Abstract
Recently, we discovered that nicotinamide riboside and nicotinic acid riboside are biosynthetic precursors of NAD+, which are utilized through two pathways consisting of distinct enzymes. In addition, we have shown that exogenously supplied nicotinamide riboside is imported into yeast cells by a dedicated transporter, and it extends replicative lifespan on high glucose medium. Here, we show that nicotinamide riboside and nicotinic acid riboside are authentic intracellular metabolites in yeast. Secreted nicotinamide riboside was detected with a biological assay, and intracellular levels of nicotinamide riboside, nicotinic acid riboside, and other NAD+ metabolites were determined by a liquid chromatography-mass spectrometry method. A biochemical genomic screen indicated that three yeast enzymes possess nicotinamide mononucleotide 5′-nucleotidase activity in vitro. Metabolic profiling of knock-out mutants established that Isn1 and Sdt1 are responsible for production of nicotinamide riboside and nicotinic acid riboside in cells. Isn1, initially classified as an IMP-specific 5′-nucleotidase, and Sdt1, initially classified as a pyrimidine 5′-nucleotidase, are additionally responsible for dephosphorylation of pyridine mononucleotides. Sdt1 overexpression is growth-inhibitory to cells in a manner that depends on its active site and correlates with reduced cellular NAD+. Expression of Isn1 protein is positively regulated by the availability of nicotinic acid and glucose. These results reveal unanticipated and highly regulated steps in NAD+ metabolism.
Introduction
NAD+ serves as a co-enzyme for hydride transfer enzymes and as a substrate of NAD+-consuming enzymes, including sirtuins, the type III protein lysine deacetylases. Whereas NAD+-dependent hydride transfer enzymes interconvert NAD+ to NADH and NADP to NADPH in redox reactions, NAD+-consuming enzymes break the bond between the ADP-ribose and the nicotinamide (Nam)2 moeities of NAD+, thereby necessitating ongoing NAD+ biosynthesis and Nam salvage (1, 2).
Yeast cells initiate Nam salvage with a specific nicotinamidase, Pnc1 (3). Nicotinic acid (NA), produced by Pnc1 or imported from the growth medium, is converted to NAD+ via the Preiss-Handler pathway, consisting of three enzymatic steps (4). The last step in Nam/NA salvage is catalyzed by the glutamine-dependent NAD+ synthetase, Qns1, which converts nicotinic acid adenine dinucleotide (NaAD) to NAD+ (5, 6). The recently discovered eukaryotic NAD+ precursors, nicotinamide riboside (NR) and nicotinic acid riboside (NAR), are each utilized through two distinct pathways. Exogenously supplied NR is transported into cells (7) and is either phosphorylated by the NR kinase, Nrk1, to form nicotinamide mononucleotide (NMN) (8) or converted to Nam by the hydrolase and phosphorylase activity of Urh1 and Pnp1, respectively (9). Finally, although NAR is poorly imported (10), it can also be utilized by Nrk1 (11) and Urh1 (10) to form nicotinic acid mononucleotide (NaMN) or NA, respectively (Fig. 1A).
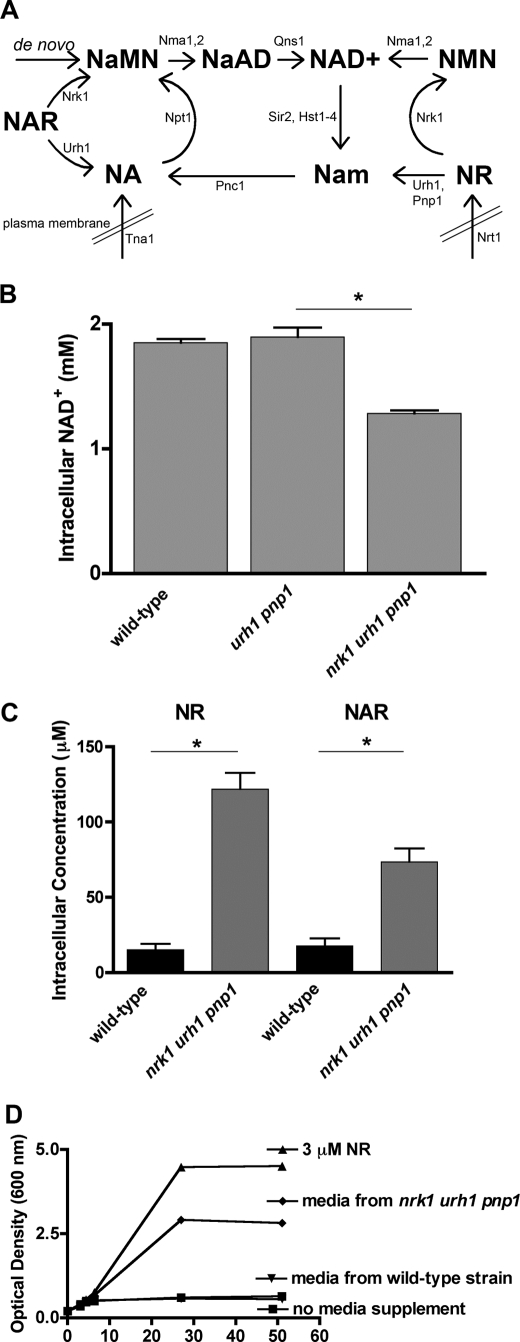
FIGURE 1.
NR and NAR are intracellular metabolites. A, model of NAD+ biosynthesis pathways. B, intracellular NAD+ concentration in wild-type yeast and in strains lacking one NR/NAR salvage pathway, urh1 pnp1, or both NR/NAR salvage pathways, nrk1 urh1 pnp1. Because cells were grown in SDC medium, which contains NA but not NR, the data suggest that cells produce NR and/or NAR at the expense of NAD+. C, LC-MS analysis of NAD+ metabolites showing substantial intracellular levels of both NR and NAR that are increased in a strain lacking NR/NAR salvage. D, bioassay using the qns1 mutant revealing that elevated levels of NR are secreted from the nrk1 urh1 pnp1 strain.
In yeast, Sir2 enzyme activity and NAD+ salvage enzymes Npt1 and Pnc1 are required for the longevity benefit of calorie restriction (CR) (12, 13). However, how CR specifically alters NAD+ metabolism is not yet clear. Competing models have suggested that CR might result in reduced Nam (13) or NADH (14) levels, which would increase Sir2 function by relief of inhibition. Alternatively, an increase in net NAD+ synthesis, which can extend lifespan in yeast (9) and which seems to occur in certain fasted (15) or calorie-restricted (16) vertebrate tissues, could be hypothesized to mediate increased Sir2 function by increasing free NAD+ over a relatively constant pool of protein-bound NAD+ co-enzyme (9). It has also been hypothesized that CR increases NAD+ biosynthetic flux without altering steady-state NAD+ levels (17). An additional study shows that CR does not result in reduced Nam, significantly reduced NADH, or increased NAD+.3 It is of interest that CR might alter NAD+ metabolic flux in ways that increase Sir2 function. However, the existing flux model (17) was proposed prior to the discovery of NMN, NR, and NAR as intermediates in yeast NAD+ metabolism, and the characterization of four gene products that function in NR and NAR metabolism to NAD+ (7–11). Clearly, to probe the mechanism of CR and NAD+ metabolic flux, the complete set of reactions must be known.
Beginning with the premise that the intracellular biogenesis of NR and NAR is the most significant remaining problem in establishing the intracellular wiring diagram for NAD+ metabolism in yeast, we hypothesized that the production of these nucleosides would be dependent on specific 5′-nucleotidases. Here, using genetic and metabolomic assays, we show that NR and NAR are normal cellular metabolites produced by the activities of 5′-nucleotidases, Isn1 and Sdt1, previously defined as IMP- (18) and pyrimidine nucleotide-specific 5′-nucleotidases (19), respectively. GAL1-driven overexpression of Sdt1 reduces intracellular NAD+ concentration and impairs growth in an active site-dependent manner. Consistent with an integral role in NAD+ metabolism and apparent regulation by CR, we find that Isn1 protein accumulation is increased by the provision of NA and decreased in response to glucose limitation. Our data, which include the first quantification of NAD+ metabolites as a function of deletion of NR/NAR salvage enzymes and the deletion of Isn1 and Sdt1, reveal complex relationships. In particular, the substantial levels of NMN and NaMN suggest that modulation of the expression and/or activities of NaMN/NMN adenylyltransferases and NaMN/NMN 5′-nucleotidases may drive pyridine nucleotide metabolism “forward” to dinucleotides or “backward” to nucleosides.
EXPERIMENTAL PROCEDURES
Saccharomyces cerevisiae Strains, Plasmids, and Media
All yeast strains used in this study were derived from BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0). Strains KB007 (sdt1) and KB006 (isn1) were prepared by a yeast deletion consortium (20). Other knock-out mutants were generated by one-step gene replacement as described (21) or obtained from genetic crosses and tetrad dissection (see supplemental Table 1). Yeast strains with chromosomally encoded tandem affinity protein (TAP) tags (22) and the plasmid for GAL1-driven overexpression of SDT1-HA (23) were obtained from Open Biosystems. The plasmid bearing SDT1-D61N was made using the QuikChange Lightning mutagenesis kit (Stratagene) using primers 5′-CAAAACCCGAACTTAAAAGTTTTCTTTTTTAATATCGACAACTGTCTC and 5′-GAGACAGTTGTCGATATTAAAAAAGAAAACTTTTAAGTTCGGGTTTTG. Media were prepared as described in an additional report,3 except when supplemented with synthetic NR (10), benzoic acid, NA or Nam, or in synthetic galactose complete (SGC) medium in which 20 g of d-galactose per liter replaced 20 g of d-glucose in synthetic dextrose complete (SDC) medium. SDC medium with reduced glucose was also utilized as indicated.
Biochemical Genomics Screen
Sixty-four pools of glutathione S-transferase fusion proteins were purified as described from 500 ml of culture (24). 10% of each pool preparation was assayed for NMN 5′-nucleotidase activity in overnight incubations (20 mm HEPES (pH 7.2), 10 mm NaCl, 5 mm 2-mercaptoethanol, 5 mm MgCl2). Nucleotides and nucleosides were resolved by HPLC on a 240 × 4.6-mm PrincetonSPHERE-60 5-μm SAX column fitted with a 4 × 10-mm guard column of the same material and particle size (Princeton Chromatography, Cranbury, NJ) using a 10-min isocratic elution in 20 mm KH2PO4.
NAD+ Metabolite Quantification
NAD+ content was assayed as described (9). Liquid chromatography mass spectrometry (LC-MS) detection of other NAD+ metabolites was performed as described in an additional report.3 All reported metabolomic assays were performed with biological triplicate samples. To assess statistical significance, standard errors of the mean were calculated, and two-tailed t tests were employed. Differences between metabolite concentrations were deemed significant at p < 0.05. Significant differences in all figures are indicated with asterisks.
NR Bioassay
A qns1 strain BY165-1D, previously described as an NR auxotroph (8), was cured of the plasmid bearing QNS1 by selection on 5-fluoroorotic acid-containing SDC medium supplemented with 10 μm NR. The plasmid-free strain was then grown for ∼8 h in SDC supplemented with 10 μm NR. This culture was diluted 1:2 in SDC without NR and allowed to grow overnight to exhaustion. In the morning, the culture was diluted to an A600 nm of 0.2 in supplemented medium conditioned by the indicated strains. Conditioned medium was supplemented 1:1 with 2× SDC to maintain glucose and other nutritional requirements. Growth of BY165-1D was followed hourly by A600 nm measurements. Growth curves with or without 3 μm NR were used as positive and negative controls.
Pnc1 Purification and Assays
Bacterial expression of PNC1 was as described (3, 25). Purification of the nontagged protein was performed by metal chelate affinity chromatography (3, 26). Peak fractions were stored in 10 mΜ Tris (pH 7.5), 150 mm NaCl, 1 mΜ MgCl2, 20% glycerol. Pnc1 assays were performed as described (3) with some modifications. Purified Pnc1 (1.2 μg) was incubated with 500 μm Nam, NMN, or NR for 30 min at 37 °C in a final volume of 500 μl containing 10 mm Tris (pH 7.5), 150 mm NaCl, 1 mΜ MgCl2. Substrates and products were analyzed by HPLC on a 240 × 4.6-mm SAX column using a gradient elution of 20 mm KH2PO4 (buffer A) to 750 mm KH2PO4 (buffer B) as mobile phase (1–3 min 100% buffer A, 3–14 min linear gradient to 100% buffer B, 14–17 min 100% buffer B, 17–27 min 100% buffer A). Detection of peaks was performed at 260 nm, and the flow rate was 1 ml/min.
Protein Expression and Overexpression Analysis
Wild-type and nrk1 urh1 pnp1 strains were transformed with plasmids pRS426GAL1, pKB07 (GAL1-SDT1-HA), and pKB10 (GAL1-SDT1-D61N-HA). These strains were grown to log phase and then stamped in 5-fold serial dilutions on SDC-ura and SGC-ura media. Galactose plates were incubated for 7 days at 28 °C. Strain KB048, constructed with a chromosomally integrated TAP-tagged Isn1 polypeptide, was grown to mid-log phase (A600 nm = 0.6–0.8). Next, 25 ml of cells were collected by centrifugation, lysed, and total cell lysates were resolved by SDS-PAGE and transferred to membranes using standard procedures. Membranes were subjected to immunoblotting with a rabbit polyclonal antibody CAB1001 directed against the TAP tag (Open Biosystems). A goat anti-rabbit horseradish peroxidase-conjugated antibody was used as a secondary.
RESULTS
NR and NAR Are Intracellular Metabolites
As shown in Fig. 1A, previous studies have defined the roles of exogenously supplied NR and NAR and defined the Nrk1-dependent and -independent pathways through which these nucleosides are utilized as NAD+ precursors (1, 7, 8, 10, 11). The vitamin properties of exogenously supplied NR include promotion of Sir2-dependent silencing and lifespan extension in glucose-replete medium (1). As shown in Fig. 1B and as demonstrated previously (1), when the Nrk1-dependent and Nrk1-independent pathways for NR and NAR salvage are deleted to produce a yeast strain of genotype nrk1 urh1 pnp1, and yeast cells are grown in standard SDC medium, which contains 3 μm NA but no NR or NAR, there is a significant diminution of intracellular NAD+. These data suggest that yeast cells may be producing NR and NAR in the absence of supplementation and maintaining their intracellular NAD+ concentration via continuous NR/NAR salvage. We therefore predicted that NR and NAR would be present in wild-type cells and would increase in the nrk1 urh1 pnp1 genotype. To test this hypothesis, we developed an LC-MS method to detect and quantify the NAD+ metabolome.3 Indeed, as shown in Fig. 1C, we measured intracellular NR and NAR levels at 15 μm and 18 μm, respectively, in wild-type cells that were grown in standard SDC medium. Noting that the standard NR supplementation concentration is 10 μm (1, 7, 8, 10, 11), the relatively high intracellular concentration of NR helps rationalize the activity of Nrt1 as a concentrative transporter (7). Moreover, in the NR/NAR-nonsalvaging genotype, nrk1 urh1 pnp1, the intracellular concentrations of NR and NAR increased dramatically to 122 μm and 73 μm, respectively.
Because the NR/NAR-nonsalvaging strain accumulates high levels of NR, we set out to determine whether NR is also an extracellularly released metabolite. To address this question, we developed a bioassay utilizing the NR auxotrophy of the qns1 mutant (8). As shown in Fig. 1A, Qns1 is required for de novo production of NAD+ as well as salvage of NA and Nam. Accordingly, we grew wild-type and the NR/NAR-nonsalvaging strain nrk1 urh1 pnp1 to stationary phase, discarded the cells, and collected the conditioned medium. After mixing conditioned medium 1:1 with 2× SDC, we assayed the growth of qns1 as an indicator of the presence of NR. We found that medium conditioned by the NR/NAR-nonsalvaging strain supported the growth of the NR auxotrophic strain qns1 (Fig. 1D), indicating the presence of NR in conditioned media. Conditioned medium diluted 2-fold with SDC has approximately two-thirds the NR activity of 3 μm NR. Thus, we estimate the extracellular concentration of NR in a late stage culture of the nrk1 urh1 pnp1 strain to be ∼4 μm. Similar results have been reported independently (27).
Analysis of the NAD+ Metabolome
Because the salvage of intracellularly produced NAR and NR has such a dramatic effect on intracellular NAD+ levels (Fig. 1B), we set out to determine the effects of knocking out NR/NAR salvage on the other NAD+ metabolites. To do this, we developed a selected reaction monitoring LC-MS method to detect the core components of the NAD+ metabolome and other nucleosides and nucleotides simultaneously. As described in an additional report, this method can separate NA, Nam, NAR, NR, the two pyridine mononucleotides, the five major pyridine dinucleotides, and other nucleosides and nucleotides, although NA and NaAD are not reliably detected in all biological samples.3 As shown in Fig. 2 and supplemental Table 2, when wild-type cells were grown in SDC, all compounds except NA were detected at concentrations of ∼1 to ∼600 μm. In addition to the 8- and 4-fold increases in NR and NAR as a function of deletion of NR/NAR salvage genes and the already reported nearly 2-fold reduction in NAD+ (1), there were several additional changes in levels of NAD+ metabolites. The level of NADPH more than doubled to ∼3 μm in the NR/NAR-nonsalvaging strain, although this remained a small fraction of the pyridine dinucleotide pool. The mononucleotides NaMN and NMN, which are produced via the activity of Nrk1, are 94 and 62% reduced compared with wild-type, whereas the level of the dinucleotide NaAD is reduced beyond the limit of detection. Interestingly, although the deletion of urh1 and pnp1 eliminates a pathway by which cells produce Nam, and deletion of nrk1 urh1 pnp1 reduces NAD+ levels and Sir2 function, the only other yeast pathway known to produce Nam (1), the nrk1 urh1 pnp1 strain increased the level of Nam by more than 5-fold. Thus, the reduced NAD+ levels and increased Nam levels of the nrk1 urh1 pnp1 strain resemble NAD+ metabolism in the pnc1 nicotinamidase mutant (3, 28) as discussed below.
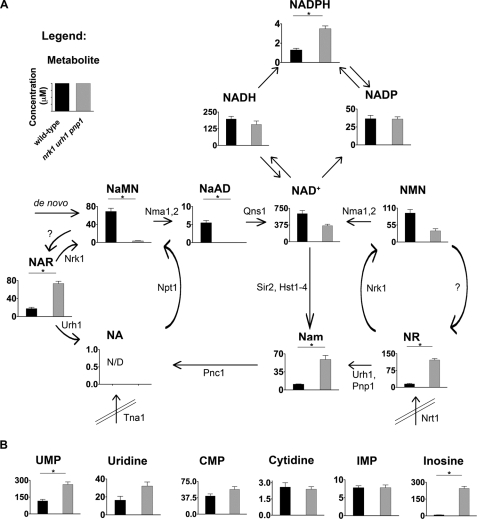
FIGURE 2.
NAD+ metabolome is dysregulated by loss of NR/NAR salvage in cells grown without NR or NAR. A, intracellular concentrations of NAD+ metabolites in wild-type (black bars) and the NR/NAR-nonsalvaging strain nrk1 urh1 pnp1 (gray bars). B, intracellular concentrations of other nucleotides and nucleosides in wild-type (black bars) and the NR/NAR-nonsalvaging strain nrk1 urh1 pnp1 (gray bars).
Concurrent with the decline in NAD+ in the NR/NAR-nonsalvaging strain, the three metabolites closest to formation of NAD+, i.e. NaMN, NaAD, and NMN, are reduced by deletion of NR/NAR salvage. We speculate that the reduced levels of NAD+ drive the equilibria of NaMN/NMN adenylyltransferase and Qns1 reactions toward product formation by homeostatic mechanisms, which remain to be uncovered. Recently, the nrk1 urh1 pnp1 mutant was shown to be resistant to the longevity benefit of CR in a chronological lifespan assay (27). Although the role and targets of NAD+ metabolism in chronological lifespan extension in yeast are unclear, the elevated Nam, reduced NAD+, and/or reduced NaMN, NaAD, and NMN observed herein may mediate the deficiency in responding to CR.
Recently, we have shown that Urh1 is more effective as an NR hydrolase than as a Urd hydrolase and that Pnp1 functions as an NR phosphorylase secondary to its known function as a purine nucleoside phosphorylase (10). Given those in vitro data, we wished to test whether deletion of NR/NAR salvage enzymes would affect the levels of a set of purine and pyrimidine nucleosides and nucleotides. In Fig. 2B we show that, consistent with roles for Pnp1 and Urh1 in the phosphorolysis and hydrolysis of Ino (29) and Urd (30), respectively, Ino levels are increased by ∼28-fold and Urd levels increased by ∼2-fold in the nrk1 urh1 pnp1 genotype. Levels of UMP are also increased in the nrk1 urh1 pnp1 strain (Fig. 2B and supplemental Table 2), potentially due to reduced Urd hydrolysis.
Isn1 and Sdt1 Are NMN 5′-Nucleotidases
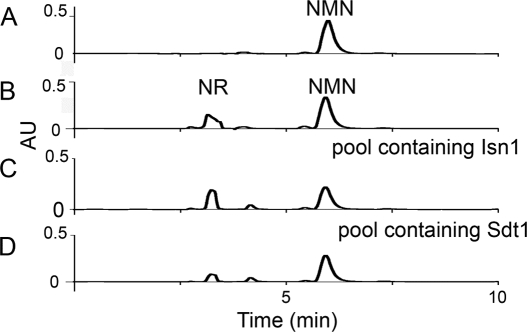
We set out to determine the enzymes responsible for NR and NAR production. Because there is no biological precedent for nucleoside synthesis de novo and because NR and NAR accumulation appear to occur at the expense of NAD+ (Figs. 1 and 2), we considered the mechanisms by which NAD+ and related compounds could be broken down to NR. Given the cellular concentrations of the pyridine mononucleotides, we hypothesized that NR and NAR are produced from NMN and NaMN via 5′-nucleotidase activities. To test this hypothesis, we screened 64 pools of glutathione S-transferase fusion proteins, which represent a substantial fraction of the yeast proteome (24, 31), for NMN 5′-nucleotidase activities. By HPLC, we found three pools of protein that produce NR from NMN in vitro (Fig. 3). The first pool contained Isn1 (Fig. 3C), previously reported as an IMP-specific 5′-nucleotidase that catalyzes the dephosphorylation of IMP to inosine (18). The second pool contained Sdt1 (Fig. 3D), described as a pyrimidine-specific 5′-nucleotidase, overexpression of which suppresses the 6-azauracil sensitivity of mutants in transcription elongation factor S-II (19). Finally, the third pool contained Phm8 (data not shown), a low phosphate-induced enzyme with no known function (32) that possesses sequence similarity to Sdt1.
FIGURE 3.
Biochemical genomic screen identifies Isn1 and Sdt1 as NMN 5′-nucleotidases. A, HPLC trace of NMN standard. B, HPLC trace of NMN + NR standards. C, positive glutathione S-transferase pool containing Isn1. D, positive glutathione S-transferase pool containing Sdt1.
To determine whether these genes are responsible for production of NR and NAR in vivo, we generated strains with single, double, and triple gene deletions. In three independent experiments (supplemental Table 4), the single deletion of isn1 produced a 60–79% reduction in NR levels. Single deletion of sdt1 reduced NR levels 70–73%. In four independent experiments, double deletion of isn1 sdt1 reduced NR levels in a range of 77% to greater than 99%. The single deletion of sdt1 also produced 73–86% reductions in NAR with respect to NAR levels in wild-type cells, whereas deletion of isn1 reduced NAR levels by 15–56% in the single mutant strain. The double deletion of sdt1 isn1 reduced NAR metabolite levels in a range of 82–100%. These data indicate that Sdt1 functions as the primary and Isn1 functions as the secondary NMN/NaMN 5′-nucleotidase in vivo. The phm8 deletion strain showed little effect of NR or NAR in the metabolite pool, leading us to conclude that Phm8 does not contribute significantly to intracellular production of NR or NAR under the growth conditions tested. Data are summarized in Fig. 4 from an experiment performed with three biological replicates.
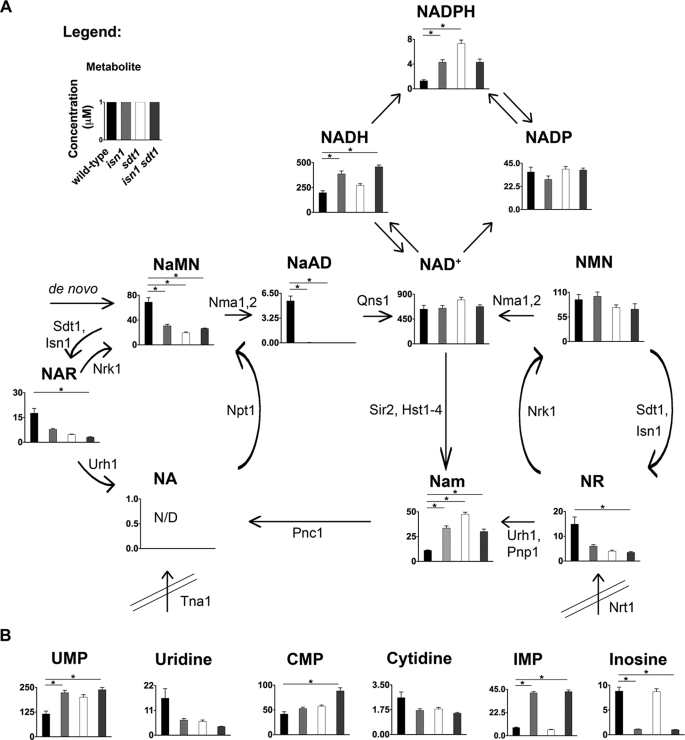
FIGURE 4.
Isn1 and Sdt1 are responsible for production of NR and NAR. A, intracellular concentrations of NAD+ metabolites in wild-type (black bars), isn1 (light gray bars), sdt1 (white bars), and isn1 sdt1 (dark gray bars). B, intracellular concentrations of other nucleotides and nucleosides in wild-type (black bars), isn1 (light gray bars), sdt1 (white bars), and isn1 sdt1 (dark gray bars).
Fig. 4 also indicates that deleting genes encoding NMN/NaMN 5′-nucleotidases dramatically alters NAD+ metabolites other than NR and NAR. For example, either single mutation increases Nam and reduces intracellular NaAD and NaMN, whereas the isn1 single mutation but not the sdt1 mutation causes a reduction in NAD+, NMN, and NADP levels.
Because Sdt1 was reported to be a UMP and CMP 5′-nucleotidase (19) and Isn1 an IMP-specific 5′-nucleotidase (18), we examined the levels of additional nucleosides and nucleotides. As shown in Fig. 4B, UMP and CMP are increased and Urd and Cyt decreased in single sdt1 and double sdt1 isn1 mutants. Similarly, IMP levels are increased and Ino levels decreased in isn1 and sdt1 isn1 double mutants. Thus, Sdt1 and Isn1 encode bifunctional enzymes. Sdt1 can now be termed a pyrimidine and pyridine-specific 5′-nucleotidase and Isn1 an IMP and pyridine-specific 5′-nucleotidase.
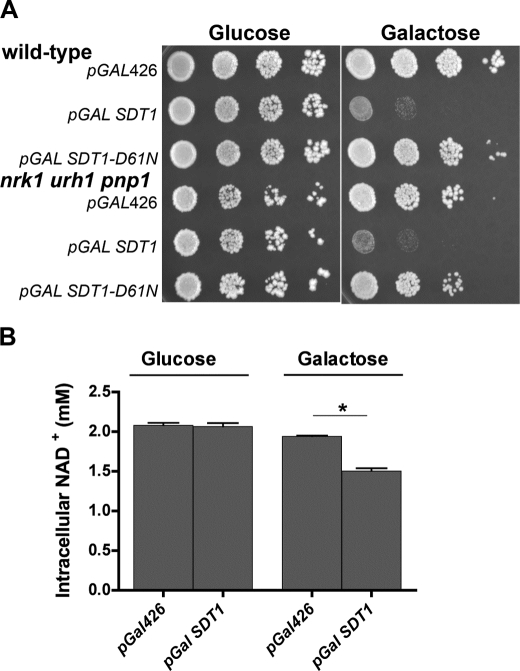
To determine the effects of aberrant regulation of Sdt1, we used a multicopy plasmid with the GAL1 promoter to drive overexpression of Sdt1 in wild-type and in the NR/NAR-nonsalvaging strain background. As shown in Fig. 5A, overexpression of Sdt1 inhibited growth in wild-type cells and inhibited growth to a slightly greater degree in the nrk1 urh1 pnp1 genotype. Sdt1 is a member of the haloalkanoic acid dehalogenase enzyme superfamily of phosphotransferases, which function as metabolite phosphatases, ATPases, and sugar-phosphate mutases (33). These enzymes possess an Asp-Xaa-Asp motif, which forms a covalent phosphorylated intermediate to the first Asp, stabilized by the general acid-base activity of the second Asp. To test whether Sdt1 overexpression inhibited growth by virtue of a mechanism independent of its enzyme activity, we generated an SDT1-D61N allele in which the carboxylate of the first Asp was converted to carboxamide. As shown in Fig. 5A, the growth-inhibitory effect of Sdt1 overexpression depends on the active site. Moreover, as shown in Fig. 5B, the galactose-dependent, plasmid-dependent toxicity of Sdt1 overexpression was accompanied by significant reduction of intracellular NAD+ levels.
FIGURE 5.
Overexpression of SDT1 inhibits growth and lowers intracellular NAD+. A, overexpression of SDT1 inhibits growth in wild-type and in the nrk1 urh1 pnp1 strain background in a manner that depends on active site residue Asp61. B, galactose-dependent overexpression of SDT1 reduces intracellular NAD+. Strains containing plasmids were grown to log phase (A600 nm = 0.8) and then stamped in 5-fold serial dilutions on SDC and SGC media.
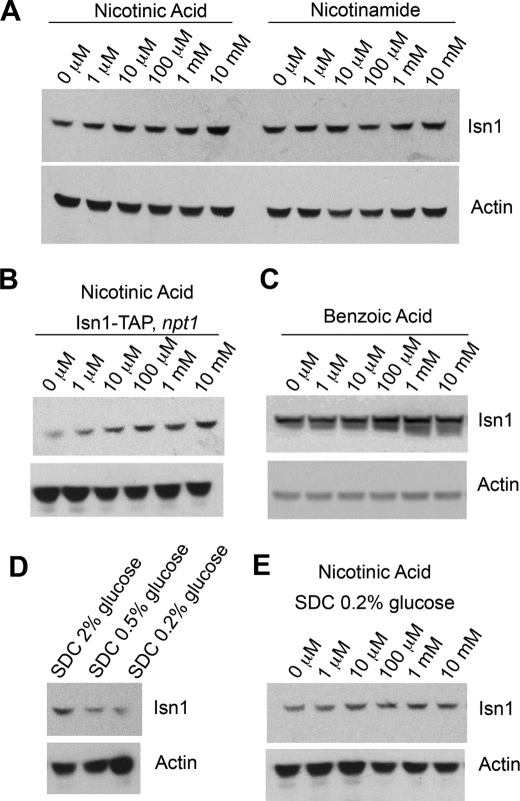
Isn1 Is Regulated by NA and Glucose
We have shown previously that the intracellular NAD+ level declines in cells grown in NR-free, NA-replete SDC medium (1), and this study shows that there is substantial NMN and NR in cells grown in NR-free, NA-replete SDC. We therefore hypothesized that one or both of the NMN/NaMN 5′-nucleotidases might be positively regulated by the availability of NA. Moreover, because IMP 5′-nucleotidase activity has been reported to be induced by glucose (34), we wished to test whether Isn1 protein might be under glucose control. A yeast strain constructed to express a chromosomally integrated TAP-tagged Isn1 polypeptide (22) was grown in vitamin-free SDC medium to which NA was added and to which Nam was added. As shown in Fig. 6A, Isn1 protein accumulates to an increasing degree with addition of NA to the culture medium. Although Nam is a biosynthetic precursor of NA, Nam addition to the medium does not increase expression of Isn1. Surprisingly, the response to NA does not depend on the presence of Npt1 (Fig. 6B), the enzyme required for NA to be incorporated into the intracellular NAD+ pool. As a further test of whether NA might possess an activity on Isn1 accumulation without incorporation into NAD+, we titrated benzoic acid, the phenyl analog of NA, into vitamin-free SDC. As shown in Fig. 6C, benzoic acid has the same effect on Isn1 accumulation as NA.
FIGURE 6.
Isn1 accumulation is positively regulated by sodium and glucose. A, Western blot analysis shows that Isn1 protein accumulates with increasing provision of NA, but not with Nam. B, Isn1 protein accumulates with provision of NA in an npt1 mutant. C, Isn1 protein accumulates with provision of benzoic acid, an NA analog that is not converted to NAD+. D, Isn1 protein in SDC (3 μm NA) decreases with glucose restriction. E, restriction of glucose to 0.2% reduces accumulation of Isn1 upon provision of NA.
To test whether the reported induction of IMP 5′-nucleotidase activity with glucose (34) might be seen at the protein level, we compared Isn1 expression in SDC (2% glucose) medium with the same medium prepared with 0.5% and 0.2% glucose. As shown in Fig. 6D, glucose restriction results in reduced Isn1 accumulation. To test whether the reduced expression of Isn1 in low glucose would preclude NA induction of Isn1 protein, we titrated NA to the TAP-tagged Isn1 strain grown in 0.2% glucose. As shown in Fig. 6E, this CR condition greatly reduces Isn1 accumulation as a function of NA.
Pnc1 Does Not Produce NaMN or NAR
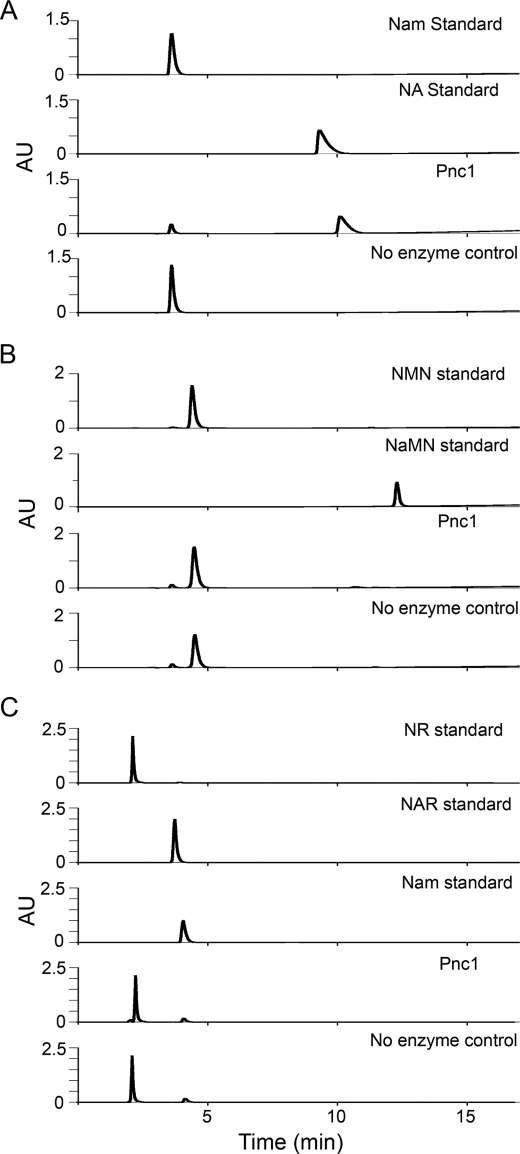
The PNC1 gene encodes a nicotinamidase that converts Nam to NA in the first step of fungal Nam salvage (3). To attempt to account for intracellular production of NAR, it was suggested that Pnc1 could produce NAR by deamidation of NR and NaMN by deamidation of NMN (27). Although there have been reports of bacterial enzyme activities that convert NMN to NaMN (35, 36), the data have been reinterpreted to explain NMN utilization without deamidation (37) and did not invoke nicotinamidase as the responsible enzyme in any case. The only data in support of Pnc1 as a source of NaMN and NAR were obtained using crude cell lysates and a kit to detect ammonia liberated from NMN or NR (27).
To determine whether the S. cerevisiae nicotinamidase Pnc1 can accommodate NMN and NR as substrates, we purified the untagged recombinant enzyme from Escherichia coli by metal chelate affinity chromatography. As shown in Fig. 7, under conditions (30 min at 37 °C) in which Pnc1 hydrolyzed 500 μm Nam to 81% completion, no NaMN was produced from NMN, nor was any NAR produced from NR. Reactions (not shown) run for 24 h showed complete conversion of Nam to NA and failed to show any evidence of NMN or NR deamidation. Indeed, as can be seen in Fig. 7C, the enzyme-independent hydrolysis of NR to Nam in neutral solution is detectable, whereas NR deamidation is not. Taken together with the genetic and metabolomic data in Fig. 4 showing that NAR production depends on Sdt1 and Isn1, a nicotinamidase-dependent route to NAR is not supported by data.
FIGURE 7.
Pnc1 does not produce NAR or NaMN. A, HPLC traces of Nam, NA, a Pnc1 reaction with Nam as substrate, and a no-enzyme control reaction show that purified Pnc1 is highly active on Nam. B, Pnc1 is inactive on NMN. C, Pnc1 is inactive on NR.
DISCUSSION
Here, we show that NR and NAR are intracellular metabolites present in wild-type cells grown in standard synthetic medium, which are increased by deletion of the NR/NAR salvage genes, nrk1 urh1 pnp1. Because SDC-grown nrk1 urh1 pnp1 cells have low NAD+ levels but maintain substantial levels of NR and NMN, although their only source of NAD+ precursor vitamins is NA, we hypothesized that NR is produced at the expense of NAD+ by an NMN 5′-nucleotidase activity. By screening for such an activity, we identified three candidate genes, two of which, ISN1 and SDT1, are essential for production of NR and NAR in vivo. The identification of Isn1 and Sdt1, previously thought to function exclusively in purine and pyrimidine metabolism, as enzymes with roles in NAD+ metabolism is reminiscent of recent discoveries with respect to Urh1 and Pnp1 (1, 10, 11). Indeed, the multifunctionality of Isn1 and Sdt1 in nucleotide-nucleoside transactions follows a recurrent theme in NAD+ biosynthesis (38). Metabolomic analysis indicates that the majority of NAD+ metabolites are altered by deletion of NR/NAR salvage enzymes, Nrk1, Urh1 and Pnp1, and producing enzymes, Isn1 and Sdt1. In addition, we found that expression of NMN/NaMN 5′-nucleotidases is highly regulated such that overexpression of Sdt1 is toxic and results in reduced intracellular NAD+, whereas Isn1 protein levels are positively regulated by NA and glucose.
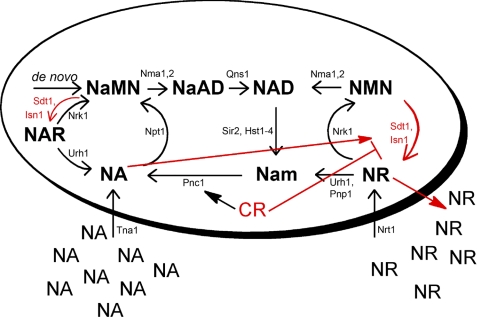
Regulation of Isn1 protein levels is among the most intriguing aspects of our discoveries of the NMN/NaMN 5′-nucleotidases. Isn1 protein accumulates in response to provision of NA, apparently in unincorporated form, because NA-dependent Isn1 accumulation occurs in an npt1 mutant and because benzoic acid, which is not a substrate for NA phosphoribosyltransferase (39), produces the same effect. However, as schematized in Fig. 8, Isn1 accumulation is reduced by glucose limitation, and the ability of NA to increase Isn1 expression is dampened by CR. This is of particular interest because it suggests a novel mechanism by which CR can control NAD+ metabolism and availability for Sir2-dependent reactions. Potentially, by retarding NAD+ catabolic pathways mediated by Isn1, CR directs NAD+ flux toward Sir2 utilization. This type of dynamic regulation of NAD+ metabolism is consistent with data in an additional report showing that the levels of intracellular NAD+ metabolites are not greatly changed by CR.3 Overall, the data indicate that in high nutrient environments, cells break down NAD+ to nonphosphorylated metabolites that are released from cells, as observed in this and another recent study (27), perhaps for reasons that have been termed altruistic (40).
FIGURE 8.
Revised model of NAD+ metabolism in yeast. New steps in NAD+ metabolism, consisting of the NMN/NaMN 5′-nucleotidase activities of Isn1 and Sdt1, are depicted in red. New regulatory steps, also depicted in red, show that Isn1 protein accumulation is promoted by NA and opposed by CR. These data suggest a novel mechanism by which CR can control NAD+ metabolism and availability for Sir2-dependent reactions: by retarding NAD+ catabolic pathways mediated by Isn1, CR may direct NAD+ flux toward Sir2 utilization.
It is also of note that the NR/NAR-nonsalvaging genotype nrk1 urh1 pnp1 has two metabolic similarities to nicotinamidase mutants, namely, low NAD+ and high Nam (3, 28). Whereas the ongoing production of NR and NAR from NMN and NaMN can explain the deficiency in NAD+, it is difficult to explain elevated Nam in the nrk1 urh1 pnp1 strain in two respects. First, the urh1 pnp1 deletions eliminate one pathway to Nam. Second, the NAD+ deficiency and the low Sir2 activity in this strain (1) should reduce the only other known pathway to produce Nam in yeast. The NR/NAR-nonsalvaging genotype is characterized by high NR and NAR levels. Whereas we have shown that NR is not a substrate and NAR not a product of Pnc1 activity, our in vivo data suggest that one or both of these pyridine nucleosides may function as an inhibitor of Pnc1, thereby leading to the accumulation of Nam.
In conclusion, the identification of Sdt1 and Isn1 as NMN/NaMN 5′-nucleotidases and the discovery of regulation of enzyme abundance by the availability of nutrients should facilitate dissection of the dynamics of NAD+ metabolism as a function of cellular age and environmental conditions.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants R37-DK046960, R01-DK079084, and T32-DK07245 and by National Science Foundation Grant MCB-0822581.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1–6.
C. Evans, K. L. Bogan, P. Song, C. F. Burant, R. T. Kennedy, and C. Brenner, manuscript in preparation.
- Nam
- nicotinamide
- NA
- nicotinic acid
- NaAD
- nicotinic acid adenine dinucleotide
- NaMN
- nicotinic acid mononucleotide
- NAR
- nicotinic acid riboside
- NMN
- nicotinamide mononucleotide
- NR
- nicotinamide riboside
- CR
- calorie restriction
- HPLC
- high pressure liquid chromatography
- LC-MS
- liquid chromatography mass spectrometry
- SDC
- synthetic dextrose complete
- SGC
- synthetic galactose complete
- TAP
- tandem affinity protein.
REFERENCES
- 1.Belenky P., Bogan K. L., Brenner C. (2007) Trends Biochem. Sci. 32, 12–19 [DOI] [PubMed] [Google Scholar]
- 2.Bogan K. L., Brenner C. (2008) Annu. Rev. Nutr. 28, 115–130 [DOI] [PubMed] [Google Scholar]
- 3.Ghislain M., Talla E., François J. M. (2002) Yeast 19, 215–224 [DOI] [PubMed] [Google Scholar]
- 4.Preiss J., Handler P. (1958) J. Biol. Chem. 233, 493–500 [PubMed] [Google Scholar]
- 5.Bieganowski P., Brenner C. (2003) J. Biol. Chem. 278, 33056–33059 [DOI] [PubMed] [Google Scholar]
- 6.Suda Y., Tachikawa H., Yokota A., Nakanishi H., Yamashita N., Miura Y., Takahashi N. (2003) Yeast 20, 995–1005 [DOI] [PubMed] [Google Scholar]
- 7.Belenky P. A., Moga T. G., Brenner C. (2008) J. Biol. Chem. 283, 8075–8079 [DOI] [PubMed] [Google Scholar]
- 8.Bieganowski P., Brenner C. (2004) Cell 117, 495–502 [DOI] [PubMed] [Google Scholar]
- 9.Belenky P., Racette F. G., Bogan K. L., McClure J. M., Smith J. S., Brenner C. (2007) Cell 129, 473–484 [DOI] [PubMed] [Google Scholar]
- 10.Belenky P., Christensen K. C., Gazzaniga F. S., Pletnev A., Brenner C. (2009) J. Biol. Chem. 284, 158–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tempel W., Rabeh W. M., Bogan K. L., Belenky P., Wojcik M., Seidle H. F., Nedyalkova L., Yang T., Sauve A. A., Park H. W., Brenner C. (2007) PLoS Biol. 5, e263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin S. J., Defossez P. A., Guarente L. (2000) Science 289, 2126–2128 [DOI] [PubMed] [Google Scholar]
- 13.Anderson R. M., Bitterman K. J., Wood J. G., Medvedik O., Sinclair D. A. (2003) Nature 423, 181–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin S. J., Ford E., Haigis M., Liszt G., Guarente L. (2004) Genes Dev. 18, 12–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodgers J. T., Lerin C., Haas W., Gygi S. P., Spiegelman B. M., Puigserver P. (2005) Nature 434, 113–118 [DOI] [PubMed] [Google Scholar]
- 16.Qin W., Yang T., Ho L., Zhao Z., Wang J., Chen L., Zhao W., Thiyagarajan M., MacGrogan D., Rodgers J. T., Puigserver P., Sadoshima J., Deng H., Pedrini S., Gandy S., Sauve A. A., Pasinetti G. M. (2006) J. Biol. Chem. 281, 21745–21754 [DOI] [PubMed] [Google Scholar]
- 17.Anderson R. M., Bitterman K. J., Wood J. G., Medvedik O., Cohen H., Lin S. S., Manchester J. K., Gordon J. I., Sinclair D. A. (2002) J. Biol. Chem. 277, 18881–18890 [DOI] [PubMed] [Google Scholar]
- 18.Itoh R., Saint-Marc C., Chaignepain S., Katahira R., Schmitter J. M., Daignan-Fornier B. (2003) BMC Biochem. 4, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakanishi T., Sekimizu K. (2002) J. Biol. Chem. 277, 22103–22106 [DOI] [PubMed] [Google Scholar]
- 20.Winzeler E. A., Shoemaker D. D., Astromoff A., Liang H., Anderson K., Andre B., Bangham R., Benito R., Boeke J. D., Bussey H., Chu A. M., Connelly C., Davis K., Dietrich F., Dow S. W., El Bakkoury M., Foury F., Friend S. H., Gentalen E., Giaever G., Hegemann J. H., Jones T., Laub M., Liao H., Liebundguth N., Lockhart D. J., Lucau-Danila A., Lussier M., M'Rabet N., Menard P., Mittmann M., Pai C., Rebischung C., Revuelta J. L., Riles L., Roberts C. J., Ross-MacDonald P., Scherens B., Snyder M., Sookhai-Mahadeo S., Storms R. K., Véronneau S., Voet M., Volckaert G., Ward T. R., Wysocki R., Yen G. S., Yu K., Zimmermann K., Philippsen P., Johnston M., Davis R. W. (1999) Science 285, 901–906 [DOI] [PubMed] [Google Scholar]
- 21.Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., Hieter P., Boeke J. D. (1998) Yeast 14, 115–132 [DOI] [PubMed] [Google Scholar]
- 22.Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., Weissman J. S. (2003) Nature 425, 737–741 [DOI] [PubMed] [Google Scholar]
- 23.Gelperin D. M., White M. A., Wilkinson M. L., Kon Y., Kung L. A., Wise K. J., Lopez-Hoyo N., Jiang L., Piccirillo S., Yu H., Gerstein M., Dumont M. E., Phizicky E. M., Snyder M., Grayhack E. J. (2005) Genes Dev. 19, 2816–2826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phizicky E. M., Martzen M. R., McCraith S. M., Spinelli S. L., Xing F., Shull N. P., Van Slyke C., Montagne R. K., Torres F. M., Fields S., Grayhack E. J. (2002) Methods Enzymol. 350, 546–559 [DOI] [PubMed] [Google Scholar]
- 25.Barski A., Cuddapah S., Cui K., Roh T. Y., Schones D. E., Wang Z., Wei G., Chepelev I., Zhao K. (2007) Cell 129, 823–837 [DOI] [PubMed] [Google Scholar]
- 26.Hu G., Taylor A. B., McAlister-Henn L., Hart P. J. (2007) Arch. Biochem. Biophys. 461, 66–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu S.-P., Kato M., Lin S.-J. (2009) J. Biol. Chem. 284, 17110–17119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sauve A. A., Moir R. D., Schramm V. L., Willis I. M. (2005) Mol. Cell 17, 595–601 [DOI] [PubMed] [Google Scholar]
- 29.Lecoq K., Belloc I., Desgranges C., Konrad M., Daignan-Fornier B. (2001) J. Bacteriol. 183, 4910–4913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kurtz J. E., Exinger F., Erbs P., Jund R. (2002) Curr. Genet. 41, 132–141 [DOI] [PubMed] [Google Scholar]
- 31.Martzen M. R., McCraith S. M., Spinelli S. L., Torres F. M., Fields S., Grayhack E. J., Phizicky E. M. (1999) Science 286, 1153–1155 [DOI] [PubMed] [Google Scholar]
- 32.Ogawa N., DeRisi J., Brown P. O. (2000) Mol. Biol. Cell 11, 4309–4321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allen K. N., Dunaway-Mariano D. (2004) Trends Biochem. Sci. 29, 495–503 [DOI] [PubMed] [Google Scholar]
- 34.Loret M. O., Pedersen L., François J. (2007) Yeast 24, 47–60 [DOI] [PubMed] [Google Scholar]
- 35.Hillyard D., Rechsteiner M., Manlapaz-Ramos P., Imperial J. S., Cruz L. J., Olivera B. M. (1981) J. Biol. Chem. 256, 8491–8497 [PubMed] [Google Scholar]
- 36.Cheng W., Roth J. (1995) J. Bacteriol. 177, 6711–6717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grose J. H., Bergthorsson U., Xu Y., Sterneckert J., Khodaverdian B., Roth J. R. (2005) J. Bacteriol. 187, 4521–4530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gazzaniga F., Stebbins R., Chang S. Z., McPeek M. A., Brenner C. (2009) Microbiol. Mol. Biol. Rev. 73, 529–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hayakawa T., Shibata K., Iwai K. (1984) Agric. Biol. Chem. 48, 445–453 [Google Scholar]
- 40.Fabrizio P., Battistella L., Vardavas R., Gattazzo C., Liou L. L., Diaspro A., Dossen J. W., Gralla E. B., Longo V. D. (2004) J. Cell Biol. 166, 1055–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.