Abstract
Polytopic membrane proteins subjected to endoplasmic reticulum (ER)-associated degradation are extracted from membranes and targeted to proteasomes for destruction. The extraction mechanism is poorly understood. One polytopic ER protein subjected to ER-associated degradation is Insig-1, a negative regulator of cholesterol synthesis. Insig-1 is rapidly degraded by proteasomes when cells are depleted of cholesterol, and its degradation is inhibited when sterols accumulate in cells. Insig-2, a functional homologue of Insig-1, is degraded slowly, and its degradation is not regulated by sterols. Here, we report that a single amino acid substitution in Insig-2, Insig-2(L210A), causes Insig-2 to be degraded in an accelerated and sterol-regulated manner similar to Insig-1. In seeking an explanation for the accelerated degradation, we found that proteasomes bind to wild type Insig-1 and mutant Insig-2(L210A) but not to wild type Insig-2, whereas the proteins are still embedded in cell membranes. This binding depends on at least two factors, ubiquitination of Insig and association with the ATPase p97/VCP complex. These data suggest that p97 recruits proteasomes to polytopic ER proteins even before they are extracted from membranes.
Introduction
Many transmembrane proteins of the endoplasmic reticulum (ER)4 are degraded via the ER-associated degradation (ERAD) pathway (1, 2). According to the current model, ERAD begins when selected membrane proteins are recognized and ubiquitinated by one of several ER-associated E3 ubiquitin ligases. Through the action of the ATPase p97/VCP complex (hereafter referred to as p97), these ubiquitinated membrane proteins are extracted from membranes and translocated into the cytosol, where they are degraded by proteasomes. The mechanism by which polytopic transmembrane proteins are unwound and extracted from membranes is unknown.
The 26 S proteasome is a protein degradation machine that consists of a 20 S catalytic core and a 19 S regulatory portion composed of 14 and 19 unique protein subunits, respectively (3, 4). Proteins subjected to ERAD are recognized by proteasomes through polyubiquitin chains that are added to these proteins by ubiquitin ligases (5). Whether proteasomes bind to ubiquitinated membrane proteins before or after their extraction from membranes is unknown.
In mammalian cells ERAD of some proteins can be regulated by small molecule metabolites. One such regulated protein is Insig-1, an ER membrane protein with six membrane-spanning helices (6). Insig-1 is an ER anchor protein that mediates feedback inhibition of cholesterol synthesis. When excess sterols accumulate in cells, Insig-1 is induced to bind to either of two ER membrane proteins; that is, Scap or 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase (6). Scap is an escort protein that carries sterol regulatory element-binding proteins (SREBPs) to the Golgi complex (7). SREBPs are membrane-bound transcription factors that must be transported to the Golgi complex for processing to release nuclear fragments that activate all of the genes required for cholesterol synthesis and uptake (8, 9). Certain SREBPs also activate the synthesis of unsaturated fatty acids (8, 9). In cholesterol-loaded cells Insig-1 binds to Scap, and this causes ER retention of Scap, thereby preventing translocation of Scap-bound SREBPs to the Golgi (10). As a result, cholesterol synthesis and uptake are reduced. Sterols also induce binding of Insig-1 to HMG-CoA reductase, which results in ubiquitination and subsequent proteasomal degradation of the reductase (11, 12). This degradation slows the production of mevalonate, a rate-limiting step in cholesterol synthesis (13). The two actions of Insig-1 serve to prevent over-accumulation of cholesterol and its sterol precursors.
We previously showed that Insig-1 itself is subject to ERAD in a reaction that is regulated by sterols and unsaturated fatty acids (14, 15). In cholesterol-depleted cells, Insig-1 does not bind to Scap or HMG-CoA reductase. Instead, Insig-1 binds to gp78, an E3 ubiquitin ligase (16). This interaction results in ubiquitination of Insig-1 (16). When cells are treated with sterols, Scap binds to Insig-1, displacing gp78 from Insig-1 (16). Consequently, ubiquitination of Insig-1 is inhibited, and Insig-1 is stabilized (16). Degradation of ubiquitinated Insig-1 requires two other proteins, Ubxd8 and p97 (15). RNAi-mediated knockdown of either of these proteins does not block ubiquitination of Insig-1, but it does prevent its degradation. When unsaturated fatty acids are added to cells, Insig-1 is stabilized even in the absence of sterols. Fatty acids do not block Insig-1 ubiquitination. Rather, they stabilize ubiquitinated Insig-1 by blocking the interaction between Insig-1 and the Ubxd8·p97 complex so that ubiquitinated Insig-1 cannot be extracted from the membranes (15).
Mammalian cells also contain a close homolog of Insig-1, namely Insig-2, which functions like Insig-1 to block cholesterol synthesis (17). Unlike Insig-1, Insig-2 does not interact with gp78 and, therefore, it is not ubiquitinated (16). As a result, Insig-2 has a much longer half-life, and its degradation is not regulated by sterols (18, 19).
In the current study we characterize a mutant Insig-2, Insig-2(L210A), which retains all the functions of Insig-2 but is degraded in a sterol-regulated manner like Insig-1. We show that proteasomes bind to ubiquitinated mutant Insig-2(L210A) and wild type Insig-1 (but not to wild type Insig-2), whereas the proteins are still embedded in ER membranes. This binding requires the p97 complex, and it appears to precede the extraction and degradation of the Insig proteins. These studies raise the possibility that proteasomes together with p97 play a direct role in the extraction of polytopic proteins from ER membranes.
EXPERIMENTAL PROCEDURES
Materials
We obtained hydroxypropyl-β-cyclodextrin from Cyclodextrin Technologies Development, arachidonic acid from NU-CHEK-PREP, INC, defatted bovine serum albumin from Roche Applied Science, Nonidet P-40 Alternative (Nonidet P-40) and digitonin from Calbiochem, Fos-choline 13 from Anatrace, MG-132 from Boston Biochem, N-Succinyl-Leu-Leu-Val-Tyr-7-amido-4-methylcoumarin (N-Suc-Leu-Leu-Val- Tyr-AMC) from Bachem. cycloheximide, sodium salt of ATP, 3× FLAG peptide, anti-c-Myc-agarose affinity gel, anti-FLAG-M2-agarose affinity gel, and rabbit polyclonal anti-FLAG from Sigma, IgG-Sepharose 6 Fast Flow, CyDye DIGE Fluor minimal labeling kit, and Deep Purple Total Protein Stain from GE Healthcare, rabbit polyclonal anti-Myc, anti-hemagglutinin (HA), anti-T7, and ReliaBLOT from Bethyl Laboratories, mouse monoclonal anti-HSV IgG and T7-tag antibody-agarose from Novagen, mouse monoclonal anti-β1 IgG, anti-α7 IgG, and anti-Rpt6 IgG from Biomol, mouse monoclonal anti-p97 IgG from BD Transduction Laboratories, and horseradish peroxidase-conjugated donkey anti-mouse and anti-rabbit IgGs (affinity-purified) from Jackson ImmunoResearch. Hybridoma cells producing IgG-9E10, a mouse monoclonal antibody against Myc tag, were obtained from the American Type Culture Collection (Manassas, VA). IgG-4H4, a mouse monoclonal antibody against hamster Scap, was generated by immunizing a mouse with a fusion protein encoding amino acids 1–767 of Scap that was epitope-tagged with 10 histidine residues at the NH2 terminus (20). Rabbit polyclonal antibodies against human gp78 (16) and human Rpt5 (21) were reported previously. Lipoprotein-deficient serum (LPDS) (22), delipidated fetal calf serum (23), sodium oleate (22), sodium mevalonate (24), and compactin (24) were prepared as described in the indicated references. SR-12813 was synthesized by the Core Medicinal Chemistry Laboratory, University of Texas Southwestern Medical Center (25). A stock solution of 10 mm sodium arachidonate-bovine serum albumin in 0.15 m NaCl (final pH 7.6) was prepared as previously described (23).
Plasmid Constructs
The following plasmids were described in the indicated references or obtained from the indicated sources. pTK-Scap and pCMV-Scap encode hamster Scap under control of the thymidine kinase (TK) and the cytomegalovirus (CMV) promoters, respectively (14). pTK-HSV-SREBP2 encodes HSV-tagged human SREBP-2 under control of the TK promoter (26). pCMV-HMGCoAR(TM1–8)-T7 encodes amino acids 1–346 of hamster HMG-CoA reductase followed by three tandem copies of the T7 epitope tag under control of the CMV promoter (11). pCMV-Myc-Ubxd8 encodes Myc-tagged human Ubxd8 under control of the CMV promoter (15). pEF1a-HA-ubiquitin (provided by Dr. Zhijian Chen, University of Texas Southwestern Medical Center) encodes amino acids 1–76 of human ubiquitin tagged with HA epitope tag at the NH2 terminus under control of the EF1a promoter (16). pCIneo-gp78 (obtained from Dr. Allan M. Weissman, National Cancer Center) encodes human gp78 under control of the CMV promoter (16). pCMV-Insig1-T7 and pCMV-Insig2-T7 encode human Insig-1 and Insig-2, respectively, each tagged with three tandem copies of the T7 epitope tag at the COOH terminus under control of the CMV promoter (16). pTK-Insig2-Myc and pCMV-Insig2-Myc encode wild type human Insig-2, each tagged with six copies of the Myc epitope tag at the COOH terminus under control of the TK and the CMV promoters, respectively (26). pCMV-FLAG-Insig1 encodes human Insig-1 with two tandem copies of a FLAG epitope tag under control of the CMV promoter (27). pCMV-Insig2-FLAG-TEV-Protein A encodes wild type human Insig-2, sequentially tagged at the COOH terminus with three tandem copies of the FLAG epitope tag, one copy of the TEV protease cleavage site, and two copies of Protein A under control of the CMV promoter. Mutants of Insig-1 or Insig-2 were generated with the QuikChange XL site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. The integrity of all plasmids was confirmed by DNA sequencing of all open reading frames.
Tissue Culture Medium
Medium A is a 1:1 mixture of Ham's F-12 medium and Dulbecco's modified Eagle's medium containing 100 units/ml penicillin and 100 μg/ml streptomycin sulfate. Medium B is medium A containing 5% (v/v) LPDS, 50 μm sodium compactin, and 50 μm sodium mevalonate. Medium C is Dulbecco's modified Eagle's medium (1 g/liter glucose) containing 10% (v/v) fetal calf serum (FCS), 100 units/ml penicillin, and 100 μg/ml streptomycin sulfate. Medium D is medium C supplemented with 5 μg/ml cholesterol (added in ethanol; final concentration, 0.05%), 1 mm sodium mevalonate, and 20 μm sodium oleate. Medium E contains IS GRO (Irvine Scientific) supplemented with 10% (v/v) FCS, 10 mm l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin sulfate. Medium F is Dulbecco's modified Eagle's medium (1 g/liter glucose) containing 10% (v/v) delipidated FCS, 50 μm sodium compactin, 50 μm sodium mevalonate, 100 units/ml penicillin, and 100 μg/ml streptomycin sulfate.
Cell Culture
Human 293S cells, a clone of HEK-293 cells adapted to suspension culture (28), were grown in monolayer at 37 °C in 5% CO2. HEK-293S/pInsig1 cells is a line of HEK-293S that stably expresses pCMV-Insig1-T7-TEV-Protein A (containing the gene encoding Zeocin resistance) (15). HEK-293S/pInsig2 and HEK-293S/pInsig2(L210A) cells are lines of HEK-293S cells that stably express pCMV-Insig2-FLAG-TEV-Protein A and pCMV-Insig2(L210A)-FLAG-TEV-Protein A, respectively. They were generated by transfecting HEK-293S cells with pCMV-Insig2-FLAG-TEV-Protein A or pCMV-Insig2(L210A)-FLAG-TEV-Protein A (both plasmids containing the Zeocin resistance gene) followed by selection with 700 μg/ml Zeocin. Monolayers of stably transfected HEK-293S cells were grown in 5% CO2 at 37 °C in medium D containing 500 μg/ml Zeocin. SRD-14/pTK-Insig1-Myc cells, a line of mutant CHO cells lacking endogenous Insig-1 that stably express pTK-Insig1-Myc (14), were grown at 37 °C in 8% CO2 in medium A supplemented with 5% LPDS and 500 μg/ml G418. SRD-13A cells, a Scap-deficient cell line derived from CHO-7 cells (29), were grown in monolayer at 37 °C in 8% CO2 in medium A supplemented with 5% FCS, 5 μg/ml cholesterol, 1 mm sodium mevalonate, and 20 μm sodium oleate. SRD-15 cells, an Insig-1/Insig-2-deficient cell line (30), were maintained in medium A supplemented with 5% LPDS, 10 μm of SR-12813, and 1 μg/ml 25-hydroxycholesterol (25-HC). Monolayers of SV-589 cells, an immortalized line of human fibroblasts expressing the SV40 large T antigen (31), were grown at 37 °C in 5% CO2 in medium C.
Transient Transfection of Cells and Immunoblot Analysis
SRD-13A, SRD-15, and SV589 cells were transiently transfected with FuGENE 6 reagent (Roche Applied Science) according to the manufacturer's protocol. Conditions of incubation after transfection are described in the figure legends. After SDS/PAGE (8 or 10% gels), the proteins were transferred to Hybond-C extra nitrocellulose filters (GE Healthcare). The filters were incubated at room temperature with one of the following primary antibodies: 0.5 μg/ml anti-HSV IgG; 1 μg/ml IgG-9E10; 0.5 μg/ml rabbit polyclonal anti-Myc, rabbit polyclonal anti-T7 and rabbit polyclonal anti-HA; 0.16 μg/ml rabbit polyclonal anti-FLAG; 5 μg/ml IgG-4H4; 1 μg/ml rabbit polyclonal anti-gp78; 1:1000 dilution of mouse monoclonal anti-p97 IgG, anti-β1 IgG, anti-α7 IgG, and anti-Rpt6 IgG; 1:5000 dilution of rabbit polyclonal anti-Rpt5. Except for Rpt5 immunoblots shown in Fig. 6A in which we used ReliaBLOT horseradish peroxidase conjugate (1:5,000 dilution) as the secondary antibody, horseradish peroxidase-conjugated donkey anti-mouse and anti-rabbit IgGs (1:5000 dilution) were used as the secondary antibody in all other immunoblot analyses. Bound antibodies were visualized by chemiluminescence (SuperSignal Substrate; Pierce) according to the manufacturer's instructions. Filters were exposed to Kodak X-Omat Blue XB-1 film at room temperature for 1–60 s.
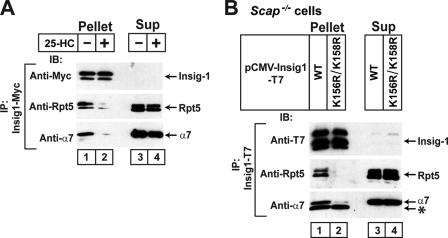
FIGURE 6.
Sterol depletion and ubiquitinated lysines required for association of Insig-1 with proteasomal subunits. A, on day 0 SRD-14/pTK-Insig1-Myc cells were set up at 7.0 × 105 cells/100-mm dish in medium A supplemented with 5% LPDS. On day 2 cells were switched to medium B. On day 3 cells were incubated with 10 μm MG-132 in the absence or presence of 1 μg/ml 25-HC. After incubation for 4 h, cells were harvested and lysed, and Insig-1 was immunoprecipitated from the cell lysate with anti-Myc. Pellets (representing 0.5 dish of cells for Insig-1 and 6.5 dishes of cells for proteasomal subunits) and supernatants (representing 0.033 dish of cells for Insig-1 and 0.43 dish of cells for proteasomal subunits) were subjected to SDS/PAGE and immunoblot analysis with IgG-9E10 (against Insig-1), anti-Rpt5, and anti-α7. IP, immunoprecipitate; IB, immunoblot; Sup, supernatant fraction. B, on day 0 Scap-deficient SRD-13A cells were set up at 1.6 × 105 cells/60-mm dish. On day 1 each dish of cells was transfected with 0.2 μg pCMV-Insig1-T7 or pCMV-Insig1(K156R/K158R)-T7 in medium A supplemented with 5% FCS. On day 4 cells were incubated with 10 μm MG-132 for 2 h and then harvested, lysed, and immunoprecipitated with anti-T7 to precipitate Insig-1. Pellets (representing 0.5 dish of cells for Insig-1 and 7.7 dish of cells for proteasome subunits) and supernatants (representing 0.033 dish of cells for Insig-1 and 0.51 dish of cells for proteasome subunits) were subjected to SDS/PAGE and immunoblot analysis with anti-T7 (against Insig-1), anti-Rpt5, and anti-α7. The asterisk (*) denotes IgG light chain.
RNA Interference (RNAi)
Duplexes of small interfering RNA (siRNA) were synthesized by Dharmacon Research (Lafayette, CO). The siRNAs were designed to target nucleotide positions 1130–1148 (relative to the codon for the initiating methionine) for human gp78 (GenBankTM accession number NM_001144) and positions 1095–1113 for human p97 (GenBankTM accession number NM_007126). SV589 cells were transfected with 400 pmol of siRNA duplexes/60-mm dish using OligofectAMINETM reagent (Invitrogen). HEK-293S/pInsig1 cells and HEK-293S/pInsig2(L210A) cells were transfected with 400 pmol of siRNA duplexes/100-mm dish using LipofectamineTM RNAiMAX reagent (Invitrogen). After transfection the cells were used for experiments as described in the legend to Fig. 7.
FIGURE 7.
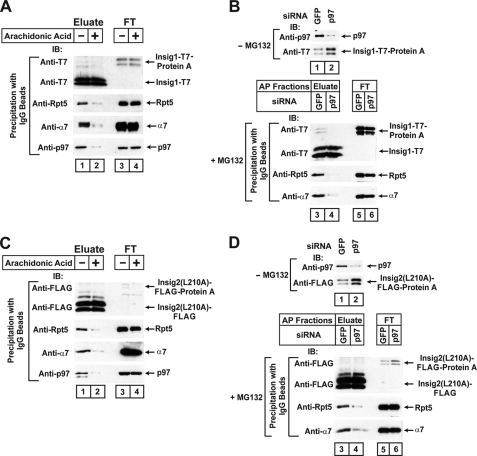
p97 is required for the interaction between proteasomes and Insig proteins. A, on day 0 HEK-293S/pInsig1 cells were set up at 5 × 105 cells/100-mm dish in medium C. On day 3 cells were switched to medium F. On day 4 cells were incubated with 10 μm MG-132 in the absence or presence of 100 μm sodium arachidonate-albumin. After incubation for 6 h, cells were harvested, and Insig-1 was precipitated from detergent-solubilized 100,000 × g membranes as described under “Experimental Procedures.” The eluate (representing 3.2 dishes of cells) and flow-through (FT) fraction (representing 0.21 dish of cells) were subjected to immunoblot analysis with anti-T7 (against Insig-1), anti-Rpt5, anti-α7, and anti-p97. IB, immunoblot. B, on day 0 HEK-293S/pInsig1 cells were set up at 4 × 105 cells/100-mm dish in medium C. On day 1 cells were transfected with 400 pmol/dish of the indicated siRNA. After incubation for 4 h, cells were switched to medium F. On day 2 dishes in lanes 3-6 received a direct addition of MG-132 at a final concentration of 10 μm. After incubation for 6 h, the cells were harvested. Aliquots of cell lysates (5 μg protein) from cells not receiving MG-132 (lanes 1 and 2) were subjected to immunoblot analysis with the indicated antibody. Cell lysates from cells receiving MG-132 (lanes 3–6) were subjected to affinity precipitation (AP) of Insig-1 with IgG-Sepharose beads, after which the eluates and flow-through (FT) fractions were subjected to immunoblot analysis with the indicated antibody as described in A. C, on day 0 HEK-293S/pInsig2(L210A) cells were set up 4 × 105 cells/100-mm dish in medium D. The interaction of proteasome subunits with Insig-2(L210A) was determined as described in A. D, on day 0 HEK-293S/pInsig2(L210A) cells were set up at 3 × 105 cells/100-mm dish in medium D. The interaction of proteasome subunits with Insig-2(L210A) was determined as described in B.
Tandem Affinity Purification and Two-dimensional Fluorescence Difference Gel Electrophoresis (Two-dimensional DIGE)
Membrane fractions from 2 liters of HEK-293S/pInsig2 cells and 4 liters of HEK-293S/pInsig2(L210A) cells were prepared as described previously (10) and solubilized with buffer A (50 mm Hepes-KOH (pH 7.6), 150 mm NaCl, and a protease inhibitor mixture containing 25 μg/ml N-acetyl-leucinal-leucinal-norleucinal, 5 μg/ml pepstatin A, 2 μg/ml aprotinin, and 10 μg/ml leupeptin) supplemented with 0.1% (w/v) Nonidet P-40. After incubation for 1 h, the insoluble material was removed by centrifugation at 100,000 × g for 1 h. All subsequent operations were carried out at 4 °C. The supernatant was incubated in a tube containing IgG-Sepharose 6 Fast Flow beads. After 16 h the beads were transferred to a Bio-Spin disposable chromatography column (Bio-Rad) and washed with 20× bead-volume of buffer A containing 0.05% Nonidet P-40 and 5 mm sodium EDTA. The beads were then incubated for 2 h with buffer A containing 0.05% Nonidet P-40, 5 mm sodium EDTA, and 50 units/ml of AcTEVTM protease (Invitrogen) followed by elution. The resulting eluate (2 ml) plus an additional 12 ml of wash with buffer A containing 0.05% Nonidet P-40 were applied 5 times to a column containing anti-FLAG-M2-agarose affinity gel followed by a wash with 20× bead-volumes of buffer A containing 0.05% Nonidet P-40. The washed beads were then transferred to a 2-ml tube and incubated for 2 h with buffer A containing 0.05% Nonidet P-40 and 0.3 mg/ml 3× FLAG peptide followed by elution. The elution procedure was repeated 3 times. The pooled eluates were concentrated ∼100-fold with an Amicon Ultra-15 Centrifugal Filter Device (10-kDa cut off; Millipore) and diluted to a final volume of 240 μl in buffer containing 0.5 mm Hepes-KOH (pH 7.6), 1.5 mm NaCl, and 0.5% Nonidet P-40. Each sample was precipitated overnight at −80 °C with 5 volumes of cold acetone and centrifuged at 20,000 × g for 30 min. The resulting pellet was dried and solubilized in 65 μl of buffer B (30 mm Tris-HCl (pH 8.8), 2 m thiourea, 7 m urea, 1% Nonidet P-40). Aliquots of the solubilized pellets (20 μl) from the HEK-293S/pInsig2 cells and HEK-293S/pInsig2(L210A) cells were each labeled with 200 pmol of the CyDye fluorochromes Cy3 and Cy5, respectively. Separate aliquots (10 μl) from each cell line were mixed together and then labeled with 200 pmol of Cy2 as the internal standard. Both labelings were done with the CyDye DIGE Fluor minimal labeling kit. Labeling was performed in the dark with a 30-min incubation, after which the reaction was quenched by the addition of 1 μl of 10 mm lysine. After incubation for 10 min, the labeled proteins were separated with the Ettan IPGphor 3 IEF System (first dimension) and 10% SDS/PAGE (second dimension) according to the manufacturer's protocol (GE Healthcare). After the second dimensional electrophoresis, fluorophore-labeled protein gels were scanned using a Typhoon TRIO+ variable mode imager (GE Healthcare) at 100-μm resolution. Spot detection and quantification were performed using the DeCyder two-dimensional differential analysis software (Version 6.5; GE Healthcare). The position of the spots that had a >3.5-fold difference in intensity between the eluates from the two cell lines were recorded. To determine the protein identity in these spots, the eluate from 8 liters of HEK-293S/pInsig2(L210A) cells was subjected to two-dimensional electrophoresis as described above and stained with the Deep Purple Total Protein Stain. Protein spots were picked by Ettan Spot Picker (GE Healthcare) and analyzed by nano-high pressure liquid chromatography-tandem mass spectrometry at the Protein Chemistry Core Facility at the University of Texas Southwestern Medical Center.
Immunoprecipitation and Protein A Affinity Purification
All operations were carried out at 4 °C. Immunoprecipitations of Insig proteins from detergent-solubilized cell extracts of SRD-14/pTK-Insig1-Myc cells and transiently transfected SRD-13A cells were performed as previously described (26) except that 1% (w/v) digitonin instead of 0.1% Nonidet P-40 was used in the experiment shown in Fig. 6A.
Protein A affinity purification of Insig proteins from stably transfected HEK-293S cells were carried out as follows. For experiments in Figs. 4, A and B, 5, 7, A and C, pooled pellets were lysed with a Dounce homogenizer in buffer C (50 mm Hepes-KOH (pH 7.6), 5 mm MgCl2, 10 mm KCl, 5 mm sodium EDTA, 5 mm EGTA, 1 mm sodium ATP, 250 mm sucrose). Cell lysates were centrifuged at 10,000 × g for 15 min, and the resulting supernatants were centrifuged at 100,000 × g for 60 min to isolate the membrane fractions. Each membrane fraction was solubilized with buffer D (50 mm Hepes-KOH (pH 7.6), 20 mm NaCl, 5 mm MgCl2, 1 mm sodium ATP, 10% (v/v) glycerol) supplemented with 0.1% Nonidet P-40. For the experiments in Figs. 7, B and D, cells were lysed directly in buffer D supplemented with 0.1% Nonidet P-40. After incubation for 1 h, the insoluble material was removed by centrifugation at 100,000 × g for 1 h. The supernatant was applied to a Bio-Spin disposable chromatography column containing IgG-Sepharose 6 Fast Flow beads, and the suspension of Sepharose beads was rotated for 16 h. The first flow-through was saved. The beads were then washed with a 30× bead-volumes of buffer D containing 0.01% Nonidet P-40 after which the bound material was eluted by incubation for 1.5 h in buffer D containing 0.01% Nonidet P-40 and 25 units/ml of AcTEV protease. The eluate (2 ml) plus an additional 1 ml of wash was concentrated 10 times to a final volume of 300 μl. One aliquot of the concentrated solution was dialyzed and used for assay of proteasomal activity (see below). A second aliquot was used for SDS/PAGE and immunoblot analysis. In some experiments the membranes were washed once by resuspension in buffer and centrifuged again at 100,000 × g for 60 min.
FIGURE 4.
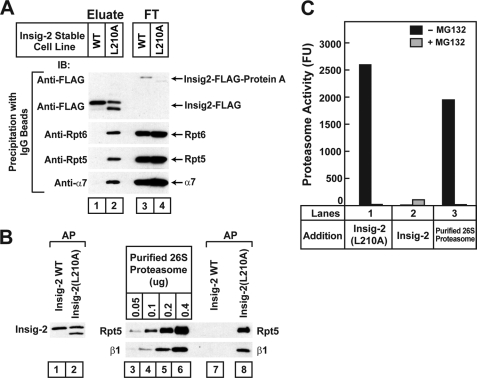
Membrane-bound Insig-2(L210A) associates with proteasomal subunits. A, on day 0 HEK-293S/pInsig2 and HEK-293S/pInsig2(L210A) cells were set up at 2.0 × 105 cells/ml and grown in suspension for 6 days in medium E at 37 °C (without CO2). On day 7 cells were treated with 10 μm MG-132 for 2 h and then harvested. A 100,000 × g membrane pellet was prepared from pooled cell pellets of HEK-293S/pInsig2 cells (0.3 liter of cells) and HEK-293S/pInsig2(L210A) cells (0.6 liter of cells). The membranes were solubilized, and Insig-2 was isolated by affinity precipitation with IgG-Sepharose beads as described under “Experimental Procedures.” Aliquots of eluates from the beads (representing 16.7% of cells) and the non-absorbed flow-through (FT) fraction (representing 0.42% of cells) were subjected to SDS/PAGE and immunoblot analysis with anti-FLAG (against Insig-2), anti-Rpt6, anti-Rpt5, and anti-α7. IB, immunoblot. B, cells were set up as described in A. 0.4 liter of HEK-293S/pInsig2 cells were mixed with 1.6 liter of HEK-293S cells. Cells pelleted from this mixture and from 2 liters of HEK-293S/pInsig2(L210A) cells were harvested, and the 100,000 × g membrane fractions were prepared. After washing once with buffer D, the membranes were solubilized, and Insig-2 was subjected to IgG-Sepharose affinity precipitation (AP) as described above. Precipitated material (0.1 liter of cells) was subjected to SDS/PAGE and immunoblot analysis with anti-FLAG (against Insig-2), anti-Rpt5, and anti-β1. The indicated amount of purified 26 S proteasomes was also subjected to SDS/PAGE and immunoblot analysis with anti-Rpt5 or anti-β1. C, shown is proteasomal activity of material coprecipitating with mutant Insig-2(L210A) but not wild type Insig-2. Each reaction was carried out as described under “Experimental Procedures” in the absence or presence of 50 μm MG-132 as indicated and one of the following sources of proteasomes; lanes 1 and 2, precipitated material from B representing 0.3 liter of the indicated cells; lane 3, 0.47 μg of purified 26 S proteasome. After incubation for 21 min at 37 °C, the amount of cleaved AMC liberated by proteasomes was quantified by fluorometry. A blank reading in reactions containing only buffer D and fluorescent substrate (507–632 fluorescence units (FU)) was subtracted from each value. Each value is the average of duplicate determinations. Similar results were obtained in two other independent experiments.
FIGURE 5.
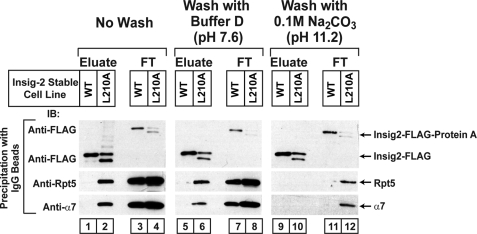
Membrane-bound Insig-2(L210A) associates with proteasomal subunits. On day 0 HEK-293S/pInsig2 and HEK-293S/pInsig2(L210A) cells were set up at 2.0 × 105 cells/ml and grown in suspension for 6 days in medium E at 37 °C (without CO2). On day 7 cells were treated with 10 μm MG-132 for 2 h and harvested. Pooled cell pellets of HEK-293S/pInsig2 cells (1 liter of cells) and HEK-293S/pInsig2(L210A) cells (2 liters of cells) were harvested, and the 100,000 × g membrane pellet was obtained as described under “Experimental Procedures.” The membrane pellet was resuspended in the indicated buffer, incubated at 4 °C for 30 min, and pelleted again at 100,000 × g. The washed membranes were detergent-solubilized and subjected to affinity precipitation with IgG-Sepharose beads to precipitate Insig-2. The eluate (representing 5.6% of cells) and flow-through (FT) fraction (representing 0.14% of cells) were subjected to SDS/PAGE and immunoblot analysis with anti-FLAG (against Insig-2), anti-Rpt5, and anti-α7. IB, immunoblot.
Purification of Proteasomes
26 S proteasomes were purified from bovine red blood cells as previously described (32).
Proteasome Activity Assay
Each concentrated, solubilized Protein A precipitate from detergent-solubilized 100,000 × g membranes (see above) was dialyzed for 44 h at 4 °C in 8 liters of buffer E (20 mm Tris-HCl (pH 7.6), 5 mm MgCl2, 1 mm sodium ATP, 1 mm 2-mercaptoethanol, 10% (v/v) glycerol) using a Slide-A-Lyzer dialysis cassette (10-kDa cut off; Pierce). Proteasome activity was assessed by measuring the hydrolysis of N-Suc-Leu-Leu-Val-Tyr-AMC as previously described (33). Each reaction contained in a final volume of 75 μl 58 mm Tris-HCl (pH 8.0), 5.6 mm dithiothreitol, 0.6 mm 2-mercaptoethanol, 13 mm MgCl2, 0.7 mm sodium ATP, 6% (v/v) glycerol, 0.1% Nonidet P-40, 50 μm N-Suc-Leu-Leu-Val-Tyr-AMC, and a source of proteasomes as described in the legend to Fig. 4C.
Ubiquitination of Insig
Immunoprecipitation and immunoblot analysis to determine the ubiquitination of Insig proteins was done as previously described (16).
RESULTS
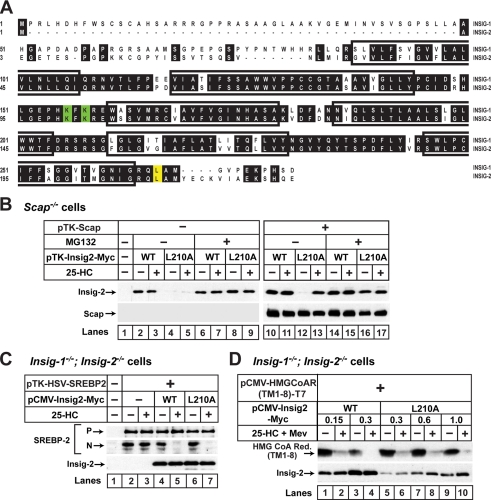
Fig. 1A compares the amino acid sequences of human Insig-1 and Insig-2 with the six putative transmembrane helices indicated. In a preliminary screen of mutagenized Insig-2 proteins, we encountered one point mutation (L210A) that caused Insig-2 to become subject to sterol-regulated ERAD, just like Insig-1. Leu-210 is located in the 18-residue cytoplasmic tail of Insig-2, indicated in yellow in Fig. 1A. Fig. 1B shows an experiment in which we compared the stability and function of wild type Insig-2 and Insig-2(L210A). We transfected cDNAs encoding wild type or mutant Insig-2 into SRD-13A cells, mutant CHO cells deficient in Scap (29). Some of the cells were subjected to cotransfection with a plasmid encoding Scap (Fig. 1B). The cells were then incubated in the presence or absence of 25-HC, an oxysterol that inhibits proteolytic activation of SREBPs by promoting binding of Insig proteins to Scap (10, 34). The amount of Insig-2 protein in these cells was then determined by immunoblot analysis. As expected, wild type Insig-2 was readily detectable, and its amount was not affected by treatment with 25-HC with or without Scap (Fig. 1B, lanes 2, 3, 10, and 11). In the absence of Scap, Insig-2(L210A) was barely visible regardless of treatment with 25-HC (Fig. 1B, lanes 4 and 5). When cells were treated with the proteasome inhibitor MG-132, the amount of Insig-2(L210A) increased to wild type levels, indicating that the low expression level was caused by accelerated degradation. When Scap was cotransfected, the amount of Insig-2(L210A) remained low in cells incubated in the absence of 25-HC, but treatment with the sterol increased the amount of Insig-2(L210A) to a level similar to that of wild type Insig-2 (Fig. 1B, lanes 12 and 13). Thus, unlike wild type Insig-2 but similar to Insig-1, the rapid degradation of Insig-2(L210A) is inhibited by sterols in a Scap-dependent manner. Importantly, the L210A mutation does not affect the ability of Insig-2 to inhibit SREBP-2 cleavage because expression of Insig-2(L210A) was as efficient as wild type Insig-2 in restoring the sterol-induced inhibition of proteolytic activation of SREBP-2 in mutant CHO cells deficient in both Insig-1 and Insig-2 (30) (Fig. 1C). Similar to wild type Insig-2, Insig-2(L210A) also accelerated the degradation of HMG-CoA reductase in these mutant CHO cells (Fig. 1D).
FIGURE 1.
L210A mutation destabilizes Insig-2 but preserves its ability to inhibit SREBP processing and to accelerate HMG-CoA reductase degradation. A, shown is amino acid sequence alignment of human Insig-1 and Insig-2. Identical residues are shown in black. Transmembrane segments (27) are boxed. Lysine residues that undergo ubiquitination in Insig-1 (14) are shown in green. The site of the L210A mutation of Insig-2 is shown in yellow. B, on day 0 Scap-deficient SRD-13A cells were set up at 3.5 × 105 cells/60-mm dish as described under “Experimental Procedures.” On day 2 each dish of cells was transfected with 0.3 μg of pTK-Insig2-Myc or 0.6 μg of pTK-Insig2(L210A)-Myc with or without 1.5 μg of pTK-Scap in medium A supplemented with 5% FCS. The total amount of DNA was adjusted to 2.1 μg/dish with pcDNA3 empty vector. After incubation for 16 h at 37 °C, the cells were switched to medium B containing 1% (w/v) hydroxypropyl-β-cyclodextrin for 1 h, after which the cells were washed twice with phosphate-buffered saline and switched to medium B in the absence or presence of 1 μg/ml 25-HC or 10 μm MG-132 as indicated. After incubation for 4 h at 37 °C, the cells were harvested and subjected to SDS/PAGE and immunoblot analysis with IgG-4H4 (against Scap) and anti-Myc (against Insig-2). C, on day 0 Insig-1/Insig-2-deficient SRD-15 cells were set up at a density of 5.0 × 105 cells/60-mm dish in medium A supplemented with 5% (v/v) LPDS. On day 1 each dish of cells was transfected with 2.5 μg of pTK-HSV-SREBP2 together with either 0.065 μg of pCMV-Insig2-Myc or 0.12 μg of pCMV-Insig2(L210A)-Myc in medium A supplemented with 5% LPDS, 5 μm compactin, and 50 μm sodium mevalonate. The total amount of DNA was adjusted to 2.62 μg/dish with pcDNA3 empty vector. After incubation for 16 h at 37 °C, the cells were switched to medium B containing 1% hydroxypropyl-β-cyclodextrin for 1 h, after which the cells were washed twice with phosphate-buffered saline and switched to medium B in the absence or presence of 1 μg/ml 25-HC. After incubation for 2 h at 37 °C, the cells were treated with 25 μg/ml N-acetyl-leucinal-leucinal-norleucinal for 2 h and then harvested, fractionated, and subjected to SDS/PAGE and immunoblot analysis with anti-HSV (against SREBP-2) and anti-Myc (against Insig-2). N and P denote the cleaved nuclear and precursor forms of SREBP-2, respectively. D, Insig-1/Insig-2 deficient SRD-15 cells were set up and transfected with 1 μg of pCMV-HMGCoAR(TM1–8)-T7 and the indicated amount of pCMV-Insig2-Myc or pCMV-Insig2(L210A)-Myc as described in D. After incubation for 16 h at 37 °C, the cells were switched to medium B containing 1% hydroxypropyl-β-cyclodextrin for 1 h, after which the cells were washed twice with phosphate-buffered saline and switched to medium B in the absence or presence of 1 μg/ml 25-HC and 10 mm sodium mevalonate (Mev). After incubation for 6 h at 37 °C, the cells were harvested and subjected to SDS/PAGE and immunoblot analysis with anti-T7 (against HMG-CoA reductase (Red.)) and anti-Myc (against Insig-2).
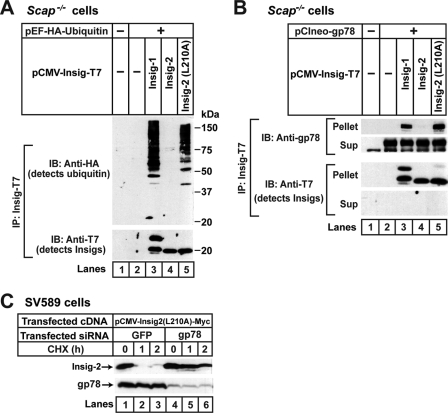
We showed previously that Insig-1 is ubiquitinated by gp78 and rapidly degraded by proteasomes in SRD-13A cells that are deficient in Scap (14, 16). In contrast to Insig-1, Insig-2 does not bind to gp78 (16), so it is neither ubiquitinated nor rapidly degraded in these cells (19). Inasmuch as Insig-2(L210A) is degraded in a way that is similar to Insig-1, we examined whether the mutant Insig-2 is ubiquitinated by gp78 in SRD-13A cells. To determine the ubiquitination of Insig proteins, we transfected SRD-13A cells with a plasmid encoding ubiquitin tagged with an epitope derived from the influenza HA protein together with a plasmid encoding epitope-tagged wild type or mutant Insig. The cells were treated with MG-132 to block degradation of ubiquitinated Insig. Insig proteins were then immunoprecipitated from the cell lysates, and the precipitates were subjected to immunoblot analysis with anti-HA to detect ubiquitinated proteins. Ubiquitinated Insig-1 and Insig-2(L210A) were visualized as high molecular weight smears in the anti-HA immunoblot (Fig. 2A, lanes 3 and 5). Ubiquitination of wild type Insig-2 was not detected (Fig. 2A, lane 4).
FIGURE 2.
Insig-2(L210A) is ubiquitinated by gp78. A, on day 0 Scap-deficient SRD-13A cells were set up at 3.5 × 105 cells/60-mm dish as described under “Experimental Procedures.” On day 2 each dish of cells was switched to medium A supplemented with 5% FCS and transfected with 0.2 μg of pEF1a-HA-ubiquitin together with one of the following plasmids: 0.1 μg pCMV-Insig1-T7, 0.3 μg pCMV-Insig2-T7, or 1.2 μg pCMV-Insig2(L210A)-T7. The total amount of DNA was adjusted to 1.4 μg/dish with pcDNA3 empty vector. After incubation for 16 h at 37 °C, the cells were incubated with 10 μm MG-132 for 30 min and harvested for preparation of detergent-solubilized cell lysates, which were subjected to immunoprecipitation with anti-T7 to precipitate Insigs. The immunoprecipitates (IP) were analyzed by immunoblot analysis (IB) with anti-HA (against ubiquitin) and anti-T7 (against Insigs). B, SRD-13A cells were set up and transfected with 0.1 μg of pCIneo-gp78 and one of the plasmids, 0.15 μg of pCMV-Insig1-T7, 0.7 μg of pCMV-Insig2-T7, and 2.1 μg of pCMV-Insig2(L210A)-T7, as described in A. The total amount of DNA was adjusted to 2.2 μg/dish with pcDNA3 empty vector. After incubation for 16 h at 37 °C, the cells were treated with 10 μm MG-132 for 2 h and then harvested, lysed, and immunoprecipitated with anti-T7 to precipitate Insigs. Pellets (representing 0.5 dish of cells) and supernatants (Sup; representing 0.05 dish of cells) were subjected to SDS/PAGE and immunoblot analysis with anti-T7 (against Insigs) and anti-gp78. C, on day 0 SV589 cells were set up at 2.0 × 105 cells/60-mm dish in medium C. On day 1, cells were transfected with 0.6 μg of pCMV-Insig2(L210A)-Myc. After incubation for 4 h at 37 °C, cells were transfected with 400 pmol/dish siRNA duplexes targeting green fluorescent protein or gp78. On day 3 cells were treated with 50 μm cycloheximide (CHX). After the indicated period of time, cells were harvested, and detergent lysates were subjected to SDS/PAGE followed by immunoblot analysis with anti-Myc (against Insig-2) and anti-gp78.
We next examined whether Insig-2(L210A) interacts with gp78. For this purpose SRD-13A cells were cotransfected with a plasmid encoding gp78 and a plasmid encoding epitope-tagged wild type or mutant Insig proteins. Insig proteins were then immunoprecipitated from cell lysates, and the amount of gp78 coimmunoprecipitated was examined by immunoblot analysis. As expected, significant amounts of gp78 were coimmunoprecipitated with Insig-1 but not Insig-2 (Fig. 2B, lanes 3 and 4). gp78 was coimmunoprecipitated with Insig-2(L210A) in an amount that was similar to that precipitated with Insig-1 (Fig. 2B, lanes 3 and 5).
To determine whether gp78 is required for degradation of Insig-2(L210A), we transfected SV589 cells, a line of immortalized human fibroblasts (31) with a plasmid encoding the mutant Insig-2 protein. The cells were subsequently transfected with duplexes of siRNA targeting either gp78 or green fluorescent protein, a control mRNA not present in the cells. The cells were then treated with cycloheximide to block the synthesis of proteins. At various times after cycloheximide treatment, cells were harvested, and the amount of Insig-2(L210A) was determined by immunoblot analysis. As shown in Fig. 2C, Insig-2(L210A) was rapidly degraded in cells transfected with the control siRNA (Fig. 2C, lanes 1–3). The transfected Insig-2(L210A) was rapidly degraded even though we did not deplete the cells of cholesterol, presumably because the amount of overexpressed mutant Insig-2 was much more than that of endogenous Scap in SV589 cells. This degradation was significantly retarded in cells receiving siRNA targeting gp78 (Fig. 2C, lanes 4–6).
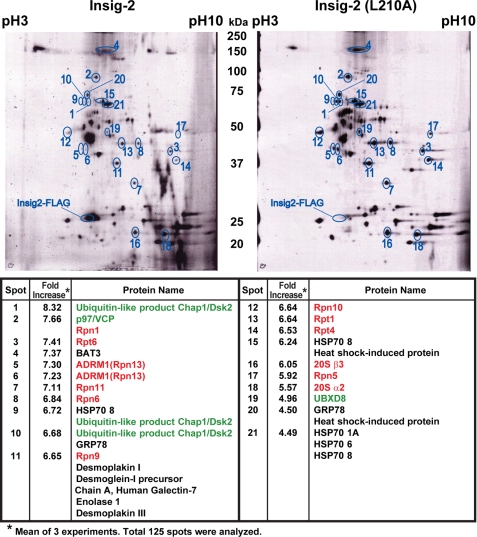
The results presented above show that the single point mutation L210A subjects Insig-2 to sterol-regulated degradation in a manner similar to wild type Insig-1 while maintaining the functions of Insig-2. To identify proteins that might participate in this degradation, we used HEK- 293S cells, a line of HEK-293 cells adapted to suspension culture (28). We stably transfected the cells with plasmids encoding Insig-2 and Insig-2(L210A), both fused with tandem Protein A and FLAG epitope tags to generate HEK-293S/pInsig2 and HEK-293S/pInsig2(L210A) cells, respectively. Membrane fractions from suspension cultures of these cells were prepared by ultracentrifugation. Wild type and mutant Insig-2 were then purified from detergent extracts of the membranes through sequential affinity purification, making use of the tandem tags. Proteins that copurified with Insigs were analyzed by two-dimensional fluorescence difference gel electrophoresis (Fig. 3). In three separate experiments we identified a total of 125 spots that copurified with Insig-2. 21 of these spots met the stringent criteria of showing in all three experiments a greater than 3.5-fold relative increase in membranes derived from the HEK-293S/pInsig2(L210A) cells as compared with cells expressing wild type Insig-2 (Fig. 3). Each of these 21 protein spots was eluted and subjected to sequencing by mass spectrometry. The majority of the peptides in each of these 21 spots corresponded to 26 S proteasome subunits (3, 4). Altogether, we detected 12 proteasome subunits or proteins that are known to associate with proteasomes, including p97 (35) and Dsk2 (36) (Fig. 3).
FIGURE 3.
Proteomic analysis of membrane-bound proteins that interact with Insig-2(L210A). On day 0 HEK-293S/pInsig2 and HEK-293S/pInsig2(L210A) cells were set up at 2.0 × 105 cells/ml and grown in suspension for 6 days in medium E at 37 °C (without CO2) as described under “Experimental Procedures.” On day 7 cells were treated with 10 μm MG-132 for 2 h and then harvested. A 100,000 × g membrane pellet was obtained and dissolved in detergent. Insig-2 and Insig-2(L210A) were isolated from this solution by tandem affinity purification. Proteins that copurified were analyzed by two-dimensional DIGE as described under “Experimental Procedures.” Three independent experiments were performed. Quantification of the intensity of the spots from one of these experiments is shown as a computer-generated image of the two-dimensional gel. Spots that had a >3.5-fold difference in intensity between the eluates from HEK-293S/pInsig2 and HEK-293S/pInsig2(L210A) cells in all three experiments are circled and numbered. Proteins in each spot were identified by mass spectrometry as described under “Experimental Procedures.” Proteasome subunits are highlighted in red; proteasome-associated proteins are highlighted in green.
To confirm the interaction between proteasome subunits and membrane-bound Insig2(L210A), we isolated wild type and mutant Insig-2, respectively, from the 100,000 × g pellet of membranes from HEK-293S/pInsig2 and HEK-293S/pInsig2(L210A) cells that had been treated with the proteasome inhibitor MG-132. After resuspending the membrane pellets in detergent, we added IgG-coupled Sepharose beads that bind to the Protein A tag fused to the Insig proteins. Wild type and mutant Insig-2 were eluted from the beads by incubation with AcTEV protease, which cleaves between the Protein A tag and Insig-2. The eluted material was subjected to immunoblot analysis with antibodies against proteasome subunits Rpt5, Rpt6, and α7 (Fig. 4A). Both wild type and mutant Insig-2 were bound and efficiently released by TEV protease from the IgG-Sepharose beads, as determined by immunoblotting against the FLAG epitope (Fig. 4A, lanes 1–4) (in these stably transfected cells, mutant Insig-2, but not the wild type protein, showed a lower molecular weight band, which was not seen in transiently transfected cells (Figs. 1, B–D); its origin is not known). The 19 S proteasome subunits Rpt5 and Rpt6 as well as the 20 S proteasome subunit α7 were coeluted with Insig-2(L210A) but not wild type Insig-2 (Fig. 4A, lanes 1 and 2). Blotting of the flow-through fractions showed that all of the cell extracts contained equal amounts of the proteasome subunits.
The results shown in Figs. 3 and 4A suggest that intact 26 S proteasomes may associate with Insig-2(L210A) on ER membranes. To confirm that the proteasomes are active, we precipitated wild type and mutant Insig-2 from 100,000 × g pellets of membranes from stably transfected HEK-293S cells that had been treated with the proteasome inhibitor MG-132 (Fig. 4B). In multiple experiments the level of precipitated wild type Insig-2 protein was consistently higher than that of the mutant Insig-2 even after treatment with MG-132. To equalize the amounts of precipitated wild type and mutant Insig-2, we prepared membranes from 0.4 liter of cells expressing wild type Insig-2 and 2 liters of cells expressing Insig-2(L210A). To equalize the amounts of total membrane protein in the precipitation reactions, we added 1.6 liters of untransfected HEK-293S cells to the 0.4 liter of HEK-293S cells expressing wild type Insig-2 to bring the total amount of cells to 2 liters. Cells were pelleted from the culture media, homogenized, and subjected to ultracentrifugation to isolate a 100,000 × g membrane pellet. Similar amounts of wild type and mutant Insig-2 were precipitated from the solubilized membranes (Fig. 4B, lanes 1 and 2). Rpt5 and β1, which are representative components of the 19 S and 20 S proteasome, respectively, were precipitated with mutant but not wild type Insig-2 (Fig. 4B, lanes 7 and 8). Calibration with purified 26 S proteasomes indicated that between 0.1 and 0.2 μg of proteasomes were coprecipitated with Insig-2(L210A) for each 0.1 liter of HEK-293S/pInsig2(L210A) cells (Fig. 4B, lanes 4, 5, and 8). After removing MG-132 by dialysis, the affinity-precipitated material from 0.3 liter of cells was assayed for proteasomal activity. Proteasomal activity corresponding to about 0.47 μg of purified 26 S proteasomes was precipitated with Insig-2(L210A) from 0.3 liter of cells, and this activity was inhibited by MG-132 in the assay buffer (Fig. 4C, lanes 1 and 3). Within experimental error, this number agrees with the immunoblots of Rpt-5 and β-1, which indicated that 0.1–0.2 μg of proteasomes was precipitated with Insig-2(L210A) from 0.1 liter of HEK-293S/pInsig2(L210A) cells (Fig. 4B).
The results presented in Figs. 3 and 4 show that active proteasomes were coprecipitated with Insig-2(L210A) in detergent lysates prepared from membrane fractions. We next examined whether the proteasomes could be released by washing the membranes with neutral or alkaline buffers. Proteasome subunits Rpt5 and α7 were coprecipitated with Insig-2(L210A) but not wild type Insig-2 when membranes were not washed (Fig. 5, lanes 1–4). Washing of membranes with a buffer at neutral pH did not affect the coprecipitation of Insig-2(L210A) with the proteasome subunits (Fig. 5, lanes 5–8). Washing of membranes with alkaline sodium carbonate buffer (pH 11), which is known to remove peripheral proteins from membranes, did not reduce the amount of Insig-2 in the precipitates (Fig. 5, lanes 2, 6, and 10), consistent with the fact that Insig-2 is an intrinsic membrane protein. However, the alkaline buffer reduced the amounts of proteasome subunits in both the flow-through fraction and the Insig-2(L210A) eluate. This indicates that the interaction of proteasomes with Insig-2(L210A), as well as other membrane proteins, is mediated by ionic forces.
Inasmuch as Insig-2(L210A) is degraded in a regulated manner that is similar to wild type Insig-1, we examined whether proteasomes also interact with Insig-1. For this purpose, we used SRD-14 cells, a mutant line of CHO cells deficient in Insig-1 (25). We stably transfected the cells with a plasmid encoding Myc epitope-tagged Insig-1 (SRD-14/pTK-Insig1-Myc cells). The amount of exogenous Insig-1 expressed in these cells is similar to that of endogenous Insig-1 in wild type CHO cells (14). These cells were treated with MG-132 in the presence or absence of 25-HC. Insig-1 was immunoprecipitated with anti-Myc from the cell lysate, and the amount of proteasome subunits coprecipitated was determined by immunoblot analysis. The two bands of Insig-1 represent translational products initiated at two different methionines, i.e. residues 1 and 37 (10). Proteasome subunits Rpt5 and α7 were coimmunoprecipitated with Insig-1 in cells incubated in the absence of 25-HC (Fig. 6A, lane 1). When cells were treated with 25-HC, the proteasome subunits no longer interacted with Insig-1 (Fig. 6A, lane 2). To confirm that ubiquitination is required for proteasomes to bind to Insig-1, we employed Insig-1(K156R/K158R), a mutant Insig-1 that is not ubiquitinated due to mutations in the two lysine residues that receive polyubiquitin chains (14). We transfected a plasmid encoding wild type Insig-1 or Insig-1(K156R/K158R) into SRD-13A cells. We used SRD-13A cells for this experiment because these cells do not express Scap, the inhibitor of Insig-1 ubiquitination (14, 16). As shown in Fig. 6B, proteasome subunits Rpt5 and α7 were coimmunoprecipitated with wild type Insig-1 but not Insig-1(K156R/K158R).
We next examined the effect of unsaturated fatty acids on the interaction between Insig-1 and proteasomes. Unsaturated fatty acids do not block ubiquitination of Insig-1, but they block its subsequent proteasomal degradation (15). Previous studies showed that unsaturated fatty acids act by blocking the binding of Insig-1 to Ubxd8, which in turn binds to p97 (15). p97 is an ATPase that interacts with proteasomes and assists in the extraction of ubiquitinated proteins from ER membranes (37). As shown in Fig. 7A, lanes 1 and 2, the amount of p97 bound to Insig-1 was markedly reduced when arachidonic acid was added to cells. The amount of the proteasome subunits Rpt5 and α7 that coprecipitated with Insig-1 was also markedly reduced in cells treated with arachidonic acid (Fig. 7A, lanes 1 and 2). Similar results were obtained when mutant Insig-2(L210A) was used instead of wild type Insig-1 (Fig. 7C).
To test the requirement for p97 directly, we used RNA interference to reduce the amount of p97. We studied HEK-293S/pInsig1 cells, a line of HEK-293-derived cells stably transfected with Insig-1. We transiently transfected the cells with duplexes of siRNA targeting p97 or green fluorescent protein as a control. The cells were cultured in serum from which all lipids had been extracted. Knockdown of p97 increased the amount of Insig-1 protein (Fig. 7B, lanes 1 and 2). To examine the interaction between proteasomes and Insig-1, we treated the cells with MG-132 to equalize the amount of Insig-1 protein, and we used immunoblots to measure the amount of proteasome subunits coprecipitated with Insig-1. As shown in Fig. 7B, knockdown of p97 markedly reduced the amount of proteasome subunits Rpt5 and α7 coprecipitated with Insig-1 (Fig. 7B, lanes 3 and 4). Similar results were obtained when mutant Insig-2(L210A) was used instead of wild type Insig-1 (Fig. 7D).
The results from Figs. 6 and 7 indicate that ubiquitination of Insig-1 (or mutant Insig-2) and recruitment of p97 to Insig-1 are both required for proteasomes to bind to Insig-1. However, it remains unclear whether ubiquitination of Insig-1 is required for its interaction with p97. Fig. 8 shows an immunoblot in which we compared the interaction between p97 and two versions of Insig-1; that is, wild type and a double lysine mutant that is not ubiquitinated (K156R/K158R) (14). When cells were cotransfected with plasmids encoding wild type Insig-1 and Ubxd8, p97 was coimmunoprecipitated (Fig. 8, lane 4). Similarly, the mutant version of Insig-1 also interacted with p97 in a Ubxd8-dependent fashion (lane 8). With both versions of Insig-1, the interaction with p97 was disrupted by the addition of arachidonate to the culture medium (lanes 5 and 9). Similar results were obtained when the corresponding lysine mutant of Insig-2(L210A), i.e. Insig-2 (K100R; K102R; L210A) was analyzed (data not shown). These results indicate that ubiquitination of Insig-1 is not required for its interaction with p97.
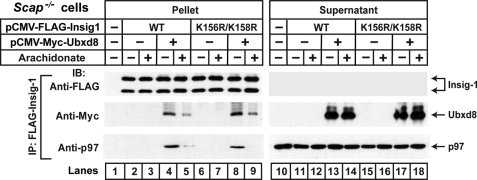
FIGURE 8.
Ubiquitination of Insig-1 is not required for its interaction with p97. On day 0 Scap-deficient SRD-13A cells were set up at 6.0 × 105 cells/60-mm dish as described under “Experimental Procedures.” On day 1 each dish of cells was transfected with 0.3 μg pCMV-Myc-Ubxd8, pCMV-FLAG-Insig1, or pCMV-FLAG-Insig1(K156R/K158R) as indicated. The total amount of transfected DNA was adjusted to 1.3 μg/dish with pcDNA3 empty vector. After incubation for 6 h each dish of cells was switched to medium A supplemented with 5% delipidated FCS. On day 2 the cells were treated for 6 h with or without 100 μm sodium arachidonate-albumin in the presence of 10 μm MG-132. The cells were then harvested and lysed with detergent, and Insig-1 was immunoprecipitated with anti-FLAG-agarose beads. Pellets (representing 0.1 dish of cells) and supernatants (representing 0.01 dish) were subjected to SDS-PAGE followed by immunoblot analysis with anti-Myc (against Ubxd8), anti-FLAG (against Insig-1), and anti-p97. IP, immunoprecipitation; IB, immunoblot. This experiment was repeated two more times with similar results. To observe arachidonate-regulated binding between Insig-1 and p97, the amount of transfected plasmids had to be adjusted to prevent excessive overexpression of Insig-1.
DISCUSSION
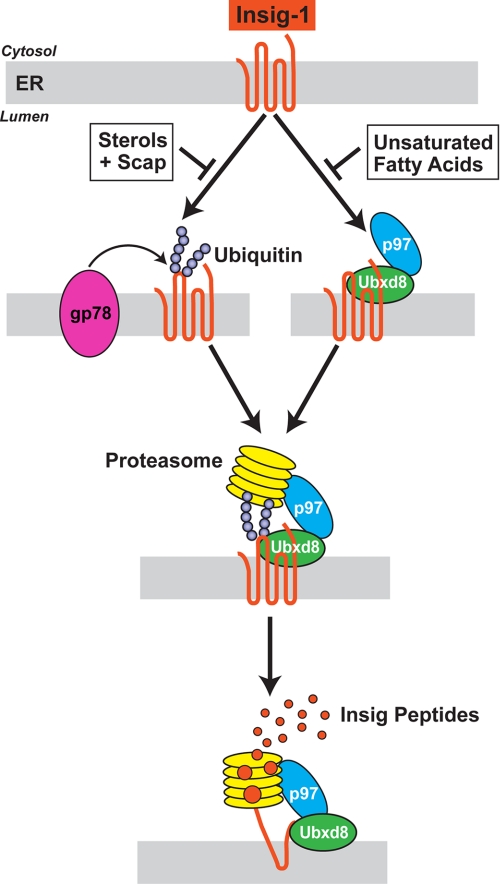
Fig. 9 shows a model for sterol- and unsaturated fatty acid-regulated ERAD of Insig-1 and mutant Insig-2(L210A). This model extends our previous model (15) by providing new insights into how proteasomes are attached to membrane-bound Insig proteins. The model is based on data gathered from experiments with either wild type Insig-1 or Insig-2(L210A). For simplicity, we frame the discussion in terms of Insig-1, recognizing that the same process applies to mutant Insig-2(L210A) but not to wild type Insig-2. In sterol- and fatty acid-depleted cells, two proteins bind to membrane-embedded Insig-1. One of these proteins, gp78, is a ubiquitin E3 ligase that attaches polyubiquitin chains to lysine 156 and lysine 158 of Insig-1, both of which are located on the first cytoplasmic loop and both of which are conserved in Insig-2 (colored green, Fig. 1A). The second protein, Ubxd8, contains a UBX domain that binds to p97 (37). We previously showed that Ubxd8 binds to Insig-1 and recruits p97 (15). The ATPase p97 has been shown by others to play a role in the extraction of membrane proteins during ERAD (1, 37). Here, we identify a new function of p97 by showing that it recruits proteasomes to ubiquitinated Insig-1 before the Insig is extracted from ER membranes, a finding that lends physiological significance to the recently reported interaction between p97 and proteasomes (35). Thereafter, the proteasomes and p97 act in concert to extract Insig-1 from the membrane in a reaction that is closely coupled to degradation (Fig. 9).
FIGURE 9.
Model for sterol and unsaturated fatty acid-regulated ERAD of Insig-1. The sequential steps in the ubiquitination of Insig-1 and its extraction from ER membranes by the proteasome are discussed in the text.
ERAD of Insig-1 is regulated by two classes of end-products, each of which inhibits one limb of the pathway shown in Fig. 9. As previously demonstrated, sterols such as cholesterol or 25-hydroxycholesterol cause Insig-1 to bind to Scap, and this precludes the binding of gp78, thereby preventing ubiquitination (16). In contrast, sterols do not inhibit binding of the Ubxd8·p97 complex to Insig-1 (15), which is consistent with the observation that this interaction does not require ubiquitination of Insig-1 (Fig. 8). Unsaturated fatty acids such as oleic acid and arachidonic acid do not block ubiquitination, but they prevent the binding of the Ubxd8·p97 complex to Insig-1 (15). Inasmuch as proteasome binding requires both ubiquitination and p97, either sterols or unsaturated fatty acids can block the degradative pathway (Fig. 9). Inasmuch as Insig-1 controls the activation of SREBPs and SREBPs increase the synthesis of both sterols and unsaturated fatty acids, the regulatory scheme outlined in Fig. 9 illustrates how both end-products can repress their own synthesis.
Although the current studies suggest that proteasomes play an active role in extracting Insig-1 from the ER membrane, they do not reveal the mechanism. An attractive hypothesis for the extraction of membrane proteins from the ER was advanced by Ploegh (38), who suggested that extraction is facilitated by splitting of the ER bilayer, due to the insertion of nonpolar lipids such as triglycerides or even cholesteryl esters. In the case of Insig-1, it is possible that Ubxd8 somehow facilitates this bilayer splitting and that arachidonic acid blocks the process by releasing Ubxd8 from the complex. This hypothesis is currently being tested in our laboratory.
A major finding in the current study is that ubiquitination of Insig-1 is not required for its interaction with p97. This is because p97 is recruited to Insig-1 by Ubxd8 whose interaction with Insig-1 does not require ubiquitination (Fig. 8). On the other hand, p97 does require ubiquitination to interact with other proteins subjected to ERAD. This list includes the class I heavy chains of the major histocompatibility complex whose interaction with p97 is mediated through Ufd1, another p97-associated adaptor protein that recognizes ubiquitinated proteins (39, 40). Thus, whether or not ubiquitination of a protein is required for its interaction with p97 depends on the adaptor protein that mediates the interaction.
A paradox in the current studies arises from the observation that the L210A mutation in the cytoplasmic tail of Insig-2 replaces a leucine that is conserved in Insig-1, reducing the overall sequence conservation in the cytoplasmic tails of the two proteins (see Fig. 1A). The paradoxical effect of this mutation in Insig-2 may be related to the neighboring insertion of four residues (YECK) that is found in Insig-2 (residues 213–216) as compared with Insig-1. We previously found that replacement of glutamic acid with alanine in the YECK sequence in Insig-2 increased its ubiquitination (19). It seems likely that a conformational aspect of this cytoplasmic tail protects Insig-2 from being recognized by gp78 for ubiquitination. Alterations to this conformation permit ubiquitination of Insig-2 but only in a sterol-depleted state. According to current ideas about Insig structure, the cytoplasmic tail in Insig-1 is only 14 residues long (18 residues in Insig-2). Whether this short sequence is sufficient to permit a direct interaction with gp78 remains to be determined.
Acknowledgments
We thank Lisa Beatty, Angela Carroll, Shomanike Head, and Ijeoma Onwuneme for invaluable help with tissue culture, Dorothy Goddard and David Thompson for excellent technical assistance, Ayako Suzuki and Masashi Yanagisawa for help with two-dimensional DIGE analysis, and Russell DeBose-Boyd for critical review of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants 2PO1HL20948 and RO1DK46181. This work was also supported by the Perot Family Foundation.
- ER
- endoplasmic reticulum
- ERAD
- ER-associated degradation
- CMV
- cytomegalovirus
- two-dimensional DIGE
- two-dimensional fluorescence difference gel electrophoresis
- FCS
- fetal calf serum
- 25-HC
- 25-hydroxycholesterol
- HMG-CoA
- 3-hydroxy-3-methylglutaryl coenzyme A
- LPDS
- lipoprotein-deficient serum
- N-Suc-Leu-Leu-Val-Tyr-AMC
- N-succinyl-Leu-Leu-Val-Tyr-7-amido-4-trifluromethylcoumarin
- siRNA
- small interfering RNA
- SREBP
- sterol regulatory element-binding proteins
- TK
- thymidine kinase
- TEV
- tobacco etch virus
- WT
- wild type
- HA
- hemagglutinin
- CHO
- Chinese hamster ovary
- HSV
- herpes simplex virus.
REFERENCES
- 1.Vembar S. S., Brodsky J. L. (2008) Nat. Rev. Mol. Cell Biol. 9, 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hampton R. Y., Garza R. M. (2009) Chem. Rev. 109, 1561–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Demartino G. N., Gillette T. G. (2007) Cell 129, 659–662 [DOI] [PubMed] [Google Scholar]
- 4.Finley D. (2009) Annu. Rev. Biochem. 78, 477–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmidt M., Hanna J., Elsasser S., Finley D. (2005) Biol. Chem. 386, 725–737 [DOI] [PubMed] [Google Scholar]
- 6.Goldstein J. L., DeBose-Boyd R. A., Brown M. S. (2006) Cell 124, 35–46 [DOI] [PubMed] [Google Scholar]
- 7.Brown M. S., Goldstein J. L. (1997) Cell 89, 331–340 [DOI] [PubMed] [Google Scholar]
- 8.Horton J. D., Goldstein J. L., Brown M. S. (2002) J. Clin. Invest. 109, 1125–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horton J. D., Shah N. A., Warrington J. A., Anderson N. N., Park S. W., Brown M. S., Goldstein J. L. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 12027–12032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang T., Espenshade P. J., Wright M. E., Yabe D., Gong Y., Aebersold R., Goldstein J. L., Brown M. S. (2002) Cell 110, 489–500 [DOI] [PubMed] [Google Scholar]
- 11.Sever N., Yang T., Brown M. S., Goldstein J. L., DeBose-Boyd R. A. (2003) Mol. Cell 11, 25–33 [DOI] [PubMed] [Google Scholar]
- 12.Sever N., Song B. L., Yabe D., Goldstein J. L., Brown M. S., DeBose-Boyd R. A. (2003) J. Biol. Chem. 278, 52479–52490 [DOI] [PubMed] [Google Scholar]
- 13.Goldstein J. L., Brown M. S. (1990) Nature 343, 425–430 [DOI] [PubMed] [Google Scholar]
- 14.Gong Y., Lee J. N., Lee P. C., Goldstein J. L., Brown M. S., Ye J. (2006) Cell Metab. 3, 15–24 [DOI] [PubMed] [Google Scholar]
- 15.Lee J. N., Zhang X., Feramisco J. D., Gong Y., Ye J. (2008) J. Biol. Chem. 283, 33772–33783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee J. N., Song B. L., DeBose-Boyd R. A., Ye J. (2006) J. Biol. Chem. 281, 39308–39315 [DOI] [PubMed] [Google Scholar]
- 17.Yabe D., Brown M. S., Goldstein J. L. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 12753–12758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee J. N., Ye J. (2004) J. Biol. Chem. 279, 45257–45265 [DOI] [PubMed] [Google Scholar]
- 19.Lee J. N., Gong Y., Zhang X., Ye J. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 4958–4963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hua X., Nohturfft A., Goldstein J. L., Brown M. S. (1996) Cell 87, 415–426 [DOI] [PubMed] [Google Scholar]
- 21.Wójcik C., DeMartino G. N. (2002) J. Biol. Chem. 277, 6188–6197 [DOI] [PubMed] [Google Scholar]
- 22.Goldstein J. L., Basu S. K., Brown M. S. (1983) Methods Enzymol. 98, 241–260 [DOI] [PubMed] [Google Scholar]
- 23.Hannah V. C., Ou J., Luong A., Goldstein J. L., Brown M. S. (2001) J. Biol. Chem. 276, 4365–4372 [DOI] [PubMed] [Google Scholar]
- 24.Brown M. S., Faust J. R., Goldstein J. L., Kaneko I., Endo A. (1978) J. Biol. Chem. 253, 1121–1128 [PubMed] [Google Scholar]
- 25.Sever N., Lee P. C., Song B. L., Rawson R. B., Debose-Boyd R. A. (2004) J. Biol. Chem. 279, 43136–43147 [DOI] [PubMed] [Google Scholar]
- 26.Radhakrishnan A., Ikeda Y., Kwon H. J., Brown M. S., Goldstein J. L. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 6511–6518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feramisco J. D., Goldstein J. L., Brown M. S. (2004) J. Biol. Chem. 279, 8487–8496 [DOI] [PubMed] [Google Scholar]
- 28.Reeves P. J., Thurmond R. L., Khorana H. G. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 11487–11492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rawson R. B., DeBose-Boyd R. A., Goldstein J. L., Brown M. S. (1999) J. Biol. Chem. 274, 28549–28556 [DOI] [PubMed] [Google Scholar]
- 30.Lee P. C., Sever N., Debose-Boyd R. A. (2005) J. Biol. Chem. 280, 25242–25249 [DOI] [PubMed] [Google Scholar]
- 31.Yamamoto T., Davis C. G., Brown M. S., Schneider W. J., Casey M. L., Goldstein J. L., Russell D. W. (1984) Cell 39, 27–38 [DOI] [PubMed] [Google Scholar]
- 32.Liu C. W., Li X., Thompson D., Wooding K., Chang T. L., Tang Z., Yu H., Thomas P. J., DeMartino G. N. (2006) Mol. Cell 24, 39–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koulich E., Li X., DeMartino G. N. (2008) Mol. Biol. Cell 19, 1072–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adams C. M., Reitz J., De Brabander J. K., Feramisco J. D., Li L., Brown M. S., Goldstein J. L. (2004) J. Biol. Chem. 279, 52772–52780 [DOI] [PubMed] [Google Scholar]
- 35.Besche H. C., Haas W., Gygi S. P., Goldberg A. L. (2009) Biochemistry 48, 2538–2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matiuhin Y., Kirkpatrick D. S., Ziv I., Kim W., Dakshinamurthy A., Kleifeld O., Gygi S. P., Reis N., Glickman M. H. (2008) Mol. Cell 32, 415–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Halawani D., Latterich M. (2006) Mol. Cell 22, 713–717 [DOI] [PubMed] [Google Scholar]
- 38.Ploegh H. L. (2007) Nature 448, 435–438 [DOI] [PubMed] [Google Scholar]
- 39.Meyer H. H., Wang Y., Warren G. (2002) EMBO J. 21, 5645–5652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Flierman D., Ye Y., Dai M., Chau V., Rapoport T. A. (2003) J. Biol. Chem. 278, 34774–34782 [DOI] [PubMed] [Google Scholar]