Abstract
Mutations in the homeobox transcription factor PITX2 result in Axenfeld-Rieger syndrome (ARS), which is associated with anterior segment dysgenesis and an increased risk of glaucoma. To understand the pathogenesis of the defects resulting from PITX2 mutations, it is essential to know the normal functions of PITX2 and its interaction with the network of proteins in the eye. Yeast two-hybrid screening was performed using a cDNA library from a human trabecular meshwork primary cell line to detect novel PITX2-interacting proteins and study their role in ARS pathogenesis. After screening of ∼1 × 106 clones, one putative interacting protein was identified named PRKC apoptosis WT1 regulator (PAWR). This interaction was further confirmed by retransformation assay in yeast cells as well as co-immunoprecipitation in ocular cells and nickel pulldown assay in vitro. PAWR is reportedly a proapoptotic protein capable of selectively inducing apoptosis primarily in cancer cells. Our analysis indicates that the homeodomain and the adjacent inhibitory domain in PITX2 interact with the C-terminal leucine zipper domain of PAWR. Endogenous PAWR and PITX2 were found to be located in the nucleus of ocular cells and to co-localize in the mesenchyme of the iridocorneal angle of the developing mouse eye, consistent with a role in the development of the anterior segment of the eye. PAWR was also found to inhibit PITX2 transcriptional activity in ocular cells. These data suggest PAWR is a novel PITX2-interacting protein that regulates PITX2 activity in ocular cells. This information sheds new light in understanding ARS and associated glaucoma pathogenesis.
Introduction
Pituitary homeobox transcription factor 2 (PITX2)2 is a member of the paired-bicoid family of homeodomain (HD) transcription factors. Members of pituitary homeobox proteins are actively involved in a wide range of developmental processes including formation of pituitary gland and hind limb and of anterior segment of the eye as well as brain morphogenesis (1–3). Expression of PITX2 is found during ocular development (4, 5). The PITX2 gene is represented by several different splicing and transcriptional isoforms. The four best characterized are PITX2A, -B, -C (5), and -D (6) These isoforms of PITX2 vary in their N termini but share common HD and C-terminal sequences. The PITX2D isoform, however, has a truncated and non-functional HD (7). These alternate transcripts (A, B, and C) encode 271, 317, and 324 amino acids, respectively (4, 8).
Numerous pathologic PITX2 mutations including missense variations, splice site alterations, and insertions/deletions have been described, producing a continuum of clinical phenotypes including Axenfeld-Rieger syndrome (ARS), iridogoniodysgenesis, and iris hypoplasia (5, 9, 10), as well as rarer cases of a Peters-like anomaly (11). Of these, ∼82% of PITX2 mutations were observed in ARS, ∼7% were observed both in iridogoniodysgenesis and in iris hypoplasia, and less than 3% were observed in Peters-like anomaly (Human Gene Mutation Database (HGMD)). ARS itself is genetically heterogeneous as Lines et al. (12) identified 39 ARS patients with mutations in PITX2 from a cohort of 91 patients having anterior segment dysgenesis and/or glaucoma. In fact, more than 50% of ARS patients do not have mutations in any of the defined loci and genes for ARS (13). ARS comprises a group of autosomal dominant clinical disorders affecting anterior eye structures derived from constituents of the embryonic neural crest (14). Classic ocular features of ARS include iridocorneal synechiae, iris hypoplasia, corectopia, polycoria, and/or prominent Schwalbe line (15, 16). Malformation of the anterior angle between iris and cornea can lead to elevated intraocular pressure and subsequent glaucomatous condition. Approximately 50% of ARS patients develop glaucoma with a great variability in age of onset, but usually in the teens (14). Systemic manifestations of ARS often include mild craniofacial dysmorphism, dental defects, and/or excessive preumbilical skin (17). Also, in rare ARS cases, congenital cardiac defects and/or hearing loss have been observed (18). Generation of mouse models lacking either one (PITX2+/−) or both (PITX2−/−) alleles caused a pleiotropic murine phenotype that overlaps with the PITX2 expression pattern and provided a good animal model for human ARS (19).
Functional analysis of different disease-causing mutants of PITX2 has been done to study the effect of mutations on protein stability (20), DNA binding capacity, transcriptional activation abilities, and subcellular localization. Moreover, mutation in the HD of PITX2 showed a shift from nuclear localization of PITX2 found normally in cells (21). The PITX2 homeobox transcription factor is part of a large network of gene regulation, which is only partially characterized at present. Mutations in FOXC1, encoding a forkhead box transcription, can also underlie ARS (22, 23). Recent work from our laboratory indicates that PITX2 and FOXC1 physically interact with each other, not only establishing both proteins into a common pathway but also showing PITX2 as a negative regulator of FOXC1 (24). Therefore, analyses of protein-protein interaction are very effective and necessary to understand the large network of gene regulation for transcription factors such as PITX2, which are involved in multiple developmental processes.
In this report, we discovered that human PRKC apoptosis WT1 regulator (PAWR) protein is a novel PITX2-interacting protein through a yeast two-hybrid (Y2H) screening. This interaction reveals a new level of regulation of PITX2 activity that connects PITX2 to apoptosis pathways in the eye.
EXPERIMENTAL PROCEDURES
Plasmid Constructs
PITX2C was cloned into the pCI mammalian expression vector as described previously (20). This pCI-PITX2C construct was used as a template for PCR of the open reading frame of PITX2C using PITX2C attB1 (5′-GGGG ACA AGT TTG TAC AAA AAA GCA GGC TTC ATG AAC TGC ATG AAA GGC C-3′) and PITX2C attB2 (5′-GGGG AC CAC TTT GTA CAA GAA AGC TGG GTA CAC GGG CCG GTC CAC TG-3′), where the underline indicates the attb1 and attb2 plasmid recombination sites in the designed primers. The PCR product containing attb1 and attB2 sites was subsequently used for recombination with pDONR (Invitrogen) to subclone PITX2C in pDONR with BP recombination kit (Invitrogen). PITX2C was further subcloned from pDONR-PITX2C into pDEST32 in-frame to the GAL4DBD by Gateway technology (Invitrogen). Human PAWR cDNA clone was purchased from Open Biosystems in pDNR vector. The open reading frame of human PAWR was amplified by PCR with 5% dimethyl sulfoxide (DMSO) using PAWR-EcoRI-2F (5′-GAA TTC ATG GCG ACC GGT GGC TAC C-3′) and PAWR-XbaI-2R (5′-TCT AGA CTA GGT CAG CTG ACC-3′), where the underline indicates restriction enzymes. The amplified PAWR open reading frame was subcloned into pcDNA4c/His Max mammalian vector and pET28a bacterial expression vector in-frame to the His6 tag. The PITX2A and its deletion clones in pCI mammalian expression vector were described previously (24). Generation of three PAWR deletion clones was done by PCR using PAWR-EcoRI-Δ1–70-F (5′-GAA TTC AAC AAC CTC CCG GGC GGC GC-3′), PAWR-EcoRI-Δ1–161-F (5′-GAA TTC TCC ACC GGC GTG GTC AAC ATC-3′), and PAWR-EcoRI-Δ1–267-F (5′-GAA TTC CTG GAA AAG AAA ATT GAA GA-3′), where the underline indicates restriction enzymes. In all cases, PAWR-XbaI-2R was used as reverse primer. Amplified products were subcloned in pET28a bacterial expression vector, and Δ1–267-PAWR was further subcloned in pcDNA4c mammalian expression vector. All vectors were sequenced to confirm that no mutations were introduced into the cDNAs. Five PITX2A HD (T68P, V83L, K88E, R90C, and R91P) and two outside the HD (L105V, N108T) mutants in pcDNA4 were subcloned in pCI to study their interaction with PAWR.
Y2H Screening
A human trabecular meshwork cDNA library fused to the GAL4AD of pEXP-AD502 (Invitrogen) was screened for proteins that interact with human PITX2C, using the ProQuest two-hybrid system (Invitrogen). The detailed method of yeast two-hybrid screening was already described previously (25).
Mammalian Cell Culture and Transfection
Maintenance of human trabecular meshwork (HTM) cells was described previously (25). SW480 cells were maintained in RPMI (Invitrogen) with 10% fetal bovine serum at 37 °C. Cells were transfected either by FuGENE (Roche Diagnostics) or by Trans IT-LT1 (Mirrus Bio) according to manufacturer's protocol. Transfected cells were subjected to immunofluorescence after 24 h and luciferase assay after 48 h of transfection.
Nickel Pulldown Assay
In Escherichia coli, His6-tagged PAWR and/or its deletion fragments were generated using pET28 based constructs containing inducible lac operator sequence. One μm isopropyl β-d-1-thiogalactopyranoside was used to induce production of the desired proteins in bacterial cells. The proteins were purified using Ni-NTA-agarose beads (Qiagen). Whole HTM cell lysates containing different PITX2 constructs were prepared using lysis buffer described previously (25) and measured thereafter by protein assay reagent (BioRad). Ni2+-agarose assay was done according to the protocol described earlier (25). Protein complexes captured on the beads were eluted in SDS-PAGE loading buffer, separated by 12% SDS-PAGE, and subjected to immunoblot analysis with antibody against the mammalian-expressed proteins.
Immunoprecipitation
Protein-G-agarose beads (Sigma) were subjected three times to 1× phosphate-buffered saline wash followed by 1-h incubation in 1% bovine serum albumin in phosphate-buffered saline at 4 °C. After incubation, the beads were centrifuged at 1000 × g and kept at 4 °C with an equal amount of radioimmune precipitation buffer as described earlier (24). HTM cells were transfected with PITX2-expressing mammalian expression vector (pcDNA-PITX2) or the empty vector (pcDNA). The cells were lysed after 48 h of transfection in the lysis buffer described previously (25). Three hundred μg of protein lysates were added into 1 ml of radioimmune precipitation buffer with a 25 μl-bed volume of bovine serum albumin-treated protein-G-agarose beads described before. The lysate was thus precleared for 1 h at 4 °C. The precleared cell lysate was incubated with 1 μg of anti-Xpress antibody overnight at 4 °C with continuous rotation. A 25-μl bed volume of bovine serum albumin-treated protein-G-agarose beads was added to the antibody-treated cell lysate and incubated for 1.5 h with continuous rotation at 4 °C followed by four washes with radioimmune precipitation buffer. Protein complexes captured on the beads were eluted in SDS-PAGE loading buffer, separated by 12% SDS-PAGE, and subjected to immunoblot analysis with antibody against human PAWR (Abcam). The input fraction represented 5% of the protein extract used for immunoprecipitation. For immunoprecipitation experiments using both PITX2 and PAWR endogenous proteins (see Fig. 1D), the input represented 10% of the protein extract used for experiment and immunoblot analysis was done using antibody against human PAWR (Abcam) and human PITX2 (Abnova).
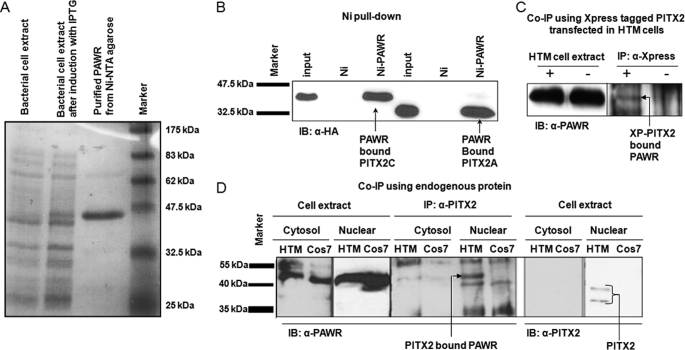
FIGURE 1.
Confirmation of interaction between PITX2 and PAWR. A, Coomassie Blue staining of 12% polyacrylamide gel showing expression of His6-tagged PAWR in bacterial cell upon induction with isopropyl β-d-1-thiogalactopyranoside (IPTG) followed by purification with Ni2+-agarose beads. B, HTM cell lysates transfected with HA-tagged PITX2A and PITX2C, were subjected to nickel pulldown assays with the His6-tagged PAWR bound to the Ni2+-NTA beads (Ni-PAWR) or the empty beads (Ni). Bound proteins were analyzed by SDS-PAGE followed by Western blot (IB) analysis using an anti-HA antibody. The inputs represent 3% of the total reaction. C, co-immunoprecipitation (Co-IP) of PITX2 and PAWR in HTM cells. pcDNA with Xpress-PITX2 (XP) (+) or empty pcDNA (−) plasmids were transfected into HTM cells. The cell lysates were immunoprecipitated with an anti-Xpress antibody and immunoblotted with an anti-PAWR antibody. The inputs represent 5% of the total reaction. D, co-immunoprecipitation of endogenous PITX2 and PAWR in HTM cells. Here both for cytosolic and for nuclear fractions, inputs represent 10% of the total reaction.
Immunofluorescence
Coverslips containing HTM, retinal ganglion cells (RGC5), non-pigmented ciliary epithelium (NPCE), and SW480 cells were subjected to fixation 24 h after transfection with Xpress-tagged pcDNA-PITX2 constructs. The methods of fixation, blocking, and antibody treatment were as described earlier (25) with slight modification to antibody concentration. The primary antibody to Xpress epitope was used in 1:500 in phosphate-buffered saline (pH 7.4) with 0.05% Triton X-100 containing 5% bovine serum albumin, whereas the primary antibody against human PAWR was used in 1:100 in the same solution described above. The images were collected using a Leica DMR immunofluorescence microscope. The no-PAWR primary antibody controls for these experiments were described in supplemental Fig. 3.
Reporter Transactivation Assay
HTM and/or SW480 cells were cultured on a 24-well tissue culture plates (4 × 104/15-mm well, 24-well plate) followed by transfection with 160 ng of PITX2 and/or PAWR mammalian expression plasmids and appropriate empty vector along with 60 ng of pGL3-bicoidBS-TK reporter (26) and 60 ng of pCMVβ transfection control vector. After 48 h of transfection, cells were harvested with 100 μl of passive lysis buffer (Promega). Subsequently, firefly luciferase activity was measured by luminometry (Turner Designs, Sunnyvale, CA) from 10 μl of protein lysate mixed with 100 μl of luciferase assay reagent (Promega), and standardized to the β-galactosidase (internal control) activity quantitated by the β-galactosidase enzyme assay system (Promega) from 75 μl of protein lysate.
Electrophoretic Mobility Shift Assay (EMSA)
Whole cell extracts from untransfected and HA-PITX2-transfected COS7 cells were treated with 32P-labeled double-stranded DNA probe containing the bicoid binding site as described previously (20). Prior to EMSA incubation and PAGE analysis, two samples were treated with increasing concentrations of purified PAWR (2 and 4 μg), eluted from Ni2+ beads.
Timed Pregnancies and Immunohistochemistry
Timed pregnancies were produced by mating C57BL/6J male and female mice from our existing colony described earlier (24) All procedures using mice were approved by the University of Michigan Committee on Use and Care of Animals and were conducted in accordance with the principles and procedures outlined in the National Institutes of Health Guidelines for the Care and Use of Experimental Animals. Embryos were fixed for 2–4 h with 4% paraformaldehyde in phosphate-buffered saline, washed and dehydrated, and embedded into paraffin. Mounted sections were deparaffinized and treated for antigen retrieval by boiling for 10 min in the citrate buffer (pH 6.0). Immunostaining was performed using standard methods. Briefly, sections were incubated with antibodies directed against PITX2 (27) and PAWR (Abcam) followed by biotinylated species-specific secondary antibodies (Jackson ImmunoResearch Laboratories). Signals were detected using tyramide signal amplification kits (PerkinElmer Life Sciences). The blocking efficiency between stains has been shown by no-PAWR primary antibody control. The lack of red stain proved that suppression of PAWR signal in between was effective, although both anti-PITX2 and anti-PAWR antibodies were raised in rabbit (supplemental Fig. 2).
RESULTS
Detection of PAWR as PITX2-interacting Protein Using Y2H Screening
A trabecular meshwork (TM) cDNA yeast two-hybrid library, created from mRNA extracted from human TM primary cell culture, was used to identify proteins that interacts with PITX2C. The TM cDNA library inserts were cloned into the plasmid pEXP-AD502 with the open reading frames fused to the GAL4 activation domain (GAL4AD) (25). Full-length PITX2C was amplified and recombined into pDONR using the BP recombination reaction protocol (Invitrogen) followed by cloning into pDEST32 fused to the GAL4 DNA binding domain (GAL4DB). Yeast cells containing three reporter genes (HIS3, URA3, and lacZ) were co-transformed with the GAL4AD-cDNA library and the GAL4DB-PITX2C plasmid.
A total of 1 × 106 clones (human TM cDNA Y2H library) were screened using PITX2C as ”bait,“ and 16 independent clones were obtained that fulfilled the criteria of interaction between gene products (supplemental Table 1). Isolation of plasmids followed by partial sequencing using vector specific primer was done for all 16 clones. Out of these 16 clones, 12 were found to match with EFEMP2, another PITX2-interacting protein (PIP), previously found in our laboratory.3 Two clones were found to be false-positive, whereas the remaining two were human PAWR cDNA sequences lacking the first 210 bp encoding the N-terminal 70 amino acids. The specificity of the interaction between PAWR and PITX2C in the yeast two-hybrid system was confirmed by retransformation of the positive cDNA clone into yeast cells, together with vectors expressing GAL4 DNA binding domain alone or with GAL4DB-PITX2C. Yeast cells containing that positive cDNA plasmid and the GAL4DB-PITX2C plasmid displayed an interaction phenotype. The empty GAL4DB was found to be negative in retransformation assay (supplemental Fig. 1).
PAWR Interacts with Both PITX2C and PITX2A Isoforms in Vitro
Interaction of PAWR and PITX2C was further confirmed by Ni2+-pulldown assay. Full-length PAWR was cloned into a bacterial expression vector (pET28a) that allowed expression of PAWR as a His6-tagged fusion protein in bacteria (E. coli). Incubation of bacterial extract with Ni-NTA-agarose beads bound His6-tagged PAWR to the column (Fig. 1A). Next, whole cell lysate prepared from HTM cells, transfected with either HA epitope-tagged PITX2C or HA epitope-tagged PITX2A (pCI vector), was incubated with either empty Ni2+ beads or Ni2+ beads containing His6-tagged PAWR fusion protein. Further, immunoblot using anti-HA antibody showed the presence of PITX2C or PITX2A in Ni2+ beads with His6-tagged PAWR (Fig. 1B).
PITX2 Interacts with PAWR in Vivo
The interaction between PITX2 and endogenous PAWR was further confirmed by immunoprecipitation experiments where HTM cells were first transfected with either Xpress-tagged PITX2 (pcDNA vector) or the empty pcDNA4 vector. HTM cells transfected with empty pcDNA4 vector served as control. Immunoprecipitation using an anti-Xpress antibody (Invitrogen) to XP-PITX2 followed by immunoblotting using the anti-PAWR antibody (Abcam) resulted in immunoprecipitated PAWR in cells transfected with Xpress-tagged PITX2 (Fig. 1C).
In another experiment, the nuclear and cytosolic fractions of untransfected HTM and COS7 cells were first tested for the presence of endogenous PITX2 and PAWR using anti-PITX2 (Abnova) and anti-PAWR (Abcam) antibody. The presence of both endogenous PITX2A and C isoforms are detected only in the nuclear fraction of HTM cells at low levels, whereas PAWR is present in both the cytosolic and the nuclear fractions of HTM and COS7 cells (Fig. 1D). The nuclear and cytosolic fractions of HTM and COS7 cells were immunoprecipitated further with previously described anti-PITX2 antibody and immunoblotted with anti-PAWR antibody. The PITX2-PAWR interaction is detected only in the nuclear fraction of HTM cells (Fig. 1D).
PITX2A Interacts through HD with PAWR
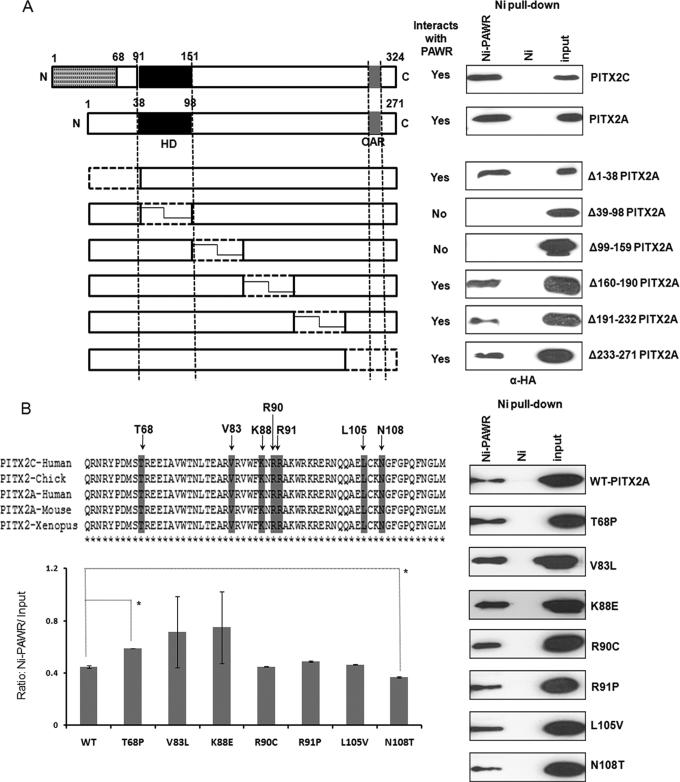
Ni2+ pulldown assays were performed again to identify the specific region of PITX2 that interacts with PAWR. Deletion constructs of PITX2 (pCI-Ha) (24) were transfected into HTM cells along with wild-type PITX2A and PITX2C (Fig. 2A). HTM cells transfected with these constructs were subjected to cell lysis followed by incubation with Ni2+ beads containing His6-tagged PAWR and subsequent PAGE and immunoblot. Except for the Δ39–98 and Δ99–159 PITX2 constructs, which lack the HD and adjacent inhibitory domain (20), respectively, all other constructs detected HA-PITX2 in the immunoblot using anti-HA antibody (Fig. 2A). These data suggested that PITX2 interacts with PAWR through its HD and adjacent C-terminal inhibitory domain.
FIGURE 2.
Identification of regions in PITX2 that interacts with PAWR. A, nickel pulldown results where HTM cells transfected with six different deletion constructs (Δ1–38, Δ39–98, Δ99–159, Δ160–190, Δ191–232, and Δ233–271) named after N-terminal locations of amino acids deleted according to the A isoform of PITX2. Thus, Δ1–38 lacks first 38 amino acids of PITX2A, and so on. Subsequent nickel pulldown assays were performed with the HTM cell lysates having different deletion constructs of PITX2 and His6-tagged PAWR bound to the Ni2+-NTA beads (Ni-PAWR) or the empty beads (Ni). Bound proteins were analyzed by SDS-PAGE followed by Western blot analysis using an anti-HA antibody. N, N terminus; C, C terminus; HD, homeodomain; OAR, orthopedia and aristaless domain. B, nickel pulldown results described above but with HTM cell lysates having seven PITX2A mutant (T68P, V83L, K88E, R90C, R91P, L105V and N108T) constructs in HA-tagged pCI vector. In both cases, inputs represent 3% of the total reaction. The upper left panel of B shows the location of mutations in conserved homeodomain and the adjacent inhibitory domain, whereas the lower left panel shows Ni-PAWR/input ratio derived from quantification of immunoblots shown in the right panel. The inputs in these experiments represent 3% of the total reaction in the form of HTM cell extracts transfected with HA-tagged PITX2A wild type (WT) and mutants. Error bars indicate S.D., whereas the asterisks indicate statistical significance in the Ni-PAWR/input ratio between WT and T68P or N108T.
ARS Mutations Residing in the PITX2 HD and Adjacent C-terminal Inhibitory Domain Have No Effect on the Interaction with PAWR
The effects of ARS-causing PITX2 mutants located in the HD and adjacent C-terminal inhibitory domain on its interaction with PAWR were determined because these regions in PITX2 interact with PAWR. HTM cells were transfected with seven PITX2 mutant constructs (pCI-HA). Five (T68P, V83L, K88E, R90C, and R91P) PITX2 HD mutations and two (L105V and N108T) inhibitory domain mutations were analyzed. Ni2+ pulldown assays were performed as described above. All mutant proteins were observed to interact with Ni2+ beads containing His6-tagged PAWR (Fig. 2B). The immunoblots were further quantified to assess the variability of PITX2 mutants binding with PAWR (Fig. 2B). Two mutants, V83L and K88E, displayed the most variability in binding to PAWR in three independent experiments. These mutants however, were not different from wild-type PITX2 in binding with PAWR on average. Two mutants displayed a significant difference in binding to PAWR. Significantly more T68P bound to PAWR than wild-type PITX2, whereas significantly less N108T bound to PAWR.
The C-terminal Leucine Zipper Domain of PAWR Interacts Both with PITX2A and with PITX2C
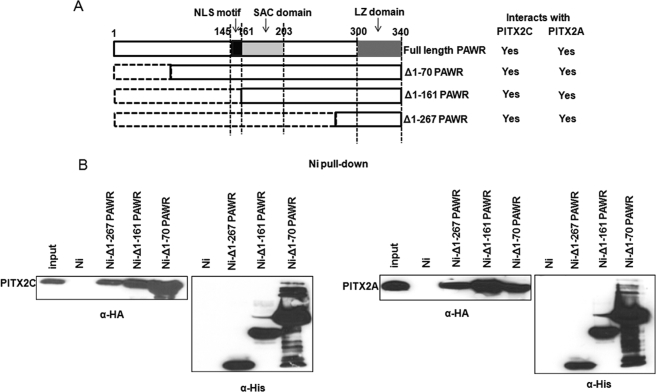
Three deletion constructs of PAWR were created to identify the region of PAWR that interacts with PITX2. These deletion constructs (Δ1–70, Δ1–161, and Δ1–267) in the pET28 vector were transformed independently in E. coli cells. His6-tagged fragments were then purified using Ni-NTA-agarose beads. Subsequently, whole cell lysates were prepared from HTM cells transfected with HA-tagged PITX2A or PITX2C and subjected to Ni2+ pulldown assay. Specific PITX2A and PITX2C bands were detected in immunoblots treated with anti-HA antibody after incubation with all three deletion fragments of PAWR (Fig. 3B). Duplicate immunoblots of the same experiment, but treated with an anti-His antibody to detect recombinant PAWR, showed the expression of correctly sized fragments of PAWR in E. coli cells (Fig. 3B). This experiment indicated that the shortest PAWR fragment containing only the C-terminal leucine zipper domain is sufficient to interact with both A and C isoforms of PITX2.
FIGURE 3.
Identification of region in PAWR that interacts with PITX2. A, schematic showing different regions and domains in PAWR and the three N-terminally deleted serial constructs (Δ1–70, Δ1–161, and Δ1–267). NLS, nuclear localization sequence; SAC, selective apoptosis in cancer. B, HTM cell lysates transfected with HA-tagged PITX2A and PITX2C were subjected to nickel pulldown assays with the His6-tagged PAWR fragments bound to the Ni2+-NTA beads (Ni-Δ1–70 PAWR, Ni-Δ1–161 PAWR, and Ni-Δ1–267 PAWR) or the empty beads (Ni). For each deletion fragment of PAWR, samples were analyzed in 12% PAGE in duplicates followed by Western blot against anti-HA to detect bound PITX2C and PITX2A as well as against anti-HIS antibody to show the different deletion fragments of PAWR expressed in E. coli cells. The inputs represent 3% of the total reaction.
PAWR Co-localizes with PITX2 in Nucleus
Immufluorescence experiments were done in untransfected HTM and RGC5. Immunofluorescence has also been done in HTM, RGC5, NPCE, and SW480 cells transfected with Xpress-tagged PITX2 to detect the distribution of PITX2 and endogenous PAWR in subcellular compartments. HTM and NPCE are cells of ocular origin (trabecular meshwork and ciliary body, respectively), whereas SW480 is a colon adenocarcinoma cell line. Because PAWR has been reported to be involved in various apoptotic and tumorigenesis processes and to be localized to the nucleus in cancer cells (28), we included SW480 cells as positive control. Cells were harvested 24 h after transfection and treated with both an anti-Xpress antibody to recombinant PITX2 and an anti-PAWR antibody to see the localization of PITX2 and PAWR in cells. Detection of Xpress-tagged PITX2 was done subsequently by anti-mouse IgG coupled with CY3 fluorescent dye (Fig. 4, red), whereas endogenous PAWR was detected by anti-rabbit IgG coupled with CY2 (green). Cells were also treated with DAPI (blue) to stain their nucleus. These data clearly showed that endogenous PAWR is localized in nucleus of untransfected HTM and RGC5 cells (Fig. 4A) and showed co-localization of endogenous PAWR and Xpress-tagged PITX2 in the nucleus of all cell types (Fig. 4B).
FIGURE 4.
Nuclear localization of PITX2 and PAWR. A and B, untransfected HTM and RGC cells (A) and HTM, RGC, NPCE, and SW480 cells (B) were transfected with Xpress-PITX2 expression vector. Endogenous PAWR (A and B) was stained with rabbit polyclonal anti-PAWR antibody followed by Cy2-conjugated anti-rabbit secondary antibody (green). Xpress-PITX2 (B) was visualized with mouse monoclonal anti-Xpress antibody followed by Cy3-conjugated mouse secondary antibody (red). All cells were stained with DAPI (blue) to detect the nucleus in cells (A and B). The merge without or with DAPI shows an overlay of red and green or red, green, and blue staining, respectively. PITX2 and PAWR are co-localized in nuclei of all cell types.
PAWR Inhibits PITX2-mediated Transcription Activation
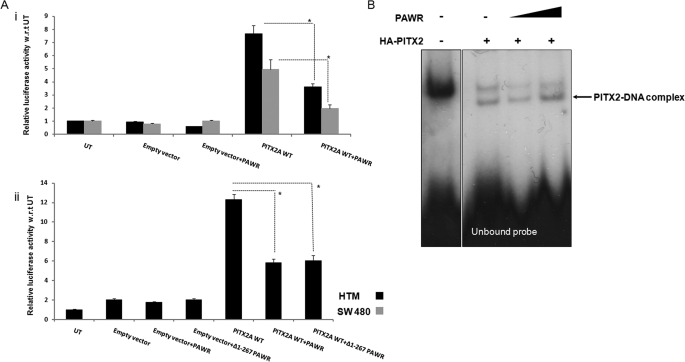
The role of PAWR in regulating PITX2 transcriptional activity was studied further by luciferase assays where a luciferase reporter driven by a TK promoter containing a PITX2 binding site (bicoidBS) upstream was used to measure PITX2 transcriptional activity. Cells expressing both PITX2 and PAWR showed more than 50% reduced PITX2 transcriptional activity than cells expressing PITX2 alone. This was observed both in HTM and in SW480 cells, and the p values are significant in both cases (HTM p = 0.003, SW480 p = 0.007) (Fig. 5A, panel i). To understand whether the C-terminal leucine zipper domain in PAWR that interacts with PITX2 is sufficient alone to decrease PITX2 activity, we subcloned the smallest deletion fragment (Δ1–267) of PAWR from pET28 vector to the mammalian expression vector (pcDNA4). The smallest fragment (Δ1–267) of PAWR (p = 0.001) was sufficient to significantly reduce PITX2 activity as did full-length PAWR (Fig. 5A, panel ii).
FIGURE 5.
PAWR mediated impairment of transcriptional activation of PITX2. A, panel i, HTM and SW480 cells were transfected with TK-bicoidBS luciferase promoter along with empty pcDNA4 or PITX2 expression vector alone or both PITX2 and PAWR expression vector. In all cases, pCMV-β-galactosidase-expressing plasmid has been taken as transfection control. Luciferase activity was measured followed by normalizing it with β-galactosidase activity, and bars were made according to relative luciferase activity along the y axis considering untransfected (UT) as 1. WT, wild type. A, panel ii, in HTM, Δ1–267 PAWR construct was used in a similar fashion to measure luciferase activity. The broken lines and asterisks designate their statistical significance (Student's t test, A, panel i, HTM p = 0.003; SW480 p = 0.007; A, panel ii, PITX2 versus PITX2 + PAWR p = 0.0006, PITX2 versus PITX2+ Δ1–267 PAWR p = 0.001). Error bars indicate S.D. B, electrophoretic mobility shift assays of whole cell lysates from transfected COS7 cells were performed using radiolabeled bicoidBS probe. Untransfected COS7 lysate is shown as a negative control exhibiting a single low mobility shifted ”background“ band. Complexes of radiolabeled bicoidBX probe bound to PITX2A showed shifted bands in the middle of the autoradiograph, with unbound probe migrating to the bottom. The last two lanes from the right were treated with increasing concentrations of purified PAWR from Ni2+ beads.
In an EMSA, lysates from COS7 cells transfected with wild-type HA-PITX2A showed a migration pattern of two bands representing PITX2A protein bound to the bicoidBS (TAATCC) DNA element (Fig. 5B). Inclusion of increasing concentrations of purified PAWR from Ni2+ beads in these EMSAs, however, displayed no difference in the band pattern (Fig. 5B). These data suggest that reduction of PITX2 transcriptional activity by PAWR is not through impairment of PITX2 DNA binding ability.
PITX2 and PAWR Co-localize in the Developing Murine Eye
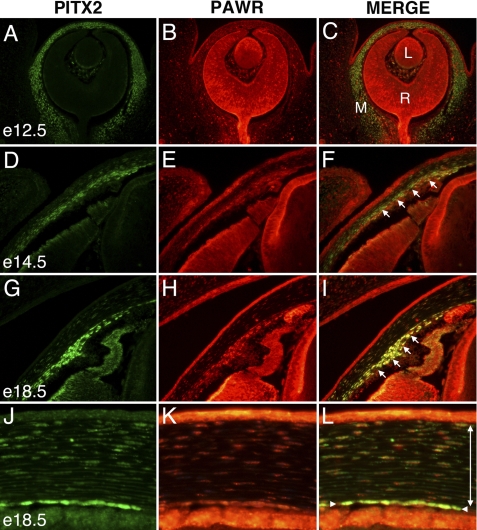
Although the expression pattern of PITX2 during eye development has been described (4), neither the expression of PAWR nor the potential co-expression of PITX2 and PAWR during eye development has been previously examined. Therefore, we used immunohistochemistry to simultaneously detect expression of PITX2 and PAWR during embryonic eye development. PITX2 is expressed throughout the periocular mesenchyme by e12.5 (Fig. 6A); expression continues at subsequent stages in emerging anterior segment structures, including the iridocorneal angle, corneal stroma, and corneal endothelium (Fig. 6, D, G, and J). PAWR is ubiquitously expressed at all time points examined (Fig. 6, B, E, H, and K). The subcellular localization is usually nuclear, as has been previously reported in cell culture (29) and in the present study, but in some structures, cytoplasmic expression is also observed (data not shown). Expression within the periocular mesenchyme was nuclear at all time points examined. Co-expression of PITX2 and PAWR is prevalent throughout the periocular mesenchyme and is particularly apparent within the mesenchyme of the presumptive iridocorneal angle, cells fated to contribute the ciliary muscles and stroma, trabecular meshwork, Schlemm canal, and iris stroma (Fig. 6, F and I). Co-expression is also prominent within the mesenchymally derived corneal stroma and corneal endothelium (Fig. 6L).
FIGURE 6.
Expresson of PITX2 and PAWR during eye development. Transverse views of an e12.5 eye (A–C), iridocorneal angles from e14.5 (D–F), and e18.5 (G–I) eyes and central cornea from e18.5 eye (J–L) are shown. Sections were co-immunostained to detect PITX2 (green) and PAWR (red). L, lens; R, retina; M, periocular mesenchyme. Single-headed arrows (F and I) indicate mesenchyme within the emerging iridocorneal angle; the single-headed arrow in L denotes the corneal endothelium, and the double-headed arrow spans the corneal stroma.
DISCUSSION
PITX2 was identified as the first candidate gene to cause ARS (5). However, knowledge of exactly how PITX2 mutations disrupt eye development leading to ARS is still lacking. Identification and characterization of new PIPs is therefore necessary to better understand the PITX2 regulatory network in the eye. In this report, we identified PAWR as a novel PIP by Y2H screening of the human trabecular meshwork cDNA library with PITX2C. Because PITX2C is the longest of the four well characterized PITX2 isoforms, we performed Y2H screening with PITX2C. Moreover, the PITX2 antibody we used for downstream experiments can detect both endogenous PITX2A isoforms and endogenous C isoforms. We later confirmed the PAWR protein-protein interaction with both the PITX2A and the C isoforms (Fig. 1, B and D). Also, our subsequent experiments proved that the PAWR interaction domain in PITX2 is common to both PITX2 A and PITX2 C isoforms (Fig. 2A).
Human PAWR is a 340-amino acid protein alternatively known as prostate apoptotic response-4 (PAR-4). The PAWR gene is ∼99 kb long and is located on chromosome 12q21 (30) with seven exons and a transcript length of 1.9 kb. Human PAR-4 was first identified in prostate cancer cells undergoing apoptosis in response to an exogenous insult (31). PAR-4 was later rediscovered in human cells as a regulator of the Wilms tumor 1 (WT1) (32), thus named as PAWR. Human PAWR is a unique proapoptotic protein that selectively induces apoptosis in cancer cells, sensitizes cells to the action of multiple apoptotic stimuli, and causes tumor regression (33) through a region harboring 59-amino acid residue starting from 145 to 203 (Universal Protein Resource (UniProt)). This region is known as selective apoptosis in cancer (SAC) cells. Other than the SAC region, PAWR has a 17-amino acid long nuclear localization sequence encompassing between the 145th and the 161st amino acid residue (Swiss-Prot accession number Q96IZ0). The best described region of PAWR is, however, the 41-amino acid long leucine zipper (LZ) domain, situated at the C-terminal end starting from the 300th to the last (340th) amino acid residue (Fig. 3A). Human PAWR interacts with numerous proteins such as WT1 (32), atypical protein kinase C (aPKC) (34), and death-associated protein-like kinase (Dlk) (35) through this LZ domain.
We performed in silico analyses to determine the expression of PAWR and PITX2. An online unification tool, The SOURCE (available for the Stanford University web site), was used and found normalized PITX2 expression in 21 different tissues, whereas 32 different tissues express PAWR. Nineteen out of 32 tissues, including the eye, heart, kidney, lung, and brain, co-expressed PITX2 and PAWR (supplemental Table 2). Co-expression of PITX2 and PAWR has also been found in embryonic tissue. We further narrowed down our search by restricting it to different eye compartments and tissues using dbEST and NEIBank. PITX2 and PAWR are co-expressed in multiple human ocular tissues including trabecular meshwork, lens, optic nerve, retinal pigment epithelium/choroid, anterior segment, ciliary body, and retina. Expressed sequence tag clones have also been found in human fetal eye for both PITX2 and PAWR, which suggests their expression during ocular development (supplemental Table 3).
We confirmed the interaction of PITX2 and PAWR in vitro (Fig. 1B) as well as in ocular cells (Fig. 1, C and D). Previous investigations have focused on studying the role of PAWR in cancer cells (28). This is the first report investigating the role of PAWR in ocular cells and in the developing mouse eye. Interestingly, PAWR is localized to the nucleus of human ocular cells (Fig. 4A) and localizes with PITX2 (Fig. 4B). In the developing mouse eye, we, for the first time, detected expression of PAWR in lens and retina at early embryonic stage (e12.5, Fig. 6B) and co-localized with PITX2 in the periocular mesenchyme and corneal endothelial cells and stroma at the later stages of development (e14.5 and e18.5). Co-localization of PAWR and PITX2 within the mesenchyme of the presumptive iridocorneal angle at e14.5 and e18.5 stages of development suggests the role of PAWR in the development of the anterior segment of the eye (Fig. 6, F and I).
Furthermore, we identified that PAWR interacts with PITX2 through its C-terminal LZ domain (Fig. 3B); the same region of PAWR is necessary to interact with WT1 and Dlk3. Because PAWR interacts with PITX2 through the PITX2 homeodomain and adjacent C-terminal inhibitory domain (Fig. 2A), we determined whether PITX2 mutations in these regions alter the ability of PITX2 to interact with PAWR. Of these seven PITX2 mutations tested (T68P, V83L, K88E, R90C, R91P, L105V, and N108T), two (T68P and N108T) had significantly altered capacity to interact with PAWR (Fig. 2B). These data are consistent with the hypothesis that altered PITX2-PAWR interactions could underlie, in part, the ARS phenotype that results from these PITX2 mutations. However, despite our best efforts, possible differences in the binding of PAWR to Ni2+ beads, transfection efficiencies of plasmid constructs, and expression of mutants could contribute to these differences in binding of PITX2 mutants to PAWR. Therefore, further exploration of the impact of PITX2 mutation on PAWR binding is warranted.
Interestingly, the PAWR-PITX2 interaction results in a reduction of PITX2 transcriptional activity in HTM cells (Fig. 5A, panel i). The LZ domain of PAWR, which interacts with PITX2, was sufficient alone to reduce PITX2 transcriptional activity in HTM cells (Fig. 5A, panel ii). This role of PAWR as a transcriptional repressor of PITX2 is consistent with previous findings where PAWR inhibited transcriptional activity of WT1, thus down-regulating expression of pro-survival proteins such as Bcl-2 (28). Our EMSA results (Fig. 5B) revealed that the down-regulation of PITX2 transactivation by PAWR is not through inhibiting the DNA binding capacity of PITX2. PAWR, by binding to PITX2, therefore appears to directly interfere with the ability of PITX2 to transactivate genes. This action might occur through prevention of the recruitment of transcriptional machinery to PITX2 target promoters or by interfering with a necessary PITX2 conformational change, previously suggested to be required for PITX2 activity (36).
Although the involvement in PAWR has already been reported in cancer-related apoptotic pathways, tumor regression, and neurodegenerative diseases including Alzheimer disease, this is the first report of its interaction with a homeobox transcription factor. Importantly, this is the first protein-protein interaction that negatively regulates PITX2 activity. Previous interactions of PITX2 with other proteins (e.g. β-catenin and LEF-1 (34)) either enhance or have no effect (e.g. FOXC1 (24)) on PITX2 transcriptional activity. In contrast, our experimental results indicate that PAWR reduces PITX2 transcriptional activity in immortalized HTM cells. This regulation of PITX2 activity through the interaction with PAWR has profound implications for PITX2 and its involvement in human disease. PITX2 is involved in numerous developmental processes including development of the anterior segment of the eye, and PITX2 mutations cause ARS. Glaucoma, the most clinically significant feature of ARS, is a neurodegenerative disease in which apoptosis of RGC results in progressive blindness. Interestingly, the LZ domain of human PAWR necessary and sufficient to interact with PITX2 and to inhibit PITX2 activity has also been found to interact with apoptosis antagonizing transcription factor (AATF) (37) and β-secretase (38), which are actively involved in another neurodegenerative disorder, Alzheimer disease. PAWR regulates β-secretase activity toward cleavage of amyloid precursor protein and production of β-amyloid in Alzheimer disease (38). Recently, the Alzheimer disease-associated protein β-amyloid has also been found to induce apoptosis in RGCs, leading to glaucoma (39). Because the ARS-associated glaucoma that results from mutations of PITX2 is also a neurodegenerative disorder, it is possible that the interaction between PITX2 and PAWR might have a role in apoptotic and/or neurodegeneration pathways involved in glaucoma. Our results indicate that further analysis of the PITX2-PAWR interaction in the eye and in ocular development and disease is warranted.
Acknowledgments
We thank May Yu for tissue culture assistance and Tim Footz and Farideh Mirzayans (all in the department of Medical Genetics, University of Alberta) for plasmid constructs and other technical assistance. We also thank Dr. Fred Berry and the members of Ocular Genetics Laboratory at the University of Alberta for helpful comments on this work and critical reading of this manuscript.
This work was supported, in whole or in part, by Operating Grant CIHRMOP77782 from the Canadian Institute of Health Research (to M. A. W.). This work was also supported by National Institutes of Health Grants EY014126 and EY007003 from the NEI (to P. J. G.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3 and supplemental Tables 1–3.
Birdi, C., and Walter, M. A., manuscript in preparation.
- PITX2
- pituitary homeobox transcription factor-2
- PIP
- PITX2-interacting proteins
- PAWR
- PRKC apoptosis WT1 regulator
- Y2H
- yeast two-hybrid
- TM
- trabecular meshwork
- HTM
- human TM
- HD
- homeodomain
- ARS
- Axenfeld-Rieger syndrome
- EMSA
- electrophoretic mobility shift assay
- LZ
- leucine zipper
- RGC
- retinal ganglion cells
- NPCE
- non-pigmented ciliary epithelium
- Ni-NTA
- nickel-nitrilotriacetic acid
- HA
- hemagglutinin
- TK
- thymidine kinase
- DAPI
- 4′,6-diamidino-2-phenylindole
- e
- embryonic day.
REFERENCES
- 1.Marcil A., Dumontier E., Chamberland M., Camper S. A., Drouin J. (2003) Development 130, 45–55 [DOI] [PubMed] [Google Scholar]
- 2.Smidt M. P., Smits S. M., Bouwmeester H., Hamers F. P., van der Linden A. J., Hellemons A. J., Graw J., Burbach J. P. (2004) Development 131, 1145–1155 [DOI] [PubMed] [Google Scholar]
- 3.Szeto D. P., Rodriguez-Esteban C., Ryan A. K., O'Connell S. M., Liu F., Kioussi C., Gleiberman A. S., Izpisúa-Belmonte J. C., Rosenfeld M. G. (1999) Genes Dev. 13, 484–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gage P. J., Camper S. A. (1997) Hum. Mol. Genet. 6, 457–464 [DOI] [PubMed] [Google Scholar]
- 5.Semina E. V., Reiter R., Leysens N. J., Alward W. L., Small K. W., Datson N. A., Siegel-Bartelt J., Bierke-Nelson D., Bitoun P., Zabel B. U., Carey J. C., Murray J. C. (1996) Nat. Genet. 14, 392–399 [DOI] [PubMed] [Google Scholar]
- 6.Cox C. J., Espinoza H. M., McWilliams B., Chappell K., Morton L., Hjalt T. A., Semina E. V., Amendt B. A. (2002) J. Biol. Chem. 277, 25001–25010 [DOI] [PubMed] [Google Scholar]
- 7.Hjalt T. A., Semina E. V. (2005) Expert Rev. Mol. Med. 7, 1–17 [DOI] [PubMed] [Google Scholar]
- 8.Arakawa H., Nakamura T., Zhadanov A. B., Fidanza V., Yano T., Bullrich F., Shimizu M., Blechman J., Mazo A., Canaani E., Croce C. M. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 4573–4578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alward W. L., Semina E. V., Kalenak J. W., Héon E., Sheth B. P., Stone E. M., Murray J. C. (1998) Am. J. Ophthalmol. 125, 98–100 [DOI] [PubMed] [Google Scholar]
- 10.Kulak S. C., Kozlowski K., Semina E. V., Pearce W. G., Walter M. A. (1998) Hum. Mol. Genet. 7, 1113–1117 [DOI] [PubMed] [Google Scholar]
- 11.Doward W., Perveen R., Lloyd I. C., Ridgway A. E., Wilson L., Black G. C. (1999) J. Med. Genet. 36, 152–155 [PMC free article] [PubMed] [Google Scholar]
- 12.Lines M. A., Kozlowski K., Kulak S. C., Allingham R. R., Héon E., Ritch R., Levin A. V., Shields M. B., Damji K. F., Newlin A., Walter M. A. (2004) Invest. Ophthalmol. Vis. Sci. 45, 828–833 [DOI] [PubMed] [Google Scholar]
- 13.Acharya M., Walter M. A. (2010) in Encyclopedia of the Eye (Besharse J., Dana R., Dartt D. A. eds) Elsevier Science Publishing Co., Inc., New York, in press [Google Scholar]
- 14.Shields M. B. (1983) Trans. Am. Ophthalmol. Soc. 81, 736–784 [PMC free article] [PubMed] [Google Scholar]
- 15.Axenfeld T. H. (1920) Klin. Monatsbl. Augenheilkd. 65, 381–382 [Google Scholar]
- 16.Rieger H. (1935) Albrecht Von Graefes Arch. Klin. Exp. Ophthalmol. 133, 602–635 [DOI] [PubMed] [Google Scholar]
- 17.Lines M. A., Kozlowski K., Walter M. A. (2002) Hum. Mol. Genet. 11, 1177–1184 [DOI] [PubMed] [Google Scholar]
- 18.Cunningham E. T., Jr., Eliott D., Miller N. R., Maumenee I. H., Green W. R. (1998) Arch. Ophthalmol. 116, 78–82 [DOI] [PubMed] [Google Scholar]
- 19.Gage P. J., Suh H., Camper S. A. (1999) Development 126, 4643–4651 [DOI] [PubMed] [Google Scholar]
- 20.Footz T., Idrees F., Acharya M., Kozlowski K., Walter M. A. (2009) Invest. Ophthalmol. Vis. Sci. 50, 2599–2606 [DOI] [PubMed] [Google Scholar]
- 21.Kozlowski K., Walter M. A. (2000) Hum. Mol. Genet. 9, 2131–2139 [DOI] [PubMed] [Google Scholar]
- 22.Mears A. J., Jordan T., Mirzayans F., Dubois S., Kume T., Parlee M., Ritch R., Koop B., Kuo W. L., Collins C., Marshall J., Gould D. B., Pearce W., Carlsson P., Enerbäck S., Morissette J., Bhattacharya S., Hogan B., Raymond V., Walter M. A. (1998) Am. J. Hum. Genet. 63, 1316–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishimura D. Y., Swiderski R. E., Alward W. L., Searby C. C., Patil S. R., Bennet S. R., Kanis A. B., Gastier J. M., Stone E. M., Sheffield V. C. (1998) Nat. Genet. 19, 140–147 [DOI] [PubMed] [Google Scholar]
- 24.Berry F. B., Lines M. A., Oas J. M., Footz T., Underhill D. A., Gage P. J., Walter M. A. (2006) Hum. Mol. Genet. 15, 905–919 [DOI] [PubMed] [Google Scholar]
- 25.Huang L., Chi J., Berry F. B., Footz T. K., Sharp M. W., Walter M. A. (2008) Invest. Ophthalmol. Vis. Sci. 49, 5243–5249 [DOI] [PubMed] [Google Scholar]
- 26.Saleem R. A., Banerjee-Basu S., Berry F. B., Baxevanis A. D., Walter M. A. (2001) Am. J. Hum. Genet. 68, 627–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hjalt T. A., Semina E. V., Amendt B. A., Murray J. C. (2000) Dev. Dyn. 218, 195–200 [DOI] [PubMed] [Google Scholar]
- 28.El-Guendy N., Rangnekar V. M. (2003) Exp. Cell Res. 283, 51–66 [DOI] [PubMed] [Google Scholar]
- 29.Roussigne M., Cayrol C., Clouaire T., Amalric F., Girard J. P. (2003) Oncogene 22, 2432–2442 [DOI] [PubMed] [Google Scholar]
- 30.Johnstone R. W., Tommerup N., Hansen C., Vissing H., Shi Y. (1998) Genomics 53, 241–243 [DOI] [PubMed] [Google Scholar]
- 31.Sells S. F., Wood D. P., Jr., Joshi-Barve S. S., Muthukumar S., Jacob R. J., Crist S. A., Humphreys S., Rangnekar V. M. (1994) Cell Growth Differ. 5, 457–466 [PubMed] [Google Scholar]
- 32.Johnstone R. W., See R. H., Sells S. F., Wang J., Muthukkumar S., Englert C., Haber D. A., Licht J. D., Sugrue S. P., Roberts T., Rangnekar V. M., Shi Y. (1996) Mol. Cell Biol. 16, 6945–6956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ranganathan P., Rangnekar V. M. (2005) Ann. N.Y. Acad. Sci. 1059, 76–85 [DOI] [PubMed] [Google Scholar]
- 34.Díaz-Meco M. T., Municio M. M., Frutos S., Sanchez P., Lozano J., Sanz L., Moscat J. (1996) Cell 86, 777–786 [DOI] [PubMed] [Google Scholar]
- 35.Kögel D., Plöttner O., Landsberg G., Christian S., Scheidtmann K. H. (1998) Oncogene 17, 2645–2654 [DOI] [PubMed] [Google Scholar]
- 36.Amendt B. A., Sutherland L. B., Russo A. F. (1999) Mol. Cell Biol. 19, 7001–7010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo Q., Xie J. (2004) J. Biol. Chem. 279, 4596–4603 [DOI] [PubMed] [Google Scholar]
- 38.Xie J., Guo Q. (2005) J. Biol. Chem. 280, 13824–13832 [DOI] [PubMed] [Google Scholar]
- 39.Guo L., Salt T. E., Luong V., Wood N., Cheung W., Maass A., Ferrari G., Russo-Marie F., Sillito A. M., Cheetham M. E., Moss S. E., Fitzke F. W., Cordeiro M. F. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 13444–13449 [DOI] [PMC free article] [PubMed] [Google Scholar]