Abstract
Recently, a few fish proteins have been described with a high homology to B-type lectins of monocotyledonous plants. Because of their mannose binding activity, they have been ascribed a role in innate immunity. By screening various fish venoms for their integrin inhibitory activity, we isolated a homologous protein from the fin stings and skin mucus of the scorpionfish (Scorpaena plumieri). This protein inhibits α1β1 integrin binding to basement membrane collagen IV. By protein chemical and spectroscopic means, we demonstrated that this fish protein, called plumieribetin, is a homotetramer and contains a high content of anti-parallel β strands, similar to the mannose-binding monocot B-lectins. It lacks both N-linked glycoconjugates and common O-glycan motifs. Despite its B-lectin-like structure, plumieribetin binds to α1β1 integrin irrespective of N-glycosylation, suggesting a direct protein-protein interaction. This interaction is independent of divalent cations. On the cellular level, plumieribetin failed to completely detach hepatocarcinoma HepG2 cells and primary arterial smooth muscle cells from the collagen IV fragment CB3. However, plumieribetin weakened the cell-collagen contacts, reduced cell spreading, and altered the actin cytoskeleton, after the compensating α2β1 integrin was blocked. The integrin inhibiting effect of plumieribetin adds a new function to the B-lectin family, which is known for pathogen defense.
Introduction
Integrins are cell surface receptors, which are expressed on cells of all metazoan animals (1). They consist of two genetically nonrelated, noncovalently associated subunits, α and β, both of which are N-glycosylated and span the plasmalemma once. The short C-terminal cytoplasmic tails of both subunits interact with cytoskeletal and signaling molecules, which are involved in integrin-mediated signaling (2, 3).
The ectodomains of both integrin subunits associate into a composite globular domain that harbors the binding site for the cognate extracellular matrix ligand (4). Matrix proteins are mainly fibrillar proteins, such as collagen (5), which constitute the scaffold structure of the interstitial connective tissue by forming supramolecular aggregates. The basement membrane is a specialized, sheet-like matrix structure that separates the connective tissue from other tissues and serves as an anchoring platform for epithelial, endothelial, muscle, and nerve cells (6, 7). It is made up of special matrix proteins, such as collagen IV, laminins, nidogen/entactin, and the proteoglycan perlecan (6, 8, 9). Some of the matrix proteins are recognized by their respective integrin receptors via the amino acid sequence RGD (10). In contrast, the collagen-binding (α1β1, α2β1, α10β1, and α11β1) and laminin-binding (α3β1, α6β1, α7β1, and α6β4) integrins require a distinct spacial structure and conformation of their ligands (11, 12). For instance, integrin α1β1 recognizes an array of positively and negatively charged amino acid residues that comes into close proximity on the surface of the collagen triple helix (13). Binding to extracellular matrix proteins not only anchors the cell to its underlying substratum but also provides environmental signals to the cell that are transmitted via a complex signaling cascade and trigger numerous cell reactions, such as cell spreading, migration, survival, proliferation, gene activation, and differentiation (1, 2). Therefore, integrin-mediated cell-matrix contacts play an essential role in tissue integrity and in various other (patho)physiological situations.
Because of their pivotal roles, integrins are targets for several naturally occurring toxins. Numerous disintegrins containing RGD sequences or homologous sequences have been identified as inhibitors of the platelet integrin αIIbβ3 in hemorrhagic venoms of snakes, leeches, and blood-sucking insects (14). Several C-type lectins have been identified as versatile toxins against platelet receptors (15). Among them, rhodocetin from the Malayan pit viper (Calloselasma rhodostoma) was characterized as a specific α2β1 integrin antagonist (16), which is able to block α2β1 integrin-mediated migration and infiltration of tumor cells (17). C-type lectins are one subclass of lectins that are characterized by a huge structural and functional variability. Other subclasses are I (immunoglobulin-like) lectins, P (mannose-phosphate-recognizing) lectins, galectins, and B (bulb)-lectins (18, 19). The B-lectins are predominantly found in monocotyledonous plants and selectively recognize mannose residues (20).
In search of integrin inhibitors, snake venoms have been a rich source. Envenomation with other animal venoms also causes local blisters, endothelial cell detachment from their subendothelial basement membrane with subsequent increase in vessel permeability, formation of erythema, and even hemorrhages (21). These symptoms indicate the presence of toxins interfering with integrin-mediated cell-matrix interactions. Some of these symptoms have been observed after envenomation by the scorpionfish (Scorpaena plumieri) (22–25) and have encouraged us to analyze this fish venom for potential integrin inhibitors.
In this study, we present a novel α1β1 integrin inhibitor from the sting glands and the mucus of the scorpionfish (S. plumieri). Structurally, it is likely to fold as a B-lectin with high homology to monocot mannose-binding B-lectins and to pufflectin from the skin and intestine of the Japanese pufferfish/Fugu fish (Takifugu rubripes) (26). In contrast to the known functions of B-lectins in defense against predators and in innate immune response against retroviral, bacterial, and metazoan parasitic pathogens, which are mediated via their mannose binding activity, the newly discovered B-lectin plumieribetin affects α1β1 integrin and thus may contribute to some of the local and systemic effects of envenomation by the scorpionfish.
EXPERIMENTAL PROCEDURES
Materials
The soluble human integrins, α1β1, α2β1, α3β1, and α7β1, were produced in transfected Schneider S2 cells and isolated as previously described (16, 27). Soluble α1β1 integrin was deglycosylated by incubation of the integrin with recombinant protein-N-glycosidase F (Roche Applied Science) in TBS,2 pH 7.4, supplemented with 2 mm MgCl2 for 13 h at 21 °C. Wild-type α1β1 integrin was isolated from human placenta as previously published (13). The production of the collagen IV fragment CB3 is described in Ref. 13, the production of collagen I is described in Ref. 16, and the production of laminin-332 from the supernatant of SCC25 cells is described in Ref. 28. Laminin-111 was a generous gift from late R. Timpl (Max-Planck-Institute for Biochemistry, Martinsried, Germany). The antiserum against plumieribetin was raised in a rabbit according to standard protocols at the animal house of the R & D Center of the Ezequiel Dias Foundation (Belo Horizonte, Brazil). The inhibitory monoclonal antibody AGF-1 directed against the integrin α1 subunit was a generous gift from Dr. H. Gardner (Biogen). Protein concentration was determined according to Lowry with self-made reagents or using the BCA kit (Pierce).
Inhibition ELISA
The inhibition ELISA with soluble integrins was performed as previously described (16). Briefly, 10 μg/ml CB3, 6 μg/ml laminin-332, and laminin-111 in 50 mm Tris/HCl, pH 7.4, 150 mm NaCl 2 mm MgCl2 (TBS/Mg) buffer and 10 μg/ml collagen I in 0.1 m acetic acid were coated onto multiwell plate at 4 °C overnight. After being washed twice, the wells were blocked with 1% BSA in TBS/Mg (BSA/TBS/Mg) buffer. The soluble integrins were added in the same buffer supplemented with 1 mm MnCl2 and the following protease inhibitors: aprotinin, leupeptin, and pepstatin (each at 2 μg/ml); 1,10-phenanthroline; and phenylmethylsulfonyl fluoride (each at 2 mm). Moreover, the incubation was carried out in the absence (positive control) and presence of S. plumieri venom (0.9 mg/ml), mucus (50 μg/ml), or gel filtration fractions thereof (50 μg/ml) or in the presence of 10 mm EDTA (negative control) for 2 h at room temperature. After nonbound integrin had been washed off with 50 mm HEPES/NaOH, pH 7.4, 150 mm NaCl, 2 mm MgCl2, 1 mm MnCl2 (HEPES/Mg/Mn) buffer, bound integrin was fixed to the wells with 2.5% glutaraldehyde solution in HEPES/Mg/Mn buffer for 10 min and quantified by ELISA using a rabbit antiserum against the human β1 integrin and a secondary anti-rabbit IgG antibody conjugated to alkaline phosphatase (both diluted 1:1000 in BSA/TBS/Mg). Conversion of p-nitro-phenyl-phosphate was measured by a multiwell plate reader (BioTek, Bad Friedrichshall, Germany) at 405 nm. After subtracting negative control blanks, relative binding was calculated by normalizing the binding signals to the signal of the positive control. The values were generally determined in duplicates.
Venom Extraction
Wild specimens of S. plumieri were fished off the coast of Espírito Santo, Brazil. The venom from the fin stings was extracted according to the batch method previously described by Schaeffer et al. (29). Prior to the extraction, the fish were immobilized by chilling to −20 °C. The dorsal and anal spines were removed, minced, and homogenized with distilled water at 4 °C. After centrifugation of the extract at 6000 × g for 15 min at 4 °C, the supernatant was immediately applied to the gel filtration separation. An aliquot was retained for the inhibition ELISA.
Isolation of the Venom Component with α1β1 Integrin Inhibiting Activity
The crude venom extract from S. plumieri was separated by gel filtration on a Sephacryl S-200 HR column (GE Life Sciences, Uppsala, Sweden). Fractions of 1.75 ml were eluted with 10 mm sodium phosphate buffer, pH 7.6, 400 mm NaCl at 5.25 ml/h and monitored for protein contents by absorbance at 280 nm. Fractions with α1β1 integrin inhibiting activity were identified by inhibition ELISA, pooled, dialyzed against water, and lyophilized. Protein-linked glycosylation was determined using the DIG glycan differentiation kit (Roche Applied Science) according to the manufacturer's instructions.
Cross-linking
Plumieribetin at 140 μg/ml in PBS (20 mm sodium phosphate, pH 7.4, 150 mm NaCl) was incubated at different concentrations of bis-(sulfosuccinimidyl) suberate (BS3) (Pierce) for 1 h at 26 °C. After stopping the reaction by adding Tris/HCl, pH 8.0, to 5 mm final concentration, the samples were analyzed by SDS-PAGE under reducing conditions.
Determination of Primary Sequence of Plumieribetin and Sequence Analysis
Approximately 2 mg of plumieribetin was dissolved in 1 ml of 0.1 m Tris/HCl, pH 8.6, 6 m guanidinium/HCl. The addition of 35 μl of β-mercaptoethanol under nitrogen and incubation at 50 °C for 4 h reduced the protein, which was alkylated with 40 μl of vinyl pyridine at 37 °C for 2 h. The reduced and vinyl-pyridinylated plumieribetin was desalted on a Vydac C4 (214TP54) column with a 0–70% gradient of acetonitrile in an aqueous 0.1% trifluoroacetic acid solution at 1 ml/min for 70 min, lyophilized, and dissolved in 0.1 ml of 8 m urea solution. After dilution with 0.9 ml 0.1% NH4HCO3, pH 7.9, the protein solution was halved and individually digested with trypsin and chymotrypsin for 4 and 3 h, respectively, at an enzyme:substrate ratio of 2%(w/w). After lyophilization, the (chymo)tryptic fragments of alkylated plumieribetin were separated on a Vydac C18 (201SP54) column in a 0–50% gradient of acetonitrile in an aqueous 0.1% trifluoroacetic acid solution at 1 ml/min for 180 min. Their amino acid sequences were determined by Edman degradation in the automatic protein sequencing system PPSQ-21A (Shimadzu, Tokyo, Japan). The almost complete protein sequence was deduced from the overlapping tryptic and chymotryptic fragments. Sequence alignment was made by FASTA.
Molecular Mass Determination by Mass Spectrometry
Samples of the purified lectin were mixed with 0.5 μl of sinapinic acid and spotted onto a Bruker AnchorChip 384-well plate and allowed to dry. The matrix-assisted laser desorption ionization time of flight analysis were performed using Bruker Daltonics mass spectrometer operated in linear mode.
CD Spectrometry
The circular dichroism spectrum of plumieribetin, dissolved in 20 mm sodium phosphate, 70 mm NaCl, pH 6.5, was recorded with an Aviv model 400 circular spectrometer (Aviv Inc., Lakewood, NJ). The molar ellipticity data were deconvoluted with the software CDNN version 2.1 utilizing a neural net trained with a set of 33 spectra.
Fabrication of Carbohydrate Microarrays
GAPS II slides (Corning) were submerged in N,N-dimethylformamide containing 2% diisopropylethylamine and 0.5 mg/ml maleimido-N-hydroxysucinimide hexanoic acid for 14 h at room temperature. Functionalized slides were washed three times with methanol, dried, and stored under an argon atmosphere. The carbohydrates were synthesized as described in Ref. 30. The oligosaccharide compounds were dissolved in PBS with one molar equivalent tris-(2-carboxyethyl) phosphine to reduce disulfides. Per spot, 1 nl of the solution was printed onto the maleimide functionalized slides using an automated printing robot. The slides were incubated in a humid chamber to complete reaction for 24 h and stored in a desiccator until usage.
Binding of Plumieribetin to Carbohydrate Microarrays
Printed carbohydrate microarrays were quenched in 0.1% (v/v) β-mercaptoethanol in PBS for 1 h, washed with PBS and blocked with 2.5% (w/v) BSA in PBS for 1 h at room temperature. Plumieribetin (10–50 μg/ml) was allowed to bind to the washed slides in incubation buffer (1% BSA in 10 mm HEPES, pH 7.4, 150 mm NaCl, 1 mm MgCl2 with 1 mm CaCl2 and 0.1% Tween 20) for 1 h at room temperature. The slides were washed and overlaid with the anti-plumieribetin serum (1:100 in incubation buffer) and subsequently with an anti-rabbit antibody Alexa 555 conjugate (1:100). For inhibition, 50 mg/ml mannan (Sigma-Aldrich) were co-incubated with the plumieribetin during the binding step. The washed slides were spun to dryness and scanned with a fluorescence microarray reader (Tecan, Maennedorf, Switzerland). The fluorescence intensities were analyzed using the GeneSpotter software (MicroDiscovery, Berlin, Germany).
Binding ELISAs
The extracellular matrix proteins were coated onto a multiwell plate as described above. Similarly, integrins were coated at 10 μg/ml in TBS/Mg buffer supplemented with 1 mm MnCl2. After being washed and blocked, the wells were incubated with a plumieribetin solution (20 μg/ml) in BSA/TBS/Mg for 2 h at room temperature. Bound plumieribetin was detected with a rabbit antiserum raised against plumieribetin and a secondary anti-rabbit antibody conjugated to alkaline phosphatase.
Reciprocally, also plumieribetin was coated at 20 μg/ml in PBS to a multiwell plate. After washing and blocking the wells, the soluble integrins dissolved in BSA/TBS/Mg were allowed to bind. Bound integrins were detected with a rabbit antiserum directed against the human integrin β1 subunit and the secondary alkaline phosphatase-conjugated anti-rabbit IgG antibodies.
Cell Adhesion Assay
Hepatocellular carcinoma cell line HepG2 and primary human arterial smooth muscle cells (huASMC) (PromoCell, Heidelberg, Germany) were grown in RPMI with 10% fetal calf serum and ASCM-medium with 5% fetal calf serum, respectively, supplemented with antibiotics. Harvested cells were plated at a density of 1.0 × 105 ml−1 onto microtiter plate wells that had been coated with 2 μg/ml CB3 overnight and blocked with 0.1% BSA in PBS for 1 h. After adhesion for 1 h in the incubator in the absence and presence of plumieribetin, nonadherent cells were washed off, and adherent cells were fluorescently labeled with 2′,7′-bis(2-carboxyethyl)-5(6)-carboxyfluorescein and fluorescein diacetate respectively (both from Molecular Probes, Leiden, The Netherlands), for 15 min. After another wash step, the fluorescent dye was released from the cells with 0.1% SDS in 30 mm Tris/HCl, pH 8.8, and fluorescently determined in an ELISA reader (BioTek) at 485-nm excitation and 535-nm emission. The signals were corrected for background values measured with 10 mm EDTA and were normalized to noninhibited controls.
Cell Imaging
Morphological changes of cells upon plumieribetin treatment were detected by scanning electron microscope and immunofluorescence microscopy of the actin cytoskeleton. Scanning electron microscope analyses of cells were performed on silicium wavers coated with CB3 (5 μg/ml) in the absence or presence of either 10 mm EDTA or rhodocetin (4 μg/ml) or rhodocetin plus plumieribetin (10 μg/ml), as described in a previous paper (17). For immunofluorescence analysis, the cells were fixed on CB3-coated chamber slides (Nunc, Wiesbaden, Germany). Their actin cytoskeleton was stained with BODIPY 581/591 phalloidin (Molecular Probes) and analyzed by confocal microscopy according to (31).
RESULTS
The Crude Venom from the Fin Stings of Scorpionfish (S. plumieri) Inhibits Integrins
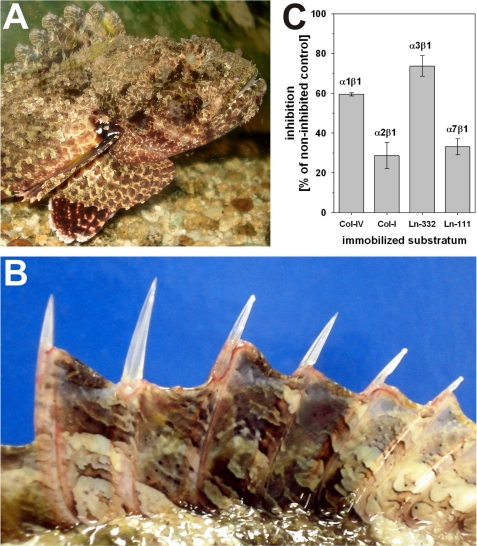
The crude venom of S. plumieri, a venomous fish (Fig. 1A) inhabiting the South Atlantic coast off the Brazilian south eastern states, was extracted from the stings of the dorsal (Fig. 1B) and anal fins. In a cell-free binding assay, binding of integrins to their respective immobilized ligands: α1β1 to the collagen IV fragment CB3, α2β1 to collagen I, α3β1 to laminin-332, and α7β1 to laminin-111, was challenged with the S. plumieri venom. The triple helical collagenous fragment CB3 harbors the high affinity binding site of α1β1 integrin within collagen IV (13), which by aggregating into a chicken wire-like supramolecular structure forms the scaffold structure of the basement membrane. As shown in Fig. 1C, the venom preferentially inhibited the interactions of α3β1 and α1β1 integrins with their cognate ligands, whereas the binding of the other two integrins, α2β1 and α7β1, to collagen I and laminin-111, respectively, more robustly withstands this venom. Although the inhibitory potential on the comparatively low affinity interaction of integrin α3β1 with laminin-332 is worthwhile, it was the exceptional preference of the venom to block specifically the avid α1β1 integrin binding to its collagen ligand to a remarkably higher extent as compared with the other collagen-binding integrin α2β1, which makes us search for the α1β1 integrin-inhibiting component of the S. plumieri venom.
FIGURE 1.
The scorpionfish (S. plumieri) (A) possesses in its fin rays (B) an integrin-inhibiting factor (C). The scorpionfish inhabits the Southern Atlantic coast of Brazil (A). Usually the fish is camouflaged in its stony environment, but in defense, it points its fin stings toward the predator. Its venom apparatus consists of 12 dorsal (B), 2 pelvic, and 3 anal fin spines. When the victim/predator steps and compresses the erected dorsal spines (B), the venom is released into the wound, resulting in severe pain and edema formation. The venom inhibits binding of the integrins, α1β1 and α3β1, to their respective ligands, collagen IV and laminin-332 (C). This was measured in a cell-free ELISA-based integrin-inhibition assay with recombinantly expressed integrin ectodomain heterodimers, called soluble integrins.
Chromatographic Separation of the S. plumieri Venom
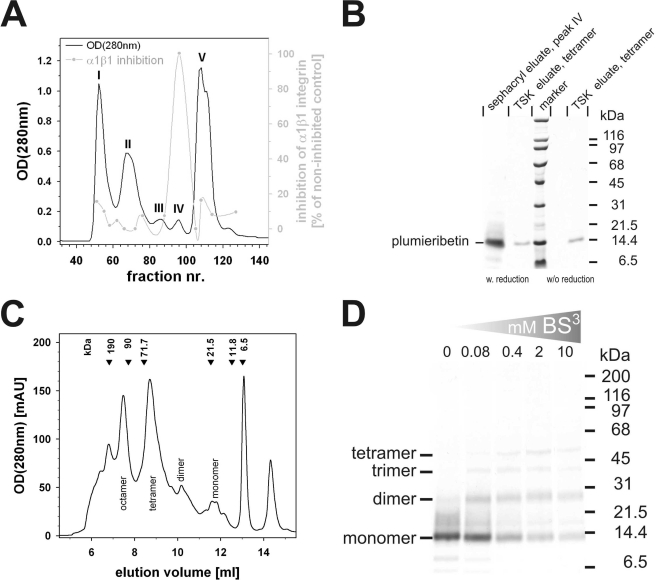
To identify the α1β1 integrin-inhibiting factor, the crude venom of S. plumieri was separated on a Sephacryl S-200 HR gel filtration column. As measured by absorbance at 280 nm, five pronounced protein peaks, numbered I–V, were eluted and tested by inhibition ELISA for their ability to interfere with α1β1 integrin binding to the collagen fragment CB3 (Fig. 2A). The peak of α1β1 inhibitory activity (Fig. 2A, gray line) coincided with peak IV, representing a molecular mass of ∼60–30 kDa. SDS-PAGE of fraction IV revealed a major protein band of ∼14 kDa (Fig. 2B). Fraction IV also contained some inhibitory activity toward α3β1 integrin binding to laminin-332. Analytic gel filtration of this fraction on a calibrated size exclusion column (TSK G 2000SWXL; TosoHaas, Stuttgart, Germany) showed several equally distanced peaks (Fig. 2C), all of which contained the same 14-kDa protein when analyzed by SDS-PAGE both without and with prior reduction of the sample (Fig. 2B). This suggested that this venom component consists of a 14-kDa monomer, which associates under physiological conditions into dimers, tetramers, or even higher aggregates, without any interchain cystine cross-linkages. Both tetrameric and octameric aggregates, as eluted from the analytical gel filtration, were equally active toward α1β1 integrin. The quaternary structure was further analyzed by chemical cross-linkage studies using BS3 as homobifunctional cross-linker molecule. Even at a very low BS3 concentration of 0.08 mm, a protein band at ∼28 kDa, indicative of a dimer, was detected (Fig. 2D). Moreover, a ladder of higher bands at ∼40 and 55 kDa was found predominantly at higher cross-linker concentration and suggested aggregation numbers of 3 and 4, in line with the gel filtration experiment. On average, 1.5 mg of this α1β1 integrin-inhibiting protein was isolated from 50 mg of venom protein (yield: 3.1%).
FIGURE 2.
Purification of the α1β1 integrin-inhibiting factor from S. plumieri venom and studies of its quaternary structure. The crude venom was separated by gel filtration on a Sephacryl S-200 HR column into five fractions, labeled I–V (A). The α1β1 integrin-inhibiting factor was eluted in fraction IV, which runs as a 14-kDa band in SDS-PAGE (B). analytical gel filtration on a calibrated TSK S2000 size exclusion column provided several peaks (C), most of which contain the same 14-kDa protein (shown by SDS-PAGE in (B)), yet in different oligomeric states (monomer, dimer, tetramer, and octamer). Calibration marker proteins with their molecular masses are indicated at the elution volumes above the chromatogram. Cross-linkage studies with the homobifunctional cross-linker BS3 proved the oligomeric nature of the α1β1 integrin-inhibiting factor in solution (D).
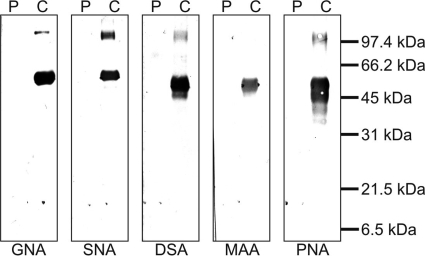
In addition to oligomerization, intense glycosylation of the protein may also change its molecular mass. Hence, after blotting to a nitrocellulose membrane, we tested the α1β1 integrin-inhibiting protein for its glycoconjugates using a panel of lectins with different capacities to recognize sugar residues typical for N- and O-glycoconjugates (Fig. 3). Although the respective positive controls (fetuin, asialofetuin, and carboxypeptidase Y) were stained properly, virtually no typical N-linked glycoconjugate residue could be detected with the lectins Galanthus nivalis agglutinin, Sambuca nigra agglutinin, and Datura stramonium agglutinin, ruling out the presence of both high mannose and complex type N-glycans. Furthermore, the negative reactions of the lectins Maackia amurensis agglutinin and peanut agglutinin, which recognize the common substituent, α(2–3)-linked sialic acid, and the core unit, galactose-β(1–3)-N-acetylgalactosamine, of the O-glycans, respectively, indicated the absence of common O-linked sugar residues within the 14-kDa protein.
FIGURE 3.
Absence of common N- and O-linked glycoconjugates on the α1β1 integrin-inhibiting factor, plumieribetin. The α1β1 integrin-inhibiting factor, plumieribetin (denoted P), along with N- and O-glycosylated control proteins (denoted C) were transferred to a nitrocellulose membrane after SDS-PAGE and stained with digoxigenin-labeled lectins and enzyme-conjugated anti-digoxigenin antibodies. The following lectins were used (target structures given in parentheses): G. nivalis agglutinin (GNA) (mannose in high mannose-type N-glycoconjugates), S. nigra agglutinin (SNA) ((2–6)-linked sialic acid in complex type N-glycoconjugates), D. stramonium agglutinin (DSA) (galactose-β(1–4)-N-acetylglucosamine as core unit of N-glycoconjugates), M. amurensis agglutinin (MAA) (α(2–6)-linked sialic acid in complex type N-glycoconjugates and O-glycans), and peanut agglutinin (PNA) (galactose-β(1–3)-N-acetylgalactosamine of O-glycans). In contrast to the controls, plumieribetin is not recognized by any of these lectins, proving the absence of any N-glycosylation and of the most common O-linked glycoconjugates.
The α1β1 Integrin-inhibiting Fraction of S. plumieri Venom Contains a Single Protein Belonging to the B-lectin Family
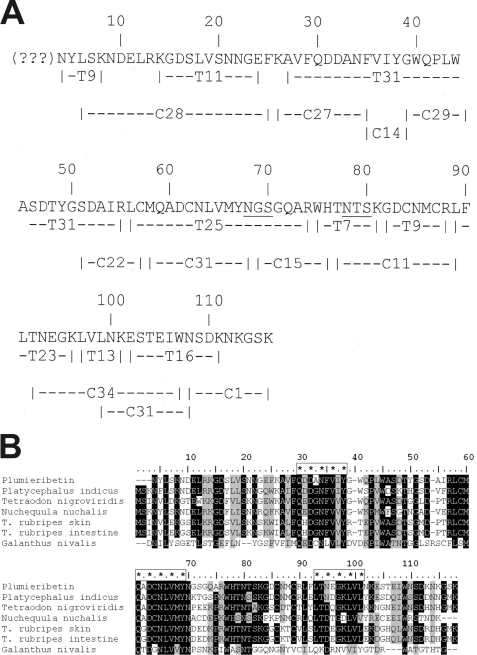
The protein of the α1β1 integrin-inhibiting fraction was sequenced by Edman degradation after reductive cleavage of disulfide bridges and pyridyl-ethylation of the reduced cysteine residues. However, the peptide chain was N-terminally blocked and inaccessible to Edman degradation. No other peptide sequences could be detected in this fraction, indicating the high purity of the isolated protein. After proteolytic fragmentation with trypsin and chymotrypsin, the fragments were sequenced and aligned to one polypeptide chain (Fig. 4A). One tryptic fragment (Fig. 4A, T9), presumably because of its N-terminal blockage, could not be sequenced. The fact that only one peptide with a blocked N terminus was found and that all other fragments could be lined up to one peptide chain indicates that the α1β1 integrin-inhibiting fraction IV contains only one protein.
FIGURE 4.
Primary structure of plumieribetin (A) and its alignment with homologous fish skin proteins and the prototypic monocot B-type lectin from snow drop (B). Tryptic and chymotryptic fragments (indexed by initial letters T and C, respectively) of plumieribetin were sequenced by Edman degradation and aligned to obtain almost the full sequence of the fish venom component. The N-terminal three amino acids could not be sequenced by Edman degradation and are indicated by question marks (A). Found by BLAST search, homologous fish proteins from P. indicus, Tetradon nigroviridis, Nuchequula nuchalis, a skin protein and an intestinal protein from T. rubripes, as well as from the monocot plant snow drop (G. nivalis) showed remarkable homology to plumieribetin (B). The sequences were aligned by the software BioEdit using the ClustalW algorithm. Similar sequence (threshold above 75%) and identical residues are shaded gray and black, respectively. The three putative mannose-binding sites are marked with enframed asterisks.
In a homology search in the protein data base, homologous proteins emerged that are found in the skin and intestine of other fish, but also homologous proteins that belong to the group of monocot lectins (Fig. 4B). The fish proteins possess higher similarities (identities as percentages are given in parentheses) to plumieribetin, such as the homologous protein from Platycephalus indicus (71.5%) and the Green puffer fish (Tetraodon nigroviridis) (63.7%) and the mucus lectin from the skin and intestine of the Japanese pufferfish (T. rubripes) (56.8%). Based on both this homology and the typical trypsin cleavage site, we deduced the N-terminal amino acids of the α1β1 integrin-inhibiting factor, which could be sequenced neither by Edman degradation nor by mass spectrometry, as likely to be MSR. Therefore, the molecular mass of the plumieribetin protein was calculated to be 13,157.67 Da. Mass spectrometric analysis of fraction IV of the Sephacryl S-200 HR gel filtration yielded a molecular mass of (14047.64 ± 3.25) Da for this protein. Therefore, its N terminus contains additional residues in contrast to other fish homologues, or it bears O-linked sugar residues, which are uncommon and not recognized by the lectins used for the glycoconjugate analysis (Fig. 3).
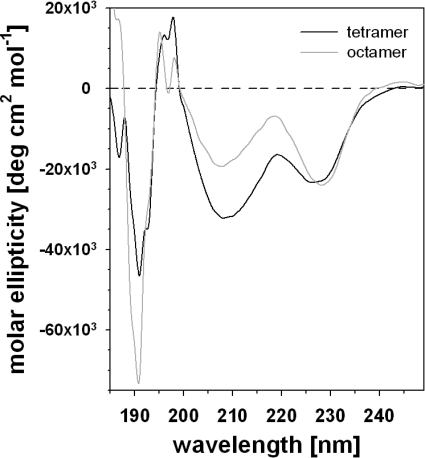
Despite the phylogenetic distance, the α1β1 integrin-inhibiting factor also has a remarkable homology to the monocot lectins, such as the bulb lectin, hence called B-lectin, from snow drop (G. nivalis) (28.4% identity) and other monocotyledonous plants. Several monocot B-lectins have been crystallized and are characterized by the β prism II fold (β barrel with a pseudo-3-fold symmetry) consisting of three four-stranded anti-parallel β-sheets, which are arranged as the three faces of a trigonal prism (20, 32–34). The circular dichroism spectra of the α1β1 integrin-inhibiting factor, both of its tetrameric and octameric form as eluted from the analytical size exclusion column (Fig. 5), resembled the circular dichroism spectrum of the monocot B-lectin of the garlic plant (35). With minima at 191, 208, and 226 nm and positive peaks at 195, 197, and 218 nm, the circular dichroism spectra indicated a high content of antiparallel β-sheets (Fig. 5). Deconvolution of both spectra revealed a high content of anti-parallel β-strands (35%) and β-turn secondary elements (19%), whereas the percentages of α-helices (7%) and parallel β-strands (3%) were very low or negligible. This secondary structure analysis and primary structure homology suggested that the α1β1 integrin inhibitor of S. plumieri folds like a B-lectin. Hence, we propose the name plumieribetin for it. It may be noted that from the three mannose-binding sites conserved in the plant B-lectins, only the first two sites seemed to be preserved in the fish lectins (Fig. 4B).
FIGURE 5.
Circular dichroism measurements of plumieribetin. Plumieribetin forms eluted from the analytical gel filtration column (Fig. 2C) as tetramer and octamer (black and gray line, respectively) were analyzed for circular dichroism. Their CD spectra indicated a high content of antiparallel β-strands.
Plumieribetin Recognizes Mannose Residues
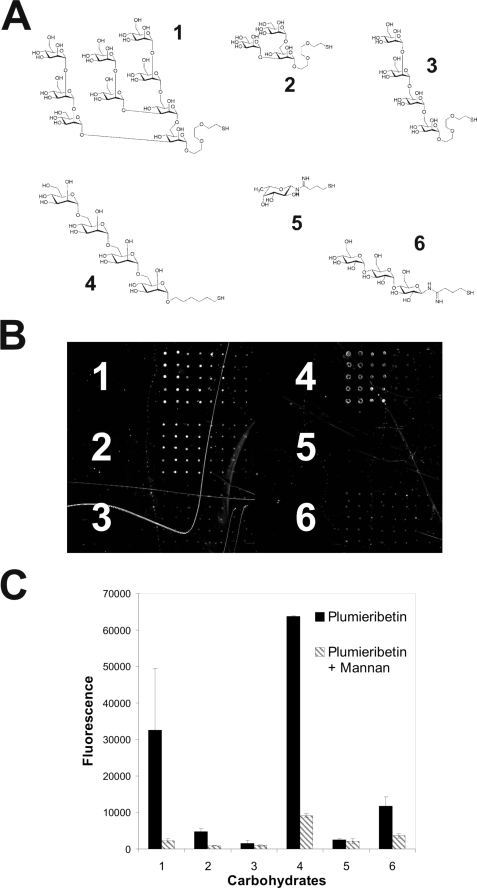
The plant B-lectins and some of the fish homologues of plumieribetin bind to mannose-containing carbohydrate structures. To test the sugar binding ability and specificity of plumieribetin, different mono- and oligosaccharides were immobilized on carbohydrate microarrays, which allow for screening sugar ligands of proteins (36). Binding of plumieribetin was detected with fluorescently labeled antibodies (Fig. 6). Plumieribetin preferentially recognized mannose-containing carbohydrate structures. Inhibition with mannan reduced binding clearly, thus demonstrating specific protein-sugar interactions. On carbohydrate microarrays, the sugar ligands are densely presented, allowing multivalent interaction with plumieribetin that leads to sufficient binding strength. However, the interaction of plumieribetin with single mannose residues was weak, and plumieribetin failed to bind to resin-linked mannose (data not shown). This is in contrast to the stronger mannose binding of pufflectin from T. rubripes (37) and the monocot B-lectins (38).
FIGURE 6.
Interaction of plumieribetin with various oligosaccharides immobilized to a carbohydrate microarray. Approximately 30 different mono- and oligosaccharides were spotted onto a microarray in different concentrations ranging from 2000, 400, 80, to 16 mm (from left to right in B), each in replicates of ten. The structures of six representative carbohydrates are shown in A: structure 1, Nonamannose; structure 2, branched trimannose; structure 3, D3 tetramannose; structure 4, tetramannan; structure 5, fucose; structure 6, maltotriose. A fluorescent image of plumieribetin binding to these selected carbohydrates on the microarray is presented in B. In C, the same carbohydrates were immobilized at 2 mm, and their plumieribetin binding was challenged with soluble mannan to test for mannose binding specificity. Bound plumieribetin was detected with rabbit antibodies against plumieribetin followed by fluorescently labeled secondary antibodies.
Plumieribetin Inhibits α1β1 Integrin Binding to Collagen IV Fragment CB3 by Direct Interaction with the Integrin
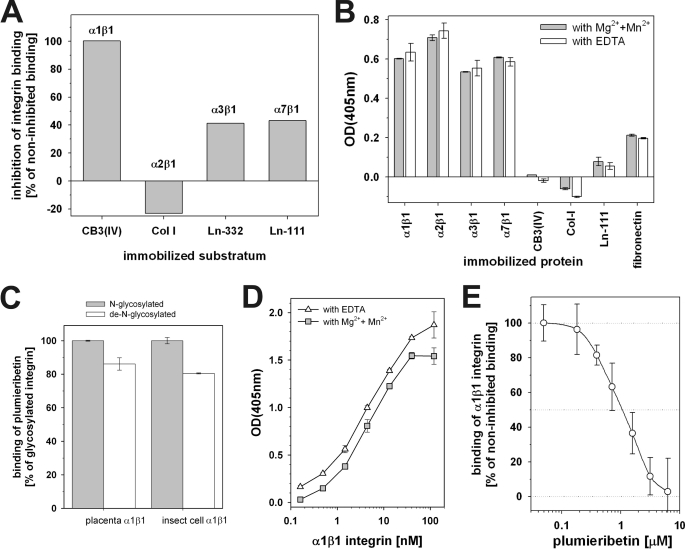
The isolated plumieribetin was assessed in the cell-free integrin inhibition assay for its activity (Fig. 7A). It entirely inhibited α1β1 integrin binding to the collagen IV fragment CB3, whereas α2β1 integrin binding was not affected at all. Both laminin-binding integrins, α3β1 and α7β1, were compromised by the fish B-lectin. To elucidate its mode of action mechanistically, we tested whether plumieribetin interacts with either the extracellular matrix proteins or with their cognate integrin receptors (Fig. 7B). In both the presence and the absence of divalent cations, Mg2+ and Mn2+ ions, which increase integrin binding activities, plumieribetin bound to all collagen- and laminin-binding integrins. In contrast, the binding signals of plumieribetin to select ECM proteins were very low and approached background. This suggests a direct interaction of plumieribetin with α1β1 integrin, which consequently inhibits interaction of the integrin with its ligand. The widespread binding of plumieribetin to all tested integrins was in contrast to its selective inhibition of α1β1 integrin binding to collagen IV fragment CB3 and its inability to block α2β1 integrin binding to collagen.
FIGURE 7.
Integrin binding and inhibiting activity of plumieribetin. A, in an inhibition ELISA, isolated plumieribetin predominantly inhibits binding of α1β1, but not α2β1 integrin to their respective collagen ligands, whereas the laminin-binding of the cognate integrin receptors is less affected. B, inhibition of integrin-ligand interaction is caused by binding of plumieribetin to the integrins but not to their binding partners of the extracellular matrix. C, interaction of plumieribetin is mediated by a protein-protein interaction, independently of N-linked glycoconjugates of the integrins. Soluble α1β1 integrin, expressed as high mannose-type N-glycoprotein in insect cells, and placenta-derived wild-type α1β1 integrin, both without (N-glycosylated) and with (de-N-glycosylated) prior treatment with PNGase F, are recognized by plumieribetin. The binding signals were normalized to the signals of the N-glycosylated form. D, independently of divalent cations, α1β1 integrin binds to plumieribetin in a saturable manner. E, by binding to α1β1 integrin, plumieribetin blocks collagen binding of the integrin effectively with an IC50 value of ∼1–2 μm.
Albeit weakly, plumieribetin binds mannose residues. Integrins are glycoproteins bearing several N-linked sugar residues. To test whether the interaction of plumieribetin with the integrin involves recognition of integrin-bound sugar epitopes, we also tested its binding to α1β1 integrin, the N-glycoconjugates of which had been cleaved off with PNGase F. Moreover, because the soluble integrins were produced in insect cells, which are unable to process the high mannose type N-glycans, we also tested plumieribetin binding to wild-type α1β1 integrin, which had been isolated from placenta and bears complex type N-glycoconjugates with less mannose residues. Both placental and insect cell-derived α1β1 integrin bound to immobilized plumieribetin irrespective of their different N-glycosylation patterns with only a slight decrease of the binding signal after removal of the glycoconjugates with PNGase F (Fig. 7C). This experiment indicates that plumieribetin interacts with α1β1 integrin via a protein-protein interaction, irrespective of the high mannose or complex type N-glycosylation of the integrin. To estimate the affinity of the interaction, immobilized plumieribetin was titrated with soluble α1β1 integrin in both the presence and the absence of divalent cations. Under both conditions, the binding signal approaches saturation with increasing concentrations of integrin, indicating a specific interaction of the binding partners (Fig. 7D). To determine the inhibitory potential of plumieribetin, its IC50 value was evaluated in an inhibition ELISA (Fig. 7E) and was in the range of 1–2 μm. Complete inhibition was achieved at plumieribetin concentrations above 5 μm.
Plumieribetin Is Also a Component of the Fish Skin Mucus
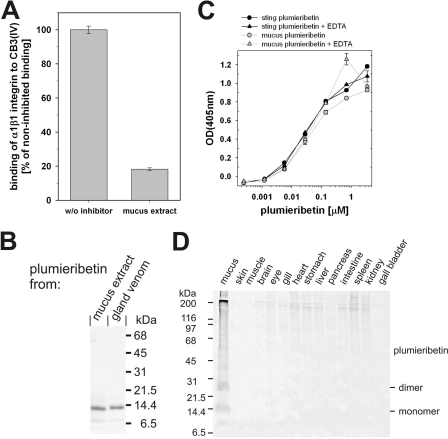
Although envenomation occurs via the stings, the venom is produced in glands located on the anterolateral grooves of the spines, which are covered by an integumental sheath. To investigate whether the compounds secreted to the skin surface also affect integrin binding, an extract of the fish mucus was tested for α1β1 integrin inhibitory activity (Fig. 8A). The mucus also contained a remarkable inhibition activity. From the mucus, a 14-kDa protein was isolated by gel filtration (Fig. 8B), although an additional ion exchange chromatography was required to achieve similar purity. Because it is recognized by the antiserum raised against plumieribetin, the 14-kDa protein from mucus is likely to be identical to the sting-derived plumieribetin. Plumieribetin, both from skin mucus and from venom, interacts with integrin α1β1 independent of divalent cations (Fig. 8C). Therefore, plumieribetin is not only found in the fin stings but also in the skin mucus. To analyze the tissue distribution of plumieribetin, different organs were dissected from two S. plumieri specimens. The proteins of these organs were extracted, separated electrophoretically, and tested for the presence of plumieribetin by immunoblot using the plumieribetin antiserum. No other fish tissue except for the stings and the skin mucus contained plumieribetin, thus corroborating a potential defensive role.
FIGURE 8.
The venom glands of the fin stings and the mucus of S. plumieri, but no other fish tissue, contain α1β1 integrin inhibitory activity. In addition to the sting-associated glands, the skin mucus of the scorpionfish strongly inhibits α1β1 integrin binding to collagen IV fragment CB3 (A). After isolation, this α1β1 integrin-inhibiting factor runs identically in SDS-PAGE to plumieribetin (B). The skin mucus-derived factor is likely identical to plumieribetin. Neither of both proteins differ from each other in their binding characteristics to α1β1 integrin, because titration of immobilized α1β1 integrin with plumieribetin yielded identical titration curves (C). Stained with an antiserum against plumieribetin, an immunoblot of various fish tissue extracts shows the restricted tissue distribution of plumieribetin in the stings and the skin mucus (D).
Plumieribetin Weakens the α1β1 Integrin-mediated Cell Interactions with Collagen IV
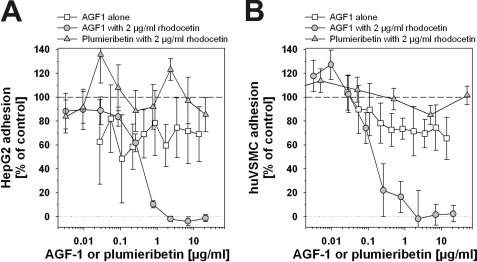
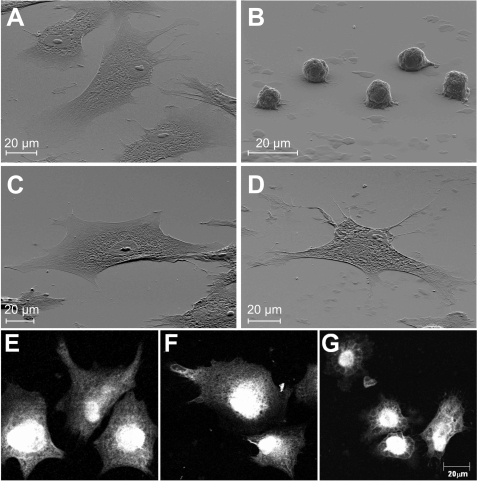
As model target cells for plumieribetin, we chose the hepatocellular carcinoma cell HepG2 and primary huASMC, which are in direct or close, respectively, contact to blood-borne venom components. HepG2 cells are anchored to collagen IV-containing basement membranes, whereas vascular smooth muscle cells are individually and completely surrounded by basement membranes. None of these structures contain laminin-332, the cognate ligand for α3β1 integrin. Both cell types express both α1β1 and α2β1 integrins as collagen receptors. The collagen IV fragment harbors binding sites for both integrins (13). Because they compensate for each other, the monoclonal antibody AGF-1 against α1β1 integrin, used as positive control, only shows a complete inhibition of HepG2 and huASMC adhesion to CB3, when given in addition to at least 2 μg/ml antagonistic α2β1 integrin blocker rhodocetin (Fig. 9). Yet, in the presence of rhodocetin, plumieribetin failed to block adhesion of either of the two cell types on the collagen IV fragment (Fig. 9). However, plumieribetin weakened the α1β1 integrin-mediated cell interactions with CB3, because the cells lose some, but not all contact points to the substratum and change their cell morphology (Fig. 10). On CB3, HuASMC spread extensively to ultrathin cells (Fig. 10A) with actin fibers running through the entire cell soma to the cell periphery (Fig. 10). Abolition of any integrin activity with EDTA resulted in nonadherent, round cells (Fig. 10B). Rhodocetin blockage of α2β1 integrin alone left the α1β1 integrin-mediated cell contacts intact and therefore did not change the ultraflat cell morphology (Fig. 10C) nor the actin cytoskeleton (Fig. 10F). Under this condition, plumieribetin inhibition of α1β1 integrin became evident, and the cells could not maintain their fully spread morphology (Fig. 10D). In addition, the organized actin stress fibers were remodeled, and the cortical actin cytoskeleton increased (Fig. 10G). Similar morphological effects were observed with HepG2 cells (data not shown). Thus, after the cell adhesive effects of the compensating collagen receptor, α2β1 integrin, are abolished, the inhibitory effect of plumieribetin on α1β1 integrin weakens the adhesion of cells to basement membrane collagen IV.
FIGURE 9.
Inhibition of α1β1 integrin-mediated adhesion of HepG2 cells (A) and huASMC (B) with a monoclonal antibody AGF-1 or plumieribetin after α2β1 integrin blockage. HepG2 cells (A) and huASMC (B) were seeded onto 2 μg/ml CB3 in the presence of AGF1 alone or rhodocetin (2 μg/ml) plus AGF-1 or rhodocetin (2 μg/ml) plus plumieribetin. Because of the compensatory activity of α2β1 integrin, monoclonal antibody AGF-1 is only active in the presence of rhodocetin and blocks cell attachment completely in an α1β1 integrin-dependent manner, although in the presence of rhodocetin, plumieribetin fails to block cell adhesion. The means and standard deviations of 8-fold determinations are shown.
FIGURE 10.
Plumieribetin induces morphological changes in α1β1 integrin-mediated cell adhesion of rhodocetin-treated huASMC. In scanning electron microscope analyses (A–D), huASMC were seeded onto CB3-coated silicium wavers in the absence (A) or presence of 10 mm EDTA (B) or rhodocetin (4 μg/ml) (C) or both rhodocetin (4 μg/ml) and plumieribetin (10 μg/ml) (D). Note, that rhodocetin alone (C) did not change cell morphology compared with the untreated cells (A), thus showing that α1β1 integrin is responsible for cell spreading after α2β1 integrin blockage. The addition of plumieribetin to rhodocetin-treated cells (D) resulted in reduced cell contacts and a more spiny or sometimes spindle-like appearance of the cells. However, plumieribetin failed to detach the cells. After immunofluorescent staining of the actin cytoskeleton (E–G), the highly organized actin stress fibers running through the entire cell were characteristic for the untreated (E) and rhodoctein-treated (F) huASMC. By weakening the α1β1 integrin-mediated cell contacts, plumieribetin reduced the actin stress fibers. Instead, the cortical actin meshwork in the cell periphery was more clearly seen (G).
DISCUSSION
The fin rays of the scorpionfish (S. plumieri) have developed into stings. In addition, glands on the grooves of these stings are able to produce a venom, which acts as a defensive weapon in cases of predator attacks on this fish. Human accidents happen when fishermen or tourists step on this fish, which hides between stones (24, 25). Like venoms of terrestrial animals, fish venoms are a complex mixture of pharmacologically highly active compounds. Envenomation is hardly ever life-threatening to humans, but the intense and strongly irradiating pain deters predators in future encounters. Additionally, local edema and erythema may arise as consequence of envenoming. The systemic symptoms of cardiotoxic and vasorelaxant effects are more severe, resulting in a drastic drop of blood pressure (22–25). Moreover, the observed increase in the epithelial permeability of the bronchi and alveoli, similar to the observed erythema and hemorrhages, indicate a loss of cell-matrix interactions (22, 23). Although we have identified α1β1 integrin as a potential target of the scorpion fish venom, the comparatively high doses of venom used in our screening experiment suggest that this novel integrin inhibitor is likely to cause local rather than systemic effects, close to the envenomation site.
When screening various venoms from different South American fish, we discovered the venom of the S. plumieri stings to have an interesting inhibition profile on integrins. By using α1β1 integrin inhibition as the target molecule, we have isolated a component from the venom and mucus of S. plumieri that strongly interferes with the α1β1 integrin-collagen IV interaction. We named this factor plumieribetin. Because no other organ produces plumieribetin, it is likely to be exclusively expressed in the sting glands and/or skin mucus, suggesting its defensive character.
Functionally, plumieribetin specifically blocks α1β1 integrin, but not α2β1 integrin, among the collagen-binding integrins. Less selectively, it also interferes with the α3β1 integrin-laminin-332 interaction but does not affect the high affinity binding of α7β1 integrin to laminin-111 (39). Although the crude venom of S. plumieri is able to disturb the low affinity interaction of α3β1 integrin with laminin-332, the isolated plumieribetin only weakly inhibited this interaction. In this study, we focused on plumieribetin and its inhibitory role on α1β1 integrin, because of its target specificity among the principal collagen-binding integrins and because of the universal abundance of collagen IV in all basement membranes (7, 9) as opposed to the more tissue-restricted expression of laminin-332 in certain basement membranes of skin, lung, and kidneys (8).
Mechanistically, plumieribetin inhibits by binding to the integrins but not to the respective matrix ligands. In contrast to its selectivity in α1β1 integrin inhibition, plumieribetin bound to all of the integrins tested. This may indicate that the interaction site of plumieribetin within the integrin comprises a region of the common integrin β1 subunit. In addition, plumieribetin binds mannose residues preferentially but weakly. Whereas other mannose-binding lectins from plants and the fish lectin pufflectin from T. rubripes were isolated by affinity chromatography on mannose columns (32, 37, 38), plumieribetin failed to bind to resin-immobilized mannose. Moreover, from the three conserved mannose-binding sites of the plant B-lectin, plumieribetin has preserved only the first two mannose-binding sites as potentially functional ones, similar to pufflectin from the Japanese pufferfish (37). The hypothesis that the soluble integrins that had been recombinantly expressed in insect cells bear high mannose-type N-glycoconjugates and thereby mediate binding to plumieribetin, was ruled out because plumieribetin recognized not only insect cell-expressed α1β1 integrin after enzymatic removal of N-glycans but also wild-type α1β1 integrin, which had been extracted from human placenta and hence bears authentic N-glycoconjugates. This suggests that plumieribetin interacts with α1β1 integrin in a direct protein-protein interaction, although we cannot rule out the possibility that its primary binding to the integrin is initiated by mannose-mediated interactions, as is seen for selectin adhesion to endothelial cells (40). These sugar-mediated interactions might be responsible for the nonselective binding to all integrins tested, inasmuch as the binding signals in Fig. 7B do not necessarily represent binding affinities but are influenced by integrin coating efficiencies or accessibility of the glycoconjugates on the immobilized integrins.
Integrin α1β1 regulates cell survival and gene activation, besides anchoring cells to the underlying basement membrane (41). It is abundantly expressed on hepatocytes and vascular smooth muscle cells. At the cellular level, plumieribetin failed to block attachment of these cell types to the collagen IV fragment CB3, even when the compensating α2β1 integrin was blocked by rhodocetin (16, 31). However, plumieribetin weakened the interaction of both cell types and resulted in rounding up of the otherwise flat cells and in rearrangement of the actin cytoskeleton. Especially, the decreased cell spreading of huVSMC indicated a reduction of force transmission to the surrounding matrix, which could aid cytolytic venom toxins in vasodilation and hypotension (42, 43). As an isolated component, plumieribetin by itself did not show any cellular effects, because inhibition of α1β1 integrin is easily compensated for by α2β1 integrin at the cellular level and, most likely, also in situ. Therefore, from our studies, we would not expect major, life-threatening pharmacological effects of plumieribetin, but rather an auxiliary or synergistic role together with other venom components. In the crude venom of S. plumieri, we could not detect any α2β1 integrin inhibiting activity. However, the integrin inhibition test with crude venom was performed in the presence of protease inhibitors. But proteases of the venom may inactivate the compensating α2β1 integrin during envenomation. We could not detect any proteolytic activity in the isolated plumieribetin preparation.
Plumieribetin binds to α1β1 integrin independently of divalent cations. This speaks against the idea that the venom component mimics the collagenous ligands and competitively blocks the collagen-binding site of the integrin. In fact, plumieribetin has a completely different structure than collagen. Structurally, plumieribetin is highly homologous to fish lectins and the B-lectins of monocotyledonous plants. Similar to the monocot B-lectins, plumieribetin shows a high content of antiparallel β-strands and assembles into higher aggregates. According to the analytical gel filtration, four or eight plumieribetin monomers form the major quaternary structure of this integrin inhibitor. In the monocot B-lectins, the monomers fold and assemble concomitantly, and the quaternary structure stabilizes the monomer folding (35). Both dimeric and tetrameric quaternary structures have been described in the group of monocot B-lectins (20, 32, 34, 44).
As yet, among the homologous fish lectins, only the pufflectin has been isolated from the skin and intestine of T. rubripes (26). All other homologous fish lectins have been deduced from genomic analysis (45) and hence lack any functional studies. However, most of these fish lectins, because of their putative mannose binding activity, have been assigned a role in innate immunity and a defense weapon against microbial and metazoan parasites (45–47). Similarly, plants use the homologous monocot B-lectins to prevent pathogenic attack, and some of them block retroviruses, such as human immunodeficiency virus and the cytomegalovirus (48). Plumieribetin with its integrin inhibiting activity adds a completely new function to this class of B-lectin.
Acknowledgment
We thank Jens Sobek (Functional Genomics Center Zurich) for printing the carbohydrate microarrays.
This work was supported by Deutsche Forschungsgemeinschaft Grants Eb177/4-2 and SFB815 (to J. A. E.), German-Brazilian Exchange Program PROBRAL of the German Academic Exchange Service Grant D/05/30352, CAPES Process 250/06, Brazilian Conselho Nacional de Desenvolvimento Cientifico e Tecnológico, Fundação de Amparo a Pesquisa do Estado de Minas Gerais Edital 24000/01, and funds from the ETH Zürich and the Schweizerischer Nationalfonds.
- TBS
- Tris-buffered saline
- ELISA
- enzyme-linked immunosorbent assay
- BSA
- bovine serum albumin
- PBS
- phosphate-buffered saline
- BS3
- bis-(sulfosuccinimidyl) suberate
- huASMC
- human arterial smooth muscle cell(s).
REFERENCES
- 1.Hynes R. O. (2002) Cell 110, 673–687 [DOI] [PubMed] [Google Scholar]
- 2.Arnaout M. A., Mahalingam B., Xiong J. P. (2005) Annu. Rev. Cell Dev. Biol. 21, 381–410 [DOI] [PubMed] [Google Scholar]
- 3.Calderwood D. A., Shattil S. J., Ginsberg M. H. (2000) J. Biol. Chem. 275, 22607–22610 [DOI] [PubMed] [Google Scholar]
- 4.Xiong J. P., Stehle T., Zhang R., Joachimiak A., Frech M., Goodman S. L., Arnaout M. A. (2002) Science 296, 151–155 [DOI] [PubMed] [Google Scholar]
- 5.Myllyharju J., Kivirikko K. I. (2004) Trends Genet. 20, 33–43 [DOI] [PubMed] [Google Scholar]
- 6.Iozzo R. V. (2005) Nat. Rev. Mol. Cell Biol. 6, 646–656 [DOI] [PubMed] [Google Scholar]
- 7.Kalluri R. (2003) Nat. Rev. Cancer 3, 422–433 [DOI] [PubMed] [Google Scholar]
- 8.Colognato H., Yurchenco P. D. (2000) Dev. Dyn. 218, 213–234 [DOI] [PubMed] [Google Scholar]
- 9.Hudson B. G., Tryggvason K., Sundaramoorthy M., Neilson E. G. (2003) N. Engl. J. Med. 348, 2543–2556 [DOI] [PubMed] [Google Scholar]
- 10.Ruoslahti E. (1996) Annu. Rev. Cell Dev. Biol. 12, 697–715 [DOI] [PubMed] [Google Scholar]
- 11.Eble J. A. (2001) Osteoarthritis Cartilage 9, S131–S140 [PubMed] [Google Scholar]
- 12.Eble J. A. (2005) Curr. Pharm. Des. 11, 867–880 [DOI] [PubMed] [Google Scholar]
- 13.Eble J. A., Golbik R., Mann K., Kühn K. (1993) EMBO J. 12, 4795–4802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calvete J. J., Marcinkiewicz C., Monleón D., Esteve V., Celda B., Juárez P., Sanz L. (2005) Toxicon 45, 1063–1074 [DOI] [PubMed] [Google Scholar]
- 15.Morita T. (2005) Toxicon 45, 1099–1114 [DOI] [PubMed] [Google Scholar]
- 16.Eble J. A., Beermann B., Hinz H. J., Schmidt-Hederich A. (2001) J. Biol. Chem. 276, 12274–12284 [DOI] [PubMed] [Google Scholar]
- 17.Rosenow F., Ossig R., Thormeyer D., Gasmann P., Schlüter K., Brunner G., Haier J., Eble J. A. (2008) Neoplasia 10, 168–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bouckaert J., Hamelryck T., Wyns L., Loris R. (1999) Curr. Opin. Struct. Biol. 9, 572–577 [DOI] [PubMed] [Google Scholar]
- 19.Rini J. M., Lobsanov Y. D. (1999) Curr. Opin. Struct. Biol. 9, 578–584 [DOI] [PubMed] [Google Scholar]
- 20.Barre A., Bourne Y., Van Damme E. J., Peumans W. J., Rougé P. (2001) Biochimie 83, 645–651 [DOI] [PubMed] [Google Scholar]
- 21.Theakston R. D., Kamiguti A. S. (2002) Toxicon 40, 579–651 [DOI] [PubMed] [Google Scholar]
- 22.Carrijo L. C., Andrich F., de Lima M. E., Cordeiro M. N., Richardson M., Figueiredo S. G. (2005) Toxicon 45, 843–850 [DOI] [PubMed] [Google Scholar]
- 23.Boletini-Santos D., Komegae E. N., Figueiredo S. G., Haddad V., Jr., Lopes-Ferreira M., Lima C. (2008) Toxicon 51, 585–596 [DOI] [PubMed] [Google Scholar]
- 24.Haddad V., Jr., Martins I. A., Makyama H. M. (2003) Toxicon 42, 79–83 [DOI] [PubMed] [Google Scholar]
- 25.Loyo J., Lugo L., Cazorla D., Acosta M. E. (2008) Invest. Clin. 49, 299–307 [PubMed] [Google Scholar]
- 26.Tsutsui S., Okamoto M., Tsasumi S., Suetake H., Kikuchi K., Suzuki Y. (2006) Comp. Biochem. Physiol. D 1, 122–127 [DOI] [PubMed] [Google Scholar]
- 27.Eble J. A., Berditchevski F. (2001) in Methods in Cell-Matrix Adhesion (Adams J. ed) pp. 223–246, Academic Press, San Diego [Google Scholar]
- 28.Eble J. A., Wucherpfennig K. W., Gauthier L., Dersch P., Krukonis E., Isberg R. R., Hemler M. E. (1998) Biochemistry 37, 10945–10955 [DOI] [PubMed] [Google Scholar]
- 29.Schaeffer R. C., Jr., Carlson R. W., Russell F. E. (1971) Toxicon 9, 69–78 [DOI] [PubMed] [Google Scholar]
- 30.Ratner D. M., Plante O. J., Seeberger P. H. (2002) Eur. J. Org. Chem. 5, 826–833 [Google Scholar]
- 31.Eble J. A., Niland S., Dennes A., Schmidt-Hederich A., Bruckner P., Brunner G. (2002) Matrix Biol. 21, 547–558 [DOI] [PubMed] [Google Scholar]
- 32.Hester G., Kaku H., Goldstein I. J., Wright C. S. (1995) Nat. Struct. Biol. 2, 472–479 [DOI] [PubMed] [Google Scholar]
- 33.Hester G., Wright C. S. (1996) J. Mol. Biol. 262, 516–531 [DOI] [PubMed] [Google Scholar]
- 34.Wood S. D., Wright L. M., Rizkallah P. J., Allen A. K., Peumans W. J., Van Damme E. J. M. (1998) Acta Crystallogr. Sect. D 55, 1264–1272 [DOI] [PubMed] [Google Scholar]
- 35.Bachhawat K., Kapoor M., Dam T. K., Surolia A. (2001) Biochemistry 40, 7291–7300 [DOI] [PubMed] [Google Scholar]
- 36.Horlacher T., Seeberger P. H. (2008) Chem. Soc. Rev. 37, 1414–1422 [DOI] [PubMed] [Google Scholar]
- 37.Tsutsui S., Tasumi S., Suetake H., Kikuchi K., Suzuki Y. (2006) Comp. Biochem. Physiol. B. 143, 514–519 [DOI] [PubMed] [Google Scholar]
- 38.Wright C. S., Kaku H., Goldstein I. J. (1990) J. Biol. Chem. 265, 1676–1677 [PubMed] [Google Scholar]
- 39.Eble J. A., Bruckner P., Mayer U. (2003) J. Biol. Chem. 278, 26488–26496 [DOI] [PubMed] [Google Scholar]
- 40.Vestweber D., Blanks J. E. (1999) Physiol. Rev. 79, 181–213 [DOI] [PubMed] [Google Scholar]
- 41.Gardner H. (2003) in I Domains in Integrins (Gullberg D. ed) pp. 25–39, Landes Bioscience, Georgetown, TX [Google Scholar]
- 42.Garnier P., Goudey-Perrière F., Breton P., Dewulf C., Petek F., Perrière C. (1995) Toxicon 33, 143–155 [DOI] [PubMed] [Google Scholar]
- 43.Low K. S., Gwee M. C., Yuen R., Gopalakrishnakone P., Khoo H. E. (1993) Toxicon 31, 1471–1478 [DOI] [PubMed] [Google Scholar]
- 44.Chandra N. R., Ramachandraiah G., Bachhawat K., Dam T. K., Surolia A., Vijayan M. (1999) J. Mol. Biol. 285, 1157–1168 [DOI] [PubMed] [Google Scholar]
- 45.Spencer R., Lumsden J. S. (2005) Vet. Immun. Immunopath. 108, 111–120 [DOI] [PubMed] [Google Scholar]
- 46.Dommett R. M., Klein N., Turner M. W. (2006) Tissue Antigens 68, 193–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsutsui S., Tasumi S., Suetake H., Suzuki Y. (2003) J. Biol. Chem. 278, 20882–20889 [DOI] [PubMed] [Google Scholar]
- 48.Balzarini J., Schols D., Neyts J., Van Damme E., Peumans W. J., De Clercq E. (1991) Antimicrob. Agents Chemother. 35, 410–416 [DOI] [PMC free article] [PubMed] [Google Scholar]