness in specific residues such as proline, glycine, tryptophan, and histidine (1, 4, 5). In addition to cationic AMPs, there are some anionic AMPs with documented activity against a range of microorganisms (6–11).
Most information on such natural antibiotics relies on amphipathic cationic AMPs. In general, their positive net charge is essential for an initial electrostatic interaction with the negative charges of the cell envelopes and phospholipid membranes of microorganisms. Through their hydrophobic portion, they may insert into the membranes and disrupt physical integrity of the cell by permeabilization, depolarization, loss of cellular contents, and consequently, lysis (4, 5).
In comparison to cationic AMPs, much less is known about how anionic AMPs work. Some studies with anionic peptides derived from the dermcidin precursor protein showed that these peptides do not permeabilize Staphylococcus aureus and Escherichia coli cytoplasmic membranes. Instead, it seems that such anionic peptides bind to S. aureus membrane via interaction with a receptor (12).
Although the membrane has been described as the main target for many AMPs, there are several reports showing that AMPs may translocate through the bacterial membrane and act on intracellular targets, leading to inhibition of specific enzymes and macromolecule (DNA, RNA, protein, and cell wall) synthesis, as well as alteration of cellular division (4, 5, 13). Examples include pyrrocoricin, which binds to the bacterial heat-shock protein DnaK and affects chaperone-assisted protein folding (14); buforin II, which interacts with RNA and DNA (15); indolicidin, which prevents RNA synthesis (16); and lactoferricin B, which inhibits DNA, RNA, and protein synthesis (17).
AMPs that interact with metal ions present yet another antimicrobial mechanism: these peptides bind to and chelate metal ions, thus preventing their uptake by microorganisms. This has been demonstrated clearly for the human neutrophil proteins calprotectin and lactoferrin (18, 19). For other peptides, such as histatins from human saliva, hepicidin from human blood and urine and poly(arginylhistidine) from the fungus Verticillium kibiense, the relationship between their metal binding and antimicrobial abilities is still unproven (20–23).
In recent years, our research group has identified several AMPs from the cattle tick Rhipicephalus (Boophilus) microplus, an important vector of veterinary pathogens (24). One AMP, corresponding to the 33–61 segment of the bovine α-hemoglobin, was isolated from tick gut contents (25). The α-hemoglobin 33–61 fragment is active against the Gram-positive bacterium Micrococcus luteus and functions by membrane disruption (26). In addition to this fragment of hemoglobin, three additional AMPs have been isolated and characterized from tick hemolymph. Two of them, a member of the invertebrate defensins (27) and ixodidin (28), were purified from hemocytes. The third AMP was purified from cell-free tick hemolymph and eggs and was designated microplusin (27, 29).
Microplusin is a 10,204-Da peptide expressed in hemocytes, fat body, female reproductive tract and eggs, as well as in BME26 embryonic cells (27, 29, 30). Its antimicrobial activity was first shown against the Gram-positive bacteria M. luteus (27). Unlike the other AMPs from R. (B.) microplus, microplusin is rather an anionic peptide at physiological pH, with an estimated pI of 5.2. Microplusin has 61% identity with hebraein, an AMP from the tick Amblyomma hebraeum, with perfect correspondence between their six cysteine residues (31). In addition to the conserved cysteine residues, both AMPs share several histidine residues at their N and C termini (31). These features have been shown for peptides from other hard ticks, such as Ixodes scapularis (GI: 67083117) and Ixodes ricinus (GI: 82494338), as well as soft ticks, such as Ornithodorus coriaceus, Ornithodorus parkeri, and Argas monolakensis (32). These peptides are between 30 and 37% identical with microplusin, suggesting that they may form a new family of AMPs.
In the current work, we investigated the antimicrobial activity spectrum of microplusin. The most sensitive microorganisms were the Gram-positive bacteria M. luteus and the yeast Cryptococcus neoformans, with an MIC50 of 0.09 μm. We have adopted M. luteus as a model to study the mechanism of action of microplusin. Surprisingly, microplusin binds copper II, and this property is related to its antimicrobial effect on M. luteus. We also determined the solution structure of microplusin and mapped its interaction with copper II.
EXPERIMENTAL PROCEDURES
Microplusin
The recombinant and native microplusin were obtained as previously described (27, 29). The elution profiles of recombinant and native peptide (separate or mixed) were compared using chromatography on an analytical C18 column (300 Å, 5 μm, 4.6 mm × 250 mm, VydacTM), using a linear gradient of 2–60% acetonitrile in 0.046% trifluoroacetic acid over 30 min at a flow rate of 1.0 ml/min. Chromatographic analyses were performed using a Shimadzu LC-10 system with an SPD-M10AV diode array detector (Shimadzu), and the column effluent was monitored by absorbance at 225 nm. Recombinant microplusin was quantified by comparison with a standard curve constructed using the peak areas of known native peptide concentrations chromatographed under the same conditions as described above.
Isotopically labeled microplusin was obtained employing the procedure described previously (33). Briefly, an overnight bacterial suspension of E. coli (strain BL-21) containing a microplusin expression construct (29) was diluted to an optical density of 0.08 at 600 nm (A600) and grown in LB broth (Invitrogen) at 37 °C. At 0.6 A600, bacteria were harvested by centrifugation at 5000 × g, during 30 min, at 4 °C and washed three times (5000 × g, 30 min, at 4 °C) with an M9 salt solution (0.1 m KH2PO4, 0.24 m Na2HPO4·7H2O, 0.04 m NaCl, 0.09 m NH4Cl, pH 7.2). After that, bacteria were resuspended in the isotopically labeled minimal medium (20% M9 salt solution, where NH4Cl was replaced by 15NH4Cl (Cambridge Isotope Laboratories, Inc.), 2% sterile [13C]glucose solution (Cambridge Isotope Laboratories, Inc.), 10% Basal Vitamins Eagle's Medium (PerkinElmer Life Sciences), 0.002 m MgSO4, and 0.01 m CaCl2) and incubated during 1 h at 37 °C to allow the clearance of unlabeled metabolites. Induction of peptide expression and purification followed the procedures described previously for microplusin (29).
Bioassays
Microorganisms
The following strains were used: Micrococcus luteus (27); Bacillus subtilis PY79 from F. Gueiros- Filho's laboratory, Chemistry Institute, São Paulo University, Brazil; B. megaterium ATCC 10778; S. aureus ATCC 6538; S. epidermidis ATCC 12228; E. coli SBS 363 (27); Enterococcus faecalis ATCC 19433; Pseudomonas aeruginosa ATCC 14502; Serratia marcescens CDC2124; Aspergillus flavus, A. fumigatus NCPF 2109, A. niger A296, and Fusarium sp., all from E. B. Bertger's laboratory, Microbiology Institute, Rio de Janeiro Federal University, Brazil; Candida albicans MDM 8 (27); Saccharomyces cerevisiae ATCC 2601; and C. neoformans T1444 (serotype A) from M. L. Rodrigues' laboratory, Institute for Microbiology P. P. Goes, Federal University of Rio de Janeiro, Brazil.
Liquid Growth Inhibition Assay
Antimicrobial activity was monitored by liquid growth inhibition as described previously (27). Peptone Broth (PB, 0.5% NaCl, 1% Peptone at pH 7.4) and Potato Dextrose Broth (Invitrogen) were used for antibacterial and antifungal assays, respectively. Briefly, bacteria or fungi were incubated with 2-fold serial dilutions of microplusin (50–0.09 μm) in a 96-well microplate at 30 °C. The microbial growth was assessed by measurements of the A595. MIC100 and MIC50 were defined as the minimal inhibitory concentration that prevents 100 and 50% of the bacterial growth, respectively. The effect of metals on peptide activity was evaluated by liquid assays using M. luteus incubated with microplusin (50–0.09 μm) in PB supplemented with a trace element solution (TES) containing: 600 μm MgCl2·6H2O, 40 μm CaCl2·2H2O, 49.9 μm FeCl3·6H2O, 5.5 μm MnCl2·2H2O, 12.4 μm ZnCl2, 2.5 μm CuCl2·6H2O, 2.5 μm CoCl2·H2O, and 2.3 μm Na2MoO4 (34) or with each component of TES. After 18 h of incubation at 30 °C, the bacterial growth was analyzed by reading of the A595. As an additional assay, M. luteus was preincubated with 0.18 μm microplusin for 24 h, and then 2-fold serial dilutions of CuCl2·6H2O (0.18–0.03 μm) were added. After another 24 h of incubation, the bacterial growth was recorded. Bacteria incubated without microplusin or incubated without copper II addition were used as controls. Experiments were repeated twice.
The copper content of PB was analyzed by Chemical Analyses Laboratories, Technological Research Institute (São Paulo, Brazil), using inductively coupled plasma optical emission spectroscopy in a Vista MPX (Varian) apparatus. The copper concentration in PB was 0.03 ± 0.008 μm.
Growth Kinetics
M. luteus harvested in log phase was incubated in the presence of 7.8 μm microplusin in LB broth at 30 °C. Aliquots were withdrawn at various time intervals (0, 4, 8, 24, and 48 h) of incubation and plated on LB agar (Invitrogen) to evaluate the number of colony-forming units (CFUs). In addition, after 24 h of incubation, bacteria were washed three times in sterile phosphate-buffered saline (136 mm NaCl, 2.6 mm KCl, 10 mm NaH2PO4, 1.8 mm KH2PO4, pH 7.4) by centrifugation (2500 × g, 5 min) and suspended in LB broth (with or without microplusin). After another 24-h incubation at 30 °C, aliquots were taken for CFU evaluation. Experiments were repeated twice.
Membrane Permeabilization Assay
M. luteus harvested in log phase was incubated in PB in the presence of 7.8 μm microplusin for 24 h at 30 °C. After that, bacteria were washed with sterile phosphate-buffered saline and treated with the reagents from the Live/Dead BacLight bacterial viability stain kit (L-7007, Invitrogen) following the manufacturer's instructions. This kit is based on the use of a mixture of two nucleic acid stains: the green fluorescent SYTO® 9 stain and propidium iodide, a red fluorescent stain. The following controls were prepared: 1) untreated bacteria, 2) bacteria treated with 100% isopropanol for 1 h, and 3) bacteria treated with 10 μm gomesin (35) for 30 min. Cells were analyzed using an Axiovert S100 fluorescence microscope (Zeiss), equipped with a Hamamatsu C5810 digital camera. Experiments were repeated three times.
Transmission Electron Microscopy
M. luteus harvested in log phase was incubated with 7.8 μm microplusin for 24 h at 30 °C. Bacteria were fixed at 4 °C with 2.5% glutaraldehyde, post-fixed in 1% OsO4 in ultrapure water, and dehydrated prior to Spurr resin (Sigma) inclusion. Ultrathin sections of 70 nm were gathered onto copper grids and stained with 0.5% uranyl acetate for 5 min, washed in ultrapure water, and stained with 0.5% lead citrate. The ultrastructure was examined on a JEOL 100 CX-II electron microscope.
Oxygen Consumption Measurements
Oxygen consumption of M. luteus was measured polarographically at 30 °C using a computer-interfaced Clark-type electrode in PB in a final volume of 1 ml. Bacteria were grown overnight at 30 °C in PB under agitation, inoculated in fresh medium, and grown until A600 of 0.2 (early log phase). Growth was stopped by cooling on ice. Bacterial cultures at A600 of 0.2 were used either immediately (time zero) or after 30 min or 3 h of further growth at 30 °C. Before oxygen consumption recordings, bacteria grown for 30 min or 3 h were diluted with PB to obtain A600 = 0.2. The electrode was first stabilized at zero oxygen consumption in 0.7 ml of fresh PB with constant stirring in the thermo-balanced chamber of a Clark-type electrode before a fixed volume of culture (0.3 ml) was transferred to the chamber. Recordings of respiration data were initiated after closing the chamber with an air-tight lid. To evaluate the immediate effect of microplusin on M. luteus respiration (“MP-treated in chamber” experimental group), 7.8 μm microplusin was added to the chamber containing M. luteus before closing it. To evaluate later effects of microplusin on respiration (“MP-treated in culture” experimental group), M. luteus was first grown in the presence of 7.8 μm microplusin for 30 min or 3 h, diluted to A600 of 0.2 in PB, and then added to the chamber. The maximum respiration rate of both microplusin-treated and untreated bacteria was stimulated by the addition of 4 μm carbonyl cyanide m-chlorophenylhydrazone (CCCP) to the chamber 9–10 min after the recording of the basal respiration was initiated. Cyanide-sensitive respiration was inhibited by the addition of 1 mm potassium cyanide to the chamber 5–6 min after addition of CCCP. Values are represented as rate of O2 consumption in nanomoles/min/A600 of 0.2 of bacterial suspension ± S.E. Experiments were repeated at least three times.
Structural Characterization
Mass Spectrometry
Molecular mass was determined by electrospray ionization mass spectrometry, using a Finnigan LCQTMDuo mass spectrometer (ThermoQuest) at a mass to charge (m/z) range of 2000. Calibration was performed as described previously (35). All samples were dissolved in 5% acetonitrile acidified with 0.2% formic acid and analyzed in a positive mode. To analyze metal-peptide interaction, microplusin was incubated with the following salts in ultrapure water during 1 h at room temperature: MgCl2·6H2O, CaCl2·2H2O, FeSO4·H2O, MnCl2·2H2O, ZnCl2, CuCl2·6H2O, CoCl2·6H2O, and Na2MoO4 at a peptide/metal molar ratio of 1:16.
Reduction and Alkylation
Microplusin was reduced and alkylated by 9 mm dithiothreitol and 17 mm iodoacetamide, respectively, following the protocol previously described (36). The purification was performed as described for production of microplusin.
CD
CD measurements were made using a Jasco-710 spectropolarimeter. CD spectra were obtained over a range of 190–260 nm, using a quartz cell of 0.5 mm path length at 22 °C. 15 μm microplusin prepared in ultrapure water was incubated with 0–50 μm CuCl2·6H2O. For each analysis, six scans were accumulated and averaged. All CD spectra were corrected by subtraction of the background and then smoothed. The CD spectra are reported as the mean residue ellipticity ([θ]) in degrees cm2 dmol−1.
NMR Spectroscopy
NMR experiments (37, 38) for 1H, 13C, and 15N chemical-shift assignments and structure determination were acquired at 298 K on microplusin dissolved in 20 mm phosphate buffer, 50 mm NaCl, and 0.02% sodium azide, at pH 5.6. Two-dimensional-NOESY (100 ms mixing time) and TOCSY (35 and 70 ms spin-lock) spectra were acquired in both 90% H2O/10% D2O and in 99.98% D2O using 0.95 mm unlabeled microplusin. A 0.52 mm 15N-labeled sample in 90% H2O (10% D2O) was used for 15N HSQC, three-dimensional 15N-edited NOESY, and 15N-edited TOCSY experiments. 0.35 mm 13C,15N-double-labeled sample was used for the three-dimensional HCCH COSY, 13C-edited NOESY, CBCA(CO)NNH, CBCANNH, HBHA(CO)NNH, H(CCCO)NH-TOCSY (13 ms), and (H)CC(CO)NH-TOCSY (13 ms) experiments. Spectra were acquired in a Bruker DRX600 spectrometer equipped with a cryo-probe head (triple-resonance experiments) or in a Bruker AVIII 800 spectrometer. Data were processed using the software Topspin (v2.0, Bruker BioSpin GmbH, Germany). Assignment was carried out using the interactive program SPARKY (v3.106, T. D. Goddard and D. G. Kneller, University of California, San Francisco).
NOE Assignment and Structure Calculation
Assignment of NOESY spectra and structure calculations were made iteratively using the program ARIA 1.2 (39, 40) with CNS 1.1 (41). Initially, we manually assigned NOEs characteristic of α-helicoidal secondary structure namely amide-amide (HN, HN i, and i+1, and HN, HN i, and i+2), Hα-amide (Hα, HN i, and i+3), (Hα, N i, and i+4), and (Hα, Hβ i, and i+3) and some unambiguous side-chain interactions. Hydrogen bonds were assigned from slowly exchanging amide-protons identified in a two-dimensional 15N HSQC spectrum following exchange into D2O. Further NOEs were assigned automatically by ARIA. For the structure calculations we also used phi and psi dihedral angles derived from CSI (42). Conversion of CSI output in dihedral restraints was done as implemented in ARIA: −65 and −35 with error estimates of 30 degrees were set, respectively, as phi and psi dihedral restraints for residues found to be in helical regions from their characteristic Hα and Cα chemical shifts (43). In the last ARIA iteration 200 structures were calculated by restrained simulated annealing, and the 15 best structures regarding total energy were refined in an explicit water box and considered as characteristic of the ensemble.
Relaxation Times and 1H–15N NOEs
15N T1 and T2 relaxation times were extracted from two series of two-dimensional 1H-15N correlated spectra (44) with relaxation delays of 12, 102, 152, 202, 302, 402, 602, 902, 2002, and 5002 ms for T1 and 6, 10, 18, 28, 34, 42, 82, 122, 162, 202, and 242 ms for T2. Relaxation times were fitted as implemented in SPARKY (v3.106). Steady-state 1H-15N NOEs were determined according to the formula NOE = I/Iref, where I is intensity of a cross-peak in a two-dimensional 1H-15N correlated spectrum with broadband 1H presaturation and Iref is the intensity in a reference spectrum recorded without presaturation (44). All experiments were performed for three 15N-labeled microplusin samples: 0.52 mm microplusin in 20 mm phosphate buffer, 50 mm NaCl, and 0.02% sodium azide, at pH 5.6, 90% H2O/10% D2O; 0.1 mm microplusin in 90% H2O/10% D2O; and 0.1 mm microplusin plus 0.1 mm CuCl2 in 90% H2O/10% D2O. Spectra were acquired in a Bruker AVIII 800 spectrometer at 298 K.
Mapping of the Interaction between Microplusin and Cu2+
A series of 1H-15N HSQC spectra with 1024 × 256 data points were acquired for an 15N-labeled microplusin sample of 0.1 mm in 90% H2O/10% D2O with increasing CuCl2 concentrations: 0, 20, 40, 60, 80, and 100 μm. Additionally, two-dimensional TOCSY spectra, spin-lock 35 ms and 1024 × 1024 data points, were acquired for two samples containing, respectively, 950 μm unlabeled microplusin in the absence or presence of 470 μm CuCl2 in 20 mm phosphate buffer, 50 mm NaCl, and 0.02% sodium azide, at pH 5.6. All spectra were compared in the absence and in presence of Cu2+.
Atomic Coordinates and NMR Restraints
The atomic coordinates and NMR restraints for the ensemble of the 15 best structures calculated for microplusin were added to the RCSB Protein Data Bank (PDB ID 2knj).
RESULTS
Microplusin Structural Studies
Recombinant Microplusin
Microplusin was produced as described previously (29) using an E. coli expression system. Native (27) and recombinant microplusin (29) have the same molecular mass (10,204 Da) and co-eluted on high-performance liquid chromatography, suggesting that both peptides have the same structure (results not shown). In addition, native and recombinant microplusin were similarly active against M. luteus, with MIC100 of 0.38 μm for the recombinant and 0.78 μm for the native peptide. To evaluate the importance of the disulfide bridges for microplusin activity, the peptide was reduced and alkylated and tested for its antimicrobial properties. The reduced alkylated microplusin did not show any activity against M. luteus in the range of concentrations tested (0.02–5 μm).
CD Spectroscopy of Microplusin
CD spectra of microplusin in water presented a positive peak at 195 nm and negative peaks at 208 and 222 nm (supplemental Fig. S1), typical of proteins in α-helical conformation.
NMR Structure of Microplusin
The sequence of the microplusin construct used in NMR experiments is shown in Fig. 1. The sequence was numbered from His 1 to His 90, following the amino acid sequence of the mature peptide. It is important to note, however, that microplusin is initially made as a precursor, with a 20 amino acid-long signal peptide prior to His 1 (27). The solution structure of microplusin was determined by multidimensional NMR spectroscopy (37, 38) with 0.50 mm 15N-labeled, 0.35 mm 13C-, 15N-labeled and 0.95 mm unlabeled peptide solution in water. In agreement with the initial CD spectra acquired, the microplusin NMR spectra are typical of α-helical structure, showing a characteristic dispersion of amide and alpha protons upfield. The assignments of distance restraints derived from NOEs were made in a semi-automated fashion. Initially, 90 H-H distances from NOEs characteristic of α-helical secondary structure, namely amide-amide (HN, HN i, i+1 and HN, HN i, i+2), Hα-amide (Hα, HN i, i+3), (Hα, N i, i+4) and (Hα, Hβ i, i+3), were manually assigned. Additionally, 40 hydrogen bonds were assigned from slowly exchanging amide protons identified in a two-dimensional 15N HSQC spectrum following exchange into D2O. The initial manual NOE assignments and slowly exchanging amide protons identified are schematically summarized in supplemental Fig. 2. We also used Hα and Cα chemical shift index analysis (42) for derivation of phi and psi dihedral angle restraints. The secondary topology of microplusin, shown in Fig. 1, is derived from the consensus analysis of chemical shift indexes, and the NOE pattern and H/D exchange.
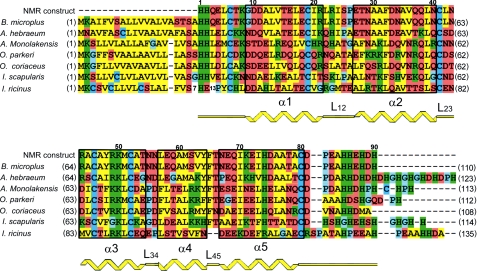
FIGURE 1.
Primary and secondary structure of microplusin-like peptides. Alignment of sequences of R. (B.) microplus microplusin and other homologous sequences from several tick species. Residue numbers over the sequences refer to R. (B.) microplus microplusin mature peptide and were used throughout this report. Numbers at left and right of the sequences indicate the positions of the first and last amino acid, respectively, in the coding sequence. Color coding: yellow are hydrophobic, green are basic, brown are acid or polar, and blue are proline, glycine, or cysteine residues. Secondary structure topology as determined by NMR data is indicated in yellow, and amino acids present in helices are boxed. GI entry codes for sequences are as follows: R. (B.) microplus 28864185, A. hebraeum 40888893, A. monolakensis 114153066, O. parkeri 149287050, O. coriaceus 172051120, I. scapularis 215501519, and I. ricinus 82494339.
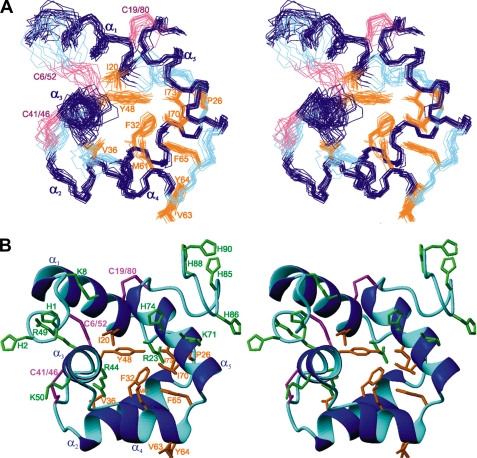
FIGURE 2.
Solution NMR structure of R. (B.) microplus microplusin (stereoview). A, superposition of the backbone atoms for the 15 lowest energy structures of microplusin, residues 6–80. α-Helices (α1–α5), loops (L12–L45), and disulfide-bridges are colored navy blue, cyan, and magenta, respectively. Selected side chains are displayed and labeled. The structural statistics are given in Table 1. B, ribbon diagram of the microplusin lowest energy structure. The coloring scheme is the same as in panel A. The N and C termini are non-convergent and were excluded from panel A for clarity.
Other manually assigned NOEs corresponded to some side-chain hydrogens with very well resolved resonances, shifted upfield (e.g. Ile-73, Ile-70, and Val-63 methyl groups), and the aromatic moieties of Phe-32, Tyr-48, Tyr-64, and Phe-65. In total, ∼110 distance restraints were assigned manually. Nevertheless, the great majority of NOE restraints were assigned automatically using the program ARIA (39, 40), yielding in total 1244 restraints. A complete count of restraints used in the final structure calculation is listed in Table 1.
TABLE 1.
Structure statistics of microplusin
| A) Number of restraints | |
| NOE restraints | 1244 |
| Intra-residual | 595 |
| Sequential | 319 |
| Medium range | 172 |
| Long range | 158 |
| Ambiguous | 179 |
| Hydrogen bonds (two restraints each) | 40 |
| Dihedral angles (φ, ψ) | 98 |
| Total | 1601 |
| B) r.m.s.d. from experimental restraints | |
| NOEs (Å) | 0.05 ± 0.02 |
| Dihedral angles (°) | 1.5 ± 0.4 |
| C) CNS potential energy (kcal mol−1) | |
| Etotal | −2240 ± 125 |
| Ebonds | 51 ± 6 |
| Eangles | 242 ± 15 |
| Eimpropers | 163 ± 18 |
| Edihedral | 557 ± 7 |
| EvdW | −235 ± 29 |
| Eelec | −3900 ± 87 |
| Enoe | 195 ± 48 |
| Ecdih | 15 ± 11 |
| D) r.m.s.d. (Å) between average structure and the ensemblea | |
| Backbone | 0.40 ± 0.07 |
| All non-H | 0.79 ± 0.12 |
| E) Ramachandran plot analysis (%)a | |
| Residues in most favored regions | 82.7 |
| Residues in additionally allowed regions | 10.5 |
| Residues in generously allowed regions | 4.3 |
| Residues in disallowed regions | 2.4 |
a For residues 6–80.
The ensemble consisting of the 15 lowest-energy structures is shown in Fig. 2 A and a ribbon representation of the lowest-energy structure is shown in Fig. 2B. This ensemble has a mean backbone root mean square deviation (r.m.s.d.) of 0.85 Å compared with the average for the well-structured regions (residues 6–80). The backbone r.m.s.d. over all residues 1–90 is 1.73 Å. Microplusin's tertiary structure is well defined by an average of 4.5 medium- or long-range NOE distance restraints per residue. Detailed structure statistics are shown in Table 1.
Microplusin consists of five α-helices. α1 comprises residues Gly-9 to Arg-21, α2 residues Glu-27 to Asn-40, α3 extends from residue Arg-44 to Thr-54, α4 from Leu-57 to Tyr-64, and α5 from Asn-67 to Cys-80. Helices α1, α2, α3, and α5 are ∼3–3.5 turns each, while α4 is shorter, with only 2 turns. These five helices are folded in three-dimensional space as a globular domain (Fig. 2). In all helices most of the hydrophobic residues (α2: Phe-32 and Val-36; α3: Ala-45 and Tyr-48; α4: Met-61; and α5: Ile-70, Ile-73, and Ala-77) are buried in the hydrophobic core of the peptide and most of the hydrophilic residues are directed toward the solvent. The six cysteines present in the primary sequence of microplusin are involved in disulfide bridges, namely between Cys-6 and Cys-52, Cys-19 and Cys-80, and Cys-41 and Cys-46. The loop regions are only slightly less convergent than the helix regions, except for the N-terminal (residues 1–5) and C-terminal (residues 81–90) regions, which are very poorly defined. Supplemental Fig. S3 shows the number of NOEs observed as well as the average local r.m.s.d. as a function of residue number. The N and C termini are characterized by only a few medium and long range NOEs (supplemental Fig. S3A), and higher local average r.m.s.d. values were obtained in these terminal regions for the corresponding NMR structures (supplemental Fig. S3B).
Backbone Dynamics of Microplusin from NMR Measurements
To assess the degree of internal mobility of microplusin, we performed 15N T1 and T2 relaxation and 1H-15N heteronuclear NOE experiments (supplemental Fig. S4). The relaxation data showed increased 15N T2 values and smaller 1H-15N heteronuclear NOEs for residues in both N and C termini (residues 1–3 and 81–90), indicating internal mobility of these regions. For residues 6 and 7 (at the beginning of helix α2) and residues 42, 43, 54, and 55 at the extremities of the helix α3, very short T2 times were obtained and are related to exchange processes on the millisecond timescale.
Similar T2 values were obtained (for the residues that are observable) using the microplusin samples at different concentrations: 0.52 (supplemental Fig. S4), 0.1, and 0.1 mm for the microplusin-Cu2+ complex (supplemental Fig. S5).
A quantitative analysis for global and internal motions was performed using the Lipari and Szabo model-free formalism (45) and the program Tensor2 (46). At 0.52 mm, average values of relaxation rates (R1 = 1.45 s−1 and R2 = 10.5 s−1) were calculated considering all the residues in secondary structure elements and from the ratio (R2/R1 = 7.3) the global correlation time for isotropic tumbling was estimated (τc = 5.7 ns) in agreement with the expected value for a 10-kDa globular protein in monomeric state. S2, the square of the order parameter, was typically between 0.8 and 0.9 for most residues, including loop regions, except for residues 1–3 and 81–90 where an S2 value between 0.60 and 0.00 and an effective correlation time (τe) in the sub-nanosecond timescale were necessary to fit the relaxation data, indicating that these regions are disordered and undergo fast movements of high amplitude. Additionally, for residues 6, 7, 42, 43, 54, and 55, exchange rates (Rex) ranging form 10 to 40 s−1 were obtained to characterize the exchange process related to the small T2 values observed.
Microplusin Binds to Metals
A growing number of studies have shown that histidine-rich AMPs are able to complex with metal ions such as copper, nickel, zinc, and iron (23, 47–50). Because microplusin has several histidine residues, 2 at the N terminus, 1 at position 74, and 4 at the C terminus (Fig. 1), we evaluated its metal-binding ability. This was done by incubating microplusin with different ions (Mg2+, Cl−, Ca2+, Fe2+, SO42−, Mn2+, Zn2+, Cu2+, Co2+, Na+, and MoO42−), followed by analysis of the mixtures by electrospray ionization-mass spectrometry, in non-denaturing conditions to preserve non-covalent interactions. For most metals, no evidence for binding could be detected, except for Cu2+ and Fe2+. The difference in molecular mass (60 Da) between microplusin alone (10,204 Da) and microplusin in the presence of copper II (10,264 Da) suggests that copper II ions binds to microplusin at a molar ratio of 1:1. Interestingly, quantitative binding was observed even at concentrations of peptide and Cu2+ in the nanomolar range. In addition to copper II, we detected a major species at 10,379 Da, suggesting that microplusin binds also to iron II, at a molar ratio of 1:3.
Mapping of the Interaction between Microplusin and Cu2+
Because the copper II-chelating ability of microplusin is crucial for its antimicrobial activity against M. luteus (see below), we further characterized the interaction between microplusin and this metal. Addition of Cu2+ from 1 to 50 μm caused only slight changes in the CD spectrum of microplusin, indicating that ion binding does not significantly alter the protein conformation (supplemental Fig. S1). Although the changes were small, [θ]222 was found to decrease hyperbolically with increasing Cu2+ concentration, reaching a plateau at ∼25 μm Cu2+ (not shown) and suggesting that copper II binding to microplusin occurs at a metal/peptide molar ratio of 1:1.
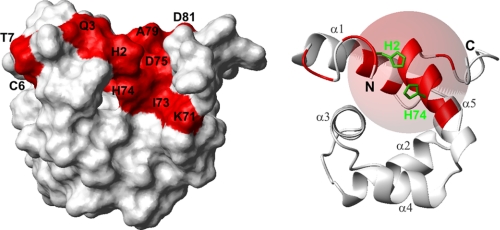
Because of the paramagnetism of Cu2+, protons in its neighborhood are expected to give no NMR signals due to fast transverse relaxation. In the microplusin-Cu2+ complex there should be a “blind region” for 1H detection around the Cu2+ with a radius of ∼8–11 Å (51). In an attempt to map the region affected by Cu2+ binding to microplusin, we observed the effect of CuCl2 titration (0–100 μm) on the 1H-15N HSQC spectrum of a 100 μm 15N-labeled microplusin sample. During the titration we observed a clear formation of the microplusin-Cu2+ complex by the appearance of a new set of signals relative to the bound form of microplusin. These new signals increase in intensity as CuCl2 is being added. On the other hand the original signals of the free microplusin decrease in intensity with increasing concentrations of CuCl2. At 100 μm CuCl2, virtually only the bound form is observed (supplemental Fig. S6), evidence for formation of a quantitative 1:1 complex. This behavior indicates slow exchange between the bound and free forms of microplusin and was observed even for amide signals that show only a small chemical shift change upon binding (e.g. Asn-67 and ∼30 Hz) (supplemental Fig. S7). Slow exchange is indicative of a strong binding with Kd usually in the nanomolar range or smaller. The 1H-15N HSQC spectrum of the microplusin-Cu2+ complex was fully assigned based on the spectrum of the free microplusin and with support from two-dimensional TOCSY spectra for microplusin alone (0.95 mm) and for microplusin in the presence of Cu2+ (0.47 mm) to identify the amino acid types. There were sixteen missing amide signals in the complex: His-2, Gln-3, Cys-6, and Thr-7 in the N-terminal region; Glu-16, Leu-17, and Ile-20 in the helix α1; Lys-71, Ile-73, His-74, Asp-75, Ala-76, Ala-77, Thr-78, and Ala-79 in the helix α5; and Asp-81 in the C-terminal region. These undetected amides were mapped in the structure of microplusin (Fig. 3). Because the termini are disordered but some residues of the N termini show the Cu2+ effect, in the structure shown in Fig. 3 we restrained His-2 and His-74 together as a model. The structured domain was not at all affected by the inclusion of this restraint. All amides under the effect of Cu2+ (blind region) are inside a sphere of ∼9-Å radius, except Cys-6 and Thr-7 (Fig. 3).
FIGURE 3.
Mapping of the microplusin binding region for copper II ions. Model structure of microplusin restraining His-2 and His-74 together. Residues not observable in a 1H-15N HSQC spectrum upon Cu2+ binding are colored red on the molecular surface of microplusin (left) and in the ribbon representation of the structure (right). A sphere of 9 Å, including almost all affected amides, has been added to the ribbon representation (right). The surface is in the same orientation as in Fig. 2.
Microplusin Mode of Action
Antimicrobial Spectra of Microplusin
To study the mode of action of microplusin we initially evaluated the activity spectrum of microplusin. We determined the MIC50, because microplusin had partial inhibitory activity for some tested microorganisms, even at the highest concentration used (50 μm). The recombinant peptide was active against almost all Gram-positive bacteria tested, except for S. aureus (Table 2). Microplusin also had a significant antifungal activity, with MIC50 below 6.25 μm for all tested filamentous fungi. However, no effect was detected for Gram-negative bacteria up to 50 μm. The most sensitive microorganisms were M. luteus and C. neoformans (MIC50 = 0.09 μm). Therefore, we adopted M. luteus as a model to study the antibacterial mechanism of microplusin.
TABLE 2.
Antimicrobial activity spectrum of recombinant microplusin
Experiments were repeated at least three times.
| Microorganisms | MIC50a |
|---|---|
| μm | |
| Gram-positive bacteria | |
| M. luteus | 0.09 |
| B. subtilis | 3.12 |
| B. megaterium | 50.0 |
| S. aureus | NDb |
| S. epidermidis | 1.56 |
| Gram-negative bacteria | |
| E. coli | ND |
| E. faecalis | ND |
| P. aeruginosa | ND |
| S. marcescens | ND |
| Filamentous fungi | |
| A. flavus | 6.25 |
| A. fumigatus | 3.12 |
| A. niger | 0.78 |
| Fusarium sp. | 6.25 |
| Yeast | |
| C. albicans | ND |
| C. neoformans | 0.09 |
| S. cerevisiae | 25.0 |
a MIC50 means minimal inhibitory concentration that inhibits 50% of the microorganism growth.
b ND, not detected up to 50 μm.
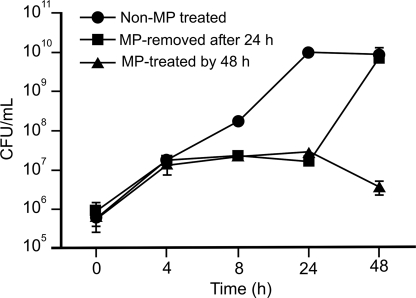
Bacteriostatic Effect of Microplusin
To ascertain whether microplusin was bacteriostatic or bactericidal, viability studies were performed (Fig. 4). Results revealed that the peptide at 7.8 μm (40× MIC100) (microplusin-treated culture, MP-treated, Fig. 4) was not lethal for M. luteus after a 48-h incubation. However, a difference in the number of CFU of approximately two orders of magnitude was observed after a 24-h incubation comparing a control culture of M. luteus (not treated with microplusin) and a microplusin-treated culture. This effect could be reversed after removing the bioactive peptide from the medium. Therefore, microplusin is bacteriostatic for M. luteus under our experimental conditions.
FIGURE 4.
Kinetics of M. luteus growth during treatment with microplusin. Recombinant microplusin (7.8 μm) was added to an exponential phase culture of M. luteus. Twenty-four hours later, bacteria were washed by centrifugation and suspended in LB, without peptide (MP-removed after 24 h) or with the peptide (MP-treated for 48 h). As a control, water was added to a bacterial culture (Non-MP treated). Aliquots were removed after 0, 4, 8, 24, and 48 h incubation at 37 °C. The number of CFUs was evaluated after an overnight incubation at 37 °C. MP stands for microplusin. The standard errors (S.E.) obtained from three CFU counts are indicated as bars. Experiments were repeated twice.
Microplusin Does Not Affect Membrane Permeability
Because several AMPs exert their effect by membrane permeabilization (4), we investigated whether the membrane might be the target of microplusin. To do this, we used two fluorochromes, SYTO® 9 and propidium iodide, which stain bacterial DNA with intact/damaged and only damaged membranes, respectively. Results showed that microplusin at 7.8 μm (40× MIC100) did not cause injury to the M. luteus membrane after a 24-h incubation (data not shown). However, bacteria treated with isopropanol or with gomesin, an AMP whose membrane-permeabilizing activity has been described (52), were damaged (data not shown). In addition, analysis by transmission electron microscopy did not show any disruption of the M. luteus membrane incubated with microplusin (data not shown).
Copper II Rescues M. luteus Growth in the Presence of Microplusin
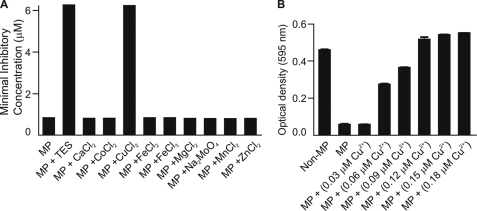
Because microplusin binds metal ions, we tested whether the addition of metals to the medium could affect its antimicrobial activity. When medium was supplemented with TES, we observed that microplusin activity against M. luteus was drastically reduced, with an increase of its MIC by 8-fold (Fig. 5A). Analysis of each component of the solution showed that addition of copper II alone reduced microplusin activity to the same level as TES (Fig. 5A). Moreover, copper II rescues bacterial growth when added after cells have been incubated with the peptide for 24 h (Fig. 5B). In this case, bacterial growth after copper II supplementation at a ratio of ∼1:1 attained an A equivalent to that found in the absence of microplusin. These results suggest that microplusin acts by chelating copper and inducing starvation for this metal. Interestingly, even though microplusin is capable of binding iron, supplementation with iron did not affect microplusin activity (Fig. 5A).
FIGURE 5.
Effect of metals on microplusin activity. A, M. luteus suspension was incubated with microplusin (50-0.09 μm) in PB supplemented with either trace element solution (TES) or separately with each component of TES. After 18 h of incubation, the MIC100 was determined by reading of the A595. B, M. luteus suspension was pre-treated with 0.18 μm microplusin for 24 h, and then serial dilutions of CuCl2·6H2O (0.18-0.03 μm) were added for an additional 24-h incubation. Optical density was determined at 595 nm. MP, microplusin. The standard error (S.E.) was obtained from three optical density measurements. Experiments were repeated twice.
Oxygen Consumption Measurements
To test whether M. luteus was indeed being starved for copper II, we evaluated the effect of microplusin on bacterial respiration. Copper II is essential for proper assembly and functioning of terminal oxidases of the electron transport chain (53). Results in Table 3 show that 7.8 μm microplusin had no immediate effect on M. luteus respiration (compare 4.02 ± 0.05 in A with 3.89 ± 0.20 nmol of O2/min/A600 0.2 in B). Similarly, respiration stimulated by the uncoupler CCCP increased to the same rate in both experimental conditions (from 4.02 ± 0.05 to 4.75 ± 0.15 in A and from 3.89 ± 0.20 to 4.60 ± 0.38 nmol of O2/min/A600 0.2 in B) and was equally inhibited by the cytochrome oxidase blocker KCN (from 4.75 ± 0.15 to 1.61 ± 0.23 in A and from 4.60 ± 0.38 to 1.44 ± 0.08 nmol of O2/min/A600 0.2 in B). To explore whether microplusin could have a delayed effect on M. luteus respiration, oxygen consumption was measured after growing bacteria in 7.8 μm microplusin for 30 min and 3 h. After adjustment of the A600 to 0.2, the rate of oxygen consumption of the control cultures grown for 30 min was the same as that observed for the control culture at time zero. However, although the rate of basal respiration of M. luteus was slightly lowered by microplusin (from 4.02 ± 0.05 in A to 3.58 ± 0.42 nmol of O2/min/A600 0.2 in C) after 30 min of exposure, stimulation of respiration by CCCP was fully impaired (compare basal respiration and respiration under CCCP stimulation in C). It is noteworthy that cyanide-resistant respiration (residual respiration after KCN addition) in the three conditions tested was intact, indicating that only cyanide-sensitive respiration was affected by chronic exposure to microplusin. At 3 h, the rate of oxygen consumption of the control cultures was greatly increased, even after adjusting the A600 to 0.2, likely reflecting the mid-exponential and more metabolically active phase of growth at this time. CCCP remained unable to stimulate respiration in microplusin-growing bacteria, but, at this time, microplusin produced further effects and considerably impaired both proliferation and basal respiration (data not shown).
TABLE 3.
Rate of O2 consumption (nmol/min/0.2 A600 nm) of bacterial suspension ± S.E.
Values in parentheses show percentage compared to basal respiration ± S.E. Experiments were repeated at least three times.
| Basala | CCCPb | KCNc | |
|---|---|---|---|
| Control (non-MP-treated) | 4.02 ± 0.05 | 4.75 ± 0.15 (118.48 ± 2.06%) | 1.61 ± 0.23 (36.29 ± 2.31%) |
| MP-treated on chamber | 3.89 ± 0.20 | 4.60 ± 0.38 (118.50 ± 4.86%) | 1.44 ± 0.08 (31.10 ± 2.64%) |
| MP-treated on culture | 3.58 ± 0.42 | 3.61 ± 0.41 (101.00 ± 0.94%) | 1.37 ± 0.05 (37.13 ± 2.52%) |
a Values for control at both time zero and 30 min of growth after adjusting A600 to 0.2.
b Addition of 4 μm CCCP.
c Addition of 1 mm potassium cyanide.
DISCUSSION
Microplusin Structure and Dynamics
We used NMR methods (37, 38) to determine a high resolution structure of microplusin in solution and to study its interaction with copper II. Fig. 1 shows the primary sequence of Rhipicephalus (Boophilus) microplus microplusin aligned to all sequences of microplusin homologs from other tick species found in GenBankTM. Comparison of these sequences shows a number of conserved aromatic/hydrophobic residues. Most of these hydrophobic residues occur in regions experimentally determined to be α-helices and occur in an i, i+3 or i, i+4, alternating manner (e.g. α2: Phe-32, Ala-35, and Leu-39), yielding amphipathic helices. In most cases these hydrophobic residues are buried, forming a hydrophobic cluster, and the hydrophilic residues are exposed to the solvent in the three-dimensional structure, with the exception of Val-63 and Tyr-64, which are exposed (Fig. 2). The beginning and end of each helix were determined by consensus analysis of chemical shift indexes, NOE pattern, and H/D exchange. The loop regions are characterized by fast H/D exchange of amide protons, absence of characteristic secondary structure NOE pattern and random-coil Cα and Hα chemical shift indexes. From the analysis of the primary sequences in Fig. 1, we observe that proline, a well known helix-breaker residue, is often found in the regions determined to be loops (e.g. L12, L34, L45, and the C terminus). Microplusin and microplusin-like peptides show a number of conserved histidines located at N and C termini as well as His-74 located in helix 5. In the solution structure of microplusin, the first five and the last ten residues of the sequence are not convergent; moreover, most of the chemical shifts of His-1, His-2, His-85, His-86, His-88, and His-90 are degenerate (except for the amide chemical shifts, which are not degenerate and were very important for differentiating among the several histidines in the mapping studies). These results suggest that both termini are disordered, and this was confirmed by measurements of 15N relaxation (supplemental Fig. S4). Nevertheless, because both N and C termini and His-74 are on the same side of the structure and the termini are very flexible, one could speculate that residues from N and C termini and His-74 may join together to form a complex with metal ions. Other conserved features in the primary sequence include six cysteines that are perfectly conserved among microplusin-like peptides. These cysteines were found to be involved in the formation of three disulfide bridges, as evidenced by mass spectroscopy (27) and by the analysis of Cβ chemical shifts (54) (39.4, 42.1, 33.4, 40.8, and 43.2 ppm for Cys-19, Cys-41, Cys-46, Cys-52, and Cys-80, respectively), which are indicative of oxidized cysteines. Preliminary structure calculations were performed without disulfide bridge constraints, but these restraints were included in further calculations because of proximity of cysteine residues in the preliminary calculations, namely between Cys-6 and Cys-52, Cys-19 and Cys-80, and Cys-41 and Cys-46. These disulfide bridges contribute to the stability of the overall fold, especially to keep the helix α1 in place, considering that fewer long range NOEs were observed for this helix (supplemental Fig. S3). The broad signals observed for Cys-6, Thr-7, and Lys-8 suggest conformational exchange in the millisecond timescale involving the Cys-6 to Cys-52 disulfide bridge, which was confirmed by 15N relaxation experiments. In support of a role for cysteines in maintaining microplusin structure, we have shown that the three disulfide bridges of microplusin are essential for its antibacterial activity. Similar roles for disulfide bridges have been described for several AMPs. For gomesin, an AMP isolated from the spider Acanthoscurria gomesiana (35), disruption of one or two of its disulfide bridges completely abolished the AMP activity (55).
The solution structure described here (Fig. 2) differs from that of other cysteine-rich AMPs, as expected from the analysis of the microplusin primary sequence and cysteine pattern, which show very low homology with other sequences previously characterized. Yet the search for homologous structures using the DALI server (56) yielded a number of structurally homologous peptides with a geometrical similarity score (Z-score) between 2.0 and 3.8. Among these structures are the C-terminal domain of the α subunit of RNA polymerase (57) (Z-score: 2.9, r.m.s.d. to microplusin: 2.6 Å, identity: 12%), and the helical domain of the α subunit of the translation initiation factor elF2 (58) (Z-score: 3.8, r.m.s.d. to microplusin: 4.6, identity: 5%). Comparison of microplusin to the complex formed by the C-terminal domain of the α subunit of RNA polymerase suggests that microplusin could also interact with DNA, but a closer inspection of the C-terminal domain of the α subunit residues involved in the interaction, namely Val-264, Arg-265, Asn-268, Asn-294, Gly-296, Lys-298, and Ser-299, indicated that these residues do not occur in structurally equivalent positions of microplusin. Thus it is likely that microplusin shares with C-terminal domain of the α subunit a similar fold but has a different function. However, further work should be done to analyze the DNA-binding capacity of microplusin.
Mapping of the Cu2+-binding Region
We showed that microplusin binds copper II and iron II and that microplusin antimicrobial activity is related to its ability to act as a copper scavenger. Due to the strong paramagnetic effect of the Cu2+ ion it is not possible to identify directly the residues involved in the coordination by conventional NMR techniques. Instead broader effects are observed in 1H-detected spectra with an expected blind region of ∼10 Å around the Cu2+ ion. Nevertheless, determining the residues under the influence of Cu2+ is still useful and could guide further structure-function studies, e.g. site-directed mutagenesis, which will prove more directly the residues responsible for Cu2+ binding.
We mapped the copper II binding region by analyzing 1H-15N HSQC (supplemental Fig. S6) and two-dimensional TOCSY spectra of microplusin alone and in complex with Cu2+. In the microplusin-Cu2+ complex we could not detect the amides from sixteen residues: His-2, Gln-3, Cys-6, and Thr-7 in the N-terminal region, Glu-16, Leu-17, and Ile-20 in the helix α1, Lys-71, Ile-73, His-74, Asp-75, Ala-76, Ala-77, Thr-78, and Ala-79 in the helix α5, and Asp-81 in the C-terminal region.
Microplusin possesses several conserved histidines at its N and C termini, and our NMR structure and dynamics studies revealed that the termini are disordered but lie on the same side of the structure. Based on the sequence conservation and on the NMR structure, our initial hypothesis was that histidines from both termini could come together to form a metal binding site. However, our results implicated several amino acids in the N-terminal region, helices α1, and α5, but none of the C-terminal histidines, which are still observable in the complex. Moreover 15N T2 measurements of the microplusin-Cu2+ complex (supplemental Fig. S5) show that the C-terminal residues are still flexible. In Fig. 3, we built a model that seems to explain our data. We performed a structure calculation, including artificial distance restraints between His-2 and His-74. The effect of these new restraints was to constrain the number of possible conformations of the flexible N-terminal region, and they did not cause any distortion in the rest of the structure. In our model structure we mapped all residues that are missing under the effect of Cu2+. Most of the residues fall into a sphere of 9-Å radius, with His-74 nearly in the center of the sphere. The restraints between His-2 and His-74 brought His-2 and Gln-3 into the sphere. Among the residues that disappear with Cu2+, Cys-6 and Thr-7 are the only mapped residues that fall outside the sphere. Signals of these two residues were already broad in the free microplusin due to exchange processes and could have disappeared because of an effect of the copper II on the exchange processes upon complex formation, or because of a summed effect on the T2 relaxation. Although we cannot directly determine the residues involved in the Cu2+ coordination, from the 16 residues mapped in our study His-74 and the N-terminal residues are the top candidates for mutagenesis studies.
Mode of Action of Microplusin
Microplusin activity against M. luteus was drastically reduced in the presence of copper II ions. Moreover, when an M. luteus suspension pre-treated with microplusin received copper II, bacterial growth was recovered. Through electrospray ionization-mass spectrometry, it was verified that microplusin forms adducts with copper II with a metal/peptide molar ratio of 1:1. Therefore, our results strongly suggest that copper binding by microplusin makes this metal unavailable for M. luteus, and this is how this AMP exerts its antibacterial effect. A nutritional effect like this has been described for the human proteins calprotectin and lactoferrin (18, 19, 59). It has been reported that calprotectin, a protein that is abundant in human neutrophils, inhibits the growth of several bacteria and fungi by zinc and manganese chelation (18, 60, 61). Interestingly, metal chelation by calprotectin was recently shown to be the mechanism behind the control of S. aureus growth in tissue abscesses, showing that metal sequestration is a physiologically relevant way to control microbial infections (18).
Another evidence in favor of a copper deprivation mechanism for microplusin comes from the measurements of M. luteus respiration. Heme-copper oxidases comprise a family of three cytochrome c oxidases and one quinol oxidase that reduce dioxygen to water (62, 63). In P. aeruginosa under copper-limiting conditions, the four heme-copper oxidases of a branched electron transport chain became defective, and respiration and aerobic growth are sustained by a cytochrome bd-type cyanide-insensitive oxidase, which lacks copper in its active site (64). Likewise, copper deprivation in the cyanobacterium Synechocystis sp. also causes an arrest of the respiratory metabolism as a result of a failure of cytochrome c oxidase function (65). Our results show that microplusin constrained both basal and CCCP-stimulated, but not cyanide-resistant respiration of Micrococcus. We propose that microplusin might affect respiration of M. luteus by removing Cu2+ from one or more of the heme-copper terminal oxidases. Once this heme-copper oxidase is defective, the respiratory chain is unable to reach the maximal electron flow capacity stimulated by the uncoupler CCCP. Additionally, the cyanide-insensitive copper-independent respiration remained apparently intact, which supports the idea that a heme-copper terminal oxidase is the component of the respiratory chain affected by the copper starvation produced by microplusin. However, as bacterial respiratory systems are usually branched to distinct heme-copper terminal oxidases (66), and there is no information regarding the type of terminal oxidase that is present in M. luteus, the specific heme-copper oxidase possibly affected by microplusin remains to be determined.
A metal depletion mode of action requires that the antimicrobial peptide be present at a concentration similar to that of the metal to be depleted. Copper depletion by microplusin is compatible with the copper content of the culture medium used in this work, which we determined to be 0.03 ± 0.008 μm. This explains why a concentration of microplusin as low as 0.09 μm can have an antibacterial effect.
Despite the scarcity of data showing the importance of copper for M. luteus metabolism, we speculate that M. luteus may be very susceptible to copper available in the culture media. Additionally, other studies by our group indicate that copper chelation by microplusin is important for its activity against the yeast C. neoformans.4 One interesting question arising from our data (Table 3), however, is why some microorganisms are insensitive to microplusin. In particular, microplusin was not active against Gram-negative bacteria, a result that is similar to what has been observed for other antimicrobial factors isolated from R. (B.) microplus (25, 27, 28). Assuming that the main antibacterial mechanism of microplusin is indeed metal chelation, there are a number of possibilities worth exploring: (i) some organisms have higher copper pools than others and, thus, are less sensitive to depletion; (ii) depending on the metabolic properties of the microorganism, copper could be more or less essential. For example, microorganisms that are capable of anaerobic energy production may tolerate better the effects of copper deprivation on the electron transport chain; and (iii) some organisms may have better copper-scavenging systems than others. If a bacterium produces a copper-scavenging molecule with higher affinity for copper than microplusin, the peptide will not have an effect. In this regard, it is known that some bacteria produce siderophores with very high affinity for iron, and this fact may be related to the absence of an iron depletion effect for microplusin (67).
Our data suggest that metal chelation may be a general mode of action for microplusin-like peptides found in other tick species (31, 32). In this regard, it is worth noting that for hebraein, an AMP of the tick Amblyomma hebraeum, which exhibits 61% identity with microplusin, deletion of its C-terminal histidine residues significantly reduced its antimicrobial activity against E. coli and the yeast Candida glabrata (31). Even though this result does not prove that hebraein is inhibiting microbial growth by metal chelation, it is certainly suggestive of this model.
Although microplusin seems to be inhibiting M. luteus because of its copper-binding capacity, we cannot discard the possibility that microplusin will have other modes of action. Indeed, some AMPs, e.g. lactoferricin (67), have been shown to function in more than one way, with modes of action varying depending on experimental conditions and the target organism. In the case of microplusin, data obtained for the protozoan Plasmodium gallinaceum suggests that it causes strong membrane permeabilization and cell death (68). This effect was specific, because under similar conditions microplusin was not toxic for human erythrocytes or for green monkey kidney (VERO) cells (69). Similarly, in the case of hebraein, deletion of its C-terminal histidine tail (and, presumably, abrogation of its metal-binding capacity) did not affect its antimicrobial effect against S. aureus (31).
One important question arising from our work is whether the metal depletion effect of microplusin is relevant for control of bacterial infections in ticks. We are addressing this question through the silencing of the microplusin gene by RNA interference in ticks infected by the natural pathogen Anaplasma marginale. In addition, the discovery that microplusin binds copper and iron provides a clue to the role of this protein in the homeostasis of these metals in ticks. In this context, the dual function of hepcidin as an antimicrobial as well as a regulator of systemic iron homeostasis in mice and humans is well known (22).
Lastly our data suggest a potential for microplusin as therapeutic agent against bacteria and fungi. Molecules that are able to sequester metals may have important roles in vivo, as demonstrated for calprotectin, which inhibits S. aureus proliferation in abscesses in mice through binding of available zinc (18). In addition, drugs that act as copper chelators may be useful for the treatment of diseases such as Wilson disease, in which patients have high levels of copper in the body (70).
Acknowledgments
We are grateful to Dr. Philippe Bulet, Prof. Maria Terêsa M. Miranda, and Prof. Martha M. Sorenson for critical reading, Susana P. Lima for technical assistance, and Cassiano Pereira for figure preparation.
This work was supported by the Fundação de Amparo a Pesquisa do Estado de São Paulo, Brazil, the Fundação de Amparo a Pesquisa do Estado do Rio de Janeiro, and the Conselho Nacional de Desenvolvimento Científico e Tecnológico, Brazil.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7.
The atomic coordinates and structure factors (code 2knj) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
F. D. Silva, D. C. P. Rossi, L. Martinez, J. D. Nosanshuk, M. L. Rodrigues, and S. Daffre, unpublished observation.
- AMP
- antimicrobial peptide
- LB
- Luria-Bertani broth
- TES
- trace element solution
- MIC
- minimal inhibitory concentration
- CFU
- colony-forming units
- CCCP
- carbonyl cyanide m-chlorophenylhydrazone
- HSQC
- heteronuclear single quantum coherence
- NOE
- nuclear Overhauser
- NOESY
- nuclear Overhauser effect spectroscopy
- r.m.s.d.
- root mean square deviation
- PB
- peptone broth
- TOCSY
- total correlation spectroscopy.
REFERENCES
- 1.Bulet P., Stöcklin R., Menin L. (2004) Immunol. Rev. 198, 169–184 [DOI] [PubMed] [Google Scholar]
- 2.Brown K. L., Hancock R. E. (2006) Curr. Opin. Immunol. 18, 24–30 [DOI] [PubMed] [Google Scholar]
- 3.Ganz T. (2005) Comb. Chem. High Throughput Screen 8, 209–217 [DOI] [PubMed] [Google Scholar]
- 4.Brogden K. A. (2005) Nat. Rev. Microbiol. 3, 238–250 [DOI] [PubMed] [Google Scholar]
- 5.Yount N. Y., Bayer A. S., Xiong Y. Q., Yeaman M. R. (2006) Biopolymers 84, 435–458 [DOI] [PubMed] [Google Scholar]
- 6.Goumon Y., Strub J. M., Moniatte M., Nullans G., Poteur L., Hubert P., Van Dorsselaer A., Aunis D., Metz-Boutigue M. H. (1996) Eur. J. Biochem. 235, 516–525 [DOI] [PubMed] [Google Scholar]
- 7.Stern A. S., Jones B. N., Shively J. E., Stein S., Undenfriend S. (1981) Proc. Natl. Acad. Sci. U.S.A. 78, 1962–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schittek B., Hipfel R., Sauer B., Bauer J., Kalbacher H., Stevanovic S., Schirle M., Schroeder K., Blin N., Meier F., Rassner G., Garbe C. (2001) Nat. Immunol. 2, 1133–1137 [DOI] [PubMed] [Google Scholar]
- 9.Lai R., Liu H., Hui Lee W., Zhang Y. (2002) Biochem. Biophys. Res. Commun. 295, 796–799 [DOI] [PubMed] [Google Scholar]
- 10.Heidari M., Hamir A., Cutlip R. C., Brogden K. A. (2002) Int. J. Antimicrob. Agents 20, 69–72 [DOI] [PubMed] [Google Scholar]
- 11.Brogden K. A., De Lucca A. J., Bland J., Elliott S. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 412–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steffen H., Rieg S., Wiedemann I., Kalbacher H., Deeg M., Sahl H. G., Peschel A., Götz F., Garbe C., Schittek B. (2006) Antimicrob. Agents Chemother. 50, 2608–2620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Otvos L., Jr. (2005) J. Pept. Sci. 11, 697–706 [DOI] [PubMed] [Google Scholar]
- 14.Kragol G., Lovas S., Varadi G., Condie B. A., Hoffmann R., Otvos L., Jr. (2001) Biochemistry 40, 3016–3026 [DOI] [PubMed] [Google Scholar]
- 15.Park C. B., Kim H. S., Kim S. C. (1998) Biochem. Biophys. Res. Commun. 244, 253–257 [DOI] [PubMed] [Google Scholar]
- 16.Hsu C. H., Chen C., Jou M. L., Lee A. Y., Lin Y. C., Yu Y. P., Huang W. T., Wu S. H. (2005) Nucleic Acids Res. 33, 4053–4064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ulvatne H., Samuelsen Ø., Haukland H. H., Krämer M., Vorland L. H. (2004) FEMS Microbiol. Lett. 237, 377–384 [DOI] [PubMed] [Google Scholar]
- 18.Corbin B. D., Seeley E. H., Raab A., Feldmann J., Miller M. R., Torres V. J., Anderson K. L., Dattilo B. M., Dunman P. M., Gerads R., Caprioli R. M., Nacken W., Chazin W. J., Skaar E. P. (2008) Science 319, 962–965 [DOI] [PubMed] [Google Scholar]
- 19.Arnold R. R., Russell J. E., Champion W. J., Brewer M., Gauthier J. J. (1982) Infect. Immun. 35, 792–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kavanagh K., Dowd S. (2004) J. Pharm. Pharmacol. 56, 285–289 [DOI] [PubMed] [Google Scholar]
- 21.Ganz T. (2006) Curr. Top. Microbiol. Immunol. 306, 183–198 [DOI] [PubMed] [Google Scholar]
- 22.Verga Falzacappa M. V., Muckenthaler M. U. (2005) Gene 364, 37–44 [DOI] [PubMed] [Google Scholar]
- 23.Nishikawa M., Ogawa K. (2004) Antimicrob. Agents Chemother. 48, 229–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sonenshine D. E. (1991) Biology of Ticks, Oxford University Press, New York [Google Scholar]
- 25.Fogaça A. C., da Silva P. I., Jr., Miranda M. T., Bianchi A. G., Miranda A., Ribolla P. E., Daffre S. (1999) J. Biol. Chem. 274, 25330–25334 [DOI] [PubMed] [Google Scholar]
- 26.Sforça M. L., Machado A., Figueredo R. C., Oyama S., Jr., Silva F. D., Miranda A., Daffre S., Miranda M. T., Spisni A., Pertinhez T. A. (2005) Biochemistry 44, 6440–6451 [DOI] [PubMed] [Google Scholar]
- 27.Fogaça A. C., Lorenzini D. M., Kaku L. M., Esteves E., Bulet P., Daffre S. (2004) Dev. Comp. Immunol. 28, 191–200 [DOI] [PubMed] [Google Scholar]
- 28.Fogaça A. C., Almeida I. C., Eberlin M. N., Tanaka A. S., Bulet P., Daffre S. (2006) Peptides 27, 667–674 [DOI] [PubMed] [Google Scholar]
- 29.Esteves E., Fogaça A. C., Maldonado R., Silva F. D., Manso P. P., Pelajo-Machado M., Valle D., Daffre S. (2009) Dev. Comp. Immunol. 33, 913–919 [DOI] [PubMed] [Google Scholar]
- 30.Esteves E., Lara F. A., Lorenzini D. M., Costa G. H., Fukuzawa A. H., Pressinotti L. N., Silva J. R., Ferro J. A., Kurtti T. J., Munderloh U. G., Daffre S. (2008) Insect Biochem. Mol. Biol. 38, 568–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lai R., Takeuchi H., Lomas L. O., Jonczy J., Rigden D. J., Rees H. H., Turner P. C. (2004) FASEB J. 18, 1447–1449 [DOI] [PubMed] [Google Scholar]
- 32.Mans B. J., Andersen J. F., Francischetti I. M., Valenzuela J. G., Schwan T. G., Pham V. M., Garfield M. K., Hammer C. H., Ribeiro J. M. (2008) Insect. Biochem. Mol. Biol. 38, 42–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marley J., Lu M., Bracken C. (2001) J. Biomol. NMR 20, 71–75 [DOI] [PubMed] [Google Scholar]
- 34.Harwood C. R., Archibald R. A. (1990) in Molecular Biology Methods for Bacillus (Harwood C. R., Cutting S. M. eds) pp. 1–26, John Wiley & Sons Ltd., New York [Google Scholar]
- 35.Silva P. I., Jr., Daffre S., Bulet P. (2000) J. Biol. Chem. 275, 33464–33470 [DOI] [PubMed] [Google Scholar]
- 36.Stone K. L., Willians K. R. (1996) in The Protocol Handbook (Walker J. M. ed) pp. 415–421, Human Press Inc., Totowa, NJ [Google Scholar]
- 37.Sattler M., Schleucher J., Griesinger C. (1999) Prog. NMR Spectrosc. 34, 93–158 [Google Scholar]
- 38.Kay L. E. (1995) Prog. Biophys. Mol. Biol. 63, 277–299 [DOI] [PubMed] [Google Scholar]
- 39.Nilges M., O'Donoghue S. (1998) Prog. NMR Spectrosc. 32, 107–139 [Google Scholar]
- 40.Linge J. P., O'Donoghue S. I., Nilges M. (2001) Methods Enzymol. 339, 71–90 [DOI] [PubMed] [Google Scholar]
- 41.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D. Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 42.Wishart D. S., Sykes B. D. (1994) J. Biomol. NMR 4, 171–180 [DOI] [PubMed] [Google Scholar]
- 43.Rezende C. A., Silva F. D., Daffre S., Pires J. R. (2009) Biomol. NMR Assign. DOI 10.1007/s12104-009-9171-7 [DOI] [PubMed] [Google Scholar]
- 44.Cavanagh J., Fairbrother W. J., Skelton N. J., Pamler A. G., III (1996) Protein NMR Spectroscopy: Principles and Practices, Academic Press, San Diego, CA [Google Scholar]
- 45.Clore G. M., Sazbo A., Bax A., Kay L. E., Driscoll P. C., Gronenborn A. M. (1990) J. Am. Chem. Soc. 112, 4989–4991 [Google Scholar]
- 46.Dosset P., Hus J. C., Blackledge M., Marion D. (2000) J. Biomol. NMR 16, 23–28 [DOI] [PubMed] [Google Scholar]
- 47.Brewer D., Lajoie G. (2000) Rapid Commun. Mass. Spectrom. 14, 1736–1745 [DOI] [PubMed] [Google Scholar]
- 48.Grogan J., McKnight C. J., Troxler R. F., Oppenheim F. G. (2001) FEBS Lett. 491, 76–80 [DOI] [PubMed] [Google Scholar]
- 49.Gusman H., Lendenmann U., Grogan J., Troxler R. F., Oppenheim F. G. (2001) Biochim. Biophys. Acta 1545, 86–95 [DOI] [PubMed] [Google Scholar]
- 50.Farnaud S., Rapisarda C., Bui T., Drake A., Cammack R., Evans R. W. (2008) Biochem. J. 413, 553–557 [DOI] [PubMed] [Google Scholar]
- 51.Arnesano F., Banci L., Piccioli M. (2005) Q. Rev. Biophys. 38, 167–219 [DOI] [PubMed] [Google Scholar]
- 52.Fázio M. A., Jouvensal L., Vovelle F., Bulet P., Miranda M. T., Daffre S., Miranda A. (2007) Biopolymers 88, 386–400 [DOI] [PubMed] [Google Scholar]
- 53.Horn D., Barrientos A. (2008) IUBMB Life 60, 421–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sharma D., Rajarathnam K. (2000) J. Biomol. NMR 18, 165–171 [DOI] [PubMed] [Google Scholar]
- 55.Fázio M. A., Oliveira V. X., Jr., Bulet P., Miranda M. T., Daffre S., Miranda A. (2006) Biopolymers 84, 205–218 [DOI] [PubMed] [Google Scholar]
- 56.Holm L., Sander C. (1998) Nucleic Acids Res. 26, 316–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Benoff B., Yang H., Lawson C. L., Parkinson G., Liu J., Blatter E., Ebright Y. W., Berman H. M., Ebright R. H. (2002) Science 297, 1562–1566 [DOI] [PubMed] [Google Scholar]
- 58.Dar A. C., Dever T. E., Sicheri F. (2005) Cell 122, 887–900 [DOI] [PubMed] [Google Scholar]
- 59.Aguila A., Herrera A. G., Morrison D., Cosgrove B., Perojo A., Montesinos I., Pérez J., Sierra G., Gemmell C. G., Brock J. H. (2001) FEMS Immunol. Med. Microbiol. 31, 145–152 [DOI] [PubMed] [Google Scholar]
- 60.Lulloff S. J., Hahn B. L., Sohnle P. G. (2004) J. Lab. Clin. Med. 144, 208–214 [DOI] [PubMed] [Google Scholar]
- 61.Sohnle P. G., Hahn B. L., Karmarkar R. (2001) J. Lab. Clin. Med. 137, 284–289 [DOI] [PubMed] [Google Scholar]
- 62.de Gier J. W., Lübben M., Reijnders W. N., Tipker C. A., Slotboom D. J., van Spanning R. J., Stouthamer A. H., van der Oost J. (1994) Mol. Microbiol. 13, 183–196 [DOI] [PubMed] [Google Scholar]
- 63.Azarkina N., Siletsky S., Borisov V., von Wachenfeldt C., Hederstedt L., Konstantinov A. A. (1999) J. Biol. Chem. 274, 32810–32817 [DOI] [PubMed] [Google Scholar]
- 64.Frangipani E., Slaveykova V. I., Reimmann C., Haas D. (2008) J. Bacteriol. 190, 6706–6717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Durán R. V., Hervás M., De La Rosa M. A., Navarro J. A. (2004) J. Biol. Chem. 279, 7229–7233 [DOI] [PubMed] [Google Scholar]
- 66.Comolli J. C., Donohue T. J. (2004) Mol. Microbiol. 51, 1193–1203 [DOI] [PubMed] [Google Scholar]
- 67.Farnaud S., Evans R. W. (2003) Mol. Immunol. 40, 395–405 [DOI] [PubMed] [Google Scholar]
- 68.Rodrigues D. G. (2005) Antimicrobial Peptides Effect on Plasmodium gallinaceum Sporozites, Master's dissertation, Universidade de São Paulo [Google Scholar]
- 69.Silva F. D. (2008) Action Mechanism of Microplusin, a Copper Chelating Peptide with Antimicrobial Activity, Doctoral dissertation, Universidade de São Paulo [Google Scholar]
- 70.Das S. K., Ray K. (2006) Nat. Clin. Pract. Neurol. 2, 482–493 [DOI] [PubMed] [Google Scholar]