Abstract
Several nascent peptides stall ribosomes during their own translation in both prokaryotes and eukaryotes. Leader peptides that induce stalling can regulate downstream gene expression. Interestingly, stalling peptides show little sequence similarity and interact with the ribosome through distinct mechanisms. To explore the scope of regulation by stalling peptides and to better understand the mechanism of stalling, we identified and characterized new examples from random libraries. We created a genetic selection that ties the life of Escherichia coli cells to stalling at a specific site. This selection relies on the natural bacterial system that rescues arrested ribosomes. We altered transfer-messenger RNA, a key component of this rescue system, to direct the completion of a necessary protein if and only if stalling occurs. We identified three classes of stalling peptides: C-terminal Pro residues, SecM-like peptides, and the novel stalling sequence FXXYXIWPP. Like the leader peptides SecM and TnaC, the FXXYXIWPP peptide induces stalling efficiently by inhibiting peptidyl transfer. The nascent peptide exit tunnel and peptidyltransferase center are implicated in this stalling event, although mutations in the ribosome affect stalling on SecM and FXXYXIWPP differently. We conclude that ribosome stalling can be caused by numerous sequences and is more common than previously believed.
Introduction
The ribosome efficiently synthesizes an enormous diversity of peptide sequences without regard to their chemical properties. This generality is not universal, however. Several polypeptides interact with the ribosome to stall their own translation, either in the elongation or termination steps (1, 2). Programmed stalling events regulate gene expression in both prokaryotes and eukaryotes (1–3) and may affect the folding of nascent polypeptides (4–6).
In two well characterized examples from Escherichia coli, ribosome stalling on a leader peptide increases the expression of a gene further downstream on the same mRNA. The secretion monitor peptide SecM, for example, regulates secA in response to changes in protein translocation activity (7). If translocation activity is low, ribosome stalling on the SecM peptide alters the secondary structure of the mRNA and up-regulates the translation of secA, a key component of the secretory machinery (8). When activity is high, the signal recognition particle/SecYEGA system binds the signal peptide sequence in SecM and pulls it from the stalled ribosome (8, 9). A second example is the regulation of tnaA, a gene required to break down tryptophan, by the leader peptide TnaC in response to Trp levels. When Trp concentrations are high, ribosome stalling on TnaC blocks a transcriptional terminator upstream of tnaA, increasing its expression (10, 11). Low tryptophan levels do not support ribosome stalling and lead to attenuation of the transcript.
Stalling on the SecM and TnaC peptides is the result of three interactions: the binding of the nascent peptide to the exit tunnel and the peptidyltransferase center, and the binding of an effector in the ribosomal A site. The peptide exit tunnel in the large ribosomal subunit is 100 Å long and 15 Å wide on average (12). Mostly made of RNA, it provides very few hydrophobic surfaces for elongating proteins to bind, accounting for their ability to pass through unhindered. A significantly constricted portion of the tunnel is formed by loops in proteins L4 and L22 (13, 14). SecM and TnaC interact with the tunnel near this constriction, using critical Trp residues 10–12 amino acids upstream of the stalling site. Ribosomal mutations that reduce stalling map to the exit tunnel, implicating A751, A2058, and U2609 in the 23 S rRNA and specific residues in the L22 protein in the stalling mechanism (15, 16). A cryo-electron microscopy study of the SecM-stalled ribosome revealed a network of conformational changes in 23 S rRNA emanating from the exit tunnel (17).
Nascent peptides also interact directly with the peptidyltransferase center (PTC)2 to induce stalling. In the case of SecM, the identity of the final six residues is critical for stalling on the FXXXXWIXXXXGIRAGP166 sequence (15). Likewise, the C-terminal Pro residue in TnaC is essential for stalling on the sequence WXXXDXXXXXXXP* (11). These amino acids must be acting within the PTC to inhibit its catalytic activity, either peptidyl transfer for SecM or peptidyl hydrolysis for TnaC. In some cases, the peptide sequence in the PTC is sufficient to induce stalling without exit tunnel interactions. A C-terminal Pro residue in the YbeL protein inhibits termination. Peptide release is especially inefficient when Pro-Stop is preceded by an Asp, Glu, or Pro residue (18).
In addition to nascent-peptide interactions with the exit tunnel and the PTC, stalling on SecM and TnaC requires a specific effector molecule to bind in the A site. This binding event is thought to create a PTC conformation that is inactive. For example, SecM stalls during elongation with unreacted Pro-tRNA bound in the A site (19, 20). Mutation of this Pro codon to Ala alleviates stalling. Likewise, TnaC stalling requires the binding of free tryptophan near the PTC (21, 22). The action of free tryptophan can be mimicked by Trp-tRNA if the tnaC stop codon is mutated to a Trp codon. Other aminoacyl-tRNAs (Phe, Met, and Pro) do not induce stalling on TnaC (11).
Although stalling peptides interact with the exit tunnel and PTC, they do so differently and share little sequence similarity. This led us to hypothesize that there are additional, unknown peptide sequences that might inhibit peptidyl transfer or hydrolysis. Here, we report the genetic identification and characterization of peptides that stall at high efficiency during elongation.
EXPERIMENTAL PROCEDURES
Library Creation
The initial 18-nt library was created by amplifying the truncated kanR gene by PCR with the forward primer CATATGGCTAGCATGAGCCATATTCAACGGGAAAC and the degenerate reverse primer CGAAAGGGTACCN18ATTACCATATTTTTGAAAAAGCCGTTTCTG. 18 random nt (6 codons) were added to the 3′-end of kanR following residue 253. To create a second library lacking stop codons in codons four through six, the random region (N18) in the degenerate primer was replaced by (NNB)3N9, where B is a mix of C, G, and T phosphoramidites. The PCR products were cloned into pBAD-KT2 (23) with NheI and BamHI, and the resulting plasmids were amplified in DH10B. The libraries were then selected in X90 ssrA::cat as described (23) in media containing 15 μg/ml kanamycin at either 25 or 37 °C. The KanR fusion sequences from surviving colonies were amplified, sequenced, and recloned to verify their activities.
Mass Spectrometry
The pGEX-3X vector was amplified with inverse PCR to create new restriction sites using the primers AGAGTAGCTAGCACGACCTTCGATCAGATCCG and AGAGTAGCATGCTTGACTGACGATCTGCCTCG. The library cassette was amplified by PCR from 12 residues upstream of the 6 random codons to the stop codon downstream, including the nucleotide sequence encoding SLQKRLFQKYGIXXXXXXGYRGSRVDRQAWLFWRMREDFQPDTD*, using the primers AGAGTAGCTAGCTCATTACAGAAACGGCTTTTTC and AGAGTAGCATGCTTTAATCTGTATCAGGCTGAAAATC. The resulting PCR products were digested with NheI and SphI and ligated to create the pGEX-WPPPSI, WPPDV*, and WPPWYR vectors.
These plasmids and pCH201 (expressing tmRNA-H encoding a His6 tag (24)) were introduced into X-90 ssrA::cat. Cultures were grown to mid-log phase and induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside for 2.5 h. The cells were then harvested and lysed in B-PER reagent (Thermo Scientific), and the cell lysate was cleared in an SS-34 rotor at 15,000 rpm for 20 min. His-tagged GST was purified on a nickel-nitrilotriacetic acid-agarose resin from the supernatant. 50 μg of protein was acetone-precipitated and digested with trypsin for 14 h at 37 °C. Tryptic fragments were purified again in a nickel-nitrilotriacetic acid slurry, and the peptides were loaded onto a reversed-phase ZipTip column, spotted onto a MALDI plate, and overlaid with an alpha-CHC matrix (Agilent Technology). Samples were analyzed with a QSTAR Pulsar QqTOF mass spectrometer.
Immunoblot Assays
Ala mutations were introduced by PCR into the pGEX-WPPPSI vector described above. X-90 ssrA::cat cells were transformed with a GST vector together with pCH201 (expressing tmRNA-H). Overnight cultures were diluted to A600 of 0.1, grown to A600 = 0.5, induced for 2 h with 1 mm isopropyl 1-thio-β-d-galactopyranoside, and pelleted. The pellets were resuspended, lysed in SDS-lysis buffer, and quantitated. Equal amounts of protein were separated by 12% SDS-PAGE, transferred to a polyvinylidene difluoride membrane, and analyzed with mouse anti-His6 and rabbit anti-GST antibodies (Cell Signaling Technology) as detected by fluorescently labeled anti-mouse and goat anti-rabbit secondary antibodies (LICOR Biosciences). Images were taken on a LICOR Odyssey IR scanner.
Cell-free Translation
Templates were prepared by PCR with the following primers: CTGTACATTAATACGACTCACTATAGGGAGATTTTATAAGGAGGAAAAAATATGCAGGGCTGGCAAGCCAC and GGTTATAATGAATTTTGCTTATTAACCAGCCAAGCTTGCCGGTC, adding the T7 promoter and a binding site for the NV1 primer. The PURExpress cell-free transcription-translation system (New England Biolabs) was used for in vitro protein synthesis. Briefly, a 0.2-pmol template was combined on ice with 2.5 μl of Solution A and 1 μl of Solution B along with either 0.5 μl of DMSO (5%) or thiostrepton (0.5 mm in 5% DMSO), and then the mixture was incubated at 37 °C for 30 min. 1 pmol of [32P]ATP-labeled NV1 primer (GGTTATAATGAATTTTGCTTATTAAC) was added, and reverse transcription was performed as described before (25). Samples were then extracted with phenol and chloroform, precipitated with ethanol, separated by 6% denaturing PAGE with C and G sequencing lanes, and visualized with a PhosphorImager (Amersham Biosciences). To detect peptidyl-tRNA in stalled complexes, [35S]Met was added to the translation reactions. Samples were analyzed by Tricine-SDS-PAGE and visualized with a PhosphorImager.
Miller Assays
The reporter plasmid was created by inserting the WPPPSI stalling sequence plus 12 upstream residues after the ninth codon of full-length lacZ (derived from pNH122 (15)). The SecM sequence FSTPVWISQAQGIRAGP was used as a control. Cells bearing a lacZ plasmid and a ribosomal mutant plasmid were induced with 1 mm isopropyl 1-thio-β-d-xgalactopyranoside at mid-log phase and analyzed for β-galactosidase activity using ortho-nitrophenyl-β-d-galactoside as described before (26).
RESULTS
A Genetic Selection for Novel Stalling Sequences
We set out to systematically identify peptide sequences like SecM and TnaC that interfere with peptidyl transfer or hydrolysis and induce ribosome stalling. To identify stalling peptides from random libraries, we modified a genetic selection that we previously developed to link ribosome stalling and rescue to the life of the cell (23). In this selection, stalled ribosomes are recognized by transfer-messenger RNA (tmRNA), a small, stable RNA found in eubacteria and a key component of a quality control system for protein synthesis (for a review, see Ref. 27). The natural function of tmRNA is to release stalled ribosomes and tag the aborted nascent peptide for destruction. Acting as a transfer RNA, tmRNA enters the empty A site of the ribosome and adds Ala to the nascent polypeptide chain. tmRNA then serves as a template, encoding a short peptide tag that is recognized by cellular proteases. After this tag is translated, the ribosome is released at a stop codon within tmRNA, and the aborted protein product is degraded. For the purposes of our selection, it is important to note that, although tmRNA was first characterized as rescuing ribosomes stalled on mRNAs lacking stop codons (28), it can also act on ribosomes stalled by nascent peptides (18, 29).
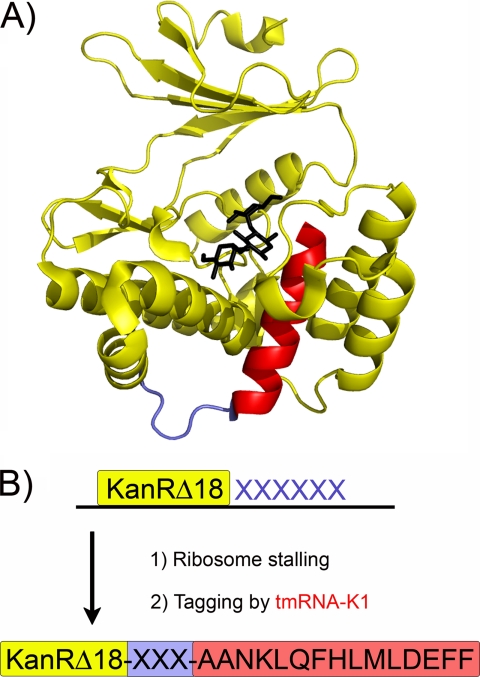
To create a genetic selection for ribosome stalling based on this ribosome rescue machinery, we altered tmRNA so that, instead of tagging proteins for proteolysis, it completes the synthesis of an essential protein, linking stalling to the life of the cell (Fig. 1). The kanamycin resistance protein (KanR) from Tn10 has a C-terminal helix of 15 amino acids that is structurally critical (30); truncation of this helix leads to loss of activity. To complement the truncated KanR protein, we changed the tmRNA template to encode the last 14 residues of KanR (ANKLQFHMLDEFF), referred to hereafter as tmRNA-K1. Together with the Ala from aminoacylated-tmRNA, these residues complete the KanR protein and restore KanR activity but only if the ribosome stalls at exactly the right site. This serves as the basis for our selection: peptide sequences that stall the ribosome at the end of a truncated KanR protein can be easily identified in random libraries, because they recruit tmRNA, complete KanR, and confer resistance to kanamycin.
FIGURE 1.
Genetic selection for sequences that induce ribosome stalling. A, structure of the Aph(3′)-IIa protein, homologous to KanR (30). Kanamycin in shown in black. The C-terminal helix (14 residues), shown in red, is essential for KanR activity. A four-residue surface-exposed loop prior to this helix is highlighted in blue. B, 18 random nucleotides (6 codons) were introduced at the C terminus of a KanR protein lacking its last 18 amino acids (yellow). If the random sequence induces stalling, tmRNA-K1 rescues the stalled ribosome and directs the synthesis of the remaining KanR residues (red). Cellular survival on kanamycin plates is therefore tied to stalling on the C terminus of KanR.
How can stalling be induced at the end of the KanR protein without interfering with the final structure and activity of KanR? Two mutations, Asn255 → Glu and Asp257 → Opal, create a Glu-Pro-(Stop) sequence that induces stalling during translational termination in a kanR gene lacking the C-terminal helix. We previously showed that expression of this truncated kanR-EP construct and the altered tmRNA, tmRNA-K1, allows cells to survive equally well on selective (15 μg/ml kanamycin) or non-selective plates at 37 °C (23). Under the same conditions, bacteria lacking the modified tmRNA-K1 gene survive at the rate of 5 colony forming units in 107 plated. These results demonstrate that the introduction of the Glu-Pro-Ala “scar” from the stalling and tagging process does not destroy KanR activity. Analysis of the crystal structure of the homologous Aph(3′)-IIa protein (30) suggests that the C-terminal helix in KanR is preceded by a surface-exposed loop of poorly conserved residues (Ile253 through Pro256). We anticipated that this loop region might tolerate a variety of sequences that induce stalling while maintaining robust KanR function.
Nucleotide sequences that induce stalling and tagging were isolated from a randomized library fused to a truncated kanR gene. 18 amino acids were deleted from KanR, including the C-terminal helix and 3 residues in the preceding loop. 18 random nucleotides (6 codons) were cloned downstream of this truncated kanR gene, beginning with residue 254. No stop codon was specified. We generated a library of 5 × 106 mutants and introduced it, together with tmRNA-K1, into an E. coli strain lacking wild-type tmRNA. We selected the mutants for survival on plates containing 15 μg/ml kanamycin at 37 °C. Roughly 1 in 104 colonies survived, suggesting that a substantial fraction of the sequences induce ribosome stalling.
Three Classes of Stalling Peptides
Although we were interested primarily in peptides that induce stalling, our selection identifies any nucleic acid sequences that elicit tagging by tmRNA. In principle, nucleotide sequences containing rare codon clusters (31, 32), secondary structures, transcriptional terminators (28), or other novel mechanisms might also survive the KanR selection. Analysis of the surviving clones, however, revealed sequences that share common features at the amino acid level. These were grouped into three classes (Table 1).
TABLE 1.
Stalling peptides isolated from the KanR selection
The variable region (six codons) is shown in bold. Class I contains Pro-stop residues at the third and fourth variable positions. Class II contains a Pro-Pro sequence at positions three and four with no nearby stop codon. Class III contains Trp-Pro-Pro at the first three positions without an adjacent stop codon.
| Class I: Pro-stop | G I S E P * (opal) |
| G I K D P * (amber) | |
| G I W P P * (opal) | |
| Class II: Pro-Pro | G I R A P P H C G T |
| G I R S P P N S G T | |
| G I L D P P G M G T | |
| Class III: Trp-Pro-Pro | G I W P P W Y R G T |
| G I W P P D V * (opal) | |
| G I W P P P S I G T |
The most common cause of stalling, found in over 90% of the clones, is inefficient termination at the sequence Pro-Stop. The Pro residue is found almost exclusively at position three of the six random codons, corresponding to native KanR residue Pro256. Although there is no significant codon bias for any particular Pro codon, the opal stop codon (UGA) is highly overrepresented (23/29 clones). There is also selection for the residue just upstream of Pro-Stop: Glu is overrepresented (16/29) and Asp, Pro, and Gly are each seen several times in the −2 position.
A second class of peptides must induce stalling during elongation, not termination. These clones contain two consecutive Pro codons, most commonly at codons three and four in the random sequence with no nearby stop codon. The majority of these clones were found by performing the selection at lower stringency, lowering the temperature to 25 °C. When tested individually, they showed poor survival at 37 °C, roughly 1–10%, much weaker than the 100% survival seen with the Pro-Stop sequences above.
A third class of clones contains the sequence Trp-Pro-Pro without a nearby stop codon (Table 1). Like the other Pro-Pro sequences, these peptides also stall during elongation rather than termination. Unlike the second class of clones, however, where the two consecutive Pro codons appear at positions three and four, here they occur at positions two and three (e.g. WPPWYR). Another difference is that WPP-containing clones survive robustly (100%) in the KanR selection at 37 °C when characterized individually. Further experiments on these sequences are described below.
The sequence Pro-Stop occurs commonly and elicits tagging at high levels; to prevent such clones from overwhelming other novel sequences, we created a second library of 18 nt (6 codons) in which codons 4 through 6 could not be stop codons. This was done by allowing only C, G, and A at the first nucleotide of these codons; this eliminates Phe, Tyr, Cys, and Trp as well. We screened an 8 × 106-member library at high stringency, obtaining colonies at rates of 0.01% survival. 21 of 23 sequenced clones contained the sequence WPPP at the first four positions (data not shown). This result confirms that WPP-containing sequences are robust inducers of stalling, particularly when coupled with a third Pro codon.
Selection of this second library at low stringency yielded higher levels of survival (0.25%). Nearly all of surviving clones fall into the second class of stalling peptides, with two consecutive Pro codons at codons three and four of the random sequence. An alignment of 46 of these sequences reveals that Arg or His are strongly preferred at the first position, with Ala, Asp, Ser, and Pro at the second position (supplemental Fig. S1). Including the constant Gly-Ile upstream, the consensus sequence becomes GI(R/H)XPPXX. These appear to be weaker versions of the SecM C-terminal sequence GIRAGP. A clone closely resembling this sequence (GIRAPP) is more active than the other members of this class and survives the KanR assay 100% at high stringency.
Stalling and Tagging Occur following WPP
The peptide sequences in class three (containing WPP) show high levels of activity in the KanR assay and stall translation during the elongation step. We chose to further characterize three sequences: WPPPSI, WPPDV*, and WPPWYR. Where does stalling occur in these sequences? Where is the tag added by tmRNA? To analyze the tagged proteins by mass spectrometry, we first transferred the stalling sequence to the C terminus of the GST protein. This full-length, stable protein served as a scaffold enabling overexpression of the stalling peptide. Some of the KanR protein context was fused to GST as well, from 12 amino acids upstream of the random hexamer through the stop codon 27 amino acids downstream. To isolate proteins tagged by tmRNA, we used a modified tmRNA encoding 6 His residues in its template sequence (tmRNA-H). GST fusions tagged by tmRNA-H were purified by affinity chromatography using nickel-nitrilotriacetic acid resin and digested with trypsin. From this tryptic digest, the C-terminal-tagged peptide was purified again with nickel-nitrilotriacetic acid resin.
The C-terminal peptide contains both the stall sequence from KanR and the tmRNA tag; determining its mass by MALDI-MS revealed the site of stalling and tagging by tmRNA. A single large peak in the mass spectra for the WPPPSI and WPPWYR C-terminal-tagged peptides corresponded to an m/z of 2041 (supplemental Fig. S2). This is the predicted result if the tmRNA tag is added after the second Pro (YGIWPPAANDH6D). The mass spectrum of the WPPDV* peptide fragment contained the same peak at 2041 together with a more abundant peak at 2255, corresponding to the peptide YGIWPPDVAANDH6D. In the WPPDV* clone, stalling occurs both after WPP and during termination at the stop codon. Peptide fingerprinting by tandem MS/MS was performed on all four of these peptides to confirm the amino acid sequence directly.
Determination of Residues Necessary and Sufficient for Stalling and Tagging
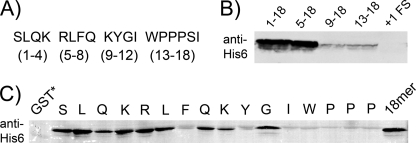
The MS data indicate that tagging occurs immediately after WPP in these three clones. What amino acids cause this stalling event? In the case of SecM and TnaC, residues essential for the highest levels of stalling are found upstream and interact with the exit tunnel. For this reason we included 12 upstream amino acids (SLQKRLFQKYGI) from KanR along with the hexamers in making the GST fusions. To assay for stalling and tagging in the GST fusions, we detected the tag added by tmRNA-H with anti-His6 antibodies (Fig. 2). High levels of tagging were detected for the full-length GST-WPPPSI fusion, referred to hereafter as 1–18 (i.e. 12 residues from KanR followed by the hexamer (Fig. 2A)). Deletion of the first four amino acids had little or no effect (Fig. 2B, 5–18), but removal of the first eight nearly eliminated tagging (Fig. 2B, 9–18). We conclude that residues upstream of the WPPPSI sequence play a critical role in high efficiency tagging. Interestingly, some minimal activity resides in the hexamer sequence alone (Fig. 2B, 13–18) with no KanR upstream sequence.
FIGURE 2.
Essential elements of a stalling peptide. A, WPPPSI and 12 upstream residues were divided into four groups for analysis. B, N-terminal deletions of the 18-mer stalling peptide were fused to the C terminus of GST. The fusions were analyzed by immunoblot for tagging by a modified tmRNA (tmRNA-H) that encodes an His6 epitope. The +1FS vector contains the same 18-mer sequence shifted into the +1 frame to test if translation of WPPPSI is necessary for stalling. C, each residue of the 18-mer was mutated individually to Ala and assayed as above. A GST-Stop construct served as a negative control and the intact full-length WPPPSI 18-mer as a positive control.
To identify how each residue contributes individually to stalling, we performed alanine scanning on the full-length stalling peptide, 1–18. Residues 1–16 were individually mutated to alanine and assayed by immunoblot (Fig. 2C). Consistent with the truncation results, mutating residues 1–4 has little or no effect on tagging. Alanine substitutions for Arg5, Leu6, Gln8, and Lys9 likewise make little difference in the level of tagging. In contrast, mutation of Phe7, Tyr10, or Ile12, or the WPP sequence dramatically decreased tagging levels. Notably, tagging was also strongly reduced by the Pro16 → Ala mutation. This is surprising because the MS data show that the third Pro in WPPPSI is not incorporated into the stalled peptide. In summary, residues in the consensus sequence FXXYXIWPPP are required for tagging.
tmRNA rescues ribosomes stalled on broken mRNA templates; perhaps these tagging events arise from RNA synthesis defects in the kanR mRNA or from nucleolytic cleavage. To prove that tagging requires translation of the peptide sequence, we created a mutant of the GST-WPPPSI fusion in which a single nucleotide is added upstream of the full-length stall peptide. The resulting +1 frameshift changes the identity of every amino acid in the stalling sequence except for Phe7 and Lys9 while retaining the same nucleotide sequence. Immunoblot analysis of this mutant revealed that tagging was completely abolished, demonstrating that tagging of the GST-WPPPSI fusion is due to the amino acid sequence and not the nucleotide sequence (Fig. 2B).
Tagging at Termination in WPPDV*
The MS data show that tagging occurs in the WPPDV* sequence both immediately after WPP and during termination. To further understand the effect on termination, we measured tagging levels for a series of GST-WPPDV* variants in the immunoblot assay (Fig. 3). Mutation of the opal stop codon (UGA) to the more efficient ochre codon (UAA) reduced tagging slightly; replacing the stop codon altogether with an Ala codon reduced it even further. We propose that the substantial tagging that remains in the WPPDVA variant represents stalling directly after the WPP as seen in the MS data.
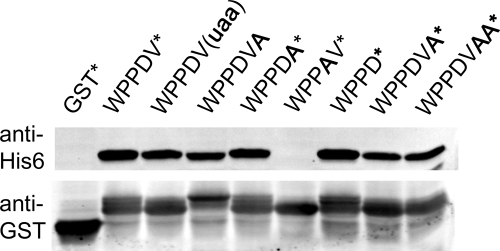
FIGURE 3.
Stalling during termination at WPPDV*. Tagging of the GST-WPPDV* fusion by tmRNA-H was monitored by anti-His6 antibodies. Mutations in bold were introduced to determine the role of spacing, stop codons, and the DV residues in this stalling event.
If the WPPDV sequence is interfering with termination, how far downstream does this effect carry? An opal stop codon immediately following WPPD tagged at the same level as the original WPPDV* sequence (Fig. 3). Moving the stop codon one or two codons downstream by inserting Ala residues, however, reduced the tagging levels to those lacking a stop codon altogether (WPPDVA). These results show that the stop codon must be only one or two codons downstream of WPPD for stalling to occur during termination.
We next examined the role of the Asp and Val residues. Val is not known to inhibit termination when found at the C terminus of proteins; indeed, the Val17 → Ala mutant showed no loss of tagging. The Asp16 → Ala mutation, however, completely alleviated tagging (WPPAV*). The Asp residue must therefore be critical for tagging after WPP as well as after WPPDV during termination. This role is consistent with the critical nature of the third Pro residue in the WPPPSI clone.
The Residue after WPP Is Critical for Tagging by tmRNA
Alanine-scanning mutagenesis of the WPPDV* and WPPPSI clones demonstrates that the residue after FXXYXIWPP plays a key role in stalling ribosomes. What amino acids besides Asp and Pro can fulfill this role? We created a library of peptide trimers following WPP in the KanR selection (WPPXXX), constrained as above to exclude stop codons. Of the clones surviving at high stringency, ∼80% contain the sequence WPPPXX and another ∼20% the sequence WPPDXX (data not shown). No selection was apparent for the final two amino acids. To perform a more quantitative analysis, we created mutants of the GST-WPPPSI fusion expressing all 20 amino acids in the position immediately following WPP. These were subjected to immunoblot analysis with tmRNA-H. Confirming the genetic data, the Pro, Asp, and Trp mutants showed high levels of tagging. The Asn mutant tagged moderately, whereas the other 16 amino acids showed much lower levels of tagging (supplemental Fig. S3).
How does the residue after FXXYXIWPP contribute to stalling and tagging? The incoming aminoacyl-tRNA does not react, yet its identity is critical. It must therefore contribute to stalling by interacting with the ribosome in the A site. During ribosome stalling on SecM, Pro-tRNA performs exactly this function (19).
But if Pro-tRNA is bound in the A site, how is the GST-WPPPSI fusion tagged by tmRNA? It should block tmRNA from entering the ribosome at all. One possibility is that ribosomes stalled on FXXYXIWPPP deplete Pro-tRNA from the available cellular pool, leading to some stalling events with empty A sites. Depletion of Pro-tRNA leads to tagging of the SecM peptide by tmRNA (19). If this is also the case with FXXYXIWPPP, then overexpression of tRNAPro should alleviate tagging. To test this hypothesis, we altered the GST-WPPPSI fusion to include one or more CCC codons. CCC is decoded by only one tRNA, Pro2, which also recognizes CCU. The original WPPPSI sequence contains neither CCC nor CCU; we altered it to include CCC at the first two Pro codons (WppPSI) or the third (WPPpSI).
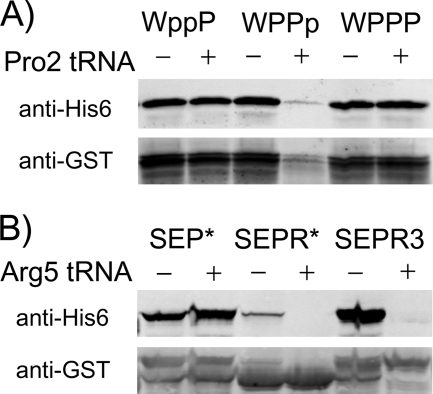
The immunoblot assay was used to visualize the tagging levels of these GST fusions with or without overexpression of Pro2 tRNA from the pRARE plasmid (Fig. 4A). Tagging of the GST-WPPPSI fusion lacking CCC codons was unaffected by overexpression of Pro2. Likewise, little or no change in tagging occurred when the first two Pro residues were encoded by CCC (WppPSI). In contrast, when the third Pro codon was CCC, tagging was sharply reduced by Pro2 tRNA overexpression. In addition to the loss of tagging, the overall expression of the GST-WPPpSI fusion was dramatically reduced. Pro2 overexpression had no effect on GST levels in WPPPSI or WppPSI fusions. These results show that depletion of the tRNA decoding the third Pro codon is necessary for tagging.
FIGURE 4.
The effect of tRNA levels on stalling and tagging. A, one or more Pro codons in the WPPPSI sequence was switched to CCC (lowercase p) so that it would only be recognized by Pro2 tRNA. An uppercase P represents a Pro codon not recognized by Pro2 tRNA. Tagging of the GST-WPPPSI fusion was monitored in the presence or absence of a plasmid (pRARE) overexpressing the Pro2 tRNA. pRARE also expresses several other rare tRNAs. B, stalling sequences containing one or three rare Arg codons (AGG) were fused to the C terminus of GST. Tagging by tmRNA-H was monitored with anti-His6 antibodies in the presence or absence of a plasmid (pRARE) overexpressing the cognate tRNA, Arg5.
The Role of Codon Usage
We anticipated at the outset of our KanR selection experiments that we might isolate stalling sequences with rare codons. Overexpression of proteins containing consecutive rare codons induces high levels of stalling and tagging by tmRNA (31, 32). The three tRNAArg isoacceptors decoding the CGG, AGA, and AGG codons are present at low levels in E. coli (33). Why do such sequences not survive the KanR selection? To address this question, we measured tmRNA tagging levels for a GST fusion construct containing SEPR* and SEPRRR encoded by the rare Arg codon AGG. SEPR* tagging was barely detectable, much lower than SEP*, whereas SEPRRR tagged at very high levels in the immunoblot assay (Fig. 4B). Tagging at both sequences was completely alleviated by overexpression of the cognate tRNA (Arg5) from the pRARE plasmid. The same SEPR* and SEPRRR sequences were then cloned in place of the randomized cassette of the KanR selection plasmid. The sequence SEP in the first three positions is known to be compatible with KanR activity; in the SEP* context it conveys 100% survival. Cells expressing SEPR* or SEPRRR sequences survived no better than an empty vector control under low stringency conditions (data not shown). These results show that tagging activity at rare codons is either insufficient or incompatible with restoring KanR function.
Direct Detection of Stalled Ribosome Complexes
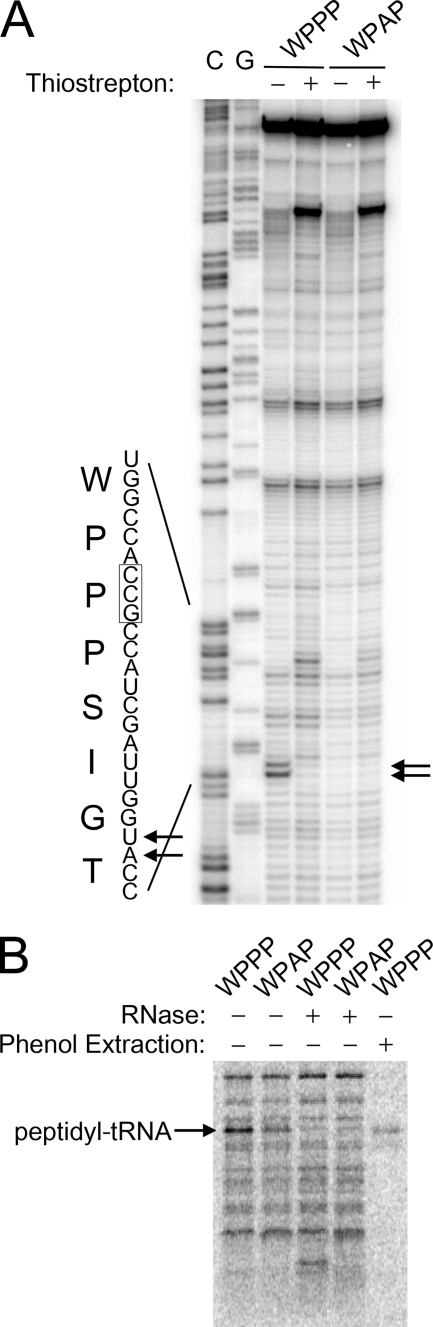
Both the KanR and the tmRNA-H immunoblot assays rely on tmRNA tagging to measure levels of ribosome stalling. To analyze stalling directly, we performed in vitro translation reactions and detected stalled ribosome complexes with toe-printing assays. Peptides corresponding to the C-terminal 64 residues of the GST fusions described above were expressed in a cell-free transcription and translation system. The protein sequence includes the full 18-mer stalling peptide, 22 residues of upstream GST sequence, and 24 residues downstream of the predicted stalling site. A radiolabeled primer was annealed to the 3′-end of the transcript and extended by reverse transcriptase. Analysis of the FXXYXIWPPP peptide translation reaction revealed that reverse transcriptase is blocked 15–16 nt downstream of the first nucleotide in the second Pro codon (Fig. 5). In contrast, no toe-print was seen in the translation of the FXXYXIWPAP peptide, consistent with the finding that mutation of the second Pro codon dramatically reduces tagging (Fig. 2C). As a control, the antibiotic thiostrepton was added to trap ribosomes in the initiation stage. The disappearance of the toe-print in the FXXYXIWPPP peptide reaction when thiostrepton is added demonstrates that the block in reverse transcription is due to stalled ribosomes and not an artifact of mRNA sequence or structure. Together with the mass spectrometry and immunoblot analyses of tmRNA tagging above, the toe-printing data demonstrate that ribosomes stall at the FXXYXIWPPP sequence with the second Pro codon in the ribosomal P site and the third Pro codon in the A site.
FIGURE 5.
Direct detection of stalled ribosome complexes. A, stalled ribosome complexes were formed by cell-free translation of a template encoding the FXXYXIWPPP sequence in a larger (64-mer) peptide. The non-stalling Ala mutant FXXYXIWPAP served as a negative control. The position of the ribosome was determined by reverse transcription of the mRNA template, and C and G sequencing lanes were run alongside. Thiostrepton was added in the fourth and sixth lanes to trap the ribosome in the initiation stage, demonstrating the observed toe-print signal in the third lane (marked by arrows) requires translation of the stalling site. The nucleotide and peptide sequence of the stalling site is shown at left. B, the [35S]Met-labeled products of the cell-free translation of the FXXYXIWPPP peptide or the non-stalling FXXYXIWPAP control were analyzed by Tricine-SDS-PAGE. Under these conditions, the peptidyl-tRNA linkage is not hydrolyzed during electrophoresis. The stalled peptidyl-tRNA disappears when treated with RNase (lane 3) but remains in the aqueous layer upon extraction with phenol (lane 5).
Ribosome stalling leads to the accumulation of peptidyl-tRNA within the ribosome. We detected this trapped peptidyl-tRNA by including [35S]methionine in the translation reaction. To prevent hydrolysis of the peptidyl-tRNA, we analyzed the products by gel electrophoresis in an acidic buffer system. Three lines of evidence support the identification of stalled peptidyl-tRNA in the FXXYXIWPPP peptide translation reaction. First, the high molecular weight band disappears upon treatment with RNase and a far smaller band appears (lanes 1 and 3). Secondly, the peptidyl-tRNA band remains in the aqueous layer following phenol extraction, whereas the other peptide bands disappear (lane 5). Finally, the stalled peptidyl-tRNA is less abundant in the FXXYXIWPAP peptide reaction (lane 2), where it is not expected to accumulate as stalling is dramatically reduced.
Ribosomal Interactions Necessary for Stalling
To better understand the interactions between the nascent peptide and the ribosome that lead to stalling, we quantified stalling levels with a series of ribosome mutants. This was done by inserting the WPPPSI 18-mer after residue nine of lacZ and assaying for the activity of β-galactosidase. Our stalling peptides were compared with SecM and a non-stalling SecM control that has an Ala substitution of the C-terminal Pro. As shown by Nakatogawa and Ito (15), the SecM peptide dramatically inhibits lacZ expression; β-galactosidase activity is 1300-fold higher in the non-stalling Pro166 → Ala mutant (Table 2). The FXXYXIWPPP peptide also reduced lacZ expression, but not as well as SecM (116 versus 8 Miller units, respectively). Mutation of the second Pro residue in FXXYXIWPPP results in 96-fold higher LacZ activity (Table 2), as expected by the reduction in tagging observed above. These results show that this selected peptide sequence induces stalling with high efficiency in a tmRNA-independent assay.
TABLE 2.
Efficiency of stalling of the SecM and FXXYXIWPPP peptides
The peptide sequences shown were inserted into the full-length lacZ gene following the ninth codon. β-Galactosidase activity is shown in Miller units along with the standard deviation. The Ala substitutions in bold are known to prevent stalling in SecM and FxxYxIWPPP.
| Stalling peptide | LacZ activity |
|---|---|
| FSTPVWISQAQGIRAGP | 8.2 ± 2.0 |
| FSTPVWISQAQGIRAGA | 10,812 ± 1,288 |
| SLQKRLFQKYGIWPPPSI | 117 ± 14 |
| SLQKRLFQKYGIWPAPSI | 11,172 ± 3,580 |
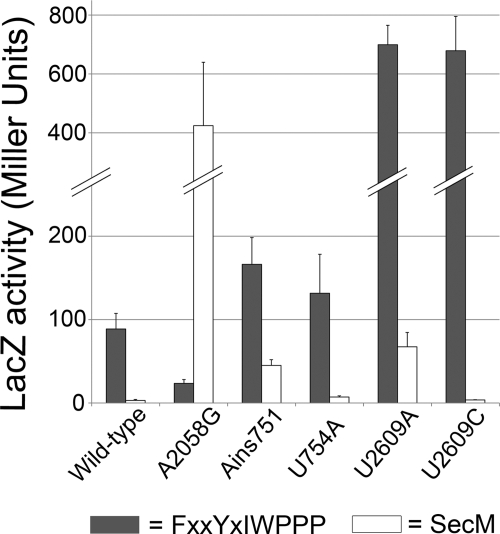
Ribosomal RNA mutations that map to the exit tunnel have been shown to affect stalling on SecM and TnaC. Does the FXXYXIWPPP peptide interact with the same ribosomal RNA nucleotides? Using β-galactosidase assays, we measured the effect of several 23 S rRNA mutations on stalling on this peptide. 23 S rRNA mutants were overexpressed in the presence of wild-type ribosomes. Stalling was reduced 7-fold by both the U2609A and U2609C mutations (Fig. 6), first studied in connection with TnaC. The U754A and A751 insertion mutations, in contrast, showed no significant effect. Surprisingly, the A2058G mutation actually increases stalling 8-fold.
FIGURE 6.
Effects of ribosome mutations on stalling on SecM and FXXYXIWPPP. The SecM stalling sequence or the WPPPSI 18-mer (see Table 2) were inserted after residue nine of the full-length lacZ gene. β-Galactosidase activity was measured for the resulting SecM (white) and FXXYXIWPPP (gray) lacZ fusions in a strain overexpressing mutant 23 S ribosomal RNA. The activity is reported in Miller units. The data represent at least three independent experiments. Error bars represent the standard deviation.
Analysis of SecM-mediated stalling with the same set of rRNA mutants yielded quite a different picture. Although the U2609A mutation reduced stalling moderately (8-fold), as it did with FXXYXIWPPP, the U2609C mutation had little or no effect. Although the A751 insertion had no effect on FXXYXIWPPP, this mutation decreased stalling on SecM 6-fold. The most striking difference, however, is that the A2058 mutation increases stalling on FXXYXIWPPP but is the most effective at reducing stalling on SecM (78-fold), consistent with the findings of Nakatogawa and Ito (15). These results show that, although stalling on FXXYXIWPPP involves some of the same rRNA nucleotides as SecM or TnaC, a unique pattern of exit tunnel interactions is required for each peptide.
DISCUSSION
We performed a genetic selection to identify novel peptides that inhibit their own synthesis. The selection is based on the ability of tmRNA to recognize and rescue stalled ribosomes. When stalling occurs at the C terminus of a truncated KanR protein, tmRNA encodes the missing amino acids to complete the protein and restore KanR activity.
Our library covered roughly 10% of the theoretical diversity of a library of random peptide hexamers. We recognize that some peptides that induce stalling were missed in our selection, because they were either too long or incompatible with the structure and activity of KanR. We were surprised that consecutive rare codons, known to induce tagging (31, 32), were not isolated in the selection. We demonstrated that tagging does occur at SEPR* and SEPRRR by immunoblot (Fig. 4), but these sequences do not support KanR rescue by tmRNA. In the case of SEPR*, tagging is probably at too low a level to support robust KanR activity. Although SEPRRR induces higher levels of tagging, the tag is probably not added at precisely the necessary site to restore the KanR protein sequence properly. Alternatively, depletion of low abundance tRNAs may be too taxing for cells. Immunoblot analysis of tagging is performed after a brief period of strong overexpression. In contrast, our genetic selection requires overexpression and tagging of KanR over long periods of cell growth and division.
The simplest and most common cause of stalling that we identified is inefficient termination at Pro-Stop sequences. Several components need to be present to cause high efficiency stalling during termination. First, the opal stop codon (UGA) was strongly preferred over the other two stop codons in the selection. UGA is the least efficient stop codon, leading to recoding events such as the programmed frameshift in the prfB gene encoding release factor 2 (34, 35). Pro-opal sequences cause strong +1 frameshifting at CCC_UGA (36) and significant levels of stalling and tagging by tmRNA (18). As seen in previous studies, the residue upstream of Pro also affects the efficiency of termination (18, 37). In particular, Glu, Asp, and Pro were overrepresented in the −2 position (e.g. Glu-Pro-opal) in our selections. These results validate our selection and demonstrate that survival in the KanR assay requires high levels of ribosome stalling.
A second set of sequences with the consensus GI(R/H)XPP show weaker activity (surviving only at low stringency). It is interesting to note that the GI residues were not part of the random hexamer library; by chance, these were the two amino acids immediately upstream. These peptides appear to be subtle variants of the SecM sequence GIRAGP166. The GIRAPP clone that matches SecM the most closely survives even at high stringency. This suggests that some alterations in this critical SecM sequence are tolerated. Mutation of Arg163 to His or replacing Ala164 with Asp, Ser, or Pro yields substantial though weaker stalling activity. These results agree with the recent findings of Yap and Bernstein (38), who showed that the GIRAGP sequence in SecM exhibits significant plasticity, with only Arg and Pro residues playing key roles.
Our third class of selectants (containing FXXYXIWPP) stall with peptidyl-tRNA in the P site. In the case of the WPPPSI clone, for example, the mass spectrometry data show that the tmRNA tag is added after WPP. Yet the Ala scanning data show that the next residue (the third Pro) is required for tagging, even though it does not react with the nascent peptide. We propose that the aminoacyl-tRNA binds and remains unreacted in the A site and that peptidyltransferase activity is inhibited by FXXYXIWPP-containing peptides. This implicates changes in the conformation of the PTC in the stalling mechanism.
The amino acid Pro plays two different roles in FXXYXIWPPP stalling. First, Pro-tRNA acts as a poor peptidyl acceptor in the A site. It fails to react with the nascent peptide. N-Alkylamino acids such as Pro have been shown to act as slow nucleophiles in the peptidyltransferase reaction (39). Using full-length tRNAs, Pavlov et al. demonstrated that the unnatural Pro-tRNAPhe reacts 23-fold slower than Phe-tRNAPhe with initiation complexes containing fMet-tRNA. They speculate that this is due to steric constraints and lower nucleophilicity. Interestingly, the rate of Pro reactivity is accelerated by the natural tRNAPro isoacceptor; Pro-tRNAPro only has a 3- to 6-fold defect. A-site bound Pro-tRNA plays a role in stalling on SecM and the 2A peptides found in viral genomes that stall at the Gly residue in the sequence D(V/I)EXNPGP (40). One possible explanation for the necessity of Pro-tRNA is that the reduced rate of peptidyl transfer to Pro gives the nascent peptide time to interact with the exit tunnel and PTC, shifting the 23 S rRNA to an inactive conformation (41).
Our results suggest that aminoacyl-tRNAs other than Pro-tRNA can induce stalling by binding in the A site. WPPD and WPPW sequences were isolated from our selections, and immunoblot analysis revealed that efficient tagging only occurs if the residue following WPP is Pro, Asp, or Trp. Although we cannot say for certain, it is probably binding of the amino acid that is critical, not the codon or tRNA. The amino acid is the key component of Pro-tRNA in SecM; the Pro analog azetidine dramatically reduces stalling (8). Likewise, the binding of free tryptophan causes stalling on TnaC (11). A second explanation for the role of the A site aminoacyl-tRNA in stalling is that amino acid binding near the PTC changes the ribosome or peptide conformation. This possibility is supported by the finding that the SecM peptide undergoes a conformational change in the tunnel upon Pro-tRNA binding in the A site (42).
If aminoacyl-tRNA is bound in the A site, how can tmRNA enter the stalled ribosomes to release them and tag the nascent peptide? SecM, TnaC, and ErmCL (another stalling leader peptide) are not tagged by tmRNA, because the A site of the ribosome is occupied (16, 19, 20, 25). Stalling on these peptides must be determined by cellular conditions to regulate gene expression; tmRNA would interfere with their biological function. But our selection and immunoblot assays rely on tagging by tmRNA to detect stalling events. We propose that overexpression of FXXYXIWPPP leads to depletion of Pro-tRNA by stalled ribosomes, creating a subset of ribosomes stalled with empty A sites that are acted on by tmRNA. Overexpression of SecM results in high levels of tagging (29, 43) for exactly this reason (19). For both SecM and FXXYXIWPPP, increasing tRNAPro levels abolishes tagging (Fig. 4). At the same time, we see that tRNA overexpression actually lowers GST levels, perhaps because stalling is more robust with the tRNA in the A site and no GST-stalled ribosomes are released by tmRNA.
The second role of Pro in FXXYXIWPPP stalling is that of a poor peptidyl donor. Peptides ending in Pro react with puromycin far slower than peptides ending in other amino acids (44, 45). It is the amino acid that inhibits peptidyl transfer, not the codon or tRNA: incorporation of Pro analogs azetidine or thiaproline restore rapid reactivity (44). In uncatalyzed reactions, however, Pro-tRNA is as reactive as other aminoacyl-tRNAs (46), suggesting that the reduced rate is not purely due to the chemistry of prolyl-esters but the interaction of the Pro residue with the ribosome. These findings suggest that the conformationally strained Pro side chain inhibits ribosome activity (45). In nature, C-terminal Pro residues inhibit termination in proteins in TnaC and in the UL4 uORF2 in the mammalian virus cytomegalovirus (47). It appears that the cyclic Pro residue interferes with conformational changes in the PTC that are required for both elongation and termination.
The WPP-containing peptides stall ribosomes robustly: in the lacZ assay, the WPPPSI 18-mer sequence reduced activity nearly 100-fold over the WPAPSI mutant. The sequence context has a great effect: upstream peptide sequences are required for high efficiency tagging. Tagging in the WPPPSI clone requires the consensus peptide sequence FXXYXIWPP. Phe7 and Tyr10 are aromatic residues that may bind rRNA in the exit tunnel. At nine residues, the FXXYXIWPP is the same length as the ErmCL peptide (MGIFSIFVI) when it stalls upon binding of erythromycin in the nascent peptide exit tunnel (25). Shortening the ErmCL peptide by deleting N-terminal residues reduces stalling significantly; this length may allow interaction with elements farther into the tunnel, such as the L22 loop (25). The sequences of these four stalling peptides are different, the only commonality being an Ile four residues from the P site in FXXYXIWPP, ErmCL, and SecM.
Analysis of stalling levels with mutant ribosomes reveals nucleotides that are required for efficient stalling on FXXYXIWPP peptides. A2058 is near the L4/L22 constriction; the A2058G mutation reduces stalling on SecM by nearly 80-fold, but it actually increases stalling on the WPPPSI clone by 8-fold. Likewise, mutation of U2609 has different effects on these three peptides. In TnaC, the U2609C mutant completely abolished stalling, whereas U2609A only affected it partially (16). SecM stalling is more reduced by the A mutant, whereas stalling on FXXYXIWPPP is reduced by either the C or A mutant equally. These data show that stalling on FXXYXIWPPP involves the same players as these other peptides, although the specifics of each interaction vary, suggesting that the peptides bind differently in the tunnel.
The plasticity of nascent peptide interactions with the PTC and exit tunnel is further highlighted by our finding that stalling on the WPPDV* clone occurs both after WPP and during termination. Presumably stalling requires upstream amino acids so the active sequence is FXXYXIWPPDx*. If specific interactions with the tunnel and PTC are lined up properly with the second Pro codon in the P site, it is difficult to imagine how they are aligned again to inhibit termination after the peptide has moved two amino acids farther into the tunnel. We believe that the simplest explanation for this is that the FXXYXIWPP sequence engenders a constrained peptide conformation that promotes interaction with the tunnel at several possible sites. This speculation is supported by the finding of Yap and Bernstein that SecM mutants containing Pro-Pro dipeptides (e.g. PPIRAGP) induce stalling even in the absence of the upstream arrest motif elements (38).
Like SecM and TnaC, FXXYXIWPP-containing sequences stall due to nascent peptide interactions in the PTC and exit tunnel, with an effector bound in the A site. Despite this common mechanism, these peptides rely on different ligands in the A site and different interactions with the exit tunnel to inhibit peptidyl transfer or hydrolysis. The lack of sequence similarity in these peptides argues that many solutions exist and that regulation of gene expression by nascent peptides may be more common than the few examples characterized so far. Further characterization of the mechanism of stalling on WPP-containing sequences and its biological significance is ongoing.
Acknowledgments
We thank Koreaki Ito and Charles Yanofsky for providing plasmids and strains, Karen Merrell for assistance with mass spectrometry, Nora Vazquez-Laslop and Alexander Mankin for assistance with toe-printing, and David Healey for comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant GM77633 (to A. B.).
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- PTC
- peptidyltransferase center
- tmRNA
- transfer-messenger RNA
- A site
- ribosomal aminoacyl-tRNA site
- P site
- ribosomal peptidyl-tRNA site
- MALDI
- matrix-assisted laser desorption ionization
- MS
- mass spectrometry
- MS/MS
- tandem MS
- nt
- nucleotide(s)
- GST
- glutathione S-transferase
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- KanR
- kanamycin resistance protein.
REFERENCES
- 1.Tenson T., Ehrenberg M. (2002) Cell 108, 591–594 [DOI] [PubMed] [Google Scholar]
- 2.Lovett P. S., Rogers E. J. (1996) Microbiol. Rev. 60, 366–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morris D. R., Geballe A. P. (2000) Mol. Cell. Biol. 20, 8635–8642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thanaraj T. A., Argos P. (1996) Protein Sci. 5, 1594–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kimchi-Sarfaty C., Oh J. M., Kim I. W., Sauna Z. E., Calcagno A. M., Ambudkar S. V., Gottesman M. M. (2007) Science 315, 525–528 [DOI] [PubMed] [Google Scholar]
- 6.Tsai C. J., Sauna Z. E., Kimchi-Sarfaty C., Ambudkar S. V., Gottesman M. M., Nussinov R. (2008) J. Mol. Biol. 383, 281–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oliver D., Norman J., Sarker S. (1998) J. Bacteriol. 180, 5240–5242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakatogawa H., Ito K. (2001) Mol. Cell 7, 185–192 [DOI] [PubMed] [Google Scholar]
- 9.Butkus M. E., Prundeanu L. B., Oliver D. B. (2003) J. Bacteriol. 185, 6719–6722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gong F., Yanofsky C. (2001) J. Biol. Chem. 276, 1974–1983 [DOI] [PubMed] [Google Scholar]
- 11.Gong F., Yanofsky C. (2002) Science 297, 1864–1867 [DOI] [PubMed] [Google Scholar]
- 12.Nissen P., Hansen J., Ban N., Moore P. B., Steitz T. A. (2000) Science 289, 920–930 [DOI] [PubMed] [Google Scholar]
- 13.Gabashvili I. S., Gregory S. T., Valle M., Grassucci R., Worbs M., Wahl M. C., Dahlberg A. E., Frank J. (2001) Mol. Cell 8, 181–188 [DOI] [PubMed] [Google Scholar]
- 14.Berisio R., Schluenzen F., Harms J., Bashan A., Auerbach T., Baram D., Yonath A. (2003) Nat. Struct. Biol. 10, 366–370 [DOI] [PubMed] [Google Scholar]
- 15.Nakatogawa H., Ito K. (2002) Cell 108, 629–636 [DOI] [PubMed] [Google Scholar]
- 16.Cruz-Vera L. R., Rajagopal S., Squires C., Yanofsky C. (2005) Mol. Cell 19, 333–343 [DOI] [PubMed] [Google Scholar]
- 17.Mitra K., Schaffitzel C., Fabiola F., Chapman M. S., Ban N., Frank J. (2006) Mol. Cell 22, 533–543 [DOI] [PubMed] [Google Scholar]
- 18.Hayes C. S., Bose B., Sauer R. T. (2002) J. Biol. Chem. 277, 33825–33832 [DOI] [PubMed] [Google Scholar]
- 19.Garza-Sánchez F., Janssen B. D., Hayes C. S. (2006) J. Biol. Chem. 281, 34258–34268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muto H., Nakatogawa H., Ito K. (2006) Mol. Cell 22, 545–552 [DOI] [PubMed] [Google Scholar]
- 21.Cruz-Vera L. R., Gong M., Yanofsky C. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 3598–3603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cruz-Vera L. R., New A., Squires C., Yanofsky C. (2007) J. Bacteriol. 189, 3140–3146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanner D. R., Dewey J. D., Miller M. R., Buskirk A. R. (2006) J. Biol. Chem. 281, 10561–10566 [DOI] [PubMed] [Google Scholar]
- 24.Hayes C. S., Bose B., Sauer R. T. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 3440–3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vazquez-Laslop N., Thum C., Mankin A. S. (2008) Mol. Cell 30, 190–202 [DOI] [PubMed] [Google Scholar]
- 26.Miller J. H. (1972) Experiments in Molecular Genetics, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 27.Moore S. D., Sauer R. T. (2007) Annu. Rev. Biochem. 76, 101–124 [DOI] [PubMed] [Google Scholar]
- 28.Keiler K. C., Waller P. R., Sauer R. T. (1996) Science 271, 990–993 [DOI] [PubMed] [Google Scholar]
- 29.Sunohara T., Jojima K., Tagami H., Inada T., Aiba H. (2004) J. Biol. Chem. 279, 15368–15375 [DOI] [PubMed] [Google Scholar]
- 30.Nurizzo D., Shewry S. C., Perlin M. H., Brown S. A., Dholakia J. N., Fuchs R. L., Deva T., Baker E. N., Smith C. A. (2003) J. Mol. Biol. 327, 491–506 [DOI] [PubMed] [Google Scholar]
- 31.Roche E. D., Sauer R. T. (1999) EMBO J. 18, 4579–4589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X., Hirano R., Tagami H., Aiba H. (2006) RNA 12, 248–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dong H., Nilsson L., Kurland C. G. (1996) J. Mol. Biol. 260, 649–663 [DOI] [PubMed] [Google Scholar]
- 34.Craigen W. J., Caskey C. T. (1986) Nature 322, 273–275 [DOI] [PubMed] [Google Scholar]
- 35.Poole E. S., Brown C. M., Tate W. P. (1995) EMBO J. 14, 151–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gurvich O. L., Baranov P. V., Zhou J., Hammer A. W., Gesteland R. F., Atkins J. F. (2003) EMBO J. 22, 5941–5950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mottagui-Tabar S., Björnsson A., Isaksson L. A. (1994) EMBO J. 13, 249–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yap M. N., Bernstein H. D. (2009) Mol. Cell 34, 201–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pavlov M. Y., Watts R. E., Tan Z., Cornish V. W., Ehrenberg M., Forster A. C. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 50–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doronina V. A., Wu C., de Felipe P., Sachs M. S., Ryan M. D., Brown J. D. (2008) Mol. Cell. Biol. 28, 4227–4239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beringer M. (2008) RNA 14, 795–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Woolhead C. A., Johnson A. E., Bernstein H. D. (2006) Mol. Cell 22, 587–598 [DOI] [PubMed] [Google Scholar]
- 43.Collier J., Bohn C., Bouloc P. (2004) J. Biol. Chem. 279, 54193–54201 [DOI] [PubMed] [Google Scholar]
- 44.Muto H., Ito K. (2008) Biochem. Biophys. Res. Commun. 366, 1043–1047 [DOI] [PubMed] [Google Scholar]
- 45.Wohlgemuth I., Brenner S., Beringer M., Rodnina M. V. (2008) J. Biol. Chem. 283, 32229–32235 [DOI] [PubMed] [Google Scholar]
- 46.Hentzen D., Mandel P., Garel J. P. (1972) Biochim. Biophys. Acta 281, 228–232 [DOI] [PubMed] [Google Scholar]
- 47.Janzen D. M., Frolova L., Geballe A. P. (2002) Mol. Cell. Biol. 22, 8562–8570 [DOI] [PMC free article] [PubMed] [Google Scholar]