Abstract
Androgen-dependent transcriptional activity by the androgen receptor (AR) and its coregulators is required for male reproductive development and function. In humans and other primates, melanoma antigen gene protein-A11 (MAGE-11) is an AR selective coregulator that increases AR transcriptional activity. Here we show that the interaction between AR and MAGE-11 is mediated by AR NH2-terminal FXXLF motif binding to a highly conserved MAGE-11 F-box in the MAGE homology domain, and is modulated by serum stimulation of mitogen-activated protein kinase phosphorylation of MAGE-11 Ser-174. The MAGE-11-dependent increase in AR transcriptional activity is mediated by a direct interaction between MAGE-11 and transcriptional intermediary factor 2 (TIF2) through the NH2-terminal region of TIF2, and by a MAGE-11 FXXIF motif interaction with an F-box-like region in activation domain 1 of TIF2. The results suggest that MAGE-11 functions as a bridging factor to recruit AR coactivators through a novel FXX(L/I)F motif-F-box interaction paradigm.
Introduction
The androgen receptor (AR)3 is a ligand-activated transcription factor that mediates the biological effects of testosterone and dihydrotestosterone (DHT). High affinity binding of androgen targets AR to the nucleus where it interacts with coregulators and androgen response element DNA associated with androgen-regulated genes. Naturally occurring functional knockouts of AR in the human population caused by AR gene mutations result in the androgen insensitivity syndrome, and demonstrate the functional importance of AR in male sex development and maturation (1). Gain-of-function AR somatic mutations identified in clinical specimens of prostate cancer support a critical role for AR during tumor progression to castration-recurrent growth (1–4). The multidomain structure of AR includes the carboxyl-terminal ligand binding domain with activation function 2 (AF2), a hydrophobic interaction surface structurally dependent on bound androgen (5). AR AF2 binds LXXLL motifs of steroid receptor p160 coactivators (SRC), but interacts preferentially with the FXXLF motif in the AR NH2 terminus (6, 7). AR activation function 1 (AF1) in the unstructured NH2-terminal region interacts with multiple coregulatory proteins (8, 9).
AR undergoes an androgen-dependent NH2- and COOH-terminal (N/C) interaction mediated by the AR NH2-terminal FXXLF motif (23FQNLF27) binding to AF2 in the ligand binding domain (5, 10–14). The ligand-dependent AR N/C interaction stabilizes AR and underlies the potency differences between testosterone and DHT (2, 11, 13–15). The AR N/C interaction coordinates transcriptional activity between AF1 and AF2, in part by competitively inhibiting the SRC/p160 coactivator LXXLL motif binding to AF2 (15). The regulatory effect of the AR N/C interaction is modulated by melanoma antigen gene protein-A11 (MAGE-11), a MAGE gene family member and AR coregulator that binds the AR NH2-terminal FXXLF motif, competes with the AR N/C interaction, and enhances AR transcriptional activity by increasing accessibility of AF2 for SRC/p160 coactivator recruitment (16).
MAGE-11 expression is limited to human and nonhuman primates (17, 18), suggesting it arose during mammalian evolution as an additional control mechanism in the AR signaling network. Post-translational modification of MAGE-11 by phosphorylation of Thr-360, and subsequent monoubiquitinylation of Lys-240 and Lys-245 within the MAGE homology domain, a highly conserved region of the MAGE gene family, is induced by epidermal growth factor (EGF) and stabilizes the interaction with AR (19). The increase in MAGE-11 levels in castration-recurrent prostate cancer during androgen deprivation therapy by DNA hypomethylation at the transcriptional start site supports the concept that MAGE-11 increases AR signaling in prostate cancer (20).
To understand the molecular mechanisms whereby MAGE-11 enhances AR transactivation, we pursued observations that MAGE-11 acts coordinately with transcriptional intermediary factor 2 (TIF2), one of the SRC/p160 coactivators implicated in AR signaling (16, 19). Previous studies have shown that TIF2 can associate with the NH2-terminal region of AR, although the sites of interaction were not well defined (8, 10, 21). We considered the possibility that MAGE-11 enhances AR activity from AF1 through a direct interaction between MAGE-11 and TIF2.
The present study provides evidence that the AR NH2-terminal FXXLF motif interacts with a highly conserved MAGE-11 F-box (residues 329–369) in the MAGE homology domain, and that the interaction is modulated by mitogen-activated protein (MAP) kinase phosphorylation of MAGE-11 Ser-174 in response to serum stimulation. MAGE-11 also interacts directly with the NH2-terminal region of TIF2, and with the TIF2 AD1 region through a MAGE-11 FXXIF motif (260FPEIF264). The results suggest AR-mediated gene transcription involves a novel protein interaction paradigm of the FXX(L/I)F motif binding to the F-box.
EXPERIMENTAL PROCEDURES
Plasmids
Expression plasmids include pCMV-AR (22), FLAG-AR (16), pCMV-AR-(1–660) (23), pCMV-ARΔ120–472 (19), VP-AR-(1–660) (6), GAL-AR-(16–36) and -(4–52) (24), pSG5-MAGE-(1–429) (pSG5-MAGE), GAL-MAGE-(2–429) (GAL-MAGE), VP-MAGE-(2–429) (VP-MAGE), FLAG-MAGE-(2–429) (FLAG-MAGE) (16), pSG5-HA-MAGE-(2–429) and -(112–429), GAL-MAGE-K236A, -K240A,K245A, and -T360A, VP-MAGE-K236A,K240A,K245A and T360A (19), GAL-TIF2.0-(1–627), 2.1-(624–1287), 2.2-(1288–1464), 2.3-(624–1287), 2.3m123-(624–1179-L644E,L645A,L693A, L694A,L748A,L749A), 2.8-(1011–1179) and 2.12-(940–1131), VP-TIF2.0-(1–627), and pSG5-TIF2 (25).
Reporter vectors include prostate-specific antigen (PSA) enhancer luciferase (PSA-Enh-Luc) (26), mouse mammary tumor virus-luciferase (MMTV-Luc), MMTVΔ-(−421–−364)-Luc, which contains a deletion of a negative response element (27–29), and 5×GAL4Luc3 (30). MMTVΔ-(−421–−364)-Luc was created by PCR mutagenesis of MMTV-Luc (31), with the XhoI/BamHI fragment inserted at the same sites of MMTV-Luc.
GST-TIF2-(1–627), -(624–1141), -(1180–1464), and –(1288–1464) were created by PCR amplifying corresponding regions of pSG5-TIF2 and inserting the EcoRI/XhoI-digested fragments into similarly digested pGEX-4T-1. FLAG-TIF2 vectors were constructed by a triple ligation: TIF2-(1–569) was PCR amplified from pSG5-TIF2 and digested with EcoRI/NdeI to obtain TIF2-(1–370); TIF2-(370–1464) was obtained by digesting the parent template with NdeI/XbaI; both fragments were ligated into EcoRI/XbaI sites of pCMV5-FLAG-b. VP-TIF2.0-I336A, Y337A, pCMV-ARΔ120–472-V33E, -K720A, -E897K, and -L26A,F27A, MAGE-11 alanine mutants, and phosphomimetics were created using QuikChange site-directed mutagenesis (Stratagene).
pSG5-HA-MAGE-(112–307), -(112–298), -(112–276), -(165–307) and -(Δ329–369) were created by PCR amplification using primers containing EcoRI/SalI ends and ligation into pSG5-HA. Additional MAGE-11 F-box deletion mutants were created by double PCR mutagenesis to create an EcoRI/XhoI MAGEΔ329–369 fragment that was inserted into the same sites of pSG5-HA to make pSG5-HA-MAGEΔ329–369, and into EcoRI/SalI sites of FLAG-b to make FLAG-MAGEΔ329–369.
GAL-MAGE-(85–205) and –(112–205) were created by PCR amplifying pSG5-MAGE and inserting EcoRI/XhoI-digested fragments into GAL0 digested with EcoRI and SalI. GAL-MAGE-(112–170) was created by PCR amplifying the parent template and ligating the EcoRI/SalI-digested fragment into the same sites of GAL0. GAL-MAGE-(251–272) wild-type and F260A,F264A mutant were created by cloning a small DNA insert into EcoRI and SalI sites of GAL0. All PCR-amplified regions were verified by DNA sequencing.
DNA Transfection
CWR-R1 prostate cancer cells were maintained and transfected using Effectene (Qiagen) as described (2, 32). CWR-R1 cells (1.6 × 105 cells/well) were transfected in 12-well plates with 0.1 μg/well of MMTV-Luc or MMTVΔ-(−421–−364)-Luc in the absence and presence of 0.1 μg of pSG5-MAGE-11 and/or 0.1 μg of pSG5-TIF2. Medium was replaced 24 h later with serum-free, phenol red-free medium in the absence and presence of DHT and/or EGF and incubated at 37 °C overnight.
For small interfering RNA (siRNA) studies, CWR-R1 cells (4 × 105/well) in 6-well plates were plated in 1 ml of antibiotic-free medium without EGF and transfected using Lipofectamine 2000 reagent (Invitrogen) in antibiotic-free medium with 0.1 μg of MMTVΔ-(−421–−364)-Luc and 0.2 μg of pSG5 or pSG5-MAGE with and without 4 nm TIF2 siRNA-3 (GAUCAGAAGUGACUAUUAA) or siCONTROL nontargeting siRNA pool (Dharmacon RNA Technologies). After 48 h at 37 °C, cells were transferred to serum-free, phenol red-free medium containing antibiotics in the absence and presence of 0.1 DHT and 0.01 μg/ml of EGF. After 24 h cells were lysed in 0.25 ml of buffer containing 1% Triton X-100, 2 mm EDTA, and 25 mm Tris phosphate, pH 7.8 (19). Luciferase activity was measured using an automated Lumistar Galaxy multiwell plate luminometer (BMG Labtech). Luciferase data are representative of at least three independent experiments, with the graphs showing the mean ± S.E.
COS cells (4 × 105/6-cm dish) were transfected in antibiotic-free medium using Lipofectamine 2000 with pSG5-MAGE, pSG5-TIF2, pCMV-AR, or empty parent vector in the absence and presence of 1 nm siRNA targeting MAGE-11, TIF2, AR, or nonspecific siCONTROL. The next day cells were placed to serum-free medium, and 24 h later extracts were analyzed by immunoblot as described below.
For inhibition of endogenous PSA mRNA expression, CWR-R1 cells (106/6-cm dish) were transfected using Lipofectamine 2000 in 2% charcoal-stripped serum medium without antibiotics with 4 nm MAGE-11 siRNA-2 (GCACUGAUCCUGCAUGCUAUU), MAGE-11 siRNA-3 (CAACUGCUCUUUGGCAUUGUU), AR siRNA-3 (UCAAAGAACUCGAUCGUAUUU), TIF2 siRNA-3, and siCONTROL nonspecific siRNA (Dharmacon RNA Technologies). After 48 h cells were treated for 6 h in fresh medium with and without 10 nm DHT and 10 ng/ml of EGF. RNA was extracted in 1 ml of TRIzol (Invitrogen) and analyzed by quantitative reverse transcription-PCR using SuperScript II reverse transcriptase (Invitrogen), 4 μg of RNA reverse transcription-PCR, CTCATCCTGTCTCGGATTGT and AGAAACAGGCTGTGCCGAC PSA primers (amplify a 189-bp fragment spanning amino acids residues 20–82), and control peptidylprolyl isomerase A primers ATCTTGTCCATGGCAAATGC and GCCTCCACAATATTCATGCC (amplify a 134-bp fragment spanning amino acids residues 97–141). Reactions were performed using a Roche LightCycler in 20 μl containing 0.4 μg of cDNA, 10 μl of 2× QuantiTect SYBR PCR Master Mix (Qiagen), and 2 μl of 2 μm primers at 95 °C for 15 min followed by 55 cycles of 94 °C for 15 s, 58 °C (PSA) or 55 °C (peptidylprolyl isomerase A) for 30 s, 72 °C for 30 s, and 79 °C for 8 s. A no template control was included in each run using standard curves for PSA and peptidylprolyl isomerase A (33).
LAPC-4 human prostate cancer cells were cultured in RPMI 1640 medium (Invitrogen) supplemented with 2 mm l-glutamine, penicillin, streptomycin, 10% fetal bovine serum, and 1 nm methyltrienolone (R1881) (34). LAPC-4 cells (5 × 105 cells/well) in 6-well plates containing 1 ml of antibiotic-free medium without R1881 were transfected the following day using Lipofectamine 2000 with 0.1 μg/well of MMTVΔ-(−421–−364)-Luc in the absence and presence of 3 nm siRNA. After 48 h at 37 °C, cells were transferred to serum-free, phenol red-free medium containing antibiotics in the absence and presence of 0.1 nm DHT and 0.1 μg/ml of EGF, and luciferase activity was measured 24 h later.
Human endometrial Ishikawa cells (105 cells/well) were transfected in 12-well plates using FuGENE 6 (Roche Applied Science) (19, 33) with 0.1 μg/well of PSA-Enh-Luc and 0.1 μg of pSG5 empty vector, wild-type, and mutant pSG5-MAGE. The next day medium was replaced with serum-free, phenol red-free medium in the absence and presence of 1 nm DHT with and without 0.1 μg/ml EGF. Twenty-four h later, luciferase activity was measured.
Monkey kidney CV1 cells (4.2 × 105/6-cm dish) were transfected using calcium phosphate (15) with 0.05 or 0.1 μg of wild-type or mutant pCMV-AR and the indicated amounts of pSG5-TIF2, pSG5-MAGE, MMTV-Luc, or PSA-Enh-Luc. Immediately after transfection and 24 h later, cells were placed in serum-free, phenol red-free medium in the absence and presence of 1 nm DHT, incubated for 24 h at 37 °C, and luciferase activity was measured.
Mammalian two-hybrid assays were performed in human HeLa epithelial cervical carcinoma cells (5 × 104 cells/well) transfected using FuGENE 6 (2) in 12-well plates with 0.1 μg/well of 5×GAL4Luc3, 0.05 μg of wild-type or mutant GAL-MAGE, GAL-TIF2, GAL-AR-(4–52), or GAL-AR-(16–36) with 0.1 μg of wild-type or mutant VP-AR-(1–660), VP-MAGE, or VP-TIF2.0. The day after transfection, cells were transferred to serum-free medium and incubated overnight at 37 °C before luciferase activity was measured.
Immunochemistry
Immunoblotting and immunoprecipitation were performed by transfecting each 10-cm dish with 2–4 dishes/group, 1.8–2.5 × 106 monkey kidney COS-1 cells and 6 × 106 human embryonic kidney 293 cells using DEAE dextran (2, 7), 2.0 × 106 CV1 cells using 450 μl of H2O, 75 μl of CaCl2, and 675 μl of 2× HBS added to 8 ml of medium/10-cm dish (2× HBS = 0.28 m NaCl, 0.05 m Hepes, and 1.5 mm Na2HP04, pH 7.1). HeLa cells (7.5 × 105) and 4.4 × 105 Ishikawa cells were transfected using 8 μl of FuGENE 6 and 160 μl of medium in 6 ml of medium. Twenty-four h after transfection, cells were transferred to serum-free, phenol red-free medium in the absence and presence of 0.1 μg/ml of EGF, 10 nm DHT, and/or 1 μm MG132, a proteosome inhibitor. After 24 h, cells were harvested in phosphate-buffered saline. For siRNA experiments, COS-1 cells (4 × 105 cells/well) plated in antibiotic-free medium in 6-well plates were transfected using Lipofectamine 2000 reagent with pSG5, pSG5-MAGE, pSG5-TIF2, or pCMV-AR in the absence and presence of siRNA.
For phosphorylation studies, COS cells (2 × 106/10-cm dish) were transfected using DEAE dextran and extracted in immunoprecipitation (IP) lysis buffer containing 0.15 m NaCl, 0.5% Nonidet P-40, 10% glycerol, 50 mm Tris-HCl, pH 7.5, Complete protease inhibitor mixture (Roche), 1 mm dithiothreitol, and 1 mm phenylmethylsulfonyl fluoride. Protein concentration was measured using the Bio-Rad assay and bovine serum albumin as standard. Cell extracts were incubated in the absence and presence of 800 IU of λ-phosphatase (New England Biolabs) for 1 h at 4 °C. For immunoblots, cells were solubilized in immunoblot (IB) lysis buffer containing 0.15 m NaCl, 1% Triton X-100, 0.1% SDS, 1% sodium deoxycholate, 0.5 mm EDTA, 50 mm Tris-HCl, pH 7.5, 1 mm phenylmethylsulfonyl fluoride, 1 mm dithiothreitol, and Complete protease inhibitor mixture (Roche). For inhibition of MAP kinase, U0126 (Promega) was added immediately after transfection to serum-containing medium. Cells were transferred 24 h later to serum-free medium with and without the same concentration of U0126 and 1 μm MG132. The next day cells were solubilized in IB lysis buffer containing phosphatase Mixture Inhibitors 1/2 (Sigma). Protein extracts were combined with 0.2 volumes of 6× sample buffer containing 10% SDS, 30% glycerol, 30% 2-mercaptoethanol, and 0.35 m Tris-HCl, pH 6.8.
Immunoprecipitation was performed using 2–4 10-cm dishes/group of transfected COS-1 or HEK293 cells harvested in phosphate-buffered saline and pooled (15). Cells were solubilized in IP lysis buffer containing 0.05 m sodium fluoride. Lysates were precleared using 0.1 ml of agarose (Sigma) by rotating for 1 h at 4 °C. Samples were transferred to 15 μl of anti-FLAG M2 affinity-agarose (Sigma) and incubated at 4 °C for 1 h or overnight. Samples were washed 3 times with IP lysis buffer, and protein was resuspended in 0.05 ml of 2× SDS sample buffer containing 3.3% SDS, 10% 2-mercaptoethanol, 10% glycerol, and 0.12 m Tris-HCl, pH 6.8. Immunoprecipitates, protein extracts, and EZ-Run Prestained Rec protein ladder (Fisher Bioreagents) were separated on 8, 10, or 12% acrylamide gels containing SDS and transferred overnight to nitrocellulose membranes at 4 °C.
Immunoblots were probed using the following antibodies: rabbit anti-HA tag (1:2500, Abcam, ab9110), mouse anti-β-actin (1:5000, Abcam), rabbit anti-GAL (1:500, Santa Cruz Biotechnology, sc-577), rabbit anti-VP16 activation domain (1:2000, Abcam, ab4809), mouse anti-FLAG M2 monoclonal antibody (1:2000, Sigma, F3165), mouse anti-TIF2 (1:275, BD Transduction Laboratories, 610985), rabbit anti-Skp1 (1:400, Abcam, ab10546), rabbit AR32 immunoglobulin G (22) (0.4 μg/ml), rabbit AR52 immunoglobulin G (5 μg/ml) (35, 36), rabbit anti-MAGE-11 peptide antibodies MagAb-(13–26), -(59–79), and -(94–108) immunoglobulin G (6–10 μg/ml) (33), and rabbit polyclonal FLAG-MAGE antibody (10 μg/ml) raised against purified baculovirus-expressed FLAG-tagged human MAGE-11. Incubations with primary antibody were performed for 1 h at room temperature or overnight at 4 °C, and with anti-mouse and anti-rabbit horseradish peroxidase-conjugated secondary IgG antibodies (1:10,000, Amersham Biosciences) at room temperature for 1 h. Signals were detected using chemiluminescence (SuperSignal West Dura Extended Duration Substrate, Pierce).
In Vitro Binding and Kinase Assays
Glutathione S-transferase (GST) affinity matrix binding assays were performed with GST-TIF2 fusion proteins and 35S-labeled MAGE-11 expressed from pSG5-MAGE-11 using the TnT T7 Quick Coupled transcription-translation system (Promega) (6, 10). GST empty vector pGEX-4T-1 and GST-TIF2 fusion proteins were expressed in XL1-Blue Escherichia coli by incubating in the presence of 1 mm isopropyl 1-thio-β-d-galactopyranoside for 3 h at 37 °C. Cell pellets were resuspended in GST binding buffer containing 0.5% Nonidet P-40, 1 mm EDTA, 0.1 m NaCl, 20 mm Tris-HCl, pH 8.0, with 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, and protease inhibitor mixture (Sigma). Protein expression was normalized by Coomassie Blue staining of a mini-gel. Fusion proteins were incubated with glutathione-Sepharose 4B beads (Amersham Biosciences) for 1.5 h at 4 °C, washed in GST binding buffer, and combined with 35S-labeled MAGE-11 prepared using 25 μCi/sample of [35S]methionine (PerkinElmer Life Sciences) and incubated for 2 h at 4 °C. Beads were washed, eluted using 2× SDS sample buffer, and analyzed on a 10% gel containing SDS.
In vitro kinase assays were performed as described (19) except using full-length recombinant active extracellular signal-regulated kinase 1 (ERK1) expressed from baculovirus in Sf9 cells (BioVision, 0.1 μg/μl, specific activity 383 nmol/min/mg) and GST-MAGE-171PPQSPQEES179, GST-MAGE-171PPQAPQEES179, and GST-APRTPGGRR ERK1 peptide substrates modeled after myelin basic protein (37). GST fusion proteins were expressed from pGEX-4T-1 by cloning short inserts into EcoRI/XhoI (MAGE-11) and EcoRI/SalI sites (ERK1 control peptide) designed to disrupt the cloning site for rapid clone identification. GST-peptide-Sepharose bead suspensions (50 μl) were washed in kinase buffer (1 mm EGTA, 0.4 mm EDTA, 5 mm MgCl2, 0.05 mm dithiothreitol, and 5 mm MOPS, pH 7.2) and incubated in kinase buffer containing 5 μl of 0.25 mm ATP and 0.16 μCi/μl of [γ-32P]ATP (10 mCi/ml, 3000 Ci/mmol, PerkinElmer Life Sciences) for 30 min at 30 °C. Samples were separated on an 8–16% gradient minigel containing SDS (Invitrogen). Dried gels were exposed to x-ray film for 16 h and rehydrated for protein staining using Coomassie Blue.
RESULTS
Dependence of AR Transactivation on MAGE-11 and TIF2
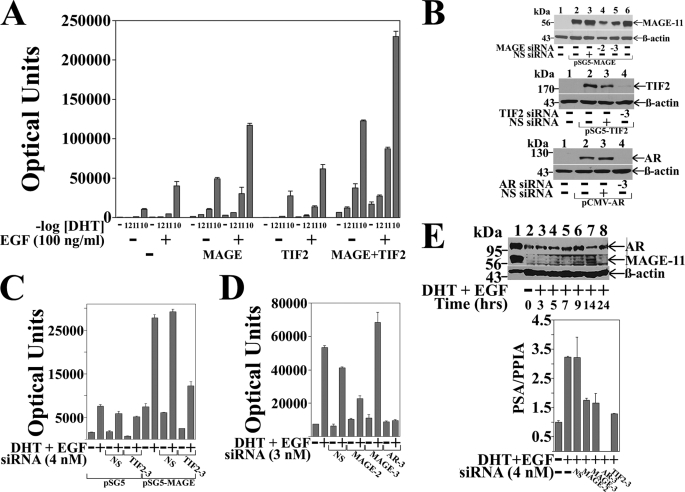
The ability of MAGE-11 to function as an AR coregulator in association with TIF2 was evaluated in CWR-R1 prostate cancer cells where the response to DHT was maximal at 0.1 nm DHT. Expression of MAGE-11 enhanced the dose-dependent increase in AR transactivation in CWR-R1 cells in response to DHT, and DHT and EGF, to a greater extent than TIF2 (Fig. 1A). Coexpression of MAGE-11 and TIF2 increased both ligand-dependent and ligand-independent AR activity to a greater extent than MAGE-11 or TIF2 alone.
FIGURE 1.
Dependence of AR transactivation on MAGE-11 and TIF2. A, CWR-R1 cells were transfected with 0.1 μg of MMTV-Luc with or without 0.1 μg of pSG5-MAGE-11 or pSG5-TIF2. Cells were incubated in the absence and presence of DHT (shown as -log DHT concentration) and 0.1 μg/ml of EGF. B, COS cells were transfected using Lipofectamine 2000 with 1 μg of pSG5 or pSG5-MAGE (top panel), 1 μg of pSG5 or pSG5-TIF2 (middle panel), and 0.5 μg of pCMV5 or pCMV-AR (bottom panel) in the absence and presence of 1 nm nonspecific (NS) siRNA, MAGE-11 siRNA-2 and -3, TIF2 siRNA-3, and AR siRNA-3. Cell extracts were analyzed by immunoblot for MAGE-11 (10 μg of protein/lane), TIF2 (40 μg/lane), AR (30 μg/lane), and β-actin. C, CWR-R1 cells were transfected with 0.1 μg of MMTVΔ-(−421–−364)-Luc, 0.2 μg of pSG5 or pSG5-MAGE in the absence and presence of 4 nm nonspecific or TIF2 siRNA-3, and incubated in the absence and presence of 0.1 nm DHT and 0.01 μg/ml of EGF. D, LAPC-4 cells were transfected with 0.1 μg of MMTVΔ-(−421–−364)-Luc and 3 nm siRNA and incubated in the absence and presence of 0.1 nm DHT and 0.1 μg/ml EGF. E, top panel, CWR-R1 cells incubated for 48 h in serum-free medium were treated for the indicated times with 0.1 nm DHT and 10 ng/ml of EGF. Cell extracts in IB lysis buffer (200 μg of protein/lane) (lanes 2–8) were analyzed by immunoblot using 10 μg/ml of MAGE-Ab-(13–26), -(59–79), and -(94–108) antibodies overnight at 4 °C, and AR32 and β-actin antibodies (33). Extracts of COS cells expressing pCMV-AR (30 μg of protein) and pSG5-MAGE (0.25 μg of protein) served as controls (lane 1). Bottom panel, CWR-R1 cells were transfected with 4 nm siRNA and incubated for 6 h with or without 10 nm DHT and 10 ng/ml of EGF. RNA was extracted and analyzed by quantitative reverse transcription-PCR as described under “Experimental Procedures” for endogenous PSA mRNA relative to peptidylprolyl isomerase A.
TIF2 siRNA-3 inhibited TIF2 expression (Fig. 1B), and the MAGE-11-dependent increase in AR transcriptional activity in CWR-R1 cells (Fig. 1C), supporting the synergistic effects of MAGE-11 and TIF2. In LAPC-4 prostate cancer cells, the DHT and EGF-dependent increase in AR transactivation was inhibited by MAGE-11 siRNA-2, but not siRNA-3 (Fig. 1D), consistent with their relative inhibitory effects on MAGE-11 expression (Fig. 1B).
In CWR-R1 cells, endogenous AR was readily detected on immunoblots (Fig. 1E), whereas endogenous MAGE-11 was evident ∼14 h after inducing quiescent cells to enter the cell cycle by treating with DHT and EGF (Fig. 1E, upper panel). The results suggest that endogenous MAGE-11 levels are low and cell cycle regulated, which may have hindered our attempts to coimmunoprecipitate endogenous MAGE-11 with AR or TIF2. However, we were able to demonstrate that siRNA knockdown of AR, MAGE-11, or TIF2 in CWR-R1 cells decreased the DHT and EGF-stimulated increase in PSA, an endogenous AR-regulated gene (Fig. 1E, lower panel). The results suggest that MAGE-11 functions coordinately with TIF2 to increase AR transactivation.
Identification and Function of a MAGE-11 F-box
The carboxyl-terminal region of MAGE-11 contains sites of EGF-dependent phosphorylation and ubiquitinylation required for MAGE-11 to interact with AR (Fig. 2A) (16, 19). Examination of the MAGE-11 sequence revealed a predicted α-helical F-box-like sequence IPEEΨ3XΨ2X2ΨXΨX7Ψ2X6ΨX8Ψ2 similar to cyclin F, where Ψ is a hydrophobic residue and X is any amino acid. Sequence similarity with the F-box of cyclin F was noted throughout the MAGE gene family (Fig. 2B). The functional significance of MAGE-11 F-box residues 329–369 was supported by the presence of the EGF-dependent Thr-360 Chk1 phosphorylation site within the F-box (underlined in Fig. 2B), because phosphorylation at Thr-360 is required for MAGE-11 to interact with AR (16, 19).
FIGURE 2.
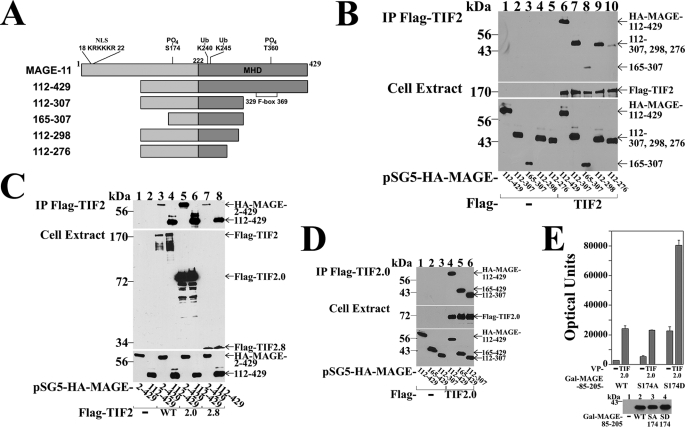
Requirement for a MAGE-11 F-box in AR transactivation. A, schematic diagram of full-length MAGE-11 showing the MAGE homology domain (MHD) with F-box residues 329–369, phosphorylation site Thr-360 within the F-box, Lys-240 and Lys-245 monoubiquitinylation (Ub) sites outside the F-box, and the NH2-terminal nuclear localization signal (NLS). B, sequence homology between MAGE-11 F-box residues 329–369, cyclin F F-box residues 35–73, and MAGE family members, with conserved acidic residues in blue and hydrophobic residues in red. C, alanine mutagenesis of the MAGE F-box at conserved hydrophobic residues. D, Ishikawa cells were transfected with pCMV-AR (10 ng/well), 0.1 μg/well of PSA-Enh-Luc, and 0.1 μg of pSG5 empty vector (—), pSG5-MAGE wild-type (WT), or the indicated F-box mutant. Cells were incubated in the absence and presence of 1 nm DHT with or without 0.1 μg/ml of EGF. Bottom panel, Ishikawa cells were transfected with 2 μg of pSG5 (—), pSG5-MAGE WT, and the indicated F-box mutant. Cells were extracted in IB lysis buffer and 30 μg of protein/lane analyzed on immunoblots probed with antibody raised against FLAG-tagged human MAGE-11 (2 μg/ml). E, FLAG-b empty vector (—) or FLAG-AR (4 μg) were expressed in COS cells with pSG5-HA-MAGE-(112–429) WT or Δ329–369 F-box deletion (0.5 μg of DNA/10-cm dish). Cells were treated in the absence and presence of 10 nm DHT and 0.1 μg/ml EGF, lysed in IP lysis buffer, and incubated with FLAG resin overnight at 4 °C. Immunoprecipitates and cell extracts (25 μg of protein/lane) were analyzed by immunoblot using FLAG and HA antibodies.
The functional properties of the MAGE-11 F-box were investigated by creating alanine point mutations at conserved hydrophobic residues (Fig. 2C) and testing their effect on AR transactivation in Ishikawa cells, a human endometrial cell line that responds to EGF (Fig. 2D). The MAGE-11 F-box mutations reduced or eliminated the MAGE-11-dependent increase in AR transcriptional activity in response to DHT, and DHT and EGF. Coimmunoprecipitation of AR with FLAG-MAGE in the absence and presence of DHT and EGF was not eliminated by the single or double mutations within the MAGE-11 F-box (data not shown), although deletion of the F-box inhibited the coimmunoprecipitation of MAGE-11 with FLAG-AR (Fig. 2E). The results suggest that a MAGE-11 F-box dependent interaction increases AR transcriptional activity.
Synergistic Effects of TIF2 on AR AF2 Activity Depend on the MAGE-11 F-box
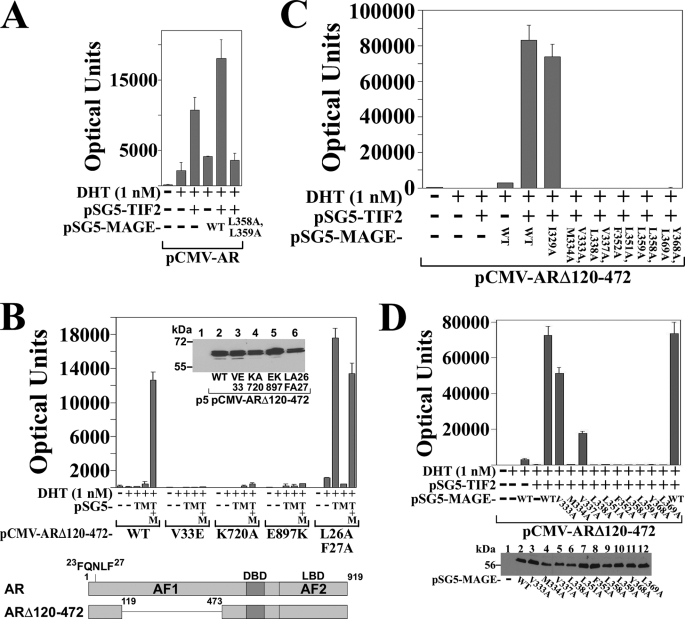
The MAGE-11 and TIF2-dependent increase AR transcriptional activity that derives from AF2 in the ligand binding domain was investigated using full-length AR and ARΔ120–472, a deletion mutant in which the AR NH2-terminal AF1 region was deleted (Fig. 3B, lower panel). ARΔ120–472 retains the AR NH2-terminal FXXLF motif (23FQNLF27) that interacts with MAGE-11 and AR AF2. Androgen-dependent transcriptional activity of ARΔ120–472 depends entirely on AF2.
FIGURE 3.
Requirement for MAGE-11 F-box in AR transactivation by TIF2. A, CV1 cells were transfected with 0.1 μg of pCMV-AR and 5 μg of MMTV-Luc, 2 μg of pSG5-TIF2, 2 μg of pSG5-MAGE wild-type (WT) or L358A/L359A F-box mutant, and incubated in the absence and presence of 1 nm DHT. B, CV1 cells were transfected with 5 μg of MMTV-Luc and 0.1 μg of pCMV-ARΔ120–472 WT or the indicated mutant, 2 μg of pSG5-TIF2 (pSG5-T) and/or 2 μg of pSG5-MAGE (pSG5-M). Cells were incubated in the absence and presence of 1 nm DHT. Inset, pCMV-ARΔ120–472 WT and mutants (5 μg) were expressed in COS cells because expression was too low in CV1 cells. Cell extracts (30 μg of protein/lane) were analyzed on immunoblots probed with AR52 antibody. Bottom panel, schematic diagram of full-length human AR amino acid residues 1–919 with the NH2-terminal FXXLF motif 23FQNLF27, activation function 1 (AF1), DNA binding domain (DBD), activation function 2 (AF2) in the ligand binding domain (LBD), and the ARΔ120–472 deletion mutant that lacks AF1. C, pCMV-ARΔ120–472 (0.1 μg) was expressed in CV1 cells with 5 μg of MMTV-Luc, 2 μg of pSG5-TIF2 and/or 2 μg of pSG5-MAGE WT or the indicated F-box mutant. Cells were incubated in the absence and presence of 1 nm DHT. D, pCMV-ARΔ120–472 (0.1 μg) was expressed in CV1 cells with 5 μg of MMTV-Luc in the absence and presence of 2 μg of pSG5-TIF2 and/or 2 μg of pSG5-MAGE WT or single residue F-box mutant. Cells were incubated in the absence and presence of 1 nm DHT. Bottom panel, CV1 cells were transfected with pSG5 (8 μg/10-cm dish) (—), pSG5-MAGE WT or the indicated single residue F-box mutant. Cell extracts in IB lysis buffer (95 μg of protein/lane) were analyzed on immunoblots probed with antibody raised against FLAG-tagged MAGE-11.
In agreement with results in Fig. 2D, MAGE-11 F-box mutants, as shown for MAGE-L358A,L359A, eliminated the MAGE-11 and TIF2-dependent increase in AR transactivation (Fig. 3A). The ability of MAGE-11 and TIF2 to increase androgen-dependent ARΔ120–472 activity (Fig. 3, B and C) demonstrates the synergistic effects of MAGE-11 and TIF2 on AF2 activity.
The dependence of ARΔ120–472 transactivation on TIF2 binding to AF2 was demonstrated by the loss of activity caused by introducing the AR AF2 K720A or E897K charge clamp mutation that prevents TIF2 binding to AF2 (Fig. 3B), and by a TIF2 LXXLL motif mutant unable to bind AF2 (data not shown) (10). Transcriptional activity of ARΔ120–472 was also eliminated by V33E, an AR mutation that inhibits AR FXXLF motif binding to MAGE-11 but not to AF2 (16) (Fig. 3B). This demonstrates that MAGE-11 binding to ARΔ120–472 is required to relieve repression of AF2 activity caused by the AR N/C interaction.
The inhibitory effect of the AR N/C interaction on AF2 transcriptional activity was also indicated by the TIF2- dependent increase in ARΔ120–472-FXXAA activity (Fig. 3B, L26A,F27A). In this case, mutations in the AR FXXLF motif inhibit binding to both AF2 and MAGE-11 (6, 16), allowing TIF2 activation of AF2. ARΔ120–472 activity in the presence of TIF2 was also inhibited by the double and most of the single mutations in the MAGE-11 F-box (Fig. 3, C and D). The results indicate that the MAGE-11 F-box is required for the synergistic effects of MAGE-11 and TIF2 on AR AF2 transcriptional activity.
MAGE-11 F-box Binds the AR FXXLF Motif and Skp1
Mammalian two-hybrid assays provide evidence that the MAGE-11 F-box interacts with the AR FXXLF motif (Fig. 4). A series of single F-box mutations in GAL-MAGE eliminated the interaction with VP-AR-(1–660), an AR NH2-terminal fragment that contains the AR FXXLF motif and DNA binding domain (Fig. 4A). The AR FXXLF motif-dependent interaction with MAGE-11 was shown by loss of the interaction with VP-AR-(1–660)-FXXAA (LFAA) as reported previously (16). Interaction of the AR FXXLF motif with MAGE-11 was also eliminated by the MAGE-T360A mutation that disrupts a Chk1 phosphorylation site within the MAGE-11 F-box, and by mutations at monoubiquitinylation sites outside the F-box region (19).
FIGURE 4.
Interaction between MAGE-11 F-box and AR FXXLF motif. A, GAL-MAGE wild-type (WT), F-box mutant, or GAL-MAGE-K236A/K240A/K245A (KA) (0.05 μg) were transfected in HeLa cells with 0.1 μg of 5×GAL4Luc3 and 0.1 μg of VP16 empty vector (—), VP-AR-(1–660) or VP-AR-(1–660)-L26A,F27A (LFAA) with a mutation in the AR FXXLF motif. Bottom panel, GAL-MAGE F-box or KA mutants, WT and GAL0 (—) (2 μg/10-cm dish) were expressed in HeLa cells. Cell extracts prepared in IB lysis buffer (150 μg of protein/lane) were analyzed on immunoblots probed using GAL antibody (1:100 dilution). B, HeLa cells were transfected with 0.1 μg of 5×GAL4Luc3, 0.05 μg of GAL-AR-(4–52), and 0.1 μg of VP16 (—), VP-MAGE WT, or the indicated mutant. Bottom panel, HeLa cells were transfected with VP-MAGE mutants (lanes 1–8), VP-MAGE WT (lane 9), and VP16 empty vector (—) (lane 10) (2 μg/10-cm dish). Cell extracts in IB lysis buffer (100 μg of protein/lane) were analyzed by immunoblot using VP16 antibody. C, HeLa cells were transfected with 0.05 μg of GAL-AR-(4–52) or GAL-AR-(16–36), 0.1 μg of 5×GAL4Luc3 and 0.1 μg of VP16 empty vector (—) or VP-MAGE WT or the indicated mutant. D, coimmunoprecipitation of endogenous Skp1 with FLAG-MAGE. FLAG empty vector (—) (3 μg), FLAG-MAGE (3 μg), and FLAG-MAGE-(112–429) (6 μg) were expressed in HEK293 cells. The next day cells were treated for 24 h with 100 ng/ml of EGF and 1 μm MG132. IP of an overnight incubation and cell extracts (35 μg of protein/lane) were analyzed using FLAG and Skp1 antibodies.
To further characterize MAGE-11 F-box binding to the AR FXXLF motif, VP-MAGE single residue F-box mutants were coexpressed with GAL-AR-(4–52) or GAL-AR-(16–36), two short AR NH2-terminal FXXLF motif peptides. VP-MAGE F-box mutations inhibited the interaction with the AR NH2-terminal peptides (Fig. 4, B and C). The interaction was also influenced by a mutation at a potential MAP kinase phosphorylation site at MAGE-11 Ser-174, which lies outside the F-box. VP-MAGE binding to GAL-AR-(4–52) or GAL-AR-(16–36) decreased with the MAGE-11 S174A mutation, but was unchanged by the S174D phosphomimetic.
MAGE-11 also interacted with Skp1 based on coimmunoprecipitation of endogenous Skp1 with FLAG-MAGE in HEK293 cells (Fig. 4D). This suggests an additional and more classical function of the MAGE-11 F-box.
The results indicate a MAGE-11 F-box interaction with the AR FXXLF motif that is influenced by phosphorylation at Thr-360 within the F-box, and potential phosphorylation at Ser-174 NH2-terminal to the F-box. MAGE-11 also interacts with Skp1, which suggests the MAGE-11 F-box serves as a binding site for Skp1, and MAGE-11 may associate with a Skp1-cullin-based E3 ubiquitin ligase complex.
Serum Stimulation of MAP Kinase Phosphorylation of MAGE-Ser-174
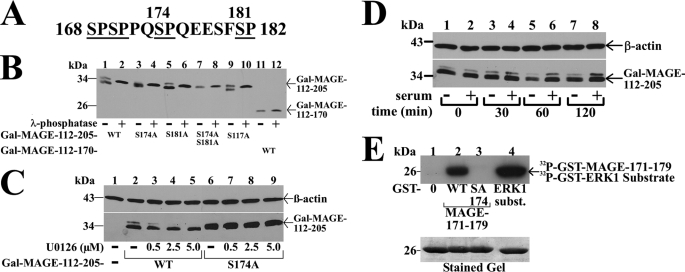
The MAGE-11 sequence flanking Ser-174 contains several potential Ser-Pro phosphorylation sites that include the MAP kinase consensus sequence 172PQSP175 (Fig. 5A). This, together with evidence that the S174A mutation inhibited MAGE-11 interaction with the AR FXXLF motif peptides, led us to investigate whether MAGE-11 is phosphorylated at Ser-174 by MAP kinase.
FIGURE 5.
Serum stimulation of MAP kinase phosphorylation of MAGE-11 Ser-174. A, MAGE-11 Ser-Pro sites Ser-168, Ser-170, Ser-174, and Ser-181 (underlined) include consensus MAP kinase site 172PQSP175. B, GAL-MAGE-(112–205) wild-type (WT) and mutants and GAL-MAGE-(112–170) (5 μg) were expressed in COS cells. Cell extracts in IP lysis buffer without sodium fluoride (35 μg) were incubated in the absence and presence of 800 IU of λ-phosphatase for 1 h at 4 °C, and the immunoblot probed with GAL antibody. C, GAL0 (—), GAL-MAGE-(112–205) WT, or S174A mutant (5 μg) were expressed in COS cells. Immediately after transfection and the next day, serum-free medium containing 1 μm MG132 and U0126 was added. Cells were extracted in IB lysis buffer containing phosphatase mixture inhibitors 1/2. Protein extracts (35 μg of protein/lane) were analyzed on immunoblots probed using GAL and β-actin antibodies. D, GAL-MAGE-(112–205) (5 μg) was expressed in COS cells incubated in the absence (lanes 1, 3, 5, and 7) or presence of 10% fetal bovine serum (lanes 2, 4, 6, and 8). Cells were harvested at 0, 30, 60, and 120 min in IB lysis buffer. Cell extracts (35 μg of protein/lane) were analyzed by immunoblots probed with GAL and β-actin antibodies. E, ERK1 in vitro phosphorylation of MAGE-11 Ser-174. In vitro kinase assay (top panel) was performed as described (19) at 30 °C for 30 min with 0.16 μCi/μl of [γ-32P]ATP, 0.1 μg of active ERK1 kinase and purified GST0 (lane 1), GST-MAGE-(171–179) WT (lane 2), and S174A mutant (lane 3), and ERK1 substrate modeled after myelin basic protein (37) (lane 4). The dried gel was exposed to x-ray film for 16 h, and rehydrated for Coomassie Blue staining to demonstrate equivalent protein loading (bottom panel).
GAL-MAGE-(112–205) is a fusion protein that contains the MAGE-(168–182) Ser-Pro region and migrates as a double band before, and single band after treatment with λ-phosphatase (Fig. 5B, lanes 1 and 2), which suggests GAL-MAGE-(112–205) is phosphorylated. Evidence that Ser-174 is a phosphorylation site was obtained from GAL-MAGE-(112–205)-S174A, which migrated as a single band of similar mobility before and after treatment with λ-phosphatase (Fig. 5B, lanes 3 and 4). GAL-MAGE-(112–205)-S181A or -S117A mutants maintained the double band migration, which was eliminated by treatment with λ-phosphatase (Fig. 5B, lanes 5 and 9). The GAL-MAGE-(112–205)-S174A,S181A double mutant (Fig. 5B, lanes 7 and 8), and GAL-MAGE-(112–170), which lacks Ser-174 (Fig. 5B, lanes 11 and 12), migrated as single bands before and after treatment with λ-phosphatase. The results point to Ser-174 as a phosphorylation site in MAGE-11. Based on the MAP kinase consensus sequence at Ser-174, and decreased binding of AR FXXLF motif peptides to MAGE-S174A but not the MAGE-S174D phosphomimetic, we investigated whether a MAP kinase inhibitor or hormone stimulation would influence phosphorylation of MAGE-Ser-174.
Increasing concentrations of U0126, a MEK1 inhibitor that prevents activation of ERK1/2 (38), eliminated the double band migration of GAL-MAGE-(112–205), but had no effect on GAL-MAGE-(112–205)-S174A (Fig. 5C). Neither treatment with EGF (0.1 μg/ml) nor dibutyryl cyclic AMP (10 mm) altered the double band migration of GAL-MAGE-(112–205) (data not shown). However, addition of 10% serum to quiescent cells caused a time-dependent increase in intensity of the slower migrating band (Fig. 5D). In addition, introducing the MAGE-S174A mutation eliminated in vitro ERK1 phosphorylation of a GST-MAGE-(171–179) fusion peptide (Fig. 5E). The results suggest that serum stimulation of ERK1 phosphorylation at MAGE-Ser-174 modulates the MAGE-11 F-box interaction with the AR FXXLF motif.
Interaction between MAGE-11 and TIF2
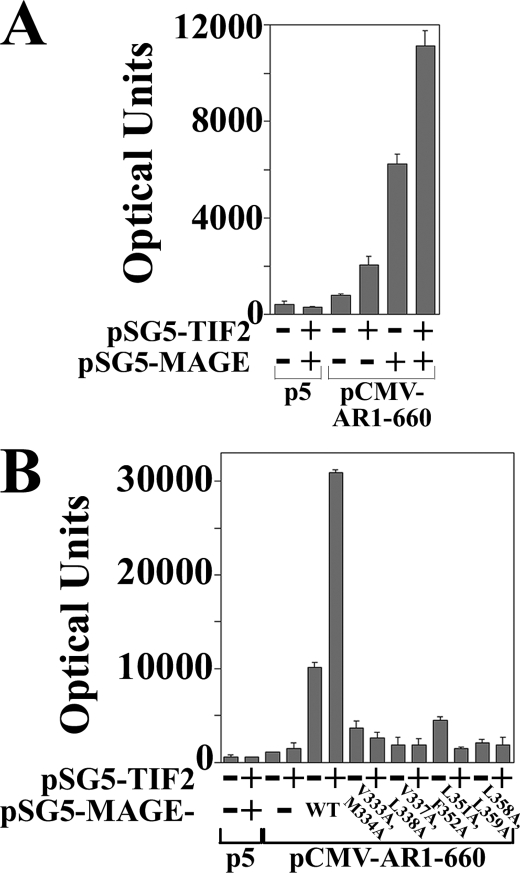
The studies suggested that MAGE-11 and TIF2 act synergistically to increase AR transactivation through AF2 (Figs. 1 and 3), but it remained unclear whether MAGE-11 could increase TIF2-dependent AR transcriptional activity independent of AF2. To address this, we made use of AR-(1–660), a constitutively active AR NH2-terminal fragment that contains AF1, but lacks the AR ligand binding domain and AF2.
The relatively low activity of AR-(1–660) increased ∼2-fold with the coexpression of TIF2 (Fig. 6A). This agrees with previous evidence that AR activation by TIF2 in the absence of MAGE-11 depends largely on TIF2 LXXLL motif binding to AR AF2 (6). AR-(1–660) activity increased to a greater extent with the expression of MAGE-11, and increased further with the coexpression of MAGE-11 and TIF2. The MAGE-11 and TIF2-dependent increase in AR-(1–660) activity was inhibited by MAGE-11 F-box mutations that interfere with MAGE-11 binding to the AR-(1–660) FXXLF motif (Fig. 6B).
FIGURE 6.
MAGE-11 and TIF2-dependent increase in AR AF1 activity. A, CV1 cells were transfected with 5 μg of PSA-Enh-Luc and 0.05 μg of pCMV5 empty vector (p5) or pCMV-AR-(1–660) in the absence and presence of 0.1 μg of pSG5-MAGE and/or 2 μg of pSG5-TIF2. B, CV1 cells were transfected with 5 μg of PSA-Enh-Luc and 0.05 μg of p5 or 0.05 μg of pCMV-AR-(1–660) in the absence and presence of 2 μg of pSG5-TIF2 and/or 1 μg of pSG5-MAGE WT or the indicated F-box mutant.
The results raised the possibility of a direct interaction between MAGE-11 and TIF2 that could increase AR transcriptional activity in the absence and presence of androgen. An interaction between endogenous MAGE-11 and TIF2 in CWR-R1 or LAPC-4 cells could not be observed, possibly due to the low level cell cycle-dependent expression of MAGE-11 (Fig. 1E). Furthermore, the association between MAGE-11 and TIF2 may be transient like AR and MAGE-11 in the presence of DHT (16). However, an interaction between TIF2 and MAGE-11 was demonstrated by coimmunoprecipitation of HA-MAGE with FLAG-TIF2 (Fig. 7A), and by the coimmunoprecipitation of TIF2 with FLAG-MAGE (Fig. 7B). A direct interaction between MAGE-11 and TIF2 was supported by in vitro affinity matrix binding studies. MAGE-11 interacted to the greatest extent with the TIF2 NH2-terminal region present in GST-TIF2.0-(1–627), and to a lesser extent with TIF2 carboxyl-terminal fragments, some of which contain AD1-(1011–1179) (Fig. 7C). The results suggest that MAGE-11 interacts directly with TIF2 to increase AR transcriptional activity.
FIGURE 7.
Interaction between MAGE-11 and TIF2. A, FLAG-b empty vector (—) (8 μg) and 5 μg of pSG5-HA-MAGE (lane 1), or 8 μg of FLAG-TIF2 and 5 μg of pSG5-HA-MAGE (lane 2) were expressed in COS cells. Cells were incubated overnight in the presence of 0.1 μg/ml EGF and 1 μm MG132, harvested in IP lysis buffer, and incubated with FLAG resin overnight at 4 °C. Immunoprecipitates and cell extracts (40 μg of protein/lane) were analyzed on immunoblots probed with HA and TIF2 antibodies. B, pSG5-TIF2 (8 μg) was expressed with 4 μg of FLAG-b empty vector (—) or 4 μg of FLAG-MAGE in COS cells. Cells were incubated overnight in the presence of 0.1 μg/ml EGF and 1 μm MG132, harvested in IP lysis buffer, and incubated with FLAG resin overnight at 4 °C. Immunoprecipitates and cell extracts (50 μg of protein/lane) were analyzed on immunoblots probed using FLAG and TIF2 antibodies. C, in vitro GST affinity matrix assays using the indicated GST-TIF2 fusion fragments expressed in E. coli and 35S-labeled MAGE-11 expressed from pSG5-MAGE-11 using the TnT T7 Quick Coupled transcription-translation system. Input lane contains 2% of the total 35S-labeled MAGE-11 used in the reactions.
MAGE-11 and TIF2 Interacting Domains
Mapping the interaction regions between MAGE-11 and TIF2 was performed initially using MAGE-11 deletion mutants (Fig. 8A). The requirement for MAGE-11 residues 112–298 was shown by coimmunoprecipitation of MAGE-(112–429), -(112–307), and -(112–298) with FLAG-TIF2, compared with weaker interactions with MAGE-(112–276) and -(165–307) (Fig. 8B). Further resolution of the MAGE-11 interaction domains for TIF2 was limited by poor expression of the smaller MAGE-11 fragments.
FIGURE 8.
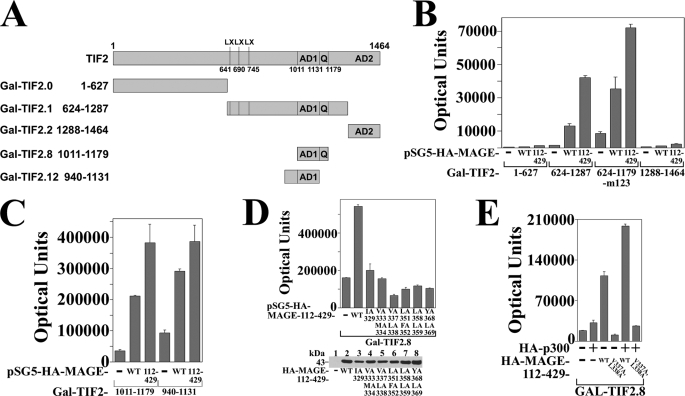
MAGE-11 and TIF2 interaction domains. A, schematic diagram of full-length MAGE-11 and deletion fragments, with nuclear localization signal (NLS, residues 18–22), monoubiquitinylation sites Lys-240 and -245, Ser-174 and Thr-360 phosphorylation sites (19), MAGE homology domain (MHD), and the F-box region. B, FLAG-b empty vector (—) (8 μg) (lanes 1–5) or 8 μg of FLAG-TIF2 (lanes 6–10) were expressed in COS cells with 0.5 μg of pSG5-HA-MAGE-(112–429), 1 μg of pSG5-HA-MAGE-(112–307), or 2 μg of pSG5-HA-MAGE-(165–307), -(112–298), and –(112–276). Cells were incubated with 0.1 μg/ml of EGF and 1 μm MG132, solubilized in IP lysis buffer and immunoprecipitated using FLAG affinity resin overnight at 4 °C. IP (upper panel) and cell extracts (lower panels) (15 μg of protein/lane for HA-MAGE, 55 μg of protein/lane for FLAG-TIF2) were analyzed on immunoblots probed using HA and FLAG antibodies. C, FLAG-b empty vector (lanes 1 and 2) (—), FLAG-TIF2 (lanes 3 and 4), FLAG-TIF2.0-(1–627), or FLAG-TIF2.8-(1011–1179) (8 μg) were expressed in COS cells with 5 μg of pSG5-HA-MAGE or 0.5 μg of pSG5-HA-MAGE-(112–429). Cells were incubated with 0.1 μg/ml of EGF and 1 μm MG132, extracted in IP lysis buffer, and incubated with FLAG affinity resin overnight at 4 °C. Immunoprecipitates (upper panel) and cell extracts (lower panels) (25 μg of protein/lane) were analyzed on immunoblots probed using FLAG and HA antibodies. D, FLAG-b empty vector (—) (8 μg) (lanes 1–3) and FLAG-TIF2.0-(1–627) (lanes 4–6) were expressed in COS cells with 1 μg of pSG5-HA-MAGE-(112–429), 2 μg of pSG5-HA-MAGE-(165–429), or 2 μg of pSG5-HA-MAGE-(112–307). Cells were incubated with 0.1 μg/ml of EGF and 1 μm MG132, extracted in IP lysis buffer, and incubated with FLAG resin for 1 h at 4 °C. Immunoprecipitated proteins (upper panel) and cell extracts (lower panels) (25 μg of protein/lane) were analyzed on immunoblots probed using HA and FLAG antibodies. E, HeLa cells were transfected with 0.05 μg of GAL-MAGE-(85–205) wild-type (WT), S174A or S174D mutant, 0.1 μg of 5×GAL4Luc3, and 0.05 μg of VP16 empty vector (—) or VP-TIF2.0-(1–627). Bottom panel, GAL0 empty vector (—) (2 μg) and 2 μg of GAL-MAGE-(85–205) WT, S174A, or S174D mutant were expressed in HeLa cells. Cell extracts (200 μg of protein/lane) were analyzed on immunoblots probed using a GAL antibody.
In agreement with the in vitro binding results (Fig. 7C), regions in TIF2 (see diagram, Fig. 9A) that interacted with MAGE-11 included TIF2 NH2-terminal 1–627 and AD1 1011–1179 residues. This was evident by the coimmunoprecipitation of HA-MAGE and HA-MAGE-(112–429) with FLAG-TIF2 (Fig. 8C, top panel, lanes 3 and 4), FLAG-TIF2.0-(1–627) (lanes 5 and 6), and FLAG-TIF2.8-(1011–1179), which contains AD1 (lanes 7 and 8). The MAGE-(112–307) region NH2-terminal to F-box residues 329–369 was sufficient to interact with the NH2-terminal region of TIF2 (Fig. 8D).
FIGURE 9.
MAGE-11 increases TIF2 AD1 activity. A, schematic diagram of GAL-TIF2 fusion proteins showing three LXXLL motifs (LX) beginning at residues 641, 690, and 745, activation domain 1 (AD1) residues 1011–1131, glutamine (Q)-rich region 1131–1179, and activation domain 2 (AD2) residues 1288–1464. B, HeLa cells were transfected with 0.1 μg of 5×GAL4Luc3 and 0.05 μg of GAL-TIF2.0-(1–627), GAL-TIF2.1-(624–1287), and GAL-TIF2.3-(624–1179)-m123, which contains L644E,L645A,L693A,L694A,L748A,L749A mutations in the LXXLL motifs, or GAL-TIF2.2-(1288–1464), and 0.1 μg of pSG5 empty vector (—), pSG5-HA-MAGE or pSG5-HA-MAGE-(112–429). C, HeLa cells were transfected with 0.1 μg/well of 5×GAL4Luc3 and 2 ng of GAL-TIF2.8-(1011–1179) or GAL-TIF2.12-(940–1131), which contain AD1 and 0.1 μg of pSG5 (—), pSG5-HA-MAGE, or pSG5-HA-MAGE-(112–429). D, GAL-TIF2.8-(1011–1179) (2 ng), which contains AD1 was expressed in HeLa cells with 0.1 μg/well of 5×GAL4Luc3, 0.1 μg of pSG5 (—), or 0.1 μg of pSG5-HA-MAGE-(112–429) wild-type (WT) or the indicated F-box mutant. Bottom panel, HeLa cells were transfected with 2 μg of pSG5, pSG5-HA-MAGE-(112–429) WT, and the indicated F-box mutant. Cell extracts (150 μg of protein/lane) were analyzed on immunoblots probed with HA antibody. E, HeLa cells were transfected with 0.1 μg/well of 5×GAL4Luc3, 2 ng of GAL-TIF2.8-(1011–1179), and 0.05 μg of pSG5 (—), pSG5-HA-p300, or pSG5-HA-MAGE-(112–429) WT or V337A/L338A F-box mutant.
Interaction between MAGE-11 and the NH2-terminal region of TIF2 was also demonstrated in mammalian two-hybrid assays using VP-TIF2.0-(1–627) and GAL-MAGE-(85–205) (Fig. 8E). The GAL-MAGE-(85–205)-S174D phosphomimetic of MAP kinase site Ser-174 had greater inherent transcriptional activity than wild-type GAL-MAGE-(85–205) or the S174A mutant. However, the fold-increase in the presence of VP-TIF2.0-(1–627) and GAL-MAGE-(85–205) or the S174A and S174D mutants, was similar relative to the VP16 empty vector controls. This suggests that phosphorylation at Ser-174 does not influence the interaction between MAGE-11 and TIF2.
Interaction between MAGE-11 and the AD1 region of TIF2 was also demonstrated by the ability of MAGE-11 to increase the transcriptional activity of GAL-TIF2 fusion proteins (Fig. 9A). Both full-length MAGE-11 and MAGE-(112–429) increased the transcriptional activity of GAL-TIF2.1-(624–1287) and GAL-TIF2.3-(624–1179)-m123, where the latter contains mutations at each of the three TIF2 LXXLL motifs (Fig. 9B). These results support the in vitro binding and coimmunoprecipitation evidence that MAGE-11 interacts directly with TIF2 independent of the TIF2 LXXLL motifs.
Evidence that transcriptional activity arising from the MAGE-11 interaction with TIF2 requires the TIF2 AD1 region was supported by the absence of a MAGE-dependent increase in GAL-TIF2.0-(1–627) activity (Fig. 9B), even though the TIF2 NH2-terminal region, which lacks AD1, interacts with MAGE-11 (see Figs. 7C and 8E). Full-length MAGE-11 and the MAGE-(112–429) fragment also increased the activity of GAL-TIF2.8-(1011–1179) and GAL-TIF2.12-(940–1131), TIF2 fragments with only AD1 in common (Fig. 9, A and C).
Mutations in the MAGE-11 F-box inhibited the ability of MAGE-11 to increase the AD1 activity of GAL-TIF2.8 (Fig. 9D). This appears to reflect a decreased transcriptional response of GAL-TIF2.8 AD1 to p300, which is known to interact with TIF2. Alone, p300 resulted in a small increase in GAL-TIF2.8 activity. In the presence of MAGE-11, p300 increased TIF2 AD1 activity in a MAGE-11 F-box-dependent manner (Fig. 9E). The results indicate that the MAGE-(165–298) region interacts with the NH2-terminal and AD1 regions of TIF2, and that the MAGE-11 F-box influences TIF2 AD1 activation by p300.
MAGE-11 FXXIF Motif
Within the MAGE-(165–298) region that interacts with TIF2 is a predicted α-helical FXXIF motif with the sequence 260FPEIF264 flanked by charged residues similar to the AR FXXLF motif sequence 23FQNLF27 (Fig. 10A). The AD1 region of TIF2 also contains a predicted α-helical sequence with spacing of hydrophobic residues similar to the F-box in cyclin F, Skp2, and MAGE-11 (Fig. 10B). We therefore investigated whether the MAGE-11 FXXIF motif contributes to the interaction with TIF2 AD1.
FIGURE 10.
FXXLF and F-box related sequences in AR, MAGE-11, TIF2, Skp1, and Skp2. A, FXXLF-like motifs include AR-(15–36) FXXLF motif 23FQNLF27 that mediates the androgen-dependent AR N/C interaction with AF2 (6) and interaction with MAGE-11 (7). MAGE-11-(252–273) FXXIF motif 260FPEIF264 interacts with the AD1 region of TIF2. TIF2-(325–346) FXXIY motif 333FSQIY337 function is unknown. Skp1-(93–114) FXXIL motif 101FELIL105 is in a region that interacts with the Skp2 F-box (43). Acidic and basic residues (blue) flank the FXXLF-like motifs (red). B, F-box related sequences have predicted α-helical structure and relatively conserved spacing of two acidic residues (blue) and multiple hydrophobic residues (red). F-box-like sequences similar to the cyclin F F-box-(35–73) include MAGE-11 F-box-(329–369) that interacts with the AR FXXLF motif, Skp2 F-box-(99–139) that interacts with a region that contains the Skp1 FXXIL motif (43), and TIF2 AD1 F-box-(1073–1112) that interacts with the MAGE-11 FXXIF motif.
Interaction between MAGE-11 and the AD1 region of TIF2 was indicated by the coimmunoprecipitation of HA-MAGE-(112–429) with FLAG-TIF2.8-(1011–1179) (Fig. 11A, top panel, lane 2). Coimmunoprecipitation of TIF2-AD1 and MAGE-11 was not eliminated by single F260A and F264A mutations in the 260FPEIF264 region of HA-MAGE-(112–429) (Fig. 11A, lanes 3 and 4) or HA-MAGE-(112–276) (data not shown). However, the MAGE-11-dependent increase in GAL-TIF2.1-(624–1287), and the increase in GAL-TIF2-(624–1179)-m123 activity, was inhibited by the F260A or F264A mutations in MAGE-11 (Fig. 11B). These same mutations also inhibited the TIF2-dependent increase in AR (Fig. 11C) and ARΔ120–472 transcriptional activity (Fig. 11D). In mammalian two-hybrid assays, interaction between GAL-MAGE-(251–272), a short fusion peptide that contains the MAGE-11 FXXIF motif, and VP-TIF2.1-(624–1287), but not VP-TIF2.2-(1288–1464) (Fig. 11E), provided evidence that the MAGE-11 FXXIF motif mediates an interaction with TIF2.
FIGURE 11.
MAGE-11 FXXIF motif-dependent interaction with TIF2. A, top panels, FLAG-b empty vector (lane 1) (—) or FLAG-TIF2.8-(1011–1179) (lanes 2–4) (8 μg) were expressed in COS cells with 0.5 μg of pSG5-HA-MAGE-(112–429) wild-type (WT) (lanes 1 and 2) or the indicated mutant (lanes 3 and 4). Cells were incubated with 0.1 μg/ml of EGF and 1 μm MG132 and immunoprecipitated overnight at 4 °C using FLAG resin. IP (upper panel) and cell extracts (25 μg of protein for HA-MAGE, 50 μg of protein for FLAG-TIF2.8, lower panels) were analyzed on immunoblots using HA and FLAG antibodies. Bottom panels, HeLa cells were transfected with 2 μg of pSG5 (—), pSG5-HA-MAGE-(2–429) WT, F260A, or F264A mutant (left panel), and pSG5-HA-MAGE-(112–429) WT, F260A, or F264A mutant, and pSG5 (—) (right panel). Cells extracts in IB lysis buffer (150 μg of protein/lane) were analyzed on immunoblots using HA antibody. B, GAL-TIF2.1-(624–1287) or GAL-TIF2.3m123 (TIF2-(624–1179)-L644E,L645A,L693A,L694A,L748A,L749A) (0.05 μg) were transfected in HeLa cells with 0.1 μg of pSG5 (—), pSG5-HA-MAGE-(112–429) WT or mutant, and 0.1 μg of 5×GAL4Luc3. C, CV1 cells were transfected with 0.1 μg of pCMV-AR and 5 μg of PSA-Enh-Luc with and without 2 μg of pSG5-TIF2 and 1 μg of WT or mutant pSG5-HA-MAGE. Cells were incubated in the absence and presence of 1 nm DHT. Bottom panel, CV1 cells were transfected with 8 μg of pSG5 (–), pSG5-HA-MAGE-(112–429) WT, F260A, or F264A mutant. Cells extracts in IB lysis buffer (200 μg of protein/lane) were analyzed on immunoblots probed with HA antibody. D, CV1 cells were transfected with 0.1 μg of pCMV-ARΔ120–472 with and without 2 μg of pSG5-TIF2, 2 μg of WT or mutant pSG5-HA-MAGE, and 5 μg of MMTV-Luc. Cells were incubated in the absence and presence of 1 nm DHT. Bottom panel, CV1 cells were transfected with 8 μg of pSG5 (—), pSG5-HA-MAGE WT, F151A, F260A, or F264A mutant. Cell extracts in IB lysis buffer (150 μg/lane) were analyzed on immunoblots using HA antibody. E, HeLa cells were transfected with 0.1 μg of 5×GAL4Luc3, 50 ng of GAL0 empty vector (—), WT or mutant GAL-MAGE-(251–272), and 0.1 μg of VP16 empty vector (—), VP-TIF2.0-(1–627), VP-TIF2.1-(624–1287), or VP-TIF2.2-(1288–1464). Bottom panel, HeLa cells were transfected with 2 μg of GAL0 (lane 1), GAL-MAGE-(251–272) WT (lane 2), or F260A,F264A mutant (lane 3). Cell extracts (100 μg of protein/lane) were analyzed on immunoblots using GAL antibody.
DISCUSSION
Interaction between MAGE-11 and TIF2
MAGE-11 increases AR transcriptional activity in part by increasing the recruitment of SRC/p160 coactivators (10, 21, 39). In the absence of MAGE-11, the AR N/C interaction competitively inhibits SRC/p160 coactivator binding to AF2, which shifts the dominant activation function from AF2 to AF1 (5) (Fig. 12A). MAGE-11 relieves the AR N/C interaction-induced repression of AR AF2 activity by binding the AR FXXLF motif and increasing AF2 accessibility for SRC/p160 coactivator binding. Here we show that MAGE-11 also interacts directly with TIF2 to increase both ligand-dependent and ligand-independent AR activity.
FIGURE 12.
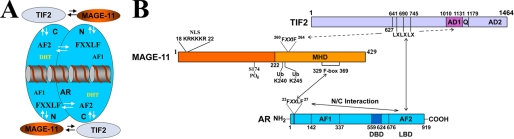
Interactions between AR, MAGE-11, and TIF2. A, schematic diagram of dynamic interactions between AR (blue), MAGE-11 (orange), and TIF2 (light blue) in the context of the AR antiparallel dimer bound to DNA. Indicated is the AR NH2-terminal (N) activation function 1 (AF1), AR carboxyl-terminal (C) activation function 2 (AF2), and the AR FXXLF motif interaction site for MAGE-11 and AF2 in the N/C interaction. B, detailed schematic diagram of interactions between MAGE-11 (orange) F-box residues 329–369 in the carboxyl-terminal MAGE homology domain (MHD) with the AR (blue) NH2-terminal FXXLF motif 23FQNLF27, which is modulated by phosphorylation in the MAGE-11 F-box at Thr-360, monoubiquitinylation (Ub) at Lys-240 and -245 (19), and serum stimulation of MAP kinase phosphorylation of MAGE-11 Ser-174 outside the F-box. AR transcriptional activity is increased by MAGE-11 F-box binding of the AR FXXLF motif, which competitively inhibits AR FXXLF motif binding to AF2 in the AR N/C interaction. This exposes AF2 in the AR ligand binding domain (LBD) for TIF2 (light blue) LXXLL (LX) motif binding. MAGE-11 also increases AR transcriptional activity through direct interactions with the TIF2 NH2-terminal region and with AD1 mediated in part by MAGE-11 FXXIF motif 260FPEIF264.
The results suggest that MAGE-11 functions as a bridging factor to stabilize and recruit SRC/p160 coactivators in a manner less dependent on AR binding of androgen. MAGE-11 interacts with the NH2-terminal region of TIF2, and with TIF2 AD1 through a MAGE-11 FXXIF motif (Fig. 12B). TIF2, SRC1, and TRAM1 each increases the activity of the AR-(1–660) NH2-terminal and the DNA binding domain fragment to a greater extent in the presence of MAGE-11. SRC/p160 coactivator interaction with the AR NH2-terminal region (10, 21) may be facilitated by their interaction with MAGE-11.
MAGE-11 Is an F-box Protein
MAGE-11 is a member of a multigene MAGE family whose functions are largely uncharacterized. MAGE- 11 is expressed in the human male and female reproductive tracts, and undergoes phosphorylation and monoubiquitinylation to interact with the AR FXXLF motif (16, 19, 33). Well characterized steroid receptor coactivator interaction motifs include the LXXLL motifs of SRC/p160 coactivators (40), FXXLF motifs of putative AR coactivators (7), bromodomains, and others (41). MAGE-11 residues 329–369 function as an F-box interaction site for the AR FXXLF motif (Fig. 12), and a MAGE-11 FXXIF motif interacts with an F-box-like sequence in the AD1 region of TIF2.
The ∼40 amino acid F-box, named for a weakly conserved hydrophobic repeat first identified in cyclin F (42), is present in the S-phase kinase-associated protein Skp2, which interacts with Skp1 in the Skp1-cullin-Skp2-F-box (SCF) E3 ubiquitin ligase complex (42–44). About 70 F-box proteins identified in humans are involved in phosphorylation-dependent ubiquitination (42, 45). F-box proteins generally lack intrinsic activity (43, 46) and are classified by their carboxyl-terminal substrate recognition motifs that recruit target proteins involved in gene regulation (44, 47–49). FBWs (FBXW) have WD40 repeats, FBLs (FBXL) have leucine-rich repeats, and FBXs (FBXO) lack these motifs or have other protein interaction motifs (45). F-box proteins include Skp2 of the SCF complex (48, 50, 51), and FBW7 (FBXW7 in humans, hCdc4 in Saccharomyces cerevisiae), a tumor suppressor that mediates the ubiquitinylation and degradation of cell cycle regulatory proteins such as c-Myc (52–54).
Based on our studies, MAGE-11 and its family members are F-box proteins that may function as components of the SCF complex. MAGE-11 would be in the FBX class of F-box proteins because it lacks a WD40 LxGH…D/N(X)5(W/F/Y)(D/N) repeat sequence (55). MAGE-11 shares size and sequence similarity with Skp2, but lacks the repeating leucine-rich β-strand and α-helix arrangement of Skp2 (56). The carboxyl-terminal position of the MAGE-11 40-amino acid F-box (amino acids 329–369) in the conserved MAGE homology domain differs from the NH2-terminal position of the F-box in components of the SCF complex. On the other hand, poxvirus ankyrin repeat proteins interact with Skp1 and have a 30-residue F-box-like sequence in the carboxyl-terminal region (57). The conserved nature of the MAGE-11 F-box across the MAGE family, the MAGE-11 F-box interaction with the AR FXXLF motif, the F-box requirement for the MAGE-11-dependent increase in GAL-TIF2 transcriptional activity, and evidence that MAGE-11 interacts with Skp1, suggest that the MAGE-11 F-box has multiple binding partners involved in domain sharing.
F-box proteins have functions other than as bait for the SCF complex (47). For example, the F-box protein MoKA is an KLF7 coregulator independent of the SCF complex and ubiquitination (58). MAGE-11 binding to the AR FXXLF motif is associated with increased AR transcriptional activity and increased degradation of AR and MAGE-11 (15, 16, 19, 59). Agonist-induced down-regulation of other steroid receptors (49, 60–63) occurs for AR through its association with MAGE-11. Like Skp2, MAGE-11 is expressed at low levels in a cell cycle-dependent manner, although MAGE-11 levels increase in human endometrium during the window of receptivity to implantation, and in castration-recurrent prostate cancer (20, 33).
Whether the phosphorylation and monoubiquitinylation-dependent MAGE-11 F-box interaction with the AR FXXLF motif involves the action of an SCF ubiquitin E3 ligase complex remains to be established. Substrates such as c-Myc and cyclin E, which bind the FBXW7 F-box protein that binds Skp1 in the SCF complex, are targeted for ubiquitinylation by phosphorylation within a phosphodegron (64). MAGE-11 is phosphorylated within the F-box by checkpoint kinase Chk1, which triggers monoubiquitinylation at lysines 240 and 245 (19), and by MAP kinase in response to serum. Monoubiquitinylation, which has been shown to mediate interactions between regulatory components of the proteosome required for transcription (65), may be required for an SCF function of MAGE-11.
MAGE-11 F-box binding to the AR FXXLF motif, and MAGE-FXXIF motif binding to an F-box-like sequence in AD1 of TIF2, suggests a novel F-box-FXX(L/I)F protein interaction paradigm. Selectivity for the MAGE-11 F-box-AR FXXLF motif interaction is supported by the lack of MAGE-11 binding to FXXLF motifs present in other AR coactivators, even though these same FXXLF motifs interact with AR AF2 (7, 16). Single amino acid mutations disrupt the MAGE-11 F-box-AR FXXLF motif and MAGE-11 FXXIF-TIF2 AD1 interactions, although these same mutations did not eliminate coimmunoprecipitation of the complex. This suggests that the protein structure outside the region to some extent compensates for single residue F-box mutations.
The FXX(L/I)F motifs in AR and MAGE-11, and the uncharacterized FXXIY motif in TIF2, are each flanked by charged residues that may facilitate protein-protein interactions by increasing the solubility of the hydrophobic region. AR FXXLF motif binding to AF2 is influenced by complementary charged clusters that surround the AF2 region of AR (24). The TIF2 FXXIY motif 333FSQIY337 and Skp1 FXXIL motif 101FELIL105 within the interaction region for Skp2 (Fig. 10A) are also flanked by charged residues (43). Although functions for the TIF2 and Skp1 FXXLF-like motifs have not yet been demonstrated, the presence of flanking charged residues support a role in mediating protein-protein interactions. The ubiquitinylation-dependent interaction between the AR FXXLF motif and the MAGE-11 F-box may be relevant to other MAGE gene family members with sequence homology within the MAGE-11 F-box.
Post-translational Regulation by Phosphorylation
Phosphorylation of steroid receptors and their coactivators has been linked to ubiquitinylation and subsequent degradation (41, 63, 66–68). AR signaling is regulated by its own phosphorylation (29, 69, 70), which includes EGF-dependent MAP kinase phosphorylation at AR Ser-515 (29), and MAP kinase phosphorylation of SRC/p160 coregulators (41, 67, 68, 71). MAP kinase signaling was also implicated in AR activity based on the inhibitory effects of U0126, a MAP kinase MEK1 inhibitor (29, 72, 73).
MAGE-11 Thr-360 phosphorylation by checkpoint kinase Chk1 within the MAGE-11 F-box is required for its interaction with the AR FXXLF motif. AR FXXLF motif binding to MAGE-11 is modulated by serum stimulation of MAP kinase phosphorylation at MAGE-Ser-174, although mutation of Ser-174 did not eliminate the ability of MAGE-11 to increase AR transcriptional activity (19). MAGE-11 Ser-174 appears to be a post-translational regulatory site phosphorylated by ERK1, based on the inhibitory effect of the S174A mutation in the context of shorter AR NH2-terminal fragments (19), and the greater transcriptional activity of GAL-MAGE-11 fusion proteins containing the S174D phosphomimetic. Conformational changes in nuclear receptor coregulators are regulated by peptidyl-prolyl isomerase 1 (Pin1), which targets phosphorylated Ser/Thr-Pro residues (74, 75). Pin1, which is highly expressed in metastatic prostate cancer, may exert regulatory effects on MAGE-11 mediated by MAP kinase phosphorylation at Ser-174.
AR Reactivation in Prostate Cancer
AR becomes reactivated during androgen deprivation therapy and prostate cancer progression to castration-recurrent growth (76, 77). This involves mechanisms that include increases in mitogen signaling, SRC/p160 coactivator levels (32), sensitivity to low levels of androgen (19, 32, 73, 78), and local tissue androgen synthesis (79). Of the coregulators that interact with AR (80), both the SRC/p160 coactivators and MAGE-11 levels increase in castration-recurrent prostate cancer (20, 81, 82). MAGE-11 mRNA levels can increase by ∼1000-fold in clinical specimens of castration-recurrent prostate cancer compared with benign prostate, and by ∼50-fold in the castration-recurrent CWR22 human prostate cancer xenograft (20). Increased expression of MAGE-11 in prostate cancer during androgen deprivation therapy is associated with DNA hypomethylation at CpG sites in the MAGE-11 promoter and may also result from increased cyclic AMP signaling. The ability of MAGE-11 to increase AR transcriptional activity by interacting with TIF2 suggests that increased levels of both coactivators contribute to AR reactivation in castration-recurrent prostate cancer.
Acknowledgments
We gratefully acknowledge the technical assistance of Aimee C. Morris and K. Michelle Cobb, and thank Frank S. French for reviewing the manuscript.
This work was supported, in whole or in part, by National Institutes of Health United States Public Health Service Grants HD16910 from the NICHD and P01-CA77739 from the NCI, a United States Department of Defense PC073586 Fellowship Award, National Institutes of Health Eunice Kennedy Shriver cooperative Agreement U54-HD35041 as part of the Specialized Cooperative Centers Program in Reproduction, and Infertility Research from the NICHD.
- AR
- androgen receptor
- MAGE-11
- melanoma antigen gene protein-A11
- TIF2
- transcriptional intermediary factor 2
- MAP kinase
- mitogen-activated protein kinase
- AD1
- activation domain 1
- DHT
- dihydrotestosterone
- AF2
- activation function 2
- SRC
- steroid receptor coactivator
- AF1
- activation function 1
- N/C
- NH2-terminal and COOH-terminal
- EGF
- epidermal growth factor
- PSA-Enh-Luc
- prostate-specific antigen enhancer luciferase
- MMTV-Luc
- mammary tumor virus luciferase
- siRNA
- small inhibitory RNA
- IP
- immunoprecipitation
- IB
- immunoblot
- ERK1
- extracellular signal-regulated kinase 1
- GST
- glutathione S-transferase
- SCF
- Skp1-cullin-F-box
- MOPS
- 4-morpholinepropanesulfonic acid
- HEK
- human embryonic kidney
- HA
- hemagglutinin.
REFERENCES
- 1.Quigley C. A., De Bellis A., Marschke K. B., el-Awady M. K., Wilson E. M., French F. S. (1995) Endocr. Rev. 16, 271–321 [DOI] [PubMed] [Google Scholar]
- 2.Askew E. B., Gampe R. T., Jr., Stanley T. B., Faggart J. L., Wilson E. M. (2007) J. Biol. Chem. 282, 25801–25816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.He B., Gampe R. T., Jr., Hnat A. T., Faggart J. L., Minges J. T., French F. S., Wilson E. M. (2006) J. Biol. Chem. 281, 6648–6663 [DOI] [PubMed] [Google Scholar]
- 4.Newmark J. R., Hardy D. O., Tonb D. C., Carter B. S., Epstein J. I., Isaacs W. B., Brown T. R., Barrack E. R. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 6319–6323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He B., Gampe R. T., Jr., Kole A. J., Hnat A. T., Stanley T. B., An G., Stewart E. L., Kalman R. I., Minges J. T., Wilson E. M. (2004) Mol. Cell 16, 425–438 [DOI] [PubMed] [Google Scholar]
- 6.He B., Kemppainen J. A., Wilson E. M. (2000) J. Biol. Chem. 275, 22986–22994 [DOI] [PubMed] [Google Scholar]
- 7.He B., Minges J. T., Lee L. W., Wilson E. M. (2002) J. Biol. Chem. 277, 10226–10235 [DOI] [PubMed] [Google Scholar]
- 8.Lavery D. N., McEwan I. J. (2008) Biochemistry 47, 3352–3359 [DOI] [PubMed] [Google Scholar]
- 9.Lavery D. N., McEwan I. J. (2008) Biochemistry 47, 3360–3369 [DOI] [PubMed] [Google Scholar]
- 10.He B., Kemppainen J. A., Voegel J. J., Gronemeyer H., Wilson E. M. (1999) J. Biol. Chem. 274, 37219–37225 [DOI] [PubMed] [Google Scholar]
- 11.Langley E., Kemppainen J. A., Wilson E. M. (1998) J. Biol. Chem. 273, 92–101 [DOI] [PubMed] [Google Scholar]
- 12.Langley E., Zhou Z. X., Wilson E. M. (1995) J. Biol. Chem. 270, 29983–29990 [DOI] [PubMed] [Google Scholar]
- 13.Wong C. I., Zhou Z. X., Sar M., Wilson E. M. (1993) J. Biol. Chem. 268, 19004–19012 [PubMed] [Google Scholar]
- 14.Zhou Z. X., Lane M. V., Kemppainen J. A., French F. S., Wilson E. M. (1995) Mol. Endocrinol. 9, 208–218 [DOI] [PubMed] [Google Scholar]
- 15.He B., Bowen N. T., Minges J. T., Wilson E. M. (2001) J. Biol. Chem. 276, 42293–42301 [DOI] [PubMed] [Google Scholar]
- 16.Bai S., He B., Wilson E. M. (2005) Mol. Cell. Biol. 25, 1238–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chomez P., De Backer O., Bertrand M., De Plaen E., Boon T., Lucas S. (2001) Cancer Res. 61, 5544–5551 [PubMed] [Google Scholar]
- 18.Rogner U. C., Wilke K., Steck E., Korn B., Poustka A. (1995) Genomics 29, 725–731 [DOI] [PubMed] [Google Scholar]
- 19.Bai S., Wilson E. M. (2008) Mol. Cell. Biol. 28, 1947–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karpf A. R., Bai S., James S. R., Mohler J. L., Wilson E. M. (2009) Mol. Cancer Res. 7, 523–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bevan C. L., Hoare S., Claessens F., Heery D. M., Parker M. G. (1999) Mol. Cell. Biol. 19, 8383–8392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lubahn D. B., Joseph D. R., Sar M., Tan J., Higgs H. N., Larson R. E., French F. S., Wilson E. M. (1988) Mol. Endocrinol. 2, 1265–1275 [DOI] [PubMed] [Google Scholar]
- 23.Simental J. A., Sar M., Lane M. V., French F. S., Wilson E. M. (1991) J. Biol. Chem. 266, 510–518 [PubMed] [Google Scholar]
- 24.He B., Wilson E. M. (2003) Mol. Cell. Biol. 23, 2135–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Voegel J. J., Heine M. J., Tini M., Vivat V., Chambon P., Gronemeyer H. (1998) EMBO J. 17, 507–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang W., Shostak Y., Tarr P., Sawyers C., Carey M. (1999) J. Biol. Chem. 274, 25756–25768 [DOI] [PubMed] [Google Scholar]
- 27.Giffin W., Torrance H., Rodda D. J., Préfontaine G. G., Pope L., Hache R. J. (1996) Nature 380, 265–268 [DOI] [PubMed] [Google Scholar]
- 28.Gottlieb T. M., Jackson S. P. (1993) Cell 72, 131–142 [DOI] [PubMed] [Google Scholar]
- 29.Ponguta L. A., Gregory C. W., French F. S., Wilson E. M. (2008) J. Biol. Chem. 283, 20989–21001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He B., Lee L. W., Minges J. T., Wilson E. M. (2002) J. Biol. Chem. 277, 25631–25639 [DOI] [PubMed] [Google Scholar]
- 31.Zhou Z. X., Sar M., Simental J. A., Lane M. V., Wilson E. M. (1994) J. Biol. Chem. 269, 13115–13123 [PubMed] [Google Scholar]
- 32.Gregory C. W., Johnson R. T., Jr., Mohler J. L., French F. S., Wilson E. M. (2001) Cancer Res. 61, 2892–2898 [PubMed] [Google Scholar]
- 33.Bai S., Grossman G., Yuan L., Lessey B. A., French F. S., Young S. L., Wilson E. M. (2008) Mol. Hum. Reprod. 14, 107–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Craft N., Shostak Y., Carey M., Sawyers C. L. (1999) Nat. Med. 5, 280–285 [DOI] [PubMed] [Google Scholar]
- 35.Quarmby V. E., Yarbrough W. G., Lubahn D. B., French F. S., Wilson E. M. (1990) Mol. Endocrinol. 4, 22–28 [DOI] [PubMed] [Google Scholar]
- 36.Tan J. A., Joseph D. R., Quarmby V. E., Lubahn D. B., Sar M., French F. S., Wilson E. M. (1988) Mol. Endocrinol. 2, 1276–1285 [DOI] [PubMed] [Google Scholar]
- 37.Erickson A. K., Payne D. M., Martino P. A., Rossomando A. J., Shabanowitz J., Weber M. J., Hunt D. F., Sturgill T. W. (1990) J. Biol. Chem. 265, 19728–19735 [PubMed] [Google Scholar]
- 38.Favata M. F., Horiuchi K. Y., Manos E. J., Daulerio A. J., Stradley D. A., Feeser W. S., Van Dyk D. E., Pitts W. J., Earl R. A., Hobbs F., Copeland R. A., Magolda R. L., Scherle P. A., Trzaskos J. M. (1998) J. Biol. Chem. 273, 18623–18632 [DOI] [PubMed] [Google Scholar]
- 39.Onate S. A., Boonyaratanakornkit V., Spencer T. E., Tsai S. Y., Tsai M. J., Edwards D. P., O'Malley B. W. (1998) J. Biol. Chem. 273, 12101–12108 [DOI] [PubMed] [Google Scholar]
- 40.Heery D. M., Kalkhoven E., Hoare S., Parker M. G. (1997) Nature 387, 733–736 [DOI] [PubMed] [Google Scholar]
- 41.Han S. J., Lonard D. M., O'Malley B. W. (2009) Trends Endocrinol. Metab. 20, 8–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bai C., Sen P., Hofmann K., Ma L., Goebl M., Harper J. W., Elledge S. J. (1996) Cell 86, 263–274 [DOI] [PubMed] [Google Scholar]
- 43.Schulman B. A., Carrano A. C., Jeffrey P. D., Bowen Z., Kinnucan E. R., Finnin M. S., Elledge S. J., Harper J. W., Pagano M., Pavletich N. P. (2000) Nature 408, 381–386 [DOI] [PubMed] [Google Scholar]
- 44.Zheng N., Schulman B. A., Song L., Miller J. J., Jeffrey P. D., Wang P., Chu C., Koepp D. M., Elledge S. J., Pagano M., Conaway R. C., Conaway J. W., Harper J. W., Pavletich N. P. (2002) Nature 416, 703–709 [DOI] [PubMed] [Google Scholar]
- 45.Jin J., Cardozo T., Lovering R. C., Elledge S. J., Pagano M., Harper J. W. (2004) Genes Dev. 18, 2573–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Westbrook T. F., Hu G., Ang X. L., Mulligan P., Pavlova N. N., Liang A., Leng Y., Maehr R., Shi Y., Harper J. W., Elledge S. J. (2008) Nature 452, 370–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hermand D. (2006) Cell Div. 1, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jackson P. K., Eldridge A. G. (2002) Mol. Cell 9, 923–925 [DOI] [PubMed] [Google Scholar]
- 49.von der Lehr N., Johansson S., Wu S., Bahram F., Castell A., Cetinkaya C., Hydbring P., Weidung I., Nakayama K., Nakayama K. I., Söderberg O., Kerppola T. K., Larsson L. G. (2003) Mol. Cell 11, 1189–1200 [DOI] [PubMed] [Google Scholar]
- 50.Craig K. L., Tyers M. (1999) Prog. Biophys. Mol. Biol. 72, 299–328 [DOI] [PubMed] [Google Scholar]
- 51.Nie L., Wu H., Sun X. H. (2008) J. Biol. Chem. 283, 684–692 [DOI] [PubMed] [Google Scholar]
- 52.Onoyama I., Nakayama K. I. (2008) Cell Cycle 7, 3307–3313 [DOI] [PubMed] [Google Scholar]
- 53.Welcker M., Clurman B. E. (2008) Nat. Rev. Cancer 8, 83–93 [DOI] [PubMed] [Google Scholar]
- 54.Yada M., Hatakeyama S., Kamura T., Nishiyama M., Tsunematsu R., Imaki H., Ishida N., Okumura F., Nakayama K., Nakayama K. I. (2004) EMBO J. 23, 2116–2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van der Voorn L., Ploegh H. L. (1992) FEBS Lett. 307, 131–134 [DOI] [PubMed] [Google Scholar]
- 56.Hao B., Zheng N., Schulman B. A., Wu G., Miller J. J., Pagano M., Pavletich N. P. (2005) Mol. Cell 20, 9–19 [DOI] [PubMed] [Google Scholar]
- 57.Sonnberg S., Seet B. T., Pawson T., Fleming S. B., Mercer A. A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 10955–10960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smaldone S., Laub F., Else C., Dragomir C., Ramirez F. (2004) Mol. Cell. Biol. 24, 1058–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chandra S., Shao J., Li J. X., Li M., Longo F. M., Diamond M. I. (2008) J. Biol. Chem. 283, 23950–23955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dennis A. P., Haq R. U., Nawaz Z. (2001) Front. Biosci. 6, D954–D959 [DOI] [PubMed] [Google Scholar]
- 61.Muratani M., Tansey W. P. (2003) Nat. Rev. Mol. Cell Biol. 4, 192–201 [DOI] [PubMed] [Google Scholar]
- 62.Verma S., Ismail A., Gao X., Fu G., Li X., O'Malley B. W., Nawaz Z. (2004) Mol. Cell. Biol. 24, 8716–8726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu R. C., Feng Q., Lonard D. M., O'Malley B. W. (2007) Cell 129, 1125–1140 [DOI] [PubMed] [Google Scholar]
- 64.Nash P., Tang X., Orlicky S., Chen Q., Gertler F. B., Mendenhall M. D., Sicheri F., Pawson T., Tyers M. (2001) Nature 414, 514–521 [DOI] [PubMed] [Google Scholar]
- 65.Archer C. T., Burdine L., Liu B., Ferdous A., Johnston S. A., Kodadek T. (2008) J. Biol. Chem. 283, 21789–21798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hoang T., Fenne I. S., Cook C., Børud B., Bakke M., Lien E. A., Mellgren G. (2004) J. Biol. Chem. 279, 49120–49130 [DOI] [PubMed] [Google Scholar]
- 67.Rowan B. G., Garrison N., Weigel N. L., O'Malley B. W. (2000) Mol. Cell. Biol. 20, 8720–8730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rowan B. G., Weigel N. L., O'Malley B. W. (2000) J. Biol. Chem. 275, 4475–4483 [DOI] [PubMed] [Google Scholar]
- 69.Gioeli D., Ficarro S. B., Kwiek J. J., Aaronson D., Hancock M., Catling A. D., White F. M., Christian R. E., Settlage R. E., Shabanowitz J., Hunt D. F., Weber M. J. (2002) J. Biol. Chem. 277, 29304–29314 [DOI] [PubMed] [Google Scholar]
- 70.Lopez G. N., Turck C. W., Schaufele F., Stallcup M. R., Kushner P. J. (2001) J. Biol. Chem. 276, 22177–22182 [DOI] [PubMed] [Google Scholar]
- 71.Zhou Z. X., Kemppainen J. A., Wilson E. M. (1995) Mol. Endocrinol. 9, 605–615 [DOI] [PubMed] [Google Scholar]
- 72.Agoulnik I. U., Bingman W. E., 3rd, Nakka M., Li W., Wang Q., Liu X. S., Brown M., Weigel N. L. (2008) Mol. Endocrinol. 22, 2420–2432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gregory C. W., Fei X., Ponguta L. A., He B., Bill H. M., French F. S., Wilson E. M. (2004) J. Biol. Chem. 279, 7119–7130 [DOI] [PubMed] [Google Scholar]
- 74.Chen S. Y., Wulf G., Zhou X. Z., Rubin M. A., Lu K. P., Balk S. P. (2006) Mol. Cell. Biol. 26, 929–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yi P., Wu R. C., Sandquist J., Wong J., Tsai S. Y., Tsai M. J., Means A. R., O'Malley B. W. (2005) Mol. Cell. Biol. 25, 9687–9699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gregory C. W., Hamil K. G., Kim D., Hall S. H., Pretlow T. G., Mohler J. L., French F. S. (1998) Cancer Res. 58, 5718–5724 [PubMed] [Google Scholar]
- 77.Yuan X., Balk S. P. (2009) Urol. Oncol. 27, 36–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Culig Z., Hobisch A., Cronauer M. V., Radmayr C., Trapman J., Hittmair A., Bartsch G., Klocker H. (1994) Cancer Res. 54, 5474–5478 [PubMed] [Google Scholar]
- 79.Mohler J. L., Gregory C. W., Ford O. H., 3rd, Kim D., Weaver C. M., Petrusz P., Wilson E. M., French F. S. (2004) Clin. Cancer Res. 10, 440–448 [DOI] [PubMed] [Google Scholar]
- 80.Heemers H. V., Tindall D. J. (2007) Endocr. Rev. 28, 778–808 [DOI] [PubMed] [Google Scholar]
- 81.Agoulnik I. U., Vaid A., Nakka M., Alvarado M., Bingman W. E., 3rd, Erdem H., Frolov A., Smith C. L., Ayala G. E., Ittmann M. M., Weigel N. L. (2006) Cancer Res. 66, 10594–10602 [DOI] [PubMed] [Google Scholar]
- 82.Gregory C. W., He B., Johnson R. T., Ford O. H., Mohler J. L., French F. S., Wilson E. M. (2001) Cancer Res. 61, 4315–4319 [PubMed] [Google Scholar]